Abstract
Fructooligosaccharide (FOS), a prebiotic well known for its health-promoting properties, can improve the human gut ecosystem most likely through changes in its microbial composition. However, the detailed mechanism(s) of action of FOS in the modulation of the gut ecosystem remain(s) obscure. Traditional methods of profiling microbes and metabolites could barely show any significant features due to the existence of large interindividual differences, but our novel microbe–metabolite correlation approach, combined with faecal immunoglobulin A (IgA) measurements, has revealed that the induction of mucosal IgA by FOS supplementation correlated with the presence of specific bacteria. Furthermore, the metabolic dynamics of butyrate, l-phenylalanine, l-lysine and tyramine were positively correlated with that of these bacteria and IgA production, whereas p-cresol was negatively correlated. Taken together, our focused intraindividual analysis with omics approaches is a powerful strategy for uncovering the gut molecular network and could provide a new vista for understanding the human gut ecosystem.
Keywords: commensal microbiota, correlation analysis, gut ecosystem, metabolite, prebiotics
1. Introduction
Our gastrointestinal tract provides residence to the intestinal microbiota, which includes both beneficial and potentially pathogenic microorganisms.1,2 It has been postulated that an imbalance in the composition of the microbiota is a risk factor in several human disorders, including inflammatory bowel disease, metabolic syndrome, allergy and cancer.3–5 Furthermore, recent reports have shown that even minor changes in the gut microbiome can have an impact on the host phenotype.6,7 Thus, the ability to shape the intestinal microbiota should have clinical importance.
The mucosal surface of the intestine is continuously exposed to an enormous variety of antigens, such as food antigens and microorganisms. Immunoglobulin A (IgA) plays important roles in the mucosal immune system, and it has been shown that the production of IgA is limited in germ-free mice compared with conventional mice.8 In addition, microbial colonization is required for secretory IgA (SIgA) production in the intestine.9,10 In studies of gnotobiotic mice, it was shown that colonization by microbes, particularly segmented filamentous bacteria, clostridia and Bacteroides species, stimulates production of IgA.11,12 However, it is still unclear what molecule(s) produced by/derived from commensal microbes induce(s) mucosal IgA production.
Prebiotics are defined as food ingredients that are non-digestible and non-absorbable in the upper gastrointestinal tract and which improve the condition of the host through selective stimulation of the growth of probiotic bacteria.13 Fructooligosaccharide (FOS) is a well-known prebiotic that has anti-tumor,14 infection-protective15 and allergy-preventive effects16 in the host through host–microbial crosstalk in the gut. The impact of FOS intake on the intestinal IgA response has been extensively studied in mouse models. The concentration of IgA in the small and large intestines is significantly increased with FOS intake.17 Furthermore, the number of B220+ IgA+ cells in Peyer's patches is significantly increased, and the level of secretory component and SIgA in the ileal gut lumen is elevated. It has also been shown that FOS intake enhances production of cytokines, such as interleukin (IL)-5, IL-6 and interferon-γ, in Peyer's patches; these cytokines can induce IgA production through their effects on CD4+ helper T cells, which further increases the amount of IgA in the mucosa.18
In humans, ulcerative colitis patients supplemented with Bifidobacterium longum and oligofructose-enriched inulin showed improvement in the clinical features of chronic inflammation,19 and daily intake of oligofructose and inulin significantly decreased Crohn's disease activity.20 Supplementation with FOS has also been shown to support the growth of Bifidobacterium species, accompanied by an increase in T lymphocytes.21 Some studies have also reported a tendency for prebiotics-treated individuals to have higher faecal SIgA levels.22,23 Although commensal microbiota have been implicated in the FOS-induced production of IgA, evaluation of the gut microbial ecosystem is not an easy task, mainly due to its highly complex composition and the large individual difference among human subjects.22
We have developed a meta-analysis platform based on a multi-dimensional profiling technique to evaluate gut environmental changes, including host–microbial crosstalk.24 In order to understand the molecular mechanisms for the induction of IgA production in human subjects by FOS supplementation through gut microbes and/or their metabolite(s), we applied the multivariate microbe–metabolite correlation analysis combined with faecal IgA secretion of the host to evaluate the inter- and intraindividual changes in the gut ecosystem occurring with FOS supplementation. Here, we show a significant correlation of faecal IgA content with microbial composition and metabolites, and further implicate the likely involvement of particular metabolites in FOS-induced IgA secretion.
2. Materials and methods
2.1. Faecal sample collection
Seven volunteers including three males and four females (20–30 year olds) from different families in Japan participated in this study (Supplementary Table S1). All subjects were informed of the purpose of this study. This study was approved by the ethical committee of RIKEN, and written consent was obtained from all subjects. All subjects had no medical history of gastrointestinal or metabolic diseases, nor had they had special dietary habits or restrictions, herbal supplements, probiotics or antibiotics within at least 1 month before the sampling. For 1 week, the volunteers ate FOS, 10 g twice a day (Meioligo W, Meiji Kabusiki Kaisya). Faecal sampling was performed at least twice during each period (before FOS intake, during FOS intake and after FOS intake). Faecal samples from each individual were immediately frozen on collection and stored at −80°C before analysis.
2.2. Measurement of faecal IgA by enzyme-linked immunosorbent assay
Faecal samples were lyophilized by using VD-800R lyophilizer (VD-800R, Tokyo, Japan) for 24 h. Freeze-dried faeces were ground with 3.0-mm Zirconia Beads (Biomedical Science, Tokyo, Japan) using a Shake Master (Biomedical Science) for 10 min. About 10 mg of faecal samples were measured and suspended into 300 μl of phosphate-buffered saline buffer containing proteinase inhibitor cocktail tablets: complete EDTA-free (Roche Applied Science, USA), and homogenized for 1 min. The homogenate was centrifuged at 17,800 × g for 15 min at 4°C, and the supernatant was collected and frozen at −20°C. The amount of faecal IgA was measured by enzyme-linked immunosorbent assay (ELISA). To measure total IgA, the wells of a Maxisorp immunoplate (Nunc, Roskilde, Denmark) were coated with goat anti-mouse IgA (A90-103A, Bethyl Laboratories, Inc., USA). After blocking of unoccupied sites on the plastic with bovine serum albumin, the test samples and standard mouse IgA (mouse reference serum; RS10-101, Bethyl Laboratories, Inc.) were added. Subsequently, bound IgA was detected by sequential incubation with horseradish peroxidase-conjugated goat anti-mouse IgA antibodies (A90-103P, Bethyl Laboratories, Inc.) (1 mg/ml), and 3,3′,5,5′-tetramethyl benzidine (TMB) (Organon Teknika, Durham, NC, USA) (1 mg/ml) and 1 : 10,000 hydrogen peroxide (H2O2, Wako Pure Chemical Industries, Japan) in sodium acetate buffer (pH 6.0). After incubation at room temperature for 15 min, colour development was stopped by adding 100 μl of 0.18 M sulphuric acid (H2SO4, Wako Pure Chemical Industries), and then the absorbance at 450 nm of each well was measured. The total IgA content of each sample was calculated by means of a standard curve.
2.3. Faecal DNA extraction
About 10 mg of freeze-dried faecal sample was suspended with mixture buffer [400 μl of 10% sodium dodecyl sulphate (SDS)/10 mM Tris–HCl, 1 mM EDTA, pH 8.0 solution, 400 μl of phenol/chloroform/isoamyl alcohol (25 : 24 : 1), and 200 μl of 3 M sodium acetate]. Faeces in mixture buffer were disrupted with 0.1 mm zirconia/silica beads (BioSpec Products, Inc., USA) by vigorous shaking (1500 rpm, for 10 min) using a Shake Master (Biomedical Science). After centrifugation at 15,000 × g for 30 min at room temperature, the DNA was purified using a phenol/chloroform/isoamyl alcohol (25 : 24 : 1) solution and was precipitated by adding ethanol and sodium acetate, and stored at −20°C.
2.4. 454 barcoded pyrosequencing of 16S rRNA gene
The V1–V2 variable region of the 16S rRNA gene was amplified by PCR using forward primer (5′-CCATCTCATCCCTGCGTGTCTCCGACTCAGNNNNNNNNNNagrgtttgatymtggctcag-3′) containing the 454 primer A and reverse primer (5′-CCTATCCCCTGTGTGCCTTGGCAGTCTCAGtgctgcctcccgtaggagt-3′) containing the 454 primer B.25 Forward primers were tagged with 10 bp unique barcode labels at the 5′ end along with the adaptor sequence (indicated in N). Each reaction mixture contained 10 pmol of each primer, 0.16 mM deoxyribonucleoside triphosphates, 25 μl of 2xEx Taq PCR buffer, 1.0 U Ex Taq polymerase (Takara Bio, Inc., Shiga), 50 ng extracted DNA and sterilized water to achieve a final volume of 50 μl. PCR program was set as follows: 96°C 2 min and 20 cycles of 96°C 30 s, 55°C 45 s, 72°C 1 min followed by 72°C for 10 min. After confirmation of the PCR product formation using agarose gel electrophoresis, purified by AMPure XP magnetic purification beads (Beckman Coulter, Inc., Brea, CA, USA), and quantified using the Quant-iT PicoGreen ds DNA Assay Kit (Life Technologies Japan, Ltd, Tokyo, Japan). Mixed samples were prepared by pooling approximately equal amounts of PCR amplicons from each sample and subjected to 454 GS FLX Titanium or 454 GS JUNIOR (Roche Applied Science) sequencing according to the manufacturer's instructions.
2.5. Analysis of 16S rRNA sequences using the QIIME pipeline
16S rRNA sequence data were processed by the quantitative insights into microbial ecology (QIIME) pipeline.26 Briefly, sequences that were less than 200 bp or greater than 1000 bp in length contained incorrect primer sequences, or contained more than 2 ambiguous bases were discarded. The remaining sequences were assigned sample aliquots based on their unique nucleotide barcodes, including error-correction. Chimeric sequences were removed using Usearch.27 Reads removed in these filters were ∼42% of all sequences (Supplementary Table S2). All filter-passed reads of the 16S V1–2 sequences were deposited in DDBJ with accession number DRA001212. Sequences were clustered into Operational Taxonomic Units (OTUs) using 97% sequence similarity. OTUs were assigned to a taxonomy using RDP classifier with the confidence level set at 0.5.28
2.6. Sample extraction for NMR measurements
Faecal samples (10 mg) were measured and suspended with extraction buffer consisting of 90% (v/v) deuterium oxide (D2O), 100 mM potassium phosphate and 1 mM 2,2-dimethyl-2-silapentane-5-sulphonate (DSS) as an internal standard. The solution was shaken at 1500 × g for 15 min at 65°C and then centrifuged at 15,000 × g for 15 min at 4°C. The supernatant was collected in an NMR tube and frozen at −20°C.29
2.7. 1D and 2D NMR measurements
All NMR spectra were recorded on a Bruker DRX-700 spectrometer operating at 1H 700.153 MHz frequency with the temperature of the NMR samples maintained at 298 K. The NMR spectra were processed using a procedure similar to that described previously.30–33 Briefly, 1H NMR data were reduced by subdividing spectra into sequential bins of 0.04 ppm designated regions between 1H chemical shifts of 0.06–9.8 ppm. 1H-13C heteronuclear single quantum coherence (HSQC) data were reduced by subdividing spectra into sequential bins of 0.3 ppm in the f1 direction and 0.03 ppm in f2 designated regions between 1H chemical shifts of 0.5–9.0 ppm and 13C chemical shifts of 40–90 ppm, respectively. After exclusion of water resonance, each region was integrated and normalized relative to the DSS intensity and analysed by principal component analysis (PCA) using the R software and plotted using the Excel software. Loading plot analysis revealed the contribution of bins to the PCA scores, and 2D spectral assignments were performed using the SpinAssign software.33
2.8. Multivariate statistical analysis
Supplementary Figure S1 provides a schematic overview of the analytical steps in this study. Multivariate statistical analysis was performed with R program environment by using an in-house program. A two-dimensional correlation map was calculated as a symmetric matrix using Spearman's rank correlation coefficient, in which an element at position (i, j) is defined as a correlation coefficient between the ith and jth positions in each set of 1D spectra of metabolites, microbial diversity of high-throughput sequencing and the amount of IgA. A more positive (negative) coefficient means the existence of a positive (negative) correlation between the ith and jth peaks or bands. Each correlation pattern was clustered using hierarchical clustering analysis (HCA). HCA was performed using a Euclidean distance and ward clustering method. The two-dimensional correlation map was drawn by the R software.29
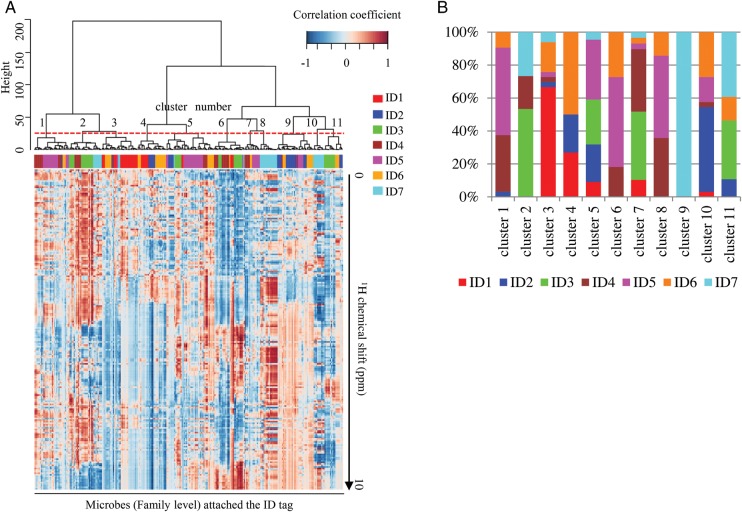
In the heat map of microbe–metabolite correlation coefficient, individual ID number was put as a tag in front of each microbe name (at the family level) to clarify the individual from whom those microbes were isolated (for example, ID4-Bacteroidaceae). Correlation coefficient between microbial composition and metabolic profile was calculated in each individual, and the data obtained from individuals were merged. Microbe–metabolite correlation pattern was clustered using HCA. HCA of the metabolic profiles in individual ID-coded microbes reveals several metabolic profile clusters that were related to different microbes. Clusters of metabolic profiles defined by cutting branches off the dendrogram. We selected clusters in which more than 70% of microbes were derived from IgA responders ID4 and ID5.
IgA–metabolite–microbe correlation network was constructed with microbes in extracted clusters, based on the multiple correlation data from ID4 and ID5 using the Cytoscape software34 (http://www.cytoscape.org/). Each node indicated IgA, a metabolite and a microbe from the result of the above analysis. Microbial node size showed the average value of relative abundance. Edges represent the correlation coefficient >0.65 between the two connected nodes in the network. Width of each edge line positively correlates with its correlation coefficient value.
3. Results and discussion
3.1. FOS effect on faecal IgA concentration is heterogeneous in different healthy human volunteers
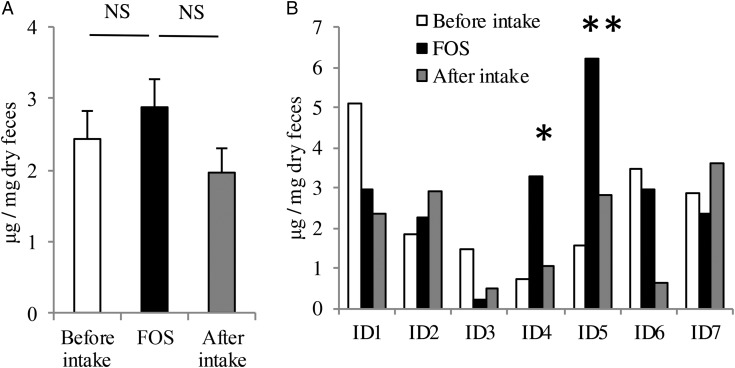
Faecal samples were collected at several time points from healthy volunteers (age = 20–30, male = 3, female = 4) before, during and after FOS intake, and the amount of faecal IgA was determined. As a group, the average faecal IgA tended to increase upon FOS intake, but the difference was not statistically significant (Fig. 1A). However, the individual faecal IgA profiles were quite heterogeneous (Fig. 1B and Supplementary Fig. S2); especially, only in volunteers ID4 and ID5, the faecal IgA levels were significantly elevated during the FOS-intake period when compared with those in other periods. Therefore, the effect of FOS on IgA induction varies widely among individuals.
Figure 1.
Effect of FOS supplementation on total faecal IgA production in human volunteers. (A) The average levels of faecal IgA in all seven volunteers before, during and after FOS supplementation. NS, no significant difference. (B) The individual average amounts of faecal IgA before, during and after FOS supplementation. Faecal sampling was performed at least twice during each period. P-values between FOS supplementation period and other periods were determined using the non-parametric Mann–Whitney U-test. *P < 0.1; **P < 0.01.
Several studies have reported that FOS increases beneficial bacteria such as Bifidobacterium, thereby stimulating the host immune system in humans.35 Another report has shown a trend toward higher SIgA concentrations following intake of a galactooligosaccharide/FOS mixture, albeit with large interindividual variations.22
3.2. Commensal microbiota composition varies greatly among individuals
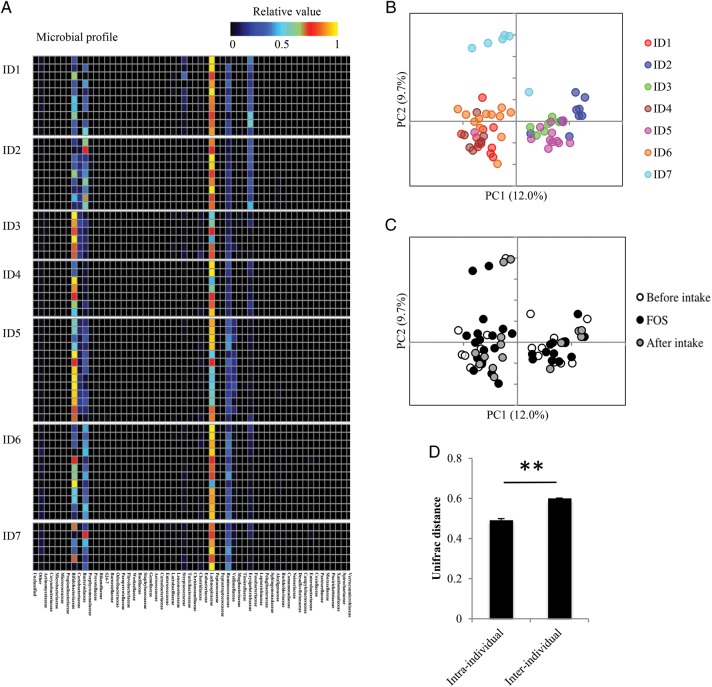
We hypothesized that the effect of FOS intake on faecal IgA levels was likely influenced by individual differences in the composition of the gut microbiota. To determine whether this was the case, PCR products of bacterial 16S rRNA-encoding genes in the faecal DNA samples were analysed using high-throughput pyrosequencing. A total of 222,863 reads were sequenced, averaging at 3537 reads per sample. Indeed, the heat maps of the individual microbial profiles at the family level looked quite different from each other (Fig. 2A). Consistently, the unweighted UniFrac PCoA of faecal microbiota clustered the faecal DNA samples from the same individuals (Fig. 2B), rather than from the samples with and without FOS intake (Fig. 2C), indicating that FOS-induced microbiota variations are overlooked because of the large interindividual variations when datasets from individuals are collectively analysed (Fig. 2D and Supplementary Fig. S3).
Figure 2.
Effect of FOS supplementation on the human microbiome. (A) Heatmap profiles of individual gut microbiome were analysed at the microbial family level. These profiles were normalized to a relative value between 0 and 1. (B) and (C) PCoA on UniFrac distance matrix from all volunteers before, during and after FOS supplementation. PCoA plots are coloured by individuals (B) and shaded by diet periods (C). (D) The comparison of Unifrac distance between intra- and interindividuals. P-values were determined using the Mann–Whitney U-test. **P < 0.01.
Our observations are consistent with a number of studies reporting individual differences in the structure of gut microbiota. Three human enterotypes can be distinguished by differences in microbial composition.36 These enterotypes are not altered by short-term diet changes.37 Consistent with this stability, FOS did not alter the major composition of the intestinal microbiota in individuals in this study (Fig. 2B and C).
3.3. Faecal metabolic profiles are distinctive among individuals
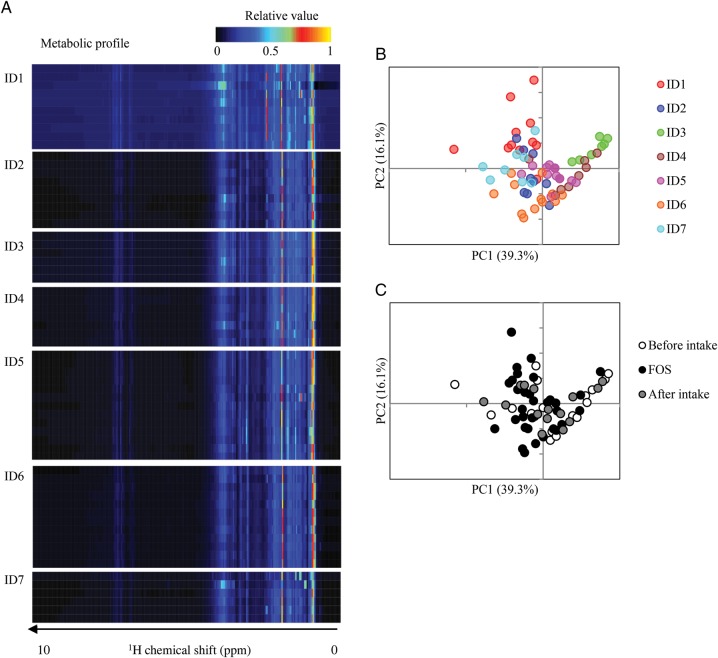
We also examined the individual faecal metabolic profiles before, during and after FOS intake, since prebiotic FOS-induced metabolic changes in the gut ecosystem could result in the modification of IgA production. To this end, faecal metabolites were extracted from the faecal samples and measured by 1H NMR spectroscopy. The heat map of the obtained 1H NMR spectra divided into bins showed similar interindividual metabolic changes upon FOS intake (Fig. 3A). Nevertheless, the PCA of these metabolic profiles showed a tendency towards individual clustering (Fig. 3B), rather than being clustered according to the presence or absence of the FOS supplementation (Fig. 3C). These observations are consistent with a previous study, reporting that 1H NMR-based metabolomics of urine and serum show individual differences.38,39
Figure 3.
Effect of FOS supplementation on faecal metabolites. (A) Heatmap profiles of individual metabolomes in this trial. Faecal metabolome profiling was performed by 1H NMR and the profiles were normalized to a relative value between 0 and 1. (B) and (C) Principal component analysis on faecal metabolome data from all volunteers before, during and after FOS supplementation. Score plots are coloured by individuals (B) and shaded by diet periods (C).
Taken together with the data on microbiota composition described above (Fig. 2), these results indicate that interindividual differences are too large for population analyses such as PCA to extract significant parameters contributing to the FOS-induced differences, and that intraindividual analysis could instead reveal the mechanisms for the effect of FOS on IgA production in humans. Thus, the individual difference in the effect of FOS on faecal IgA concentration could be due to the individual differences in their gut microbiota and/or metabolites.
3.4. Intraindividual correlation analyses among commensal microbiota composition, faecal metabolic profiles and faecal IgA concentration reveal microbial and metabolic changes related to enhanced faecal IgA levels
To evaluate intraindividually the impact of FOS intake on gut microbiota composition in the absence of the interindividual differences, we performed individual PCA (Supplementary Fig. S4A). The data revealed that FOS intake caused changes in microbiota composition in each individual, but that these changes were different from individual to individual. We also evaluated the influence of FOS intake on the individual faecal metabolic profile, by performing PCA individually (Supplementary Fig. S4B). Similar to the changes observed in the microbiota, FOS also induced faecal metabolic changes in each individual.
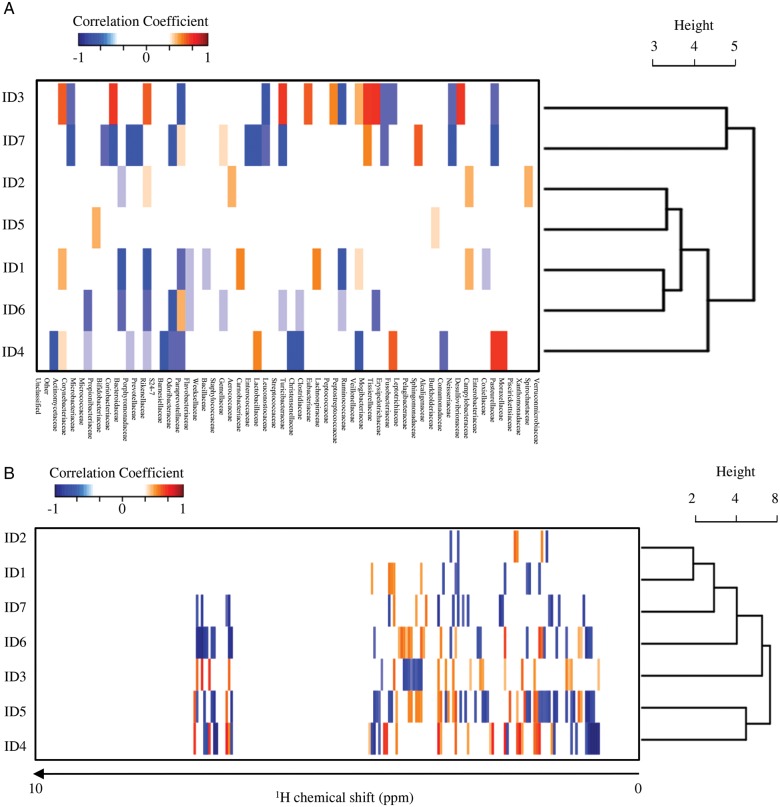
In order to obtain more integrated information about the effect of FOS intake on IgA production, we next performed intraindividual two-dimensional correlation analyses to link the commensal microbial variation, the faecal metabolic profile and the amount of faecal IgA. The coefficient of correlation for each individual gut environmental factor was calculated using the Spearman's rank correlation coefficient, and the correlative information was clustered individually by HCA to seek the covariation factor(s) that related to the induction of faecal IgA production (Fig. 4). The microbe–IgA correlation analysis showed that the commensal microbiota composition co-varied strongly with the individual faecal IgA production profile (Fig. 4A). HCA on the microbe–IgA correlation patterns from seven volunteers showed that the profile from ID3 and ID7 was strikingly distinct from the others. This might result from an apparently abnormal IgA profile in ID3 in that the faecal IgA levels were extremely low during and after FOS intake (Supplementary Fig. S2). In addition, microbiota composition in ID7 is a different from the other individuals (Fig. 2B). In the metabolite–IgA correlation analysis, we found that the correlation patterns were divided into two groups; ID4 and ID5 were grouped together and separated from the others (Fig. 4B). As described above, the amount of IgA was significantly increased during FOS intake only in ID4 and ID5, suggesting that the change in metabolites common to ID4 and ID5, but not to the other individuals, may contribute to their FOS-induced increase in faecal IgA. 1H chemical shifts revealed by this analysis to be common in ID4 and ID5 are listed in Supplementary Table S3.
Figure 4.
Individual ecological dynamics in the gut during FOS supplementation. (A) Correlation coefficients between faecal IgA–microbiome profile and (B) faecal IgA–faecal metabolome were calculated by Spearman's rank correlation coefficient using the R software (http://www.r-project.org/). A correlation matrix is represented by negative correlation (R < −0.65; blue) and positive correlation (R > 0.65; red). The correlation similarity among individuals was further clustered by HCA.
To analyse the relationship between microbiota and faecal metabolites before, during and after FOS intake, we performed microbe–metabolite correlation analysis (Fig. 5A). Characteristic correlation patterns were represented as the phenolic region (around 6.5–8.5 ppm). Based on the HCA, the correlation patterns divided the microbiota into 11 bacterial clusters. Faecal bacteria from each individual contributed differently to these clusters (Fig. 5B); intriguingly, Clusters 1, 6 and 8 consisted of the microbes derived predominantly from ID4 and ID5 (>70%). The result showed that these microbes were closely related to Rikenellaceae, Bifidobacteriaceae, S24-7, Peptostreptococcaceae, Barnesiellaceae, Mogibacteriaceae, Pasteurellaceae, Veillonelaceae, Coriobacteriaceae, Clostridiaceae, Alcaligenaceae, Desulfovibrionaceae and Pasteurellaceae at the family level (Supplementary Table S3). Major OTUs in these microbial families were assigned using BLAST against non-redundant database (Supplementary Table S4). Supplementary Table S3 lists the 1H chemical shifts of metabolites correlated with these clusters which were commonly seen in ID4 and ID5.
Figure 5.
Identification of the gut microbes correlated with faecal IgA induction by FOS supplementation. (A) Correlation coefficients between gut microbiome–faecal metabolome were calculated by Spearman's rank correlation coefficient. The 11 clusters based on the correlation similarity among microbes (the cut-off height of 30, which was determined so that the number of leaf nodes in a cluster is over 10, shown as a dotted line) were further clustered by HCA. (B) Composition of the individually derived microbes in each cluster.
3.5. Assignment of the candidate metabolites involved in the induction of faecal IgA production
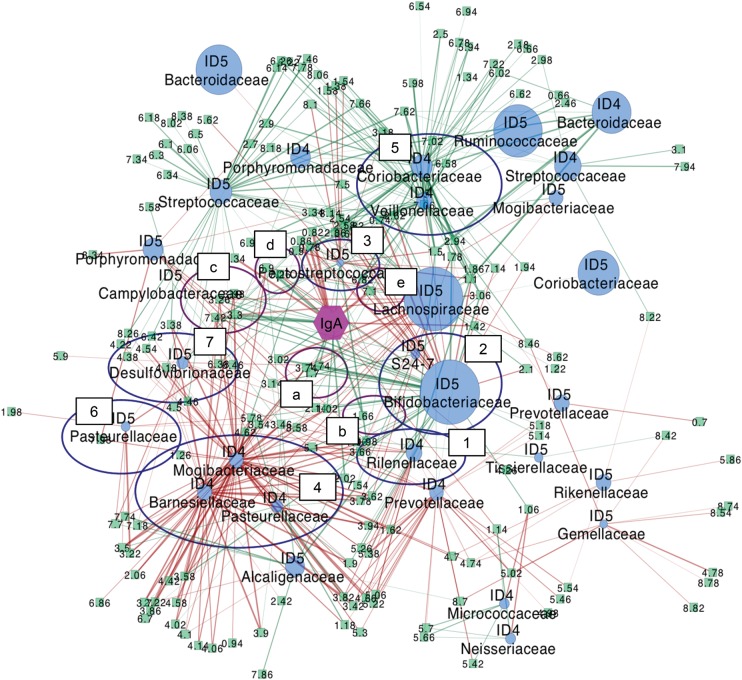
Comparison of metabolites correlated with IgA and microbes shared in ID4 and ID5 (Supplementary Table S3) revealed that some of the 1H chemical shifts were commonly extracted from both the microbe–metabolite correlation and the metabolite–IgA correlation. These metabolites were assigned by two-dimensional J-resolved (2D-J) spectroscopy, 1H-13C HSQC and total correlation spectroscopy (TOCSY),40 as well as according to the literature41–43 and the PRIME website44 (Supplementary Table S5). Metabolites that showed a positive correlation with increased IgA were identified as l-phenylalanine (3.30 and 7.42 ppm), l-lysine (1.70 and 3.02 ppm), tyramine (6.90 ppm) and butyrate (2.14 ppm); and p-cresol (6.82 and 7.10 ppm) was negatively correlated with IgA levels. The molecular network around IgA based on these results is shown in Fig. 6. Taken together, our results suggest that these bacteria in clusters are directly or indirectly involved in the metabolism of these molecules, an activity that may ultimately result in the up-regulation of faecal IgA production in the gut of ID4 and ID5.
Figure 6.
Molecular network of the gut ecosystem supplemented with FOS. This network was constructed based on the multiple correlation data from ID4 and ID5 using the Cytoscape software34 (http://www.cytoscape.org/). Green rectangles, blue spheres and purple hexagon indicate metabolites, microbes and IgA, respectively. Sphere size of microbial nodes corresponds to their average relative abundance. Positive and negative correlations are shown as green and red lines, respectively. Purple and blue circles indicate metabolites and microbes, respectively, which highly correlate with faecal IgA abundance. Metabolite a: l-lysine, b: butyrate, c: l-phenylalanine, d: tyramine, e: p-cresol. Microbe 1: ID4 Rikenellaceae, 2: ID5 Bifidobacteriaceae, S24-7, 3: ID5 Peptostreptococcaceae, 4: ID4 Barnesiellaceae, Mogibacteriaceae, Pasteurellaceae and ID5 Clostridiaceae, 5: ID4 Veillonelaceae, Coriobacteriaceae, 6: ID5 Pasteurellaceae, 7:ID5 Desulfovibrionaceae.
In this study, a positive correlation of l-lysine, l-phenylalanine, tyramine and butyrate, and a negative correlation of p-cresol, were found with the FOS intake-induced increase in faecal IgA, using the combination of microbe–metabolite and metabolite–IgA correlation analyses (Fig. 6). Consistent with our observations, it has been reported using in vitro experiments that human intestinal contents with live commensal microbes metabolize FOS into butyrate and tyramine.45 Bifidobacterium preferentially utilize FOS as an energy source to produce lactate and acetate. Bifidobacterium themselves do not generally produce butyrate,46 but other commensal microbes can produce butyrate from acetate and lactate produced by Bifidobacterium.47,48 It has also been reported that both lactate and acetate can be converted to butyrate by human faecal microbes in vitro.49
Tyramine is a decarboxylation product of tyrosine, and two types of tyrosine metabolic pathways exist in commensal microbes. One pathway produces p-cresol,35,50 while the other produces tyramine.51,52 Our data showed that tyramine and p-cresol were positively and negatively correlated with IgA production, respectively, in FOS responders ID4 and ID5, suggesting that FOS intake may impact the microbiota composition to change the tyrosine metabolic pathways in favour of tyramine production in the gut ecosystem of FOS responders.
It has been reported that an important function of phenylalanine is to up-regulate the expression of GTP cyclohydrolase I, which is the first and rate-controlling enzyme for the synthesis of tetrahydrobiopterin, an essential cofactor for nitric oxide synthase (NOS). Consequently, an adequate intake of dietary phenylalanine is required to maintain a sufficient provision of tetrahydrobiopterin for the production of NO by inducible NOS (iNOS) in activated macrophages and other leucocytes.53 Moreover, an inadequate intake of dietary lysine reduces antibody responses and cell-mediated immunity.54 By sharing the same transport systems with arginine, the availability of dietary or extracellular lysine can modulate the entry of arginine into leucocytes and NO synthesis by iNOS.53 iNOS is expressed mainly in macrophages and dendritic cells, which produce NO upon stimulation with cytokines and bacterial components. Recent investigations have demonstrated that IgA production is decreased in iNOS−/− mice.55 Therefore, these amino acids may be involved in the induction of IgA production through NO production.
3.6. Conclusion
In the present study, our intraindividual, multiple time point correlation analyses showed that enhancement of faecal IgA levels by FOS supplementation correlates with changes in the intestinal metabolites, likely produced by commensal microbes. Our study also have confirmed that the human intestinal microbiota possess large individual variations that make it difficult to detect significant but small changes commonly induced in individuals by an intervention, such as FOS intake in this study. This fact emphasizes the importance of intraindividual analyses, such as those employed in our study, for analysing changes in the gut microbiota in the human population. Consistent with this notion, a similar approach has recently been taken to reveal human gut environmental changes in response to short-term dietary intervention.56 Collectively, we propose that the multiple omics approach on the repeated intraindividual time-course sampling should be appropriate for uncovering the gut environmental changes upon intake of probiotics and prebiotics, as well as foods and drugs. In contrast, when repeated intraindividual samples are not available, these approaches may not detect significant differences/changes unless data from a large scale population are collected.
In conclusion, our study has demonstrated that intraindividual multiple time-point correlation analyses are useful for understanding the FOS intake-induced increase in faecal IgA concentration. Since an enormous amount of biological data can be obtained with recent innovations in analytical hardware, such as the pyrosequencer, NMR and mass spectrometry, data-mining techniques are becoming more and more important. Application of our multiple correlation approach for analysing the individual gut ecosystems will enable us to better understand the whole picture of the impact of prebiotics, such as FOS and probiotics, on our gut ecosystem.
Authors' contributions
S.F. and H.O. conceived and designed the experiments. T.K., S.F., A.F., W.S. and M.H. performed the experiments. T.K. and S.F. analysed the data. J.K. provided analytical tool and database. T.K., S.F. and H.O. wrote the paper.
Supplementary Data
Supplementary Data are available at www.dnaresearch.oxfordjournals.org.
Funding
This study was supported in part by Grants-in-Aid for Scientific Research (B) (24380072 to S.F. and 21390155 to H.O.), Scientific Research on Innovative Areas (24117524 to S.F. and 20113003 to H.O.) and Challenging Exploratory Research (24658129 to S.F.) from the Ministry of Education, Culture, Sports, Science and Technology of Japan. This work was also supported in part by grants from the Institute for Fermentation, Osaka (S.F.), the Mishima Kaiun Memorial Foundation (S.F.), the Takeda Science Foundation (S.F.) and the Uehara Memorial Foundation (S.F.).
Supplementary Material
Acknowledgements
We thank Yumi Chiba, Chikako Uetake, Hideaki Shima (Laboratory for Intestinal Ecosystem, RIKEN IMS-RCAI) and Yuuri Tsuboi (Environmental Metabolic Analysis Research Team, RIKEN CSRS) for technical support. We also appreciate Dr Peter D. Burrows (Department of Microbiology, University of Alabama at Birmingham, Birmingham) for critical reading of the manuscript and helpful comments on it.
Conflict of Interest statement: none declared.
References
- 1.Guarner F., Malagelada J.R. Gut flora in health and disease. Lancet. 2003;361:512–9. doi: 10.1016/S0140-6736(03)12489-0. [DOI] [PubMed] [Google Scholar]
- 2.Turnbaugh P.J., Ley R.E., Mahowald M.A., Magrini V., Mardis E.R., Gordon J.I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–31. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 3.Nagalingam N.A., Lynch S.V. Role of the microbiota in inflammatory bowel diseases. Inflammatory Bowel Dis. 2011;18:968–84. doi: 10.1002/ibd.21866. [DOI] [PubMed] [Google Scholar]
- 4.Vijay-Kumar M., Aitken J.D., Carvalho F.A., et al. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science. 2010;328:228–31. doi: 10.1126/science.1179721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tappenden K.A., Deutsch A.S. The physiological relevance of the intestinal microbiota—contributions to human health. J. Am. Coll. Nutr. 2007;26:679S–83S. doi: 10.1080/07315724.2007.10719647. [DOI] [PubMed] [Google Scholar]
- 6.Holmes E., Nicholson J. Variation in gut microbiota strongly influences individual rodent phenotypes. Toxicol. Sci. 2005;87:1–2. doi: 10.1093/toxsci/kfi259. [DOI] [PubMed] [Google Scholar]
- 7.Rohde C.M., Wells D.F., Robosky L.C., et al. Metabonomic evaluation of Schaedler altered microflora rats. Chem. Res. Toxicol. 2007;20:1388–92. doi: 10.1021/tx700184u. [DOI] [PubMed] [Google Scholar]
- 8.Shroff K.E., Meslin K., Cebra J.J. Commensal enteric bacteria engender a self-limiting humoral mucosal immune response while permanently colonizing the gut. Infect. Immun. 1995;63:3904–13. doi: 10.1128/iai.63.10.3904-3913.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Macpherson A.J., Hunziker L., McCoy K., Lamarre A. IgA responses in the intestinal mucosa against pathogenic and non-pathogenic microorganisms. Microbes Infect. 2001;3:1021–35. doi: 10.1016/s1286-4579(01)01460-5. [DOI] [PubMed] [Google Scholar]
- 10.Hapfelmeier S., Lawson M.A., Slack E., et al. Reversible microbial colonization of germ-free mice reveals the dynamics of IgA immune responses. Science. 2010;328:1705–9. doi: 10.1126/science.1188454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Umesaki Y., Setoyama H. Structure of the intestinal flora responsible for development of the gut immune system in a rodent model. Microbes Infect. 2000;2:1343–51. doi: 10.1016/s1286-4579(00)01288-0. [DOI] [PubMed] [Google Scholar]
- 12.Yanagibashi T., Hosono A., Oyama A., et al. Bacteroides induce higher IgA production than Lactobacillus by increasing activation-induced cytidine deaminase expression in B cells in murine Peyer's patches. Biosci. Biotechnol. Biochem. 2009;73:372–7. doi: 10.1271/bbb.80612. [DOI] [PubMed] [Google Scholar]
- 13.Gibson G.R., Roberfroid M.B. Dietary modulation of the human colonic microbiota: introducing the concept of prebiotics. J. Nutr. 1995;125:1401–12. doi: 10.1093/jn/125.6.1401. [DOI] [PubMed] [Google Scholar]
- 14.Pierre F., Perrin P., Champ M., Bornet F., Meflah K., Menanteau J. Short-chain fructo-oligosaccharides reduce the occurrence of colon tumors and develop gut-associated lymphoid tissue in Min mice. Cancer Res. 1997;57:225–8. [PubMed] [Google Scholar]
- 15.Cazzola M., Tompkins T.A., Matera M.G. Immunomodulatory impact of a synbiotic in T(h)1 and T(h)2 models of infection. Ther. Adv. Respir. Dis. 2010;4:259–70. doi: 10.1177/1753465810379009. [DOI] [PubMed] [Google Scholar]
- 16.Fujitani S., Ueno K., Kamiya T., et al. Increased number of CCR4-positive cells in the duodenum of ovalbumin-induced food allergy model Nc/jic mice and antiallergic activity of fructooligosaccharides. Allergol Int. 2007;56:131–8. doi: 10.2332/allergolint.O-06-450. [DOI] [PubMed] [Google Scholar]
- 17.Nakamura Y., Nosaka S., Suzuki M., et al. Dietary fructooligosaccharides up-regulate immunoglobulin A response and polymeric immunoglobulin receptor expression in intestines of infant mice. Clin. Exp. Immunol. 2004;137:52–8. doi: 10.1111/j.1365-2249.2004.02487.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hosono A., Ozawa A., Kato R., et al. Dietary fructooligosaccharides induce immunoregulation of intestinal IgA secretion by murine Peyer's patch cells. Biosci. Biotechnol. Biochem. 2003;67:758–64. doi: 10.1271/bbb.67.758. [DOI] [PubMed] [Google Scholar]
- 19.Furrie E. A molecular revolution in the study of intestinal microflora. Gut. 2006;55:141–3. doi: 10.1136/gut.2005.081695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lindsay J.O., Whelan K., Stagg A.J., et al. Clinical, microbiological, and immunological effects of fructo-oligosaccharide in patients with Crohn's disease. Gut. 2006;55:348–55. doi: 10.1136/gut.2005.074971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guigoz Y., Rochat F., Perruisseau-Carrier G., Rochat I., Schiffrin E.J. Effects of oligosaccharide on the faecal flora and non-specific immune system in elderly people. Nutr. Res. 2002;22:13–25. [Google Scholar]
- 22.Bakker-Zierikzee A.M., Tol E.A., Kroes H., Alles M.S., Kok F.J., Bindels J.G. Faecal SIgA secretion in infants fed on pre- or probiotic infant formula. Pediatr. Allergy Immunol. 2006;17:134–40. doi: 10.1111/j.1399-3038.2005.00370.x. [DOI] [PubMed] [Google Scholar]
- 23.Scholtens P.A., Alliet P., Raes M., et al. Fecal secretory immunoglobulin A is increased in healthy infants who receive a formula with short-chain galacto-oligosaccharides and long-chain fructo-oligosaccharides. J. Nutr. 2008;138:1141–7. doi: 10.1093/jn/138.6.1141. [DOI] [PubMed] [Google Scholar]
- 24.Date Y., Nakanishi Y., Fukuda S., et al. New monitoring approach for metabolic dynamics in microbial ecosystems using stable-isotope-labeling technologies. J. Biosci. Bioeng. 2010;110:87–93. doi: 10.1016/j.jbiosc.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 25.Kim S.W., Suda W., Kim S., et al. Robustness of gut microbiota of healthy adults in response to probiotic intervention revealed by high-throughput pyrosequencing. DNA Res. 2013;20:241–53. doi: 10.1093/dnares/dst006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Caporaso J.G., Kuczynski J., Stombaugh J., et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7:335–6. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Edgar R.C., Haas B.J., Clemente J.C., Quince C., Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Q., Garrity G.M., Tiedje J.M., Cole J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007;73:5261–7. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fukuda S., Toh H., Hase K., et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature. 2011;469:543–7. doi: 10.1038/nature09646. [DOI] [PubMed] [Google Scholar]
- 30.Tian C., Chikayama E., Tsuboi Y., et al. Top-down phenomics of Arabidopsis thaliana: metabolic profiling by one- and two-dimensional nuclear magnetic resonance spectroscopy and transcriptome analysis of albino mutants. J. Biol. Chem. 2007;282:18532–41. doi: 10.1074/jbc.M700549200. [DOI] [PubMed] [Google Scholar]
- 31.Fukuda S., Nakanishi Y., Chikayama E., Ohno H., Hino T., Kikuchi J. Evaluation and characterization of bacterial metabolic dynamics with a novel profiling technique, real-time metabolotyping. PloS ONE. 2009;4:e4893. doi: 10.1371/journal.pone.0004893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kikuchi J., Shinozaki K., Hirayama T. Stable isotope labeling of Arabidopsis thaliana for an NMR-based metabolomics approach. Plant Cell Physiol. 2004;45:1099–104. doi: 10.1093/pcp/pch117. [DOI] [PubMed] [Google Scholar]
- 33.Akiyama K., Chikayama E., Yuasa H., et al. PRIMe: a web site that assembles tools for metabolomics and transcriptomics. In Silico Biol. 2008;8:339–45. [PubMed] [Google Scholar]
- 34.Shannon P., Markiel A., Ozier O., et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith E.A., Macfarlane G.T. Formation of phenolic and indolic compounds by anaerobic bacteria in the human large intestine. Microb. Ecol. 1997;33:180–8. doi: 10.1007/s002489900020. [DOI] [PubMed] [Google Scholar]
- 36.Arumugam M., Raes J., Pelletier E., et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–80. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu G.D., Chen J., Hoffmann C., et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–8. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walsh M.C., Brennan L., Malthouse J.P., Roche H.M., Gibney M.J. Effect of acute dietary standardization on the urinary, plasma, and salivary metabolomic profiles of healthy humans. Am. J. Clin. Nutr. 2006;84:531–9. doi: 10.1093/ajcn/84.3.531. [DOI] [PubMed] [Google Scholar]
- 39.Kochhar S., Jacobs D.M., Ramadan Z., Berruex F., Fuerholz A., Fay L.B. Probing gender-specific metabolism differences in humans by nuclear magnetic resonance-based metabonomics. Anal. Biochem. 2006;352:274–81. doi: 10.1016/j.ab.2006.02.033. [DOI] [PubMed] [Google Scholar]
- 40.Bax A., Davis D.G. MLEV-17-based two-dimensional homonuclear magnetization transfer spectroscopy. J. Magn. Reson. 1985;65:355–60. [Google Scholar]
- 41.Saric J., Wang Y., Li J., et al. Species variation in the fecal metabolome gives insight into differential gastrointestinal function. J. Proteome Res. 2008;7:352–60. doi: 10.1021/pr070340k. [DOI] [PubMed] [Google Scholar]
- 42.Nicholson J.K., Foxall P.J., Spraul M., Farrant R.D., Lindon J.C. 750 MHz 1H and 1H-13C NMR spectroscopy of human blood plasma. Anal. Chem. 1995;67:793–811. doi: 10.1021/ac00101a004. [DOI] [PubMed] [Google Scholar]
- 43.Martin F.P., Sprenger N., Yap I.K., et al. Panorganismal gut microbiome-host metabolic crosstalk. J. Proteome Res. 2009;8:2090–105. doi: 10.1021/pr801068x. [DOI] [PubMed] [Google Scholar]
- 44.Chikayama E., Sekiyama Y., Okamoto M., et al. Statistical indices for simultaneous large-scale metabolite detections for a single NMR spectrum. Anal. Chem. 2010;82:1653–8. doi: 10.1021/ac9022023. [DOI] [PubMed] [Google Scholar]
- 45.Makelainen H., Forssten S., Saarinen M., Stowell J., Rautonen N., Ouwehand A.C. Xylo-oligosaccharides enhance the growth of bifidobacteria and Bifidobacterium lactis in a simulated colon model. Beneficial Microb. 2010;1:81–91. doi: 10.3920/BM2009.0025. [DOI] [PubMed] [Google Scholar]
- 46.Salminen S., Bouley C., Boutron-Ruault M.C., et al. Functional food science and gastrointestinal physiology and function. Br. J. Nutr. 1998;80(Suppl 1):S147–71. doi: 10.1079/bjn19980108. [DOI] [PubMed] [Google Scholar]
- 47.Duncan S.H., Louis P., Flint H.J. Lactate-utilizing bacteria, isolated from human feces, that produce butyrate as a major fermentation product. Appl. Environ. Microbiol. 2004;70:5810–7. doi: 10.1128/AEM.70.10.5810-5817.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Belenguer A., Duncan S.H., Calder A.G., et al. Two routes of metabolic cross-feeding between Bifidobacterium adolescentis and butyrate-producing anaerobes from the human gut. Appl. Environ. Microbiol. 2006;72:3593–9. doi: 10.1128/AEM.72.5.3593-3599.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Morrison D.J., Mackay W.G., Edwards C.A., Preston T., Dodson B., Weaver L.T. Butyrate production from oligofructose fermentation by the human faecal flora: what is the contribution of extracellular acetate and lactate? Br. J. Nutr. 2006;96:570–7. [PubMed] [Google Scholar]
- 50.De Preter V., Geboes K., Verbrugghe K., et al. The in vivo use of the stable isotope-labelled biomarkers lactose-[15N]ureide and [2H4]tyrosine to assess the effects of pro- and prebiotics on the intestinal flora of healthy human volunteers. Br. J. Nutr. 2004;92:439–46. doi: 10.1079/bjn20041228. [DOI] [PubMed] [Google Scholar]
- 51.Coton M., Fernandez M., Trip H., et al. Characterization of the tyramine-producing pathway in Sporolactobacillus sp. P3J. Microbiology. 2011;157(Pt 6):1841–9. doi: 10.1099/mic.0.046367-0. [DOI] [PubMed] [Google Scholar]
- 52.Gale E.F. Production of amines by bacteria: the decarboxylation of amino-acids by organisms of the groups Clostridium and Proteus with an addendum by G. L. Brown, F. C. MacIntosh and P. Bruce White. Biochem. J. 1941;35:66–80. doi: 10.1042/bj0350066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shi W., Meininger C.J., Haynes T.E., Hatakeyama K., Wu G. Regulation of tetrahydrobiopterin synthesis and bioavailability in endothelial cells. Cell Biochem. Biophys. 2004;41:415–34. doi: 10.1385/CBB:41:3:415. [DOI] [PubMed] [Google Scholar]
- 54.Chen C., Sander J.E., Dale N.M. The effect of dietary lysine deficiency on the immune response to Newcastle disease vaccination in chickens. Avian Dis. 2003;47:1346–51. doi: 10.1637/7008. [DOI] [PubMed] [Google Scholar]
- 55.Tezuka H., Abe Y., Iwata M., et al. Regulation of IgA production by naturally occurring TNF/iNOS-producing dendritic cells. Nature. 2007;448:929–33. doi: 10.1038/nature06033. [DOI] [PubMed] [Google Scholar]
- 56.David L.A., Maurice C.F., Carmody R.N., et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–63. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.