Abstract
The steroid hormone ecdysone directs the massive destruction of obsolete larval tissues during Drosophila metamorphosis, providing a model system for defining the molecular mechanisms of steroid-regulated programmed cell death. Although earlier studies have identified an ecdysone triggered genetic cascade that immediately precedes larval tissue cell death, no death regulatory genes have been functionally linked to this death response. We show here that ecdysone-induced expression of the death activator genes reaper (rpr) and head involution defective (hid) is required for destruction of the larval midgut and salivary glands during metamorphosis, with hid playing a primary role in the salivary glands and rpr and hid acting in a redundant manner in the midguts. We also identify the Drosophila inhibitor of apoptosis 1 as a survival factor in the larval cell death pathway, delaying death until its inhibitory effect is overcome by rpr and hid. This study reveals functional interactions between rpr and hid in Drosophila cell death responses and provides evidence that the precise timing of larval tissue cell death during metamorphosis is achieved through a steroid-triggered shift in the balance between the Drosophila inhibitor of apoptosis 1 and the rpr and hid death activators.
Keywords: steroid hormone, autophagy
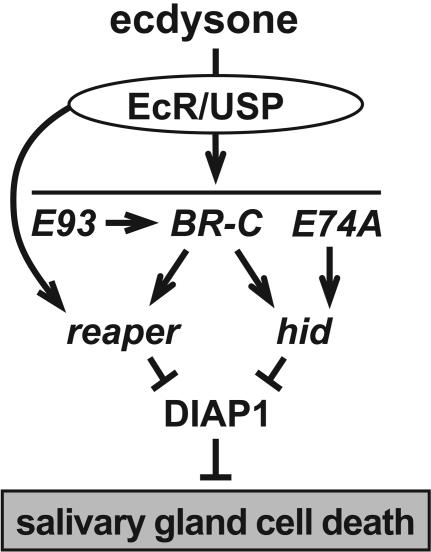
Small lipophilic hormones provide a critical signal for directing the appropriate patterns of programmed cell death during animal development, acting through members of the nuclear receptor family of ligand-regulated transcription factors. In frogs, thyroid hormone signals the destruction of the tadpole tail and remodeling of the intestine as the animal progresses from a juvenile to adult form (1–3). Similarly, steroid hormones regulate mammalian apoptotic pathways, including the glucocorticoid-induced apoptosis of immature thymocytes and mature T cells (4–6). In Drosophila, the steroid hormone ecdysone acts as a key signal to trigger the stage-specific destruction of obsolete larval tissues during metamorphosis. A high titer pulse of ecdysone at the end of larval development signals puparium formation and the destruction of the larval midgut as an adult gut forms around the dying cells (7, 8). A second ecdysone pulse, ≈10 h after puparium formation, triggers adult head eversion, marking the prepupal-to-pupal transition and signaling the rapid destruction of the larval salivary glands. Destruction of the larval midguts and salivary glands is accompanied by classic hallmarks of apoptosis, including acridine orange staining, DNA fragmentation, and caspase activation, although these larval tissues undergo a distinct form of programmed cell death referred to as autophagy, characterized by the formation of intracellular autophagic vesicles (9–11). Drosophila larval tissue cell death is foreshadowed by the coordinate transcriptional induction of two key death activator genes, reaper (rpr) and head involution defective (hid) (9). Ecdysone directly induces rpr transcription in doomed larval salivary glands (12). This effect is augmented by the ecdysone-induction of three transcription factor-encoding genes, BR-C (Broad-Complex), E74A, and E93, that, in turn, are required for appropriate rpr and hid expression and salivary gland cell death (12–14) (see Fig. 5). A similar steroid-triggered regulatory cascade is operative in doomed midgut cells, although E74 plays no apparent role in this pathway (15).
Fig. 5.
A model for the steroid regulation of salivary gland cell death. Ecdysone functions through the heterodimeric ecdysone receptor (EcR)/Ultraspiracle (USP) receptor to directly induce primary response transcription factors, E93, BR-C, and E74A, which are required for proper rpr and hid induction. Ecdysone can also regulate rpr directly through an ecdysone response element in its promoter. reaper and hid act together in a combinatorial manner to overcome the inhibitory effect of DIAP1, triggering salivary gland cell death.
Functions for rpr and hid are defined by Df(3L)H99, a chromosomal deficiency that removes these genes along with a third death activator gene, grim, resulting in a complete block in embryonic programmed cell death (16–18). The protein products of these genes bind to the Drosophila inhibitor of apoptosis 1 (DIAP1), releasing its complex with caspases and thereby initiating a proteolytic cascade that culminates in cell death (19–23). The death activators also down-regulate DIAP1 protein levels through at least two mechanisms involving a general suppression of protein synthesis (24, 25) and polyubiquitination and degradation (24–30). The mammalian death activators Smac/Diablo and Omi/HtrA2 appear to act in the same manner as rpr, hid, and grim, inhibiting the action of death repressors, such as survivin and X-linked IAP (inhibitor of apoptosis), demonstrating that this pathway has been conserved through evolution (31–36).
Although the timing of ecdysone-induced rpr and hid expression correlates with the onset of larval midgut and salivary gland cell death, no functional studies have linked any death regulatory genes to the destruction of larval tissues during metamorphosis. rpr expression appears to accurately foreshadow cell death at other stages of development and is sufficient to drive programmed cell death (16, 37); however, hid can be expressed in cells that are fated to survive, consistent with posttranslational mechanisms for modifying Hid activity (38, 39). Similarly, no effects are seen on nurse cell apoptosis in Df(3L)H99 germline clones despite the observation that rpr, hid, grim, and diap1 are expressed in doomed nurse cells during Drosophila oogenesis (40). Recent analysis of a rpr-null mutant revealed no effects on larval midgut or salivary gland cell death, although significant defects were observed in the central nervous system, raising the possibility that rpr plays no role in larval tissue destruction (41). A possible independent or combinatorial role for hid in these pathways has been difficult to ascertain because the available genetic tools, Df(3L)H99 and hid-null mutations, lead to early lethality (16, 18, 42). Moreover, the embryonic lethality of diap1 mutants has prevented an understanding of its possible roles during metamorphosis (19–21).
In this study, we exploit heat-inducible RNA interference (RNAi) as a means of disrupting gene function at later stages of development (43). We show that hid is required for larval salivary gland cell death and demonstrate that rpr and hid act together in a redundant manner to direct efficient destruction of the larval midguts and salivary glands. diap1 is required to block premature programmed cell death during larval development. In contrast, diap2, which is expressed immediately preceding salivary gland cell death, appears to play no independent role in these pathways (9, 44). This study provides a functional link between the genetic hierarchy that leads to rpr and hid induction and the destruction of larval tissues during Drosophila metamorphosis. This work also suggests that the integration and modulation of death activator and death repressor levels is vital for determining the proper timing of larval tissue cell death.
Materials and Methods
Supporting Information. Additional methods and details on the P element transformants for RNAi can be found as Supporting Materials and Methods and Fig. 6, which are published as supporting information on the PNAS web site.
Developmental Staging and Induction of Double-Stranded RNA (dsRNA) Expression. Third-instar larvae were staged by rearing on food containing 0.05% bromophenol blue (45). For larval midgut investigations, light-blue-gut, third-instar larvae were transferred to a 1.5-ml microcentrifuge tube plugged with cotton and subjected to a 30-min heat treatment in a 37.5°C water bath at 8 and 4 h before puparium formation; RNA was isolated from dissected larval midguts at 0 or 3 h after puparium formation (APF) for Northern blot hybridization. For salivary gland studies, control and hs-hid-RNAi-30 prepupae were transferred to a 1.5-ml microcentrifuge tube and subjected to a sequential 30-min heat treatment in a 37.5°C water bath at 4 and 8 h APF. Salivary glands were dissected at the onset of head eversion, or 2 h later, for Northern blot hybridization. To study effects on salivary gland cell death, prepupae were subjected to the same heat-shock regime and salivary glands were dissected at 8 h after head eversion or analyzed by GFP fluorescence (with 34B-GAL4 and UAS-GFP) 12 h after head aversion. For induction of diap1 or diap2 dsRNA expression, dark-blue-gut, mid-third-instar larvae were transferred to a 1.5-ml microcentrifuge tube plugged with cotton, subjected to a 30-min heat treatment at 37.5°C in a water bath, and allowed to recover for 2 or 4 h at 25°C before total RNA was isolated for Northern blot hybridization. An identical heat treatment followed by 6 h of recovery at 25°C was used to study the phenotypes resulting from diap1 or diap2 dsRNA expression. hs-diap2-RNAi-35 animals were also subjected to a 30-min 37.5°C heat treatment at either 3–5 h after egg lay or 36–48 h after egg lay to test for effects on viability.
Immunohistochemistry. Dark-blue-gut, w1118 control and hs-diap1-RNAi-11, third-instar larvae were subjected to a 30-min 37.5°C heat treatment. These animals were allowed to recover at 25°C for 6 h; they were then dissected and fixed with 4% formaldehyde, 1× PBST (PBS plus 0.1% Tween 20), and three volumes of heptane for 30 min at room temperature. Samples were washed four times in 1× PBST and blocked in 1× PBST/4% normal goat serum (NGS) for 2 h at room temperature. The samples were stained with antibodies directed against either DIAP1 (a generous gift from B. Hay, California Institute of Technology, Pasadena) at 1:400 dilution or against active caspase-3 (Cell Signaling Technology, Beverly, MA) at 1:1,000 in 1× PBST/4% NGS overnight at 4°C. Samples were washed four times in 1× PBST and stained with either Cy3 donkey anti-mouse secondary antibody (The Jackson Laboratory) for DIAP1 or Cy3 donkey anti-rabbit secondary antibody (The Jackson Laboratory) for active caspase-3, at 1:400 in 1× PBST/4% NGS for 2 h at room temperature. Samples were mounted in VECTAShield (Vector Laboratories) and images were captured as a Z-series by using a Bio-Rad 1024 confocal laser scanning microscope.
Results
hid Function Is Required for Efficient Larval Salivary Gland Cell Death. Transgenic flies were generated that carry the heat-inducible hsp70 (heat-shock protein 70) promoter upstream from a tandem inverted repeat of hid coding sequences. Several independent lines were established, one of which, hs-hid-RNAi-30, was used for studies of hid function. As an initial test of the efficiency of RNAi, we examined the levels of endogenous hid mRNA and cuticle preparations from embryos expressing hid dsRNA (Fig. 6). Whereas hid mRNA is readily detectable in heat-treated control embryos, this level is significantly reduced upon expression of hid dsRNA (Fig. 6). Consistent with this reduced level of hid expression, we see head involution defects in these embryos that phenocopy a hid-null mutant (42). The penetrance of head defects in the null mutant (70%, n = 154), however, is higher than that seen with RNAi (45%, n = 96). These observations indicate that the expression of hid dsRNA results in a strong loss-of-function hid mutant phenotype.
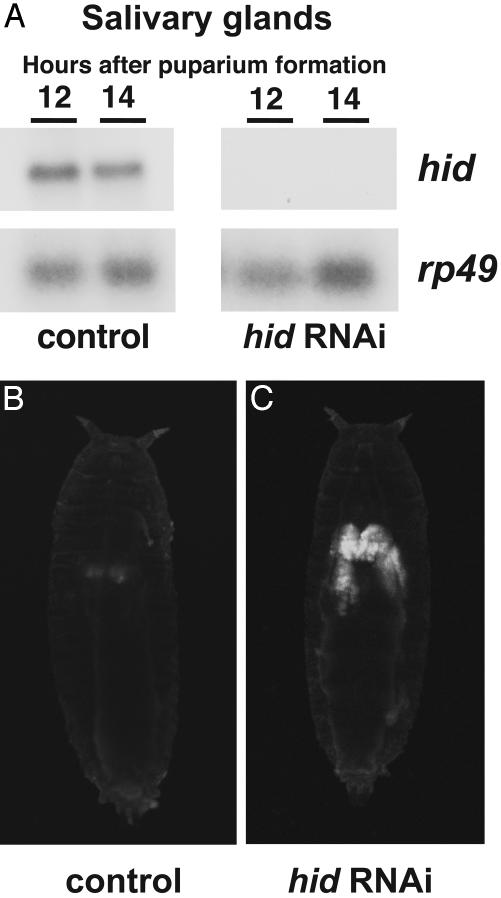
To determine a possible role for hid in larval salivary gland cell death, control w1118 and hs-hid-RNAi-30 transformants were subjected to sequential 30-min heat treatments at 4 and 8 h APF. RNA was isolated from salivary glands of animals either at head eversion or 2 h later, ≈12 or 14 h APF, and analyzed by Northern blot hybridization. As expected, the levels of endogenous hid mRNA are greatly reduced upon expression of hid dsRNA (Fig. 1A). To determine the functional consequences of this reduction in hid expression, we scored for persistent larval salivary glands by either dissection or direct visualization of salivary glands by using the salivary gland-specific 34B-GAL4 driver in combination with a UAS-GFP reporter. Only residual GFP is detected in control 34B-GAL4, UAS-GFP pupae at 24 h APF, consistent with the normal completion of salivary gland cell death (Fig. 1B), whereas persistent larval salivary glands are present in pupae that express hid dsRNA (Fig. 1C). Dissection of heat-treated hs-hid-RNAi-30 pupae at 8 h after head eversion (≈20 h APF) revealed that 27% (n = 37) had persistent glands, compared with 0% (n = 31) in controls. A similar result was observed by scoring for GFP expression (25%, n = 28). We conclude that a severe reduction in hid expression results in a partial block in larval salivary gland cell death.
Fig. 1.
Expression of hid dsRNA results in persistent larval salivary glands. (A) Control w1118 and hs-hid-RNAi-30 prepupae were subjected to two sequential heat treatments at 4 and 8 h APF. Animals were allowed to recover until head eversion, ≈12 and 14 h APF, after which RNA was extracted from salivary glands for Northern blot hybridization. Hybridization to detect rp49 was used as a control for loading and transfer. (B and C) Control w; 34B-GAL4/+; UAS-GFP/+ (B) and w; 34B-GAL4/+; UAS-GFP/hs-hid-RNAi-30 (C) prepupae were subjected to the same heat-treatment regimen and scored at 24 h APF for GFP expression in the larval salivary glands. Although programmed cell death of control salivary glands is complete at this stage with only minimal residual fluorescence (B), salivary glands remain intact after expression of hid dsRNA (C).
Consistent with an earlier study (41), Df(3L)XR38/Df(3L)H99 mutant animals that lack rpr but have normal hid function display essentially normal salivary gland cell death, with only 4.2% (n = 24) displaying persistent larval salivary glands at 20 h APF (8 h after head eversion). Expression of hid dsRNA in this genetic background, however, increased this number to 65% (n = 23), significantly higher than that seen with hid RNAi alone. This result cannot be attributed to the reduced dose of hid in the Df(3L)XR38/Df(3L)H99 mutant background, because heat-treated Df(3L)H99, hs-hid-RNAi-30/+ pupae displayed a 24% frequency of persistent salivary glands at 20 h APF (n = 17), similar to the result obtained with hid RNAi alone. Taken together, these observations indicate that rpr and hid act together to drive efficient larval salivary gland cell death during the onset of metamorphosis.
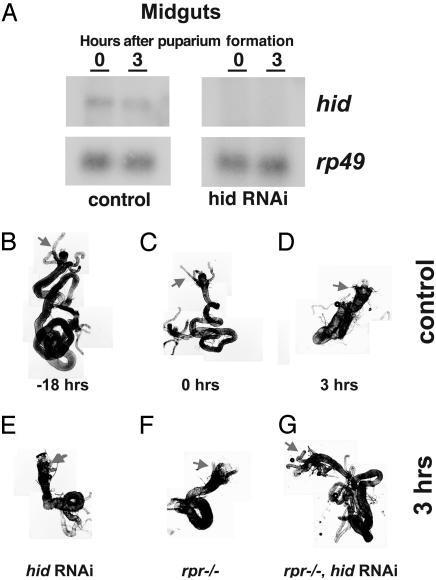
rpr and hid Function in a Redundant Manner to Activate Cell Death in Larval Midguts. To reduce the level of hid expression in larval midguts, control w1118 and hs-hid-RNAi-30 transformants were subjected to two sequential 30-min heat treatments at 8 and 4 h before puparium formation. RNA was isolated from larval midguts at 0 and 3 h APF, the times when hid is normally induced in this tissue (9). Northern blot hybridization confirmed that the levels of endogenous hid mRNA are significantly reduced under these conditions (Fig. 2A). Dissection of these animals at 3 h APF revealed that neither reducing hid function with hs-hid-RNAi-30 (4.4%, n = 45) nor the removal of rpr function with the Df(3L)XR38/Df(3L)H99 chromosomal deficiencies (0%, n = 20) had a significant effect on larval midgut cell death (Fig. 2, compare E and F with D). In contrast, 80% (n = 20) of heat-treated Df(3L)XR38/Df(3L)H99, hs-hid-RNAi-30 animals, which lack rpr activity and have reduced hid function, display persistent larval midguts (Fig. 2G). These larval midguts have failed to contract by 3 h APF and retain their gastric caeca (Fig. 2G, arrow), similar to the midguts of control mid-third-instar larvae (Fig. 2B). Midgut contraction normally begins at puparium formation (Fig. 2C), coinciding with the onset of cell death, and is complete by 3 h APF, when the gastric caeca are no longer present and the midgut is highly contracted (Fig. 2D) (9). These observations indicate that rpr and hid are functionally redundant in this tissue, acting in a cooperative manner to direct larval midgut cell death.
Fig. 2.
Expression of hid dsRNA in a rpr-null mutant background leads to persistent larval midguts. (A) Control w1118 and hs-hid-RNAi-30 third-instar larvae were subjected to two sequential heat treatments at 8 and 4 h before puparium formation, and RNA was isolated from larval midguts dissected at either 0 or 3 h APF for Northern blot hybridization. Hybridization to detect rp49 was used as a control for loading and transfer (B–G). Larval midguts were dissected from a control w1118 mid-third-instar larva (B), newly formed prepupa (C), or 3 h APF (D) to depict the normal time course of cell death. Shortened gastric caeca (arrows) are evident at puparium formation, along with an overall contraction in the length of the midgut (C). This contraction is complete by 3 h APF, and gastric caeca are no longer present (D). Neither expression of hid dsRNA (E) nor a rpr-null mutation (F) has an effect on larval midgut cell death when examined at 3 h APF, whereas expression of hid dsRNA in a rpr-null mutant background leads to an efficient block in the destruction of this tissue (G).
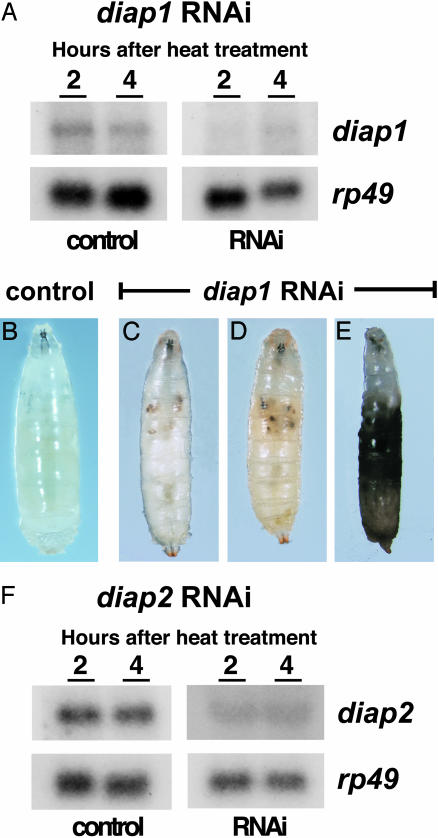
diap1 Is Required for Third-Instar Larval Viability. diap1 is highly expressed in late third-instar larvae and significantly reduced at 6 h APF, with low constant levels of expression in both the larval midgut and salivary glands throughout the onset of metamorphosis (data not shown). To address possible roles for this death repressor in preventing premature larval tissue cell death, we reduced diap1 expression during larval development by inducible RNAi. Transgenic flies were established that carry the hsp70 promoter upstream from a tandem inverted repeat of diap1 coding sequences; one line, hs-diap1-RNAi-11, was selected for studies of diap1 function. Heat-induced diap1 dsRNA expression in mid-third-instar larvae (≈18 h before puparium formation) led to a significant reduction in the levels of endogenous diap1 transcripts (Fig. 3A). Some larvae stopped moving as early as 2 h after heat treatment, with necrotic patches often appearing in their larval tissues. By 6 h, most larvae exhibited visible signs of death (76%, n = 100), from relatively restricted death of larval tissues (Fig. 3 C and D) to widespread necrosis (Fig. 3E), compared with heat-treated, control third-instar larvae (Fig. 3B). Although the remaining animals formed morphologically normal prepupae, they all underwent massive death before head eversion, leaving only an empty pupal case.
Fig. 3.
Expression of diap1 dsRNA leads to larval lethality. (A) Control w1118 and hs-diap1-RNAi-11 mid-third-instar larvae were heat treated, and total RNA was isolated 2 or 4 h later for Northern blot analysis. Six hours after heat treatment, control larvae (B) exhibited no signs of premature cell death, whereas larvae expressing diap1 dsRNA display patches of death in larval tissues (C and D) or overall necrosis (E). Control and hs-diap2-RNAi-35 mid-third-instar larvae were subjected to the same heat-treatment regimen as hs-diap1-RNAi-11 animals, and total RNA was isolated 2 or 4 h later for Northern blot analysis. Expression of diap2 dsRNA leads to reduced levels of endogenous diap2 mRNA (F).
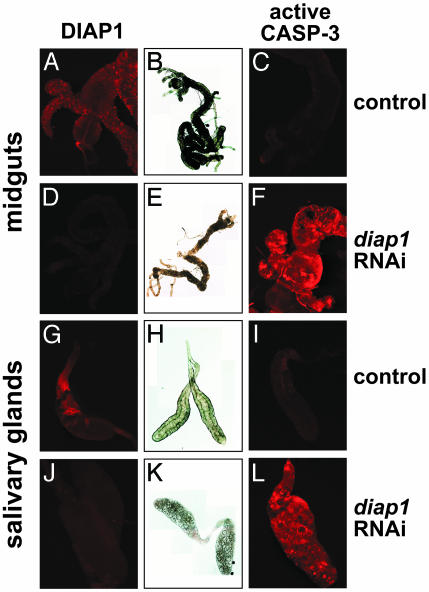
diap1 Is Required to Suppress Premature Destruction of Larval Tissues. To determine the consequences of reducing diap1 function on larval tissue cell death, hs-diap1-RNAi-11 third-instar larvae were heat-treated at ≈18 h before puparium formation and dissected 6 h later. As expected, the levels of DIAP1 protein are significantly reduced under these conditions (Fig. 4, compare D and J with A and G). This observation is consistent with the reduced levels of diap1 mRNA under these conditions (Fig. 3A) and the short half-life of DIAP1 protein (24). The larval midguts and salivary glands of heat-treated hs-diap1-RNAi-11 larvae resemble those undergoing cell death. The larval midgut is contracted with a significantly reduced proventriculus and gastric caeca and an overall degraded morphology (Fig. 4E), similar to that normally seen 2 h APF (9) and contrasting dramatically with the healthy morphology of heat-treated control larval midguts (Fig. 4B). Similarly, the larval salivary glands of heat treated hs-diap1-RNAi-11 larvae are severely degraded (Fig. 4K), resembling the morphology of wild-type glands dissected from 14.5-h pupae (9). In contrast, salivary glands dissected from heat-treated control larvae display a tight cellular structure and apparent lumen in the center of the gland (Fig. 4H). Consistent with these morphological indications of premature cell death, those tissues that lack diap1 function are selectively stained with an antibody directed against active caspase-3, which detects the Drosophila DrICE effector caspase, indicating they are undergoing cell death (Fig. 4, compare F and L with C and I) (46).
Fig. 4.
Expression of diap1 dsRNA leads to premature larval midgut and salivary gland cell death. Control w1118 and hs-diap1-RNAi-11 mid-third-instar larvae were subjected to heat treatment and allowed to recover for 6 h. Larval midguts and salivary glands were then dissected from these animals and examined by light microscopy (B, E, H, and K), immunostaining to detect DIAP1 (A, D, G, and J), or immunostaining to detect active caspase-3 (C, F, I, and L). Physical signs of cell death (E and K) and caspase activation (F and L) are only seen when DIAP1 levels are reduced by RNAi (D and J).
diap2 Has No Apparent Independent Role in the Regulation of Cell Death. Previous studies have shown that a brief burst of diap2 transcription precedes rpr and hid induction in doomed larval salivary glands (9), with expression that parallels that of diap1 throughout late larval and prepupal stages (data not shown). To determine whether diap2 contributes to the regulation of larval tissue cell death, we used inducible RNAi to inhibit diap2 function during the onset of metamorphosis. Transgenic flies were established that carry the hsp70 promoter upstream from a tandem inverted repeat of diap2 coding sequences, and one line, hs-diap2-RNAi-35, was selected for studies of diap2 function. Heat-induced diap2 dsRNA expression in mid-third-instar larvae led to a significant reduction in the levels of endogenous diap2 transcripts (Fig. 3F). To test for a role for diap2 in preventing premature larval salivary gland cell death, we heat-treated hs-diap2-RNAi-35 animals at 4 and 8 h APF and dissected salivary glands at 10 and 12 h APF. All salivary glands examined died on time (n = 15 animals), and all hs-diap2-RNAi-35 animals exposed to this sequential heat treatment survived to adulthood (n = 50). No effects on viability were seen in heat-treated hs-diap2-RNAi-35 embryos or first- and second-instar larvae (data not shown). In addition, a heat-shock regime identical to that used to inactivate diap1 during third-instar larval development (Fig. 3C–E) resulted in no effects on hs-diap2-RNAi-35 animals. These observations suggest that diap2 has no significant independent role in the regulation of Drosophila programmed cell death.
Discussion
rpr and hid Cooperate to Destroy Larval Tissues During Metamorphosis. Our understanding of the functional interactions between rpr, hid, and grim has been hampered by a paucity of genetic tools in this region. Extensive screens for lethal mutations within the Df(3L)H99 interval resulted in the recovery of only hid alleles, suggesting that the three death activators might functionally compensate for one another and indicating that more directed efforts would be required to recover rpr and grim mutations (16, 18, 42). Reversion of a P element 225 kb proximal to rpr resulted in the Df(3L)XR38 deficiency. This deletion, in combination with Df(3L)H99, leads to the selective elimination of rpr, although it remains a possibility that other unidentified modifiers of the death response could be affected by these deletions (41). Interestingly, these rpr-null mutants develop into morphologically normal adults with a highly enlarged central nervous system, indicating a relatively restricted requirement for rpr during development (41). Characterization of hid mutants and Df(3L)X25 mutants, which lack grim and hid, have implied functional interactions between the death activators for embryonic head morphogenesis and programmed cell death of neurons that express crustacean cardioactive peptide (47, 48). This model is further supported by a gain-of-function study showing that targeted expression of both rpr and hid is required for the death of midline cells in the embryonic nervous system, suggesting that these genes act in a cooperative and dose-dependent manner in this cell type (49). A loss-of-function analysis to support this model, however, has not been possible because of the difficulty of recombining hid alleles onto the Df(3L)XR38 rpr mutant chromosome. Here, we used inducible RNAi to selectively inactivate hid function, both in the presence and absence of rpr, and showed that these genes act together in a combinatorial manner to drive the death of larval tissues during metamorphosis. This study provides the first direct genetic evidence that rpr and hid are functionally redundant for programmed cell death; it also supports the results of Peterson et al. (41) regarding the absence of larval tissue cell death in rpr mutants and indicates that this response requires the simultaneous induction of hid. The cooperativity and dose-dependency of death activator function is consistent with the ability of either rpr or hid to drive programmed cell death in the adult eye (16–18, 44) and their interchangeable role in inactivating the diap1 death inhibitor both in vivo and in vitro (19–21).
We find that 27% of hid RNAi animals display persistent larval salivary glands, in contrast to the almost complete absence of this phenotype in rpr-null mutants. This observation supports the earlier proposal that, at least in some cells, hid is a more potent activator of programmed cell death than rpr (18, 41). Our results are also consistent with the observation that E74A mutant salivary glands, which display an almost complete absence of hid transcription but normal levels of rpr, fail to die in ≈20% of the pupae examined 24 h APF (12). Similarly, BR-C mutant salivary glands fail to express both rpr and hid and display highly penetrant persistent larval salivary glands (12, 50, 51).
In contrast to our results in larval salivary glands, larval midgut cell death occurs essentially normally in the presence of hid dsRNA. Both hid RNAi and the rpr-null mutation are required to see penetrant death defects in this tissue, indicative of complete functional redundancy. This observation supports the proposal that the ecdysone-induced death program in the larval midgut has differences from that activated by the hormone in doomed salivary glands (15).
diap1 Acts as a Survival Factor to Prevent Premature Cell Death During Postembryonic Development. With the exception of Drosophila adult eye development, functions for IAP death inhibitors at later stages of development have been undefined. Our studies indicate an essential role for diap1 in holding back the death response during postembryonic development, restricting it to the appropriate space and time. We propose that the reduced levels of diap1 expression seen in larval tissues during the early stages of metamorphosis prime the system for their destruction. The appropriate spatial and temporal patterns of ecdysone-induced rpr and hid expression then tip the balance in these cells, signaling their elimination.
It is interesting to note that visible necrotic patches appear rapidly in third-instar larvae that express diap1 dsRNA (Fig. 3C–E). The distinction between apoptosis and necrosis was established in early studies of dying cells, based on morphology, arguing that the two pathways are distinct and involve different regulatory factors (52). More recent studies, however, have uncovered common features shared by these death responses (53, 54). For example, studies of neutrophil cell death showed that X-linked IAP is a target of calpain, a key component of the necrotic pathway (55), and that calpain inhibitors not only protect against necrosis in some neuronal cells but also inhibit apoptosis (56). Future studies should provide a better understanding of how IAPs may function at the intersection between necrotic and programmed cell death responses.
Earlier work showed that diap2 is expressed in a brief burst, immediately before the induction of rpr and hid in doomed larval salivary glands, suggesting that it may contribute to the regulation of cell death in this tissue (9). The studies of diap2 RNAi presented here, however, do not support this model. This conclusion is consistent with a complementary study, in which premature ectopic overexpression of diap2 was shown to have no effect on larval salivary gland cell death (10). Thus, although diap2 was originally identified by its ability to block rpr- or hid-induced programmed cell death in the adult eye, its ability to exert this function must be cell-type specific (44). Rather, we propose that diap1 acts as the primary inhibitor of cell death in larval salivary glands. It is possible that diap2 could contribute to biological pathways other than programmed cell death. For example, RNAi studies of survivin function have uncovered a role in cell cycle regulation (57). The characterization of specific diap2 mutations should provide a better understanding of its functions during Drosophila development.
A Steroid-Triggered Regulatory Cascade Directs the Stage-Specific Destruction of Larval Tissues During Metamorphosis. The steroid-triggered destruction of larval tissues during Drosophila metamorphosis provides one of the best systems for understanding the temporal and spatial control of programmed cell death. Several transcription factors that are induced directly by ecdysone have been linked to the regulation of key death genes in these tissues, defining a genetic pathway for the control of programmed cell death (12, 14, 15). This cascade has been most extensively characterized in the larval salivary gland, in which numerous death regulators are induced just before the destruction of this tissue (58, 59). What has been missing from these studies, however, is a functional link between the death regulators and the destruction of larval tissues during metamorphosis. Indeed, the recent report that rpr-null mutants have no effect on larval midgut and salivary gland cell death raised the possibility that an alternate pathway might be operative, as appears to be the case in nurse cell apoptosis (40, 41). The results presented here complete the pathway, showing that ecdysone-induced rpr and hid expression, mediated by EcR (ecdysone receptor), E93, BR-C, and E74A, is required for the stage-specific destruction of the larval midgut and salivary glands (Fig. 5). Moreover, this study shows that diap1 is required to hold back the death response, adding a new critical player to the pathway. Further studies of Drosophila metamorphosis should provide a better understanding of the regulation of programmed cell death during development as well as a model system for understanding how steroid hormones control apoptotic responses in vertebrate organisms.
Supplementary Material
Acknowledgments
We thank B. Hay for the DIAP1 antibodies, the Bloomington Stock Center for stocks used in this study, K. White for providing the hid and rpr mutants, A. Bashirullah for critical reading of this manuscript, and S. King-Jones for help with image processing. This study was supported by National Institutes of Health Grant R01 GM60954 (to C.S.T.). C.S.T. is an Investigator with the Howard Hughes Medical Institute.
Abbreviations: rpr, reaper; hid, head involution defective; dsRNA, double-stranded RNA; RNAi, RNA interference; APF, after puparium formation; PBST, PBS plus 0.1% Tween 20; BR-C, Broad-Complex; DIAP1, Drosophila inhibitor of apoptosis 1.
References
- 1.Yoshizato, K. (1996) in Cell Death and Histolysis in Amphibian Tail Metamorphosis, eds. Gilbert, L. I., Tata, J. R. & Atkinson, B. G. (Academic, New York), pp. 647–671.
- 2.Tata, J. R. (1966) Dev. Biol. 13, 77–94. [DOI] [PubMed] [Google Scholar]
- 3.Shi, Y. B., Wong, J., Puzianowska-Kuznicka, M. & Stolow, M. A. (1996) BioEssays 18, 391–399. [DOI] [PubMed] [Google Scholar]
- 4.Evans-Storms, R. B. & Cidlowski, J. A. (1995) J. Steroid Biochem. Mol. Biol. 53, 1–8. [DOI] [PubMed] [Google Scholar]
- 5.Winoto, A. & Littman, D. R. (2002). Cell 109, Suppl., S57–S66. [DOI] [PubMed] [Google Scholar]
- 6.Smith, S. W., McLaughlin, K. A. & Osborne, B. A. (1995) Curr. Top Microbiol. Immunol. 200, 147–162. [DOI] [PubMed] [Google Scholar]
- 7.Robertson, C. W. (1936) J. Morphol. 59, 351–399. [Google Scholar]
- 8.Bodenstein, D. (1965) in The Postembryonic Development of Drosophila, ed. Demerec, M. (Hafner, New York), pp. 275–367.
- 9.Jiang, C., Baehrecke, E. H. & Thummel, C. S. (1997) Development (London) 124, 4673–4683. [DOI] [PubMed] [Google Scholar]
- 10.Lee, C. Y. & Baehrecke, E. H. (2001) Development (London) 128, 1443–1455. [DOI] [PubMed] [Google Scholar]
- 11.Martin, D. N. & Baehrecke, E. H. (2004) Development (London) 131, 275–284. [DOI] [PubMed] [Google Scholar]
- 12.Jiang, C., Lamblin, A.-F., Steller, H. & Thummel, C. S. (2000) Mol. Cell 5, 445–455. [DOI] [PubMed] [Google Scholar]
- 13.Lee, C. Y., Wendel, D. P., Reid, P., Lam, G., Thummel, C. S. & Baehrecke, E. H. (2000) Mol. Cell 6, 433–443. [DOI] [PubMed] [Google Scholar]
- 14.Lee, C. Y., Simon, C. R., Woodard, C. T. & Baehrecke, E. H. (2002) Dev. Biol. 252, 138–148. [DOI] [PubMed] [Google Scholar]
- 15.Lee, C. Y., Cooksey, B. A. & Baehrecke, E. H. (2002) Dev. Biol. 250, 101–111. [DOI] [PubMed] [Google Scholar]
- 16.White, K., Grether, M. E., Abrams, J. M., Young, L., Farrell, K. & Steller, H. (1994) Science 264, 677–683. [DOI] [PubMed] [Google Scholar]
- 17.Chen, P., Nordstrom, W., Gish, B. & Abrams, J. M. (1996) Genes Dev. 10, 1773–1782. [DOI] [PubMed] [Google Scholar]
- 18.Grether, M. E., Abrams, J. M., Agapite, J., White, K. & Steller, H. (1995) Genes Dev. 9, 1694–1708. [DOI] [PubMed] [Google Scholar]
- 19.Lisi, S., Mazzon, I. & White, K. (2000) Genetics 154, 669–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goyal, L., McCall, K., Agapite, J., Hartwieg, E. & Steller, H. (2000) EMBO J. 19, 589–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang, S. L., Hawkins, C. J., Yoo, S. J., Muller, H. A. & Hay, B. A. (1999) Cell 98, 453–463. [DOI] [PubMed] [Google Scholar]
- 22.Vucic, D., Kaiser, W. J. & Miller, L. K. (1998) Mol. Cell Biol. 18, 3300–3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vucic, D., Kaiser, W. J., Harvey, A. J. & Miller, L. K. (1997) Proc. Natl. Acad. Sci. USA 94, 10183–10188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoo, S. J., Huh, J. R., Muro, I., Yu, H., Wang, L., Wang, S. L., Feldman, R. M., Clem, R. J., Muller, H. A. & Hay, B. A. (2002) Nat. Cell Biol. 4, 416–424. [DOI] [PubMed] [Google Scholar]
- 25.Holley, C. L., Olson, M. R., Colon-Ramos, D. A. & Kornbluth, S. (2002) Nat. Cell Biol. 4, 439–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hays, R., Wickline, L. & Cagan, R. (2002) Nat. Cell Biol. 4, 425–431. [DOI] [PubMed] [Google Scholar]
- 27.Ditzel, M., Wilson, R., Tenev, T., Zachariou, A., Paul, A., Deas, E. & Meier, P. (2003) Nat. Cell Biol. 5, 467–473. [DOI] [PubMed] [Google Scholar]
- 28.Ryoo, H. D., Bergmann, A., Gonen, H., Ciechanover, A. & Steller, H. (2002) Nat. Cell Biol. 4, 432–438. [DOI] [PubMed] [Google Scholar]
- 29.Wilson, R., Goyal, L., Ditzel, M., Zachariou, A., Baker, D. A., Agapite, J., Steller, H. & Meier, P. (2002) Nat. Cell Biol. 4, 445–450. [DOI] [PubMed] [Google Scholar]
- 30.Wing, J. P., Schreader, B. A., Yokokura, T., Wang, Y., Andrews, P. S., Huseinovic, N., Dong, C. K., Ogdahl, J. L., Schwartz, L. M., White, K. & Nambu, J. R. (2002) Nat. Cell Biol. 4, 451–456. [DOI] [PubMed] [Google Scholar]
- 31.Goyal, L. (2001) Cell 104, 805–808. [DOI] [PubMed] [Google Scholar]
- 32.Martin, S. J. (2002) Cell 109, 793–796. [DOI] [PubMed] [Google Scholar]
- 33.Liu, Z., Sun, C., Olejniczak, E. T., Meadows, R. P., Betz, S. F., Oost, T., Herrmann, J., Wu, J. C. & Fesik, S. W. (2000) Nature 408, 1004–1008. [DOI] [PubMed] [Google Scholar]
- 34.Suzuki, Y., Imai, Y., Nakayama, H., Takahashi, K., Takio, K. & Takahashi, R. (2001) Mol. Cell 8, 613–621. [DOI] [PubMed] [Google Scholar]
- 35.Wu, G., Chai, J., Suber, T. L., Wu, J. W., Du, C., Wang, X. & Shi, Y. (2000) Nature 408, 1008–1012. [DOI] [PubMed] [Google Scholar]
- 36.Wu, J. W., Cocina, A. E., Chai, J., Hay, B. A. & Shi, Y. (2001) Mol. Cell 8, 95–104. [DOI] [PubMed] [Google Scholar]
- 37.White, K., Tahaoglu, E. & Steller, H. (1996) Science 271, 805–807. [DOI] [PubMed] [Google Scholar]
- 38.Bergmann, A., Agapite, J., McCall, K. & Steller, H. (1998) Cell 95, 331–341. [DOI] [PubMed] [Google Scholar]
- 39.Kurada, P. & White, K. (1998) Cell 95, 319–329. [DOI] [PubMed] [Google Scholar]
- 40.Foley, K. & Cooley, L. (1998) Development (London) 125, 1075–1082. [DOI] [PubMed] [Google Scholar]
- 41.Peterson, C., Carney, G. E., Taylor, B. J. & White, K. (2002) Development (London) 129, 1467–1476. [DOI] [PubMed] [Google Scholar]
- 42.Abbott, M. K. & Lengyel, J. A. (1991) Genetics 129, 783–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lam, G. & Thummel, C. S. (2000) Curr. Biol. 10, 957–963. [DOI] [PubMed] [Google Scholar]
- 44.Hay, B. A., Wassarman, D. A. & Rubin, G. M. (1995) Cell 83, 1253–1262. [DOI] [PubMed] [Google Scholar]
- 45.Andres, A. J. & Thummel, C. S. (1994) in Methods for Quantitative Analysis of Transcription in Larvae and Prepupae, eds. Goldstein, L. S. B. & Fyrberg, E. A. (Academic, New York), pp. 565–573. [DOI] [PubMed]
- 46.Yu, S. Y., Yoo, S. J., Yang, L., Zapata, C., Srinivasan, A., Hay, B. A. & Baker, N. E. (2002) Development (London) 129, 3269–3278. [DOI] [PubMed] [Google Scholar]
- 47.Nassif, C., Daniel, A., Lengyel, J. A. & Hartenstein, V. (1998) Dev. Biol. 197, 170–186. [DOI] [PubMed] [Google Scholar]
- 48.Draizen, T. A., Ewer, J. & Robinow, S. (1999) J. Neurobiol. 38, 455–465. [DOI] [PubMed] [Google Scholar]
- 49.Zhou, L., Schnitzler, A., Agapite, J., Schwartz, L. M., Steller, H. & Nambu, J. R. (1997) Proc. Natl. Acad. Sci. USA 94, 5131–5136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Restifo, L. L. & White, K. (1992) Roux's Arch. Dev. Biol. 201, 221–234. [DOI] [PubMed] [Google Scholar]
- 51.Zhimulev, I. F., Belyaeva, E. S., Mazina, O. M. & Balasov, M. L. (1995) Eur. J. Entomol. 92, 263–270. [Google Scholar]
- 52.Kerr, J. F., Wyllie, A. H. & Currie, A. R. (1972) Brit. J. Cancer 26, 239–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Proskuryakov, S. Y., Konoplyannikov, A. G. & Gabai, V. L. (2003) Exp. Cell Res. 283, 1–16. [DOI] [PubMed] [Google Scholar]
- 54.Syntichaki, P. & Tavernarakis, N. (2002) EMBO Rep. 3, 604–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kobayashi, S., Yamashita, K., Takeoka, T., Ohtsuki, T., Suzuki, Y., Takahashi, R., Yamamoto, K., Kaufmann, S. H., Uchiyama, T., Sasada, M. & Takahashi, A. (2002) J. Biol. Chem. 277, 33968–33977. [DOI] [PubMed] [Google Scholar]
- 56.Wang, K. K. (2000) Trends Neurosci. 23, 20–26. [DOI] [PubMed] [Google Scholar]
- 57.Li, F., Ackermann, E. J., Bennett, C. F., Rothermel, A. L., Plescia, J., Tognin, S., Villa, A., Marchisio, P. C. & Altieri, D. C. (1999) Nat. Cell Biol. 1, 461–466. [DOI] [PubMed] [Google Scholar]
- 58.Gorski, S. M., Chittaranjan, S., Pleasance, E. D., Freeman, J. D., Anderson, C. L., Varhol, R. J., Coughlin, S. M., Zuyderduyn, S. D., Jones, S. J. & Marra, M. A. (2003) Curr. Biol. 13, 358–363. [DOI] [PubMed] [Google Scholar]
- 59.Lee, C. Y., Clough, E. A., Yellon, P., Teslovich, T. M., Stephan, D. A. & Baehrecke, E. H. (2003) Curr. Biol. 13, 350–357. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.