Abstract
Background and Aims
The putative FASCICLIN-LIKE ARABINOGALACTAN PROTEIN 4 (At-FLA4) locus of Arabidopsis thaliana has previously been shown to be required for the normal growth of wild-type roots in response to moderately elevated salinity. However, the genetic and physiological pathway that connects At-FLA4 and normal root growth remains to be elucidated.
Methods
The radial swelling phenotype of At-fla4 was modulated with growth regulators and their inhibitors. The relationship of At-FLA4 to abscisic acid (ABA) signalling was analysed by probing marker gene expression and the observation of the At-fla4 phenotype in combination with ABA signalling mutants.
Key Results
Application of ABA suppresses the non-redundant role of At-FLA4 in the salt response. At-FLA4 positively regulates the response to low ABA concentration in roots and is required for the normal expression of ABA- and abiotic stress-induced genes. The At-fla4 phenotype is enhanced in the At-abi4 background, while two genetic suppressors of ABA-induced gene expression are required for salt oversensitivity of At-fla4. Salt oversensitivity in At-fla4 is suppressed by the CYP707A inhibitor abscinazole E2B, and salt oversensitivity in At-fla4 roots is phenocopied by chemical inhibition of ABA biosynthesis.
Conclusions
The predicted lipid-anchored glycoprotein At-FLA4 positively regulates cell wall biosynthesis and root growth by modulating ABA signalling.
Keywords: Fasciclin, arabinogalactan protein, root growth, cell wall, abscisic acid, plant cell wall signalling, Arabidopsis thaliana, At-FLA4
INTRODUCTION
Among the biopolymers of the plant cell wall, the vast group of hydroxyproline-rich glycoproteins (Showalter et al., 2010) comprises lightly glycosylated proline-rich proteins, moderately glycosylated extensins and highly glycosylated arabinogalactan-proteins (AGPs). The latter, structurally highly diverse family of glycoproteins has been implicated to have a wide range of biological roles; however, there is still an overall lack of understanding of the biophysical and biochemical mode of action of any individual AGP (Seifert and Roberts, 2007; Ellis et al., 2010; Tan et al., 2012).
A sub-group of AGPs, specified by the presence of one or two copies of a fasciclin domain (Fas1), have been termed fasciclin-like AGPs or FLAs (Johnson et al., 2003). The Fas1 domain was named after glycoproteins found in the axonal fascicles of grasshoppers (Bastiani et al., 1987); however, Fas1-containing proteins occur across all phyla (Moody and Williamson, 2013). Fas1 proteins mediate the adhesion between cells and their matrix, e.g. in bacterial biofilms (Moody and Williamson, 2013), or in animal cells, where Fas1 proteins such as periostin and transforming growth factor-β-induced matrix protein (βIG-h3) interact with extracellular matrix receptors of the integrin type (Gillan et al., 2002; Kim et al., 2002). Like many other Fas1-containing proteins, most FLAs localize to the outer leaflet of the plasma membrane via a lipid moiety – specifically a glycosylphosphatidylinositol (GPI) anchor – and associate with putative membrane nanodomains also known as lipid rafts (Borner et al., 2003). The biological roles of FLAs in plant growth and development follow the theme of an involvement in cell wall deposition. The Arabidopsis thaliana FLA11 and FLA12 genes are preferentially expressed in secondary cell wall- (SCW) forming cells (Ito et al., 2005; Persson et al., 2005), similar to their orthologues from various other plant species (Dahiya et al., 2006; MacMillan et al., 2010; Liu et al., 2013). Supporting a role in SCWs, the At-fla11/ At-fla12 double mutant displays a reduction in cellulose content accompanied by reduced tensile strength and tensile modulus of elasticity. This suggests an effect of FLAs both on cellulose deposition and on cell wall matrix integrity (MacMillan et al., 2010). The authors of this paper propose a direct biomechanical role for At-FLA11 and At-FLA12 proteins in the cell wall matrix and also consider a role in signalling that might influence cellulose deposition. Indeed, there is accumulating evidence for FLAs to modulate signalling upstream of cell wall polymer biosynthesis. Overexpression of the cotton orthologue of At-FLA12, named Gh-FLA1 (Liu et al., 2013), in transgenic cotton, results in an increased rate of fibre initiation and elongation and causes an upregulation of a suite of genes related to cell wall biosynthesis including other FLA genes, whereas antisense suppression has the opposite effect (Huang et al., 2013). This gain-of-function phenotype shows that FLAs can control the transcriptional programme for cell wall formation, which is best explained by a function in signalling.
The At-fla4 mutant of A. thaliana, also named salt overly sensitive 5 (sos5), highlights a non-redundant role for At-FLA4 in root growth under salt stress. The root of At-fla4 shows a drastic reduction of elongation growth combined with radial swelling of the elongation zone. Cell walls appear abnormally thin in At-fla4, apparently lacking the middle lamella (Shi et al., 2003). Support for the possibility that At-FLA4 is involved in a pathway upstream of cell wall deposition comes from the At-fei1 At-fei2 double mutant, that lacks two leucine-rich repeat receptor-like kinases (LRR-RLKs) resulting in salt oversensitivity just like At-fla4, and abnormal cellulose deposition. The At-fei1 At-fei2 double mutant non-additively interacts with At-fla4, suggesting that At-FEI1 and At-FEI2 act redundantly and At-FLA4 might act in the same genetic pathway. Moreover, the phenotype of both At-fei1 At-fei2 and At-fla4 is suppressed by α-aminoisobutyric acid (AIB), a structural analogue of the ethylene precursor 1-aminocyclopropane-1-carboxylic acid (ACC), and the cytoplasmic domain of FEI2 interacts with several ACC synthase (ACS) proteins, leading to the hypothesis that At-FLA4 and the At-FEI loci might act in a linear genetic pathway that depends on ACC but not on ethylene signalling, upstream of cellulose deposition (Xu et al., 2008).
To better understand the genetic pathway linking At-FLA4 with salt tolerance and root growth, we used chemical and genetic tools to test the possible involvement of stress-and growth-related signalling pathways. We found that abscisic acid (ABA) suppresses the At-fla4 mutant phenotype and that ABA signalling is affected by the At-FLA4 locus. We propose that At-FLA4 might act on ABA signal transduction upstream of cell wall deposition.
MATERIALS AND METHODS
Growth conditions and inhibitor treatments
Arabidopsis thaliana ecotype Col gl wild type and the At-fla4 mutant (sos5-1) were kindly provided by Jian-Kang Zhu (University of California, Riverside, CA, USA). Mutants At-cpl1-1, At-cpl3-1 and At-sad1-1 (ecotype C24), At-abi4-1 and At-abi5-1 (ecotype Col) were available in our department and, like all mutant combinations, were confirmed by sequencing. Growth conditions were as previously described (Blaukopf et al., 2011). For phenotypic observation, ten seedlings were manually transferred to test media containing standard medium alone or also including the indicated additives, and were examined using a dissecting microscope (Leica EZ4 HD). Unless otherwise stated, figures show representative individuals 48 h after transfer to control or test medium. Abscisic acid, pyrabactin, fluridone [1-methyl-3-phenyl-5-(α,α,α-trifluoro-m-tolyl)-4-pyridone] and standard chemicals were obtained from Sigma-Aldrich (Vienna, Austria). Abscinazole E2B was kindly provided by Yasushi Todoroki. Stock solutions were prepared in dimethylsulfoxide (DMSO), except for ABA that was dissolved in 0·01 m NaOH.
Quantitative real-time PCR (qRT-PCR)
Samples were treated in biological triplicates. For each biological replicate, 150 seedlings were grown on a nylon mesh (20 µm mesh size; Prosep, Belgium) for 5 d and were transferred to standard medium with or without 100 mm NaCl and incubated for 40 min. Roots were removed from the seedlings, frozen in liquid nitrogen, ground in a ball mill (Retsch, Germany) for 2 min and RNA was extracted using peqGOLD Trifast (Peqlab, Germany) according to the manufacturer's instructions. The RNA concentration was measured using a Nano Drop 2000c Spectrophotometer (Thermo Scientific, USA). For each sample, 1 µg of total RNA was reverse-transcribed with oligo(dT) primers using a first-strand cDNA synthesis kit (Thermo Scientific, USA) according to the manufacturer's instructions. Real-time PCR was performed using Solis BioDyne 5 × HOT FIREPol EvaGreen qPCR Mix Plus (no ROX) (Medibena, Austria), and a CFX96™ Real-Time PCR Detection System (Bio-Rad, USA) was used for detection. Information on the oligonucleotides used can be found in Supplementary Data Table S1. For real-time PCR, the following program was used: 95·0 °C for 15 min, 40 cycles of 95·0 °C for 10 s, 55·0 °C for 30 s, 72·0 °C for 30 s. Each biological replicate was analysed in technical duplicates. The average technical error was 0·5(±1) Ct values. Technical outliers were identified when the ΔCt between technical replicates was >2·5, and the higher Ct value was removed. The remaining technical replicates were averaged and the ΔCt (Ct test – Ct UBQ5) was calculated. The effect of genotypes and treatments was tested by subjecting the ΔCt values of three biological replicates to a two-sided t-test. Test values ≤0·05 and values ≤0·01 are indicated with lower case and uppercase letters, respectively, in the figures as specified in the legend of Fig. 1. To compare the expression levels of different probes graphically, we subtracted the average ΔCt (of three biological replicates) from the series average (average of all samples) to obtain –ΔΔCt. Note that in this way relatively high values indicate relatively high expression levels.
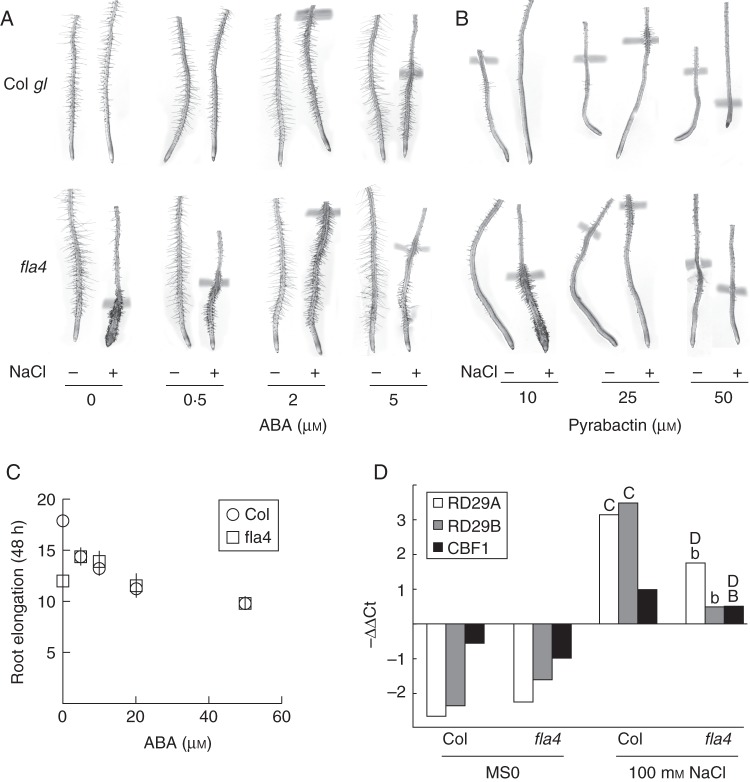
Fig. 1.
Synergistic effect of At-FLA4 and ABA signaling. The At-fla4 root phenotype is suppressed by (A) ABA and (B) pyrabactin. (C) The effect of ABA on root length requires At-FLA4. Root length measured after 48 h on media containing different concentrations of ABA (n = 20, ±confidence interval, α = 0·01). (D) The effect of salt on the expression of ABA-responsive transcripts in roots depends on At-FLA4. The indicated pairs were tested for statistically significant differences, and t-test values ≤0·05 and values ≤0·01 are indicated with lower case and uppercase letters, respectively, in the figures. A: Col vs. At-fla4 on standard medium (MS0), B: Col vs. At-fla4 on 100 mm NaCl, C: Col MS0 vs. Col NaCl, D: At-fla4 MS0 vs. At-fla4 NaCl.
For this study, the relative mRNA levels of At-FLA4 and six At-FLA loci were analysed. The FLA loci were selected for their domain structure being similar to that of At-FLA4 (At-FLA1, At-FLA2, and At-FLA8), for displaying global expression profiles closely correlating with At-FLA4 (At-FLA8 and At-FLA1; Bjœrn Ost Hansen, unpublished data), for previously showing slight upregulation by ABA [At-FLA13 and AtFLA2 (Matsui et al., 2008)] or for generally high expression in roots [At-FLA2, AtFLA7, At-FLA8 and AT-FLA9; (Johnson et al., 2003)].
RESULTS
ABA suppresses the At-fla4 phenotype and At-FLA4 is required for the ABA-mediated stress response
To define further the physiological process that is controlled by At-FLA4, we tested the effect of growth regulators and other bioactive compounds on the At-fla4 mutant. As previously described (Shi et al., 2003; Xu et al., 2008), At-fla4 mutants grown in the presence of 100 mm NaCl display a dramatic short root phenotype and radial swelling of the root elongation zone (Fig. 1A). The addition of different growth regulators and compounds affects the At-fla4 phenotype to varying degrees (data not shown); however, ABA at between 0·5 and 2 µm partially and at 5 µm fully suppresses the NaCl-induced phenotype of At-fla4 (Fig. 1A). At this concentration, the wild-type and At-fla4 roots become indistinguishable. Pyrabactin, a synthetic inhibitor of seed germination (Zhao et al., 2007) acting as a selective agonist of ABA perception (Park et al., 2009), suppresses the At-fla4 root phenotype at a concentration of 25 µm (Fig. 1B). Moreover, pyrabactin inhibits root elongation and root hair growth in the presence and absence of NaCl; however, At-fla4 appears less sensitive to this inhibition than the wild type. At-FLA4 is not only necessary for normal root growth on 100 mm NaCl- or 4 % sucrose-containing medium (Xu et al., 2008). Also on NaCl-free medium containing 1 % sucrose, At-fla4 roots are significantly (P < 0·001) shorter than those of the wild type (Fig. 1C) and more radially expanded compared with the wild type, giving the appearance of relatively dense root hairs initiating closer to the root tip (Fig. 1A). Application of ABA causes a dose-dependent decrease of root elongation in the wild type. By contrast, the elongation of At-fla4 roots is not negatively affected by up to 10 µm ABA; in fact, between zero and 5 µm ABA, there is a significant (P < 0·001) increase in At-fla4 root length. On the one hand this means that the suppression of the At-fla4 phenotype by ABA is not restricted to high salt treatment. On the other hand, this observation also suggests that At-FLA4 acts in the inhibition of root elongation by relatively low doses of ABA.
The mRNA levels of the abiotic stress-induced, ABA-regulated genes At-RD29A, At-RD29B and At-CBF1 in At-fla4 are not significantly different from those of the wild type 40 min after transfer to NaCl-free control medium. After 40 min treatment with 100 mm NaCl, the three marker genes are upregulated in both genotypes. However, the transcript levels of all three genes are significantly lower in At-fla4 compared with the wild type (Fig. 1C). In summary, ABA suppresses the role of At-FLA4 for root growth and salt tolerance and At-FLA4 positively regulates responses to ABA and salt in roots.
The At-fla4 phenotype is modulated by ABA response mutants
To demonstrate that the role of At-FLA4 in salt tolerance and root growth is mediated by genetic regulators of the ABA response, we isolated double mutants between At-fla4 and ABA-insensitive and ABA-oversensitive mutants. The At-ABI4 and At-ABI5 loci encode positive transcriptional regulators of the ABA response acting during seed germination and other ABA-dependent processes (Finkelstein, 1994; Dai et al., 2013; Wind et al., 2013). However, At-abi4 At-fla4 and At-abi5 At-fla4 roots are fully responsive to 5 µM ABA with respect to the suppression of the At-fla4 phenotype (Fig. 2), which means that this ABA effect does not depend on the function of At-ABI4 or At-ABI5. Neither At-abi4 nor At-abi5 displays obvious root growth phenotypes on NaCl-containing or control medium. By contrast, the At-abi4 At-fla4 double mutant appears phenotypically abnormal on NaCl-free medium, displaying a relatively short root and epidermal bulging (Fig. 2). This suggests a genetic synergy of At-FLA4 and At-ABI4 in root growth.
Fig. 2.
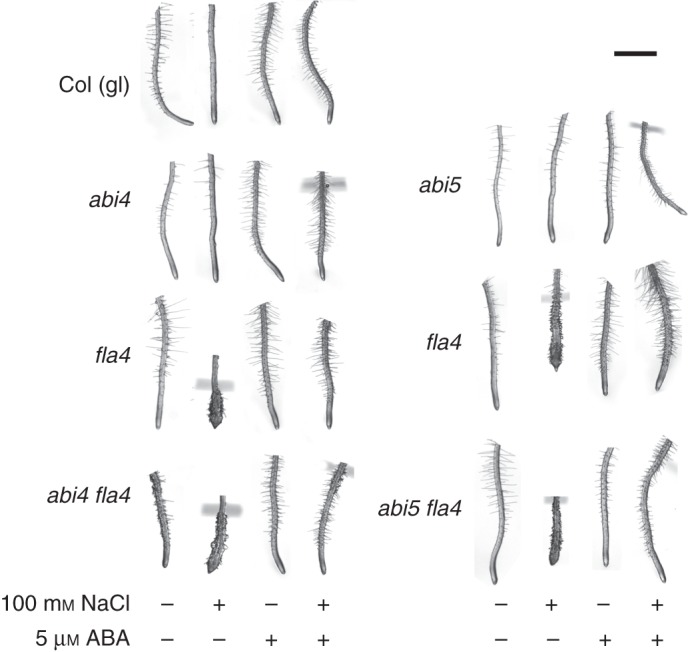
Genetic interaction between At-FLA4 and positive ABA response regulators At-ABI4 and At-ABI5. Note that the abi4 fla4 double mutant shows abnormal root growth on NaCl-free medium. Scale bar = 1 mm.
The C-TERMINAL DOMAIN PHOSPHATASE LOCI 1 and -3 [At-CPL1 (Koiwa et al., 2002; Xiong et al., 2002a) and At-CPL3 (Koiwa et al., 2002)] and SUPERSENSITIVE TO ABA AND DROUGHT [At-SAD1 (Xiong et al., 2001)] encode negative regulators of ABA-induced mRNA species. In the presence and absence of 100 mm NaCl, root morphology in double mutants At-cpl1 At-fla4 and At-sad1 At-fla4 is comparable with that of the wild type (Fig. 3). By contrast, the At-cpl3 At-fla4 double mutant displays salt oversensitivity comparable with that of the At-fla4 single mutant. This indicates that At-CPL1 and At-SAD1 are required for the genetic regulation of salt sensitivity by At-FLA4. Taken together, the non-redundant role of At-FLA4 in root growth is enhanced by the positive ABA regulator At-ABI4 and suppressed by the negative ABA regulators At-CPL1 and At-SAD1.
Fig. 3.
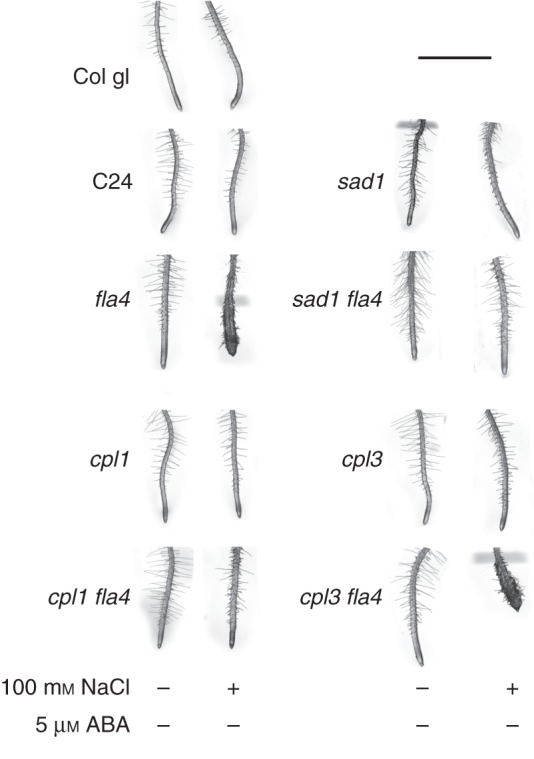
Genetic interaction between At-FLA4 and ABA repressors At-CPL1, At-CPL3 and At-SAD1. Conditions are the same as for Fig. 2. Note that on 100 mm NaCl-containing medium, the At-cpl1 At-fla4 and the At-sad1 At-fla4 double mutants are comparable with the wild type, while the At-cpl3 At-fla4 double mutant is comparable with the At-fla4 single mutant. Scale bar = 2 mm.
Involvement of ABA metabolism in the role of At-FLA4 in root growth
For the investigation of the potential interaction between At-FLA4 and ABA metabolism, we used fluridone to inhibit carotenoid biosynthesis, an early step in ABA biosynthesis. Fluridone in the absence of NaCl reduces root elongation and enhances radial expansion in the wild type and to a visibly higher degree in At-fla4 (Fig. 4A). In combination with NaCl, fluridone induces dramatic root swelling in the wild type that strikingly resembles the At-fla4 phenotype. However, under the same conditions, At-fla4 mutants show a more severe phenotype compared with the wild type, suggesting that At-FLA4 action might be synergistic with but independent of ABA biosynthesis (Fig. 4A). The specific CYP707A inhibitor abscinazole E2B that interferes with ABA 8'-hydroxylase (Okazaki et al., 2012) suppresses salt oversensitivity in At-fla4. The subtle At-fla4 root phenotype on NaCl-free medium is also suppressed by this compound (Fig. 4B). Hence with respect to At-FLA4 function in roots, ABA biosynthesis has a synergistic effect while ABA catabolism has an antagonistic effect.
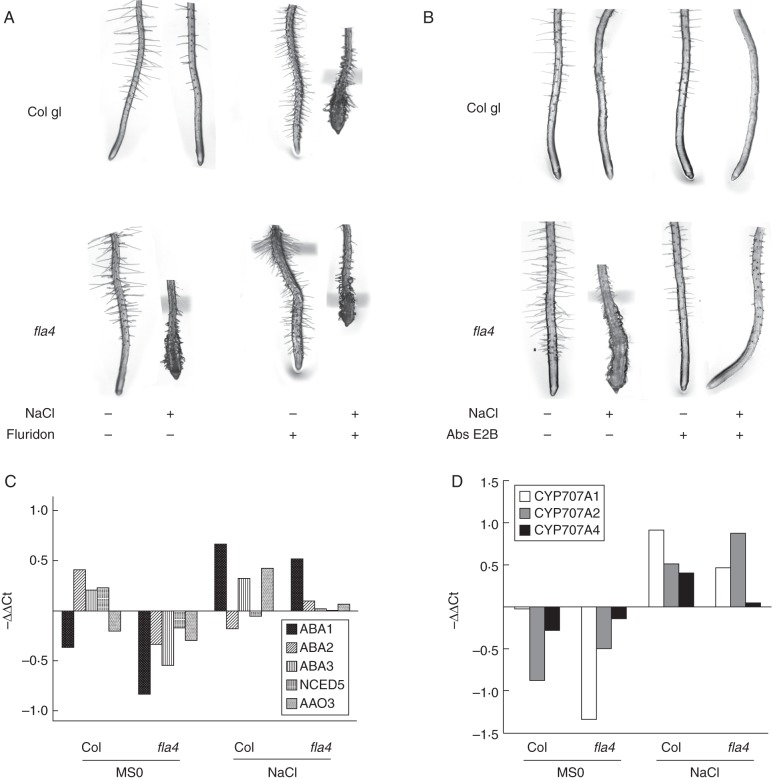
Fig. 4.
Interaction of At-FLA4 with ABA metabolism. (A) Inhibition of ABA biosynthesis with fluridone phenocopies the At-fla4 phenotype. (B) Inhibition of ABA catabolism with abscinazole E2B suppresses the At-fla4 phenotype. Expression of genes involved in ABA (C) biosynthesis and (D) catabolism is not significantly different between Col and At-fla4.
To investigate the possible involvement of At-FLA4 as a regulator of ABA metabolism, we measured the relative transcript level of genes involved in this process (Nambara and Marion-Poll, 2005) in At-fla4 mutants compared with wild-type roots before and after 40 min exposure to 100 mm NaCl. None of five ABA biosynthetic loci (At-ABA1, At-ABA2, At-ABA3, At-NCED5 and At-AAO3) or three ABA catabolic loci (At-CYP707A1, At-CYP707A2 and At-CYP707A4) displays a significant difference between At-fla4 mutant and wild-type roots. However, the transcript level of all five tested biosynthetic loci is reduced in the mutant compared with the wild type under salt-free conditions, and after salt treatment At-ABA1, At-ABA3 and At-AAO3 are reduced in At-fla4 compared with the wild type (Fig. 4C). By contrast, two of the three tested ABA catabolic loci are insignificantly upregulated in At-fla4 compared with the wild type under salt-free conditions. After salt exposure, only At-CYP707A2 is upregulated, while At-CYP707A1 and -4 are downregulated in At-fla4 compared with the wild type (Fig. 4D). Taken together, we do not observe a clear effect of At-FLA4 on ABA metabolism at the individual transcript level. Nevertheless, the locus might contribute to achieving the appropriate balance between ABA catabolic and biosynthetic gene expression. Consistent with the possibility that At-FLA4 stimulates ABA biosynthesis and downregulates ABA catabolism, we observe that At-fla4 loss of function and ABA biosynthesis inhibition are synergistic, and chemical inhibition of ABA catabolism phenotypically suppresses At-fla4.
ABA does not affect FLA transcript levels in roots
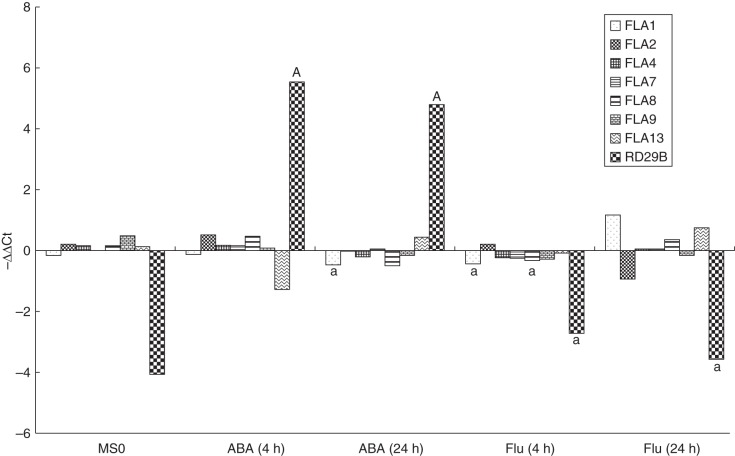
To test the effect of ABA on FLA4 expression, we compared the level of At-FLA4 transcript in roots treated with 5 µm ABA or 5 µm fluridone for 4 and 24 h with untreated controls. While At-RD29B mRNA is significantly (P < 0·01) upregulated by ABA treatment, the level of At-FLA4 transcript barely varies between the treatments. To test whether other FLA loci might be induced by ABA, to compensate for the loss of At-FLA4, we selected At-FLA1, At-FLA2, At-FLA7, At-FLA8, At-FLA9 and At-FLA13 genes based on considerations outlined in the Materials and Methods. Compared with At-RD29B that displays a difference of approx. 10 Ct units between untreated and ABA-treated roots, the fluctuations of the selected FLA genes are very moderate, the largest albeit statistically insignificant difference of 1·29 Ct units seen after 4 h ABA treatment being shown by At-FLA13 (Fig. 5). Of the seven tested FLA genes, At-FLA1 shows the only statistically significant (P < 0·05) difference between ABA-treated and non-treated roots. With a value of 0·29 Ct units, this corresponds to a very subtle, probably irrelevant decrease. Likewise, fluridone treatment causes only weak alterations in the expression level in any of the tested FLA genes, with At-FLA1 and At-FLA8 being expressed at a slightly lower level after 4 h of treatment. Hence under our test conditions, ABA does not influence the RNA level of At-FLA1, At-FLA2, AtFLA4, At-FLA7, At-FLA8, At-FLA9 and At-FLA13.
Fig. 5.
Effect of ABA (5 µM) and fluridone (5 µm) on the transcript level of selected At-FLA genes and At-RD29B. To mark significant differences between standard medium (MS0) and media containing ABA or fluridone, t-test values ≤ 0·05 and ≤0·01 are indicated with lower case a and uppercase A, respectively.
DISCUSSION
The actions of At-FLA4 and ABA are synergistic
In this study, multiple lines of evidence support the view that At-FLA4 acts in synergy with ABA signalling (Fig. 6). (1) Externally applied ABA suppresses At-fla4, both its salt-oversensitive phenotype and its root elongation phenotype, under salt-free conditions. (2) Root elongation in the At-fla4 mutant shows reduced sensitivity towards inhibition by ABA, and ABA-regulated transcripts are downregulated in salt-treated At-fla4 compared with the wild type. (3) The ABA-insensitive At-abi4-1 mutation enhances At-fla4 under NaCl-free conditions and the ABA-oversensitive At-cpl1-1 and At-sad1-1 alleles suppress At-fla4. (4) The At-fla4 phenotype is phenocopied by inhibition of ABA biosynthesis and suppressed by the inhibition of ABA degradation.
Fig. 6.
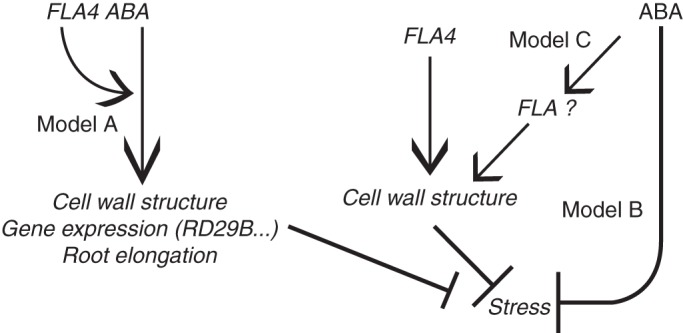
Genetic model for the interaction of At-FLA4 with ABA. For an explanation, see the Discussion.
The central observation presented in this study is the phenotypic suppression of At-fla4 by ABA and pyrabactin. The latter is a selective agonist of the PYR1/PYL-type ABA receptor and does not interact with other potential ABA receptor types (Park et al., 2009; Klingler et al., 2010). The ‘core signalling pathway’ consists of PYRABACTIN RESISTANT 1/PYR1 LIKE (At-PYR1/PYL) proteins that are induced by ABA to bind to type 2C protein phosphatases (PP2Cs) such as At-ABI1, thereby releasing them from inhibiting SNF1-related protein kinases (SnRK2s). The de-repressed SnRK2s in turn phosphorylate and activate a number of substrates including membrane channels and transcription factors such as At-ABI4 and At-ABI5 (Hubbard et al., 2010). Consequently, activation of the ABA ‘core signalling pathway’ is sufficient to suppress the At-fla4 salt-oversensitive phenotype. Although the effect of At-ABI4 on ABA sensitivity of the root is only partial (data not shown), the enhancement of the At-fla4 phenotype on NaCl-free medium suggests that At-FLA4 and At-ABI4 are synergistic. Consistently, two negative regulators of ABA-induced gene expression, At-CPL1 and At-SAD1, are antagonistic with respect to At-FLA4. The At-CPL1, At-CPL3 and At-SAD1 loci were identified as negative regulators of the stress response and ABA signalling, and their loss-of-function mutant alleles display overaccumulation of At-RD29A transcript (Xiong et al., 2001, 2002a; Koiwa et al., 2002). While various CPLs might negatively regulate RNA polymerase II by dephosphorylating its C-terminus, At-CPL1 is essential for normal micro RNA biogenesis, and genetic defects in this process lead to ABA hypersensitivity (Manavella et al., 2012). Another ABA-hypersensitive locus that antagonizes At-FLA4 is At-SAD1, encoding the SM-like 5 protein [LSM5 (Xiong et al., 2001)]. The At-SAD1/LSM5 protein is a component of both the LSM1–LSM7 and the LSM2–LSM8 complex that act in mRNA degradation and pre-mRNA splicing, respectively (Perea-Resa et al., 2012). The suppression of At-fla4 by At-cpl1 and At-sad1 suggests that sufficient accumulation of ABA-induced mRNA species is required for At-FLA4 to fulfil its role. The lack of suppression of At-fla4 by At-cpl3 might be explained by genetic redundancy of At-CPL3 in the root. It might also indicate that the set of mRNAs repressed by At-CPL3 is not identical to the set of At-CPL1- and At-SAD1-regulated mRNAs that are downregulated in salt-treated At-fla4 and which might be involved in the At-fla4 phenotype.
The reduced level of At-RD29A, At-RD29B and At-CBF1 in the At-fla4 background upon salt treatment is consistent with a positive role for At-FLA4 in ABA signalling because normal induction of these transcripts depends on ABA signalling (Xiong et al., 2002b; Knight et al., 2004). Initially, we expected an increased level of abiotic stress response in the At-fla4 mutant compared with the wild type. After genetic and chemical inhibition of cellulose biosynthesis, transcriptional signatures indicative of jasmonic acid- and ethylene-regulated biotic and wounding stress can be observed (Ellis and Turner, 2001; Guan and Nothnagel, 2004; Hamann et al., 2009). Such responses might, however, result from lesions in a constitutive mutant or be caused by long-term inhibitor treatment. By analysing roots exposed to NaCl for 40 min, we tested a time point when root morphology was not visibly damaged but when the transcriptional salt stress response was already activated, as indicated by induction of At-RD29A, At-RD29B and At-CBF1. We also analysed several other genes that respond to cell wall inhibitors such as isoxaben (Hamann et al., 2009) and β-glucosyl Yariv reagent (Guan and Nothnagel, 2004). OXIDATIVE SIGNAL INDUCIBLE 1 (Rentel et al., 2004) and RESPIRATORY BURST OXYDASE HOMOLOG D (Ma et al., 2012) and several other genes were not significantly changed by 40 min NaCl treatment or between the mutant and wild type. At-ACS6, At-MYB15, At-MUR4 and At3g11280 were induced by NaCl; however, none of these loci showed a significant effect of At-fla4 (data not shown). By contrast, all tested ABA-dependent genes displayed a significant repression in At-fla4, suggesting that the effect of At-FLA4 on stress signalling might be specific for ABA.
Fluridone inhibits the biosynthesis of carotenoid precursors for ABA (Nambara and Marion-Poll, 2005) and it causes salt oversensitivity closely resembling that of At-fla4. Interestingly, root elongation defects were previously observed in At-aba1 and At-aba2 mutants (Barrero et al., 2005; Lin et al., 2007); however, residual ABA that is detected in aba mutants suggests the existence of an alternative ABA biosynthesis pathway supporting essential growth processes (Barrero et al., 2005). Fluridone might more fully deplete the endogenous ABA, leading to the observed growth defects. The apparent additive effect of At-fla4 and fluridone, however, indicates that although At-FLA4 and ABA biosynthesis are synergistic with respect to root growth and salt sensitivity, they act in separate pathways. Accordingly, At-FLA4 does not act by affecting ABA biosynthesis. However, this hypothesis requires genetic confirmation to exclude side effects of fluridone.
The suppression of At-fla4 by the CYP707A-specific inhibitor abscinazole E2B (Okazaki et al., 2012) is consistent with our view that ABA and At-FLA4 are synergistic, because inhibition of CYP707A that functions as an ABA 8'-hydroxylase in planta is expected to block the main catabolic pathway for ABA (Nambara and Marion-Poll, 2005). Our observation indicates that in At-fla4, the intracellular ABA level or sensitivity are below a threshold level to counterbalance salt stress sufficiently and that the ABA level is raised by blocking ABA degradation. However, the expression level of seven ABA biosynthetic genes and three catabolic genes is not significantly different between At-fla4 and the wild type, which argues against the alteration of the endogenous ABA level dependent on At-FLA4. Nevertheless, we cannot exclude cell type-specific alterations of ABA metabolism in At-fla4.
How does ABA suppress At-fla4?
Taken together, we suggest that At-FLA4 and ABA signalling act in genetic synergy, leading to suppression of At-fla4 by ABA and to a reduced ABA response in At-fla4. We envisage three alternative models for this effect. First, At-FLA4 might positively modulate an ABA-dependent effect on root elongation, gene expression and cell wall structure (Fig. 6, Model A). According to this view, the loss of At-FLA4 function is expected to compromise the final output of ABA signalling. External ABA supply corrects this defect by exceeding a threshold required for normal cell growth under salt-free conditions and for tolerance of salt. This hypothesis also predicts that besides known ABA responses such as At-RD29A expression and inhibition of root elongation, processes required for cell wall structural integrity exist downstream of At-FLA4-modulated ABA signalling. This prediction is consistent with our observation that depletion of ABA by fluridone phenocopies At-fla4 under salt stress.
The two alternative hypotheses suggest that At-FLA4 acts primarily in an ABA-independent pathway upstream of cell wall structure (Fig. 6, Models B and C). Consequently, the primary effect of the loss of At-FLA4 function is a damaged cell wall subsequently leading to cellular stress responsible for the apparent phenotypic changes. In one variant of the model, ABA counteracts the indirect consequences of cell wall damage initiated by the At-fla4 mutation (Fig. 6, Model B), possibly by the induction of salt export (Shi and Zhu, 2002) or antioxidants (Ding et al., 2013). In a second variant, ABA activates genes that make At-FLA4 redundant. The induction of alternative FLA genes by ABA might provide a simple mechanism for this scenario (Fig. 6, Model C). However, although many FLA loci are differentially regulated by ABA, in most cases a repression of FLA mRNA by ABA is observed. In one study, At-FLA1, At-FLA2 and At-FLA8 mRNAs are downregulated by ABA (Johnson et al., 2003). Likewise, the genetic manipulation of ABA levels affects the transcript levels of 11 of the 21 FLA genes in the A. thaliana genome (Okamoto et al., 2010) and also under these conditions FLA genes are exclusively downregulated by ABA. Finally, in our study that focuses on roots, there is no apparent response of a set of seven At-FLA loci to ABA application and depletion. Besides the possibility of post-transcriptional regulation that we have not tested, these observations make an ABA-induced suppression of At-fla4 by alternative FLAs unlikely.
At present we cannot exclude any of the proposed scenarios of how At-fla4 and ABA might interact; however, neither Model C nor Model B (Fig. 6) accounts for the reduced level of ABA-regulated transcripts in At-fla4 or for its reduced response to low concentrations of ABA, whereas Model A explains both the suppression of the At-fla4 phenotype by ABA and the suppression of ABA responses in At-fla4. To support Model A further, we envisage a novel mechanism of how FLA proteins might influence ABA signalling.
How FLAs might modulate ABA signalling
To explain our observations, we favour a model in which At-FLA4 positively modulates ABA signalling (Fig. 6, Model A) and hypothesize how the At-FLA4 protein might be mechanistically involved in this process. In one plausible scenario, At-FLA4 might regulate the early events in the ‘core signalling pathway’ of ABA signal transduction. The crucial step in ABA perception requires the suppressive effect of PP2Cs on SnRK2s to be released by the ABA-bound form of PYR1/PYL proteins (Hubbard et al., 2010). We speculate that the physical interaction between PP2Cs and one or more of their partners in this complex might be modulated by factors in addition to ABA. Intriguingly, the PP2C At-ABI1 and another PP2C protein have recently been identified in detergent-resistant lipid rafts (Demir et al., 2013). FLA proteins are typically associated with the outer face of lipid rafts in the plasma membrane via their GPI anchor (Borner et al., 2003; Kierszniowska et al., 2009; Demir et al., 2013). Hence FLAs and ABI1 might assemble in the same plasma membrane sub-domain. However, if At-FLA4 indeed physically interacts with the cytosolic At-ABI1, such an interaction can only be indirect. A protein group that is in a position to connect At-FLA4 and At-ABI1 physically are receptor-like kinases, many of which have recently been detected in lipid rafts (Shahollari et al., 2004; Demir et al., 2013). Furthermore, the At-FEI1 and At-FEI2 LRR-RLKs have been speculated to bind to At-FLA4 and mediate its action (Xu et al., 2008). Interestingly, both At-FEI2 and At-ABI1 physically interact with different forms of ACS. At-ABI1 interacts with At-ACS2 and At-ACS6 (Ludwikow et al., 2009), while At-FEI2 interacts with At-ACS5 and At-ACS9 but not with At-ACS2 in yeast two-hybrid assays (Xu et al., 2008). Although such assays do not necessarily reflect in vivo binding, ACS isoforms forming a complex with At-ABI1 and At-FEI2 in principle might physically link At-FLA4 and ABA signalling in this speculative scenario. Thereby, this putative complex might connect ABA signalling with the previously postulated ethylene-independent action of ACC on elongation growth (Xu et al., 2008; Tsang et al., 2011).
At present, direct evidence for a physical interaction between FLAs and receptor-like proteins on the one hand and between ABI1 and membrane proteins on the other hand is lacking. The physical interaction between Fas1 domain-containing mammalian glycoproteins βIG-h3 and periostin with integrin-type cell surface receptors is revealed in adhesion assays using intact cells spreading on coated glass cover slips (Kim et al., 2000, 2002; Gillan et al., 2002). Such cell adhesion assays are not available for plant cells, but other approaches such as co-immunoprecipitation and Förster resonance energy transfer (FRET) might be more promising to identify physical interactors of FLA proteins.
Overall, we conclude that At-FLA4 regulates normal root growth via an ABA-depedent signalling pathway that might be upstream of cell wall biosynthesis.
SUPPLEMENTARY DATA
ACKNOWLEDGEMENTS
We thank Isabel Damm (University of Applied Sciences, Wr. Neustadt) for assistance with RT-PCR, and Jose M. Estevez (University of Buenos Aires, Argentinia) for critical reading of the manuscript. We gratefully acknowledge the generous gift of abscinazoles by Yasushi Todoroki (Shizuoka University, Japan). Bjœrn Ost Hansen (Max Planck Institute for Molecular Plant Physiology, Golm, Germany) provided an RNaseq-based co-expression matrix for At-FLA4. This work was supported by the Austrian Science Fund (P21782-B12, I1182-B22). H.X. was supported by the China Scholarship Council. T.A. was supported by The Council of Higher Education (PhD Research Scholarship), Turkey. G.J.S planned and conducted the research and wrote the manuscript; H.X. and T.A. conducted the research and critically read the manuscript.
LITERATURE CITED
- Barrero JM, Piqueras P, Gonzalez-Guzman M, et al. A mutational analysis of the ABA1 gene of Arabidopsis thaliana highlights the involvement of ABA in vegetative development. Journal of Experimental Botany. 2005;56:2071–2083. doi: 10.1093/jxb/eri206. [DOI] [PubMed] [Google Scholar]
- Bastiani MJ, Harrelson AL, Snow PM, Goodman CS. Expression of fasciclin I and II glycoproteins on subsets of axon pathways during neuronal development in the grasshopper. Cell. 1987;48:745–755. doi: 10.1016/0092-8674(87)90072-9. [DOI] [PubMed] [Google Scholar]
- Blaukopf C, Krol MZ, Seifert GJ. New insights into the control of cell growth. Methods in Molecular Biology. 2011;715:221–244. doi: 10.1007/978-1-61779-008-9_16. [DOI] [PubMed] [Google Scholar]
- Borner GH, Lilley KS, Stevens TJ, Dupree P. Identification of glycosylphosphatidylinositol-anchored proteins in arabidopsis. A proteomic and genomic analysis. Plant Physiology. 2003;132:568–577. doi: 10.1104/pp.103.021170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahiya P, Findlay K, Roberts K, McCann MC. A fasciclin-domain containing gene, ZeFLA11, is expressed exclusively in xylem elements that have reticulate wall thickenings in the stem vascular system of Zinnia elegans cv Envy. Planta. 2006;223:1281–1291. doi: 10.1007/s00425-005-0177-9. [DOI] [PubMed] [Google Scholar]
- Dai M, Xue Q, McCray T, et al. The PP6 phosphatase regulates ABI5 phosphorylation and abscisic acid signaling in Arabidopsis. The Plant Cell. 2013;25:517–34. doi: 10.1105/tpc.112.105767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demir F, Horntrich C, Blachutzik JO, et al. Arabidopsis nanodomain-delimited ABA signaling pathway regulates the anion channel SLAH3. Proceedings of the National Academy of Sciences, USA. 2013;110:8296–82301. doi: 10.1073/pnas.1211667110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Cao J, Ni L, Zhu Y, Zhang A, Tan M, Jiang M. ZmCPK11 is involved in abscisic acid-induced antioxidant defence and functions upstream of ZmMPK5 in abscisic acid signalling in maize. Journal of Experimental Botany. 2013;64:871–884. doi: 10.1093/jxb/ers366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis C, Turner JG. The Arabidopsis mutant cev1 has constitutively active jasmonate and ethylene signal pathways and enhanced resistance to pathogens. The Plant Cell. 2001;13:1025–1033. doi: 10.1105/tpc.13.5.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis M, Egelund J, Schultz CJ, Bacic A. Arabinogalactan-proteins: key regulators at the cell surface? Plant Physiology. 2010;153:403–419. doi: 10.1104/pp.110.156000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR. Mutations at two new Arabidopsis ABA response loci are similar to the abi3 mutations. The Plant Journal. 1994;5:765–771. [Google Scholar]
- Gillan L, Matei D, Fishman DA, Gerbin CS, Karlan BY, Chang DD. Periostin secreted by epithelial ovarian carcinoma is a ligand for alpha(V)beta(3) and alpha(V)beta(5) integrins and promotes cell motility. Cancer Research. 2002;62:5358–5364. [PubMed] [Google Scholar]
- Guan Y, Nothnagel EA. Binding of arabinogalactan proteins by Yariv phenylglycoside triggers wound-like responses in arabidopsis cell cultures. Plant Physiology. 2004;135:1346–1366. doi: 10.1104/pp.104.039370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamann T, Bennett M, Mansfield J, Somerville C. Identification of cell-wall stress as a hexose-dependent and osmosensitive regulator of plant responses. The Plant Journal. 2009;57:1015–1026. doi: 10.1111/j.1365-313X.2008.03744.x. [DOI] [PubMed] [Google Scholar]
- Huang GQ, Gong SY, Xu WL, et al. A fasciclin-like arabinogalactan protein, GhFLA1, is involved in fiber initiation and elongation of cotton. Plant Physiology. 2013;161:1278–1290. doi: 10.1104/pp.112.203760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard KE, Nishimura N, Hitomi K, Getzoff ED, Schroeder JI. Early abscisic acid signal transduction mechanisms: newly discovered components and newly emerging questions. Genes and Development. 2010;24:1695–1708. doi: 10.1101/gad.1953910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Suzuki Y, Miyamoto K, Ueda J, Yamaguchi I. AtFLA11, a fasciclin-like arabinogalactan-protein, specifically localized in sclerenchyma cells. Bioscience, Biotechnology, and Biochemistry. 2005;69:1963–1969. doi: 10.1271/bbb.69.1963. [DOI] [PubMed] [Google Scholar]
- Johnson KL, Jones BJ, Bacic A, Schultz CJ. The fasciclin-like arabinogalactan proteins of arabidopsis. A multigene family of putative cell adhesion molecules. Plant Physiology. 2003;133:1911–1925. doi: 10.1104/pp.103.031237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kierszniowska S, Seiwert B, Schulze WX. Definition of Arabidopsis sterol-rich membrane microdomains by differential treatment with methyl-beta-cyclodextrin and quantitative proteomics. Molecular and Cellular Proteomics. 2009;8:612–623. doi: 10.1074/mcp.M800346-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JE, Kim SJ, Lee BH, Park RW, Kim KS, Kim IS. Identification of motifs for cell adhesion within the repeated domains of transforming growth factor-beta-induced gene, betaig-h3. Journal of Biological Chemistry. 2000;275:30907–30915. doi: 10.1074/jbc.M002752200. [DOI] [PubMed] [Google Scholar]
- Kim JE, Jeong HW, Nam JO, et al. Identification of motifs in the fasciclin domains of the transforming growth factor-beta-induced matrix protein betaig-h3 that interact with the alphavbeta5 integrin. Journal of Biological Chemistry. 2002;277:46159–46165. doi: 10.1074/jbc.M207055200. [DOI] [PubMed] [Google Scholar]
- Klingler JP, Batelli G, Zhu JK. ABA receptors: the START of a new paradigm in phytohormone signalling. Journal of Experimental Botany. 2010;61:3199–3210. doi: 10.1093/jxb/erq151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight H, Zarka DG, Okamoto H, Thomashow MF, Knight MR. Abscisic acid induces CBF gene transcription and subsequent induction of cold-regulated genes via the CRT promoter element. Plant Physiology. 2004;135:1710–1717. doi: 10.1104/pp.104.043562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koiwa H, Barb AW, Xiong L, et al. C-terminal domain phosphatase-like family members (AtCPLs) differentially regulate Arabidopsis thaliana abiotic stress signaling, growth, and development. Proceedings of the National Academy of Sciences, USA. 2002;99:10893–10898. doi: 10.1073/pnas.112276199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin PC, Hwang SG, Endo A, Okamoto M, Koshiba T, Cheng WH. Ectopic expression of ABSCISIC ACID 2/GLUCOSE INSENSITIVE 1 in Arabidopsis promotes seed dormancy and stress tolerance. Plant Physiology. 2007;143:745–758. doi: 10.1104/pp.106.084103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Shi R, Wang X, et al. Characterization and expression analysis of a fiber differentially expressed fasciclin-like arabinogalactan protein gene in Sea Island cotton fibers. PLoS One. 2013;8:e70185. doi: 10.1371/journal.pone.0070185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwikow A, Kierzek D, Gallois P, Zeef L, Sadowski J. Gene expression profiling of ozone-treated Arabidopsis abi1td insertional mutant: protein phosphatase 2C ABI1 modulates biosynthesis ratio of ABA and ethylene. Planta. 2009;230:1003–1017. doi: 10.1007/s00425-009-1001-8. [DOI] [PubMed] [Google Scholar]
- Ma L, Zhang H, Sun L, et al. NADPH oxidase AtrbohD and AtrbohF function in ROS-dependent regulation of Na(+)/K(+)homeostasis in Arabidopsis under salt stress. Journal of Experimental Botany. 2012;63:305–317. doi: 10.1093/jxb/err280. [DOI] [PubMed] [Google Scholar]
- MacMillan CP, Mansfield SD, Stachurski ZH, Evans R, Southerton SG. Fasciclin-like arabinogalactan proteins: specialization for stem biomechanics and cell wall architecture in Arabidopsis and Eucalyptus. The Plant Journal. 2010;62:689–703. doi: 10.1111/j.1365-313X.2010.04181.x. [DOI] [PubMed] [Google Scholar]
- Manavella PA, Hagmann J, Ott F, et al. Fast-forward genetics identifies plant CPL phosphatases as regulators of miRNA processing factor HYL1. Cell. 2012;151:859–870. doi: 10.1016/j.cell.2012.09.039. [DOI] [PubMed] [Google Scholar]
- Matsui A, Ishida J, Morosawa T, et al. Arabidopsis transcriptome analysis under drought, cold, high-salinity and ABA treatment conditions using a tiling array. Plant and Cell Physiology. 2008;49:1135–1149. doi: 10.1093/pcp/pcn101. [DOI] [PubMed] [Google Scholar]
- Moody RG, Williamson MP. Structure and function of a bacterial Fasciclin I Domain Protein elucidates function of related cell adhesion proteins such as TGFBIp and periostin. FEBS Open Bio. 2013;3:71–77. doi: 10.1016/j.fob.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambara E, Marion-Poll A. Abscisic acid biosynthesis and catabolism. Annual Review of Plant Biology. 2005;56:165–185. doi: 10.1146/annurev.arplant.56.032604.144046. [DOI] [PubMed] [Google Scholar]
- Okamoto M, Tatematsu K, Matsui A, et al. Genome-wide analysis of endogenous abscisic acid-mediated transcription in dry and imbibed seeds of Arabidopsis using tiling arrays. The Plant Journal. 2010;62:39–51. doi: 10.1111/j.1365-313X.2010.04135.x. [DOI] [PubMed] [Google Scholar]
- Okazaki M, Kittikorn M, Ueno K, et al. Abscinazole-E2B, a practical and selective inhibitor of ABA 8'-hydroxylase CYP707A. Bioorganic and Medicinal Chemistry. 2012;20:3162–3172. doi: 10.1016/j.bmc.2012.03.068. [DOI] [PubMed] [Google Scholar]
- Park SY, Fung P, Nishimura N, et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science. 2009;324:1068–1071. doi: 10.1126/science.1173041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YB, Cosgrove DJ. A revised architecture of primary cell walls based on biomechanical changes induced by substrate-specific endoglucanases. Plant Physiology. 2012;158:1933–1943. doi: 10.1104/pp.111.192880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perea-Resa C, Hernandez-Verdeja T, Lopez-Cobollo R, del Mar Castellano M, Salinas J. LSM proteins provide accurate splicing and decay of selected transcripts to ensure normal Arabidopsis development. The Plant Cell. 2012;24:4930–4947. doi: 10.1105/tpc.112.103697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson S, Wei H, Milne J, Page GP, Somerville CR. Identification of genes required for cellulose synthesis by regression analysis of public microarray data sets. Proceedings of the National Academy of Sciences, USA. 2005;102:8633–8638. doi: 10.1073/pnas.0503392102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rentel MC, Lecourieux D, Ouaked F, et al. OXI1 kinase is necessary for oxidative burst-mediated signalling in Arabidopsis. Nature. 2004;427:858–861. doi: 10.1038/nature02353. [DOI] [PubMed] [Google Scholar]
- Seifert GJ, Roberts K. The biology of arabinogalactan proteins. Annual Review of Plant Biology. 2007;58:137–161. doi: 10.1146/annurev.arplant.58.032806.103801. [DOI] [PubMed] [Google Scholar]
- Shahollari B, Peskan-Berghofer T, Oelmuller R. Receptor kinases with leucine-rich repeats are enriched in Triton X-100 insoluble plasma membrane microdomains from plants. Physiologia Plantarum. 2004;122:397–403. [Google Scholar]
- Shi H, Zhu JK. Regulation of expression of the vacuolar Na+/H+ antiporter gene AtNHX1 by salt stress and abscisic acid. Plant Molecular Biology. 2002;50:543–550. doi: 10.1023/a:1019859319617. [DOI] [PubMed] [Google Scholar]
- Shi H, Kim Y, Guo Y, Stevenson B, Zhu JK. The arabidopsis SOS5 locus encodes a putative cell surface adhesion protein and is required for normal cell expansion. The Plant Cell. 2003;15:19–32. doi: 10.1105/tpc.007872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Showalter AM, Keppler B, Lichtenberg J, Gu D, Welch LR. A bioinformatics approach to the identification, classification, and analysis of hydroxyproline-rich glycoproteins. Plant Physiology. 2010;153:485–513. doi: 10.1104/pp.110.156554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L, Showalter AM, Egelund J, Hernandez-Sanchez A, Doblin MS, Bacic A. Arabinogalactan-proteins and the research challenges for these enigmatic plant cell surface proteoglycans. Frontiers in Plant Science. 2012;3:140. doi: 10.3389/fpls.2012.00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang DL, Edmond C, Harrington JL, Nuhse TS. Cell wall integrity controls root elongation via a general 1-aminocyclopropane-1-carboxylic acid-dependent, ethylene-independent pathway. Plant Physiology. 2011;156:596–604. doi: 10.1104/pp.111.175372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wind JJ, Peviani A, Snel B, Hanson J, Smeekens SC. ABI4: versatile activator and repressor. Trends in Plant Science. 2013;18:125–132. doi: 10.1016/j.tplants.2012.10.004. [DOI] [PubMed] [Google Scholar]
- Xiong L, Gong Z, Rock CD, et al. Modulation of abscisic acid signal transduction and biosynthesis by an Sm-like protein in Arabidopsis. Developmental Cell. 2001;1:771–781. doi: 10.1016/s1534-5807(01)00087-9. [DOI] [PubMed] [Google Scholar]
- Xiong L, Lee H, Ishitani M, et al. Repression of stress-responsive genes by FIERY2, a novel transcriptional regulator in Arabidopsis. Proceedings of the National Academy of Sciences, USA. 2002a;99:10899–10904. doi: 10.1073/pnas.162111599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L, Lee H, Ishitani M, Zhu JK. Regulation of osmotic stress-responsive gene expression by the LOS6/ABA1 locus in Arabidopsis. Journal of Biological Chemistry. 2002b;277:8588–8596. doi: 10.1074/jbc.M109275200. [DOI] [PubMed] [Google Scholar]
- Xu SL, Rahman A, Baskin TI, Kieber JJ. Two leucine-rich repeat receptor kinases mediate signaling, linking cell wall biosynthesis and ACC synthase in Arabidopsis. The Plant Cell. 2008;20:3065–3079. doi: 10.1105/tpc.108.063354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Chow TF, Puckrin RS, et al. Chemical genetic interrogation of natural variation uncovers a molecule that is glycoactivated. Nature Chemical Biology. 2007;3:716–721. doi: 10.1038/nchembio.2007.32. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.