Abstract
Background and Aims
In Arabidopsis thaliana, the degree of methylesterification (DM) of homogalacturonans (HGs), the main pectic constituent of the cell wall, can be modified by pectin methylesterases (PMEs). In all organisms, two types of protein structure have been reported for PMEs: group 1 and group 2. In group 2 PMEs, the active part (PME domain, Pfam01095) is preceded by an N-terminal extension (PRO part), which shows similarities to PME inhibitors (PMEI domain, Pfam04043). This PRO part mediates retention of unprocessed group 2 PMEs in the Golgi apparatus, thus regulating PME activity through a post-translational mechanism. This study investigated the roles of a subtilisin-type serine protease (SBT) in the processing of a PME isoform.
Methods
Using a combination of functional genomics, biochemistry and proteomic approaches, the role of a specific SBT in the processing of a group 2 PME was assessed together with its consequences for plant development.
Key Results
A group 2 PME, AtPME17 (At2g45220), was identified, which was highly co-expressed, both spatially and temporally, with AtSBT3.5 (At1g32940), a subtilisin-type serine protease (subtilase, SBT), during root development. PME activity was modified in roots of knockout mutants for both proteins with consequent effects on root growth. This suggested a role for SBT3.5 in the processing of PME17 in planta. Using transient expression in Nicotiana benthamiana, it was indeed shown that SBT3.5 can process PME17 at a specific single processing motif, releasing a mature isoform in the apoplasm.
Conclusions
By revealing the potential role of SBT3.5 in the processing of PME17, this study brings new evidence of the complexity of the regulation of PMEs in plants, and highlights the need for identifying specific PME–SBT pairs.
Keywords: Arabidopsis thaliana, co-expression, pectin, pectin methylesterase, PME, subtilase, SBT, post-translational modification, protein processing, gene expression, plant cell walls, subtilisin-like serine protease
INTRODUCTION
Pectins are a family of highly complex cell-wall polysaccharides with several applications in the food industry. In plants, multiple biological functions have been attributed to pectins, most of them related to cell-wall mechanical properties. Pectins can be considered as multiblock co-polymers. The simplest and the most abundant of these blocks is homogalacturonan (HG), an unbranched polymer of α-(1–4) linked d-galacturonic acid residues. HG is synthesized in the Golgi apparatus in a fully methylesterified form and subsequently selectively de-methylesterified in the cell wall by pectin methylesterases (PMEs), which constitute a gene family of 66 members in Arabidopsis (Pelloux et al., 2007). Apoplastic PME activity is itself post-translationally controlled through a 1:1 interaction with specific pectin methylesterase inhibitors (PMEIs; Juge, 2006).
Over recent years, the PME–PMEI-mediated control of the degree of methylesterification (DM) of HG has been shown to play a central role in plant development and in response to stresses. For instance, using reverse genetics approaches, a role for PME and PMEI was shown in plant–pathogen interactions (Hewezi et al., 2008; Osorio et al., 2008; Raiola et al., 2011), the control of pollen development and pollen tube growth (Jiang et al., 2005; Francis et al., 2006), the modulation of stem mechanical properties (Hongo et al., 2012), the control of seed mucilage extrusion (Saez-Aguayo et al., 2013; Voiniciuc et al., 2013), radicle emergence at the onset of germination (Müller et al., 2013), the subsequent regulation of etiolated hypocotyl elongation (Derbyshire et al., 2007; Pelletier et al., 2010) and the control of primordia emergence at the shoot apical meristem (Peaucelle et al., 2008, 2011a, b). For the last of these, a clear relationship was shown between auxin signalling and the control of PME activity modulating the cell-wall physical properties at the shoot apical meristem, thus enabling proper primordia formation (Braybrook and Peaucelle, 2013). Despite this increasing wealth of data concerning the functions of some Arabidopsis PME isoforms in planta, much remains to be discovered with regard to their substrate specificity, mode of action and regulation. This notably includes a better understanding of the role of pH in the modulation of the activity of a given PME isoform, the identification of specific PME–PMEI pairs, and lastly the determination of the role of protein processing in the release of active PME isoforms.
PME protein sequence analysis shows that PMEs can be classified in two subgroups (1 and 2). Group 2 PMEs indeed contain, in addition to the catalytic domain (PME domain, Pfam01095, IPR000070), an N-terminal extension (PRO part, PMEI domain, Pfam04043, IPR006501) showing similarities to PMEI. Group 1 PMEs do not have the PRO region, whereas PMEs from group 2 can contain one to three PMEI domains. Cleavage of the PMEI domain(s) of group 2 PMEs, which is required for activation and secretion of PMEs, occurs at a conserved R(R/K)LL processing site, with a preference towards RRLL motifs (Bosch et al., 2005; Dorokhov et al., 2006; Wolf et al., 2009; Weber et al., 2013). This might involve subtilases (SBTs), serine proteases from the S8 family (Pfam00082). Two subgroups of SBTs can be identified: S8A, subtilisins; and S8B, kexins (Schaller et al., 2012). In plants, no proteins have been identified in the S8B subfamily thus far, while the S8A subfamily is large, comprising 56 members in Arabidopsis (Beers et al., 2004; Rautengarten et al., 2005). While SBTs were previously shown to play a role in immune priming during plant–pathogen interactions (Ramírez et al., 2013), the processing of peptide hormones (Matos et al., 2008; Srivastava et al., 2008, 2009), the differentiation of stomata and epidermis (Berger and Altmann, 2000; Tanaka et al., 2001; Xing et al., 2013), seed development (D’Erfurth et al., 2012), germination (Rautengarten et al., 2008) and cell death (Chichkova et al., 2010), the identification of their physiological substrates and roles remains a challenge.
There are several lines of evidence linking PMEs and SBTs. PME activity is enhanced in seeds of AtSBT1.7 loss-of-function mutants. As a consequence of increased PME activity in the mutants, the DM is reduced in seed mucilage, mucilage fails to be released upon hydration and the efficiency of germination is reduced under low water conditions (Rautengarten et al., 2008; Saez-Aguayo et al., 2013). Owing to the protease activity of SBTs, the observed changes could be related to a degradative function of this SBT isoform in the wild-type context (Hamilton et al., 2003; Schaller et al., 2012). However, SBTs were also shown to be involved in the processing of group 2 PMEs. First, site-directed mutagenesis of the dibasic motifs R(R/K)LL between the PMEI and PME domains led to the retention of PMEs in the Golgi apparatus. The processing of group 2 PMEs would therefore be a prerequisite for the secretion of active isoforms to the apoplasm. A role of SBTs in the process was proposed when AtSBT6.1 (Site-1-protease, S1P) was shown to interact with PMEs in co-immunoprecipitation experiments and to co-localize with unprocessed PME proteins in the Golgi apparatus (Wolf et al., 2009). Furthermore, in atsbt6.1 mutants PME processing was impaired. However, Golgi-resident S1P is only distantly related to most other SBTs that are secreted, questioning the roles of other SBT isoforms in PME processing and the localization of the processing itself. The interaction between SBTs and group 2 PMEs could occur in the late Golgi, thus mediating the export of only the active and processed PMEs into the cell wall (Wolf et al., 2009). Some analyses have indeed shown that peptides matching the PRO part of group 2 PMEs are rarely recovered in the cell-wall proteome (Al-Qsous et al., 2004; Boudart et al., 2005; Feiz et al., 2006; Irshad et al., 2008; Minic et al., 2009). However, as other data indicate the presence of both SBTs and unprocessed group 2 PMEs in the wall (Boudart et al., 2005; Feiz et al., 2006; Irshad et al., 2008; Minic et al., 2009; Mareck et al., 2012), PME processing and activation could occur inside or outside of the cell depending on developmental stages and/or the specific balance between SBT and group 2 PME pools. Specific co-expression was observed for individual members of the PME and SBT gene families in Arabidopsis tissues, developmental stages or in response to biotic and abiotic stresses, suggesting that AtSBT6.1 may not be the sole SBT involved in the secretion and activation of PMEs.
Using transcriptome data mining, we identified AtSBT3.5 as being strongly co-expressed with AtPME17, a group 2 PME, during development and in response to various stresses. Real-time quantitative PCR (RT-qPCR) analysis and promoter GUS fusions confirmed the overlapping expression patterns of both genes during root development. Using knockout (KO) mutants for both genes, we further showed that the encoded proteins were absent in cell-wall-enriched extracts and that both PME activity and root growth were impaired. Co-expression of AtSBT3.5 and tagged versions of AtPME17 in Nicotiana benthamiana confirmed the ability of SBT3.5 to release processed PME17 in the apoplasm. Our results provide evidence that processing of PMEs involves, depending on the tissues considered, specifically co-expressed PME–SBT pairs.
MATERIALS AND METHODS
Plant material and growth conditions
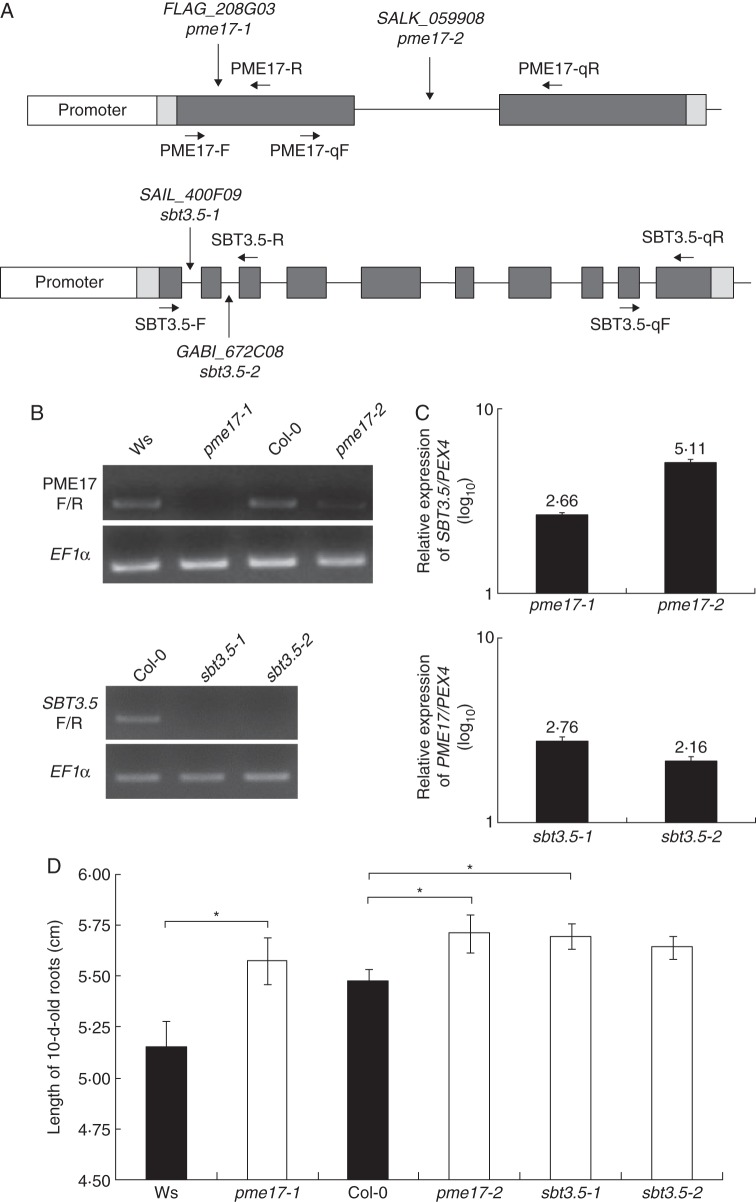
Homozygous pme17–1, pme17–2, sbt3.5–1 and sbt3.5–2 mutants were isolated from FLAG (INRA, Versailles, France), SALK (SIGnAL, USA), SAIL (Syngenta, Basel, Switzerland) and GABI (CeBiTec, Bielefeld, Germany) T-DNA insertion collections, using gene-specific forward and reverse primers and T-DNA left border specific primers (Supplementary Data Table S1).
Arabidopsis thaliana plants (wild-types, mutants and prom:GUS lines) from ecotypes Col-0 and Ws were grown on 0·5× MS solid media (Duchefa, Cat. No. M0221.0001) containing 1 % sucrose and 0·05 % MES monohydrate at pH 5·8. Seeds were treated for 3 d at 4 °C to synchronize germination, and placed in a phytotronic chamber (16-h photoperiod at 120 μmoL m–2 s–1 and 22 °C constant temperature) for in vitro seedling growth. Plants grown on soil were placed in a phytotronic chamber (16-h photoperiod at 100 μmoL m–2 s–1, 70 % relative humidity and 23 °C/19 °C day/night temperature). Transfer to the chamber is referred to as t = 0 for all experiments. Seedlings were harvested at 10 d for RNA and protein extractions and at various time points (1, 2, 3, 4, 7 and 10 d) to determine the activity of the promoters. Various organs were harvested from adult plants for RNA extraction. For root length measurements, 90 seedlings were analysed using ImageJ software (http://rsbweb.nih.gov/ij/) and the NeuronJ plugin, for each of the three biological replicates, and data were statistically analysed using the parametric Student's test (Statistica v9.1, StatSoft, Tulsa, OK, USA). To determine the germination rate, non-sterilized seeds were sown on nutrient-free media, cold-treated for 3 d and transferred to the growth chamber as already mentioned for seedling growth. Germination was followed from 24 to 72 h. Data shown are the means with standard errors (SE) of four replicates, with 30 seeds per replicate. Statistical analyses were performed using a non-parametric Mann–Whitney test with the Statistica software (Statistica v9.1, StatSoft).
Total RNA extraction, cDNA synthesis and gene expression analysis
In-vitro-grown seedlings (10-d-old roots and leaves) and organs from plants grown on soil [young and old leaves, stem, flowers buds, siliques from 3 to 8 and 9 to 17 d after fertilization (DAF) and mature seeds] were dissected and immediately placed in liquid nitrogen. Total RNA was extracted from 100 mg tissue, using TRIzol® reagent (Invitrogen, Carlsbad, CA, USA; Cat. No. 15596–026), according to the manufacturer's recommendations. Genomic DNA was removed using Turbo DNA-free™ kit (Ambion, Austin, TX, USA; Cat. No. AM1907), according to the manufacturer's protocol. cDNA synthesis was performed using 4 μg of RNA, 50 μm oligo (dT)20 and the SuperScript™ III First-Strand Synthesis SuperMix (Invitrogen; Cat. No. 18080–400), using manufacturer's protocol. Semi-quantitative and RT-qPCR analyses were performed on 1/20 diluted cDNA. For RT-qPCR, the LightCycler® 480 SYBR Green I Master (Roche, Indianapolis, IN, USA; Cat. No. 04887352001) was used in 384-well plates in the LightCycler® 480 Real-Time PCR System (Roche). The CT values for each sample (crossing threshold values are the number of PCR cycles required for the accumulated fluorescence signal to cross a threshold above the background) were acquired with the LightCycler 480 software (Roche) using the second derivative maximum method. Primers used are shown in Supplementary Data Table S1 (see also Fig. 4A). Stably expressed reference genes (PEX4, CLA, TIP41, At4g26410 and APT1), selected using GeNorm software (Vandesompele et al., 2002), were used as internal controls to calculate relative expression of target genes, according to the method described by Gutierrez et al. (2009).
Fig. 4.
Characterization of T-DNA insertions lines for PME17 and SBT3.5. (A) Localization of T-DNA insertions in PME17 (top) and SBT3.5 (bottom) genomic DNA sequences. Promoter, 5′-UTR and 3′-UTR, and exons are represented in white, light grey and dark grey bars, respectively. Introns are represented as a black line. Primers F/R and qF/qR were used for semi-quantitative PCR and qPCR analyses, respectively. (B) Semi-quantitative PCR on cDNA from 10-d-old roots of wild-type and mutant plants are shown for PME17 (top) and SBT3.5 (bottom). PME17F/R and SBT3.5F/R primers, flanking the insertion site for pme17–1 and sbt3.5–1/sbt3.5–2, respectively, were used. EF1α is shown as an internal positive control. (C) Relative expression of SBT3.5 in pme17 mutants (top) and PME17 in sbt3.5 mutants (bottom) was quantified in 10-d-old roots using references genes PEX4, CLA and At4g26410. Similar variations were observed with the three references genes, but only the results obtained with PEX4 are shown. (D) Length of 10-d-old roots for wild-type and mutant plants. Data represent the means in ± SE of three independent experiments (n = 90). Significant differences were determined with parametric Student's test (*P < 0·05).
Promoter amplification, plant transformation and GUS staining
1·5 kb upstream of the AtPME17 5′-untranslated region (5′-UTR) were amplified from arabidopsis Col-0 genomic DNA using the Phusion® Taq polymerase (Finnzymes, Waltham, MA, USA; Cat. No. F-540L) and specific forward and reverse primers (Supplementary Data Table S1). The amplified fragment was recombined into pENTR™/D-TOPO® entry vector (Invitrogen; Cat. No. K2400–20) using attL1 and attL2 recombination sites. After sequencing, the promoter was recombined upstream of the GUS coding sequence into the destination vector pKGWFS7,1 (Gent, http://www.psb.ugent.be/), using LR clonase (Invitrogen; Cat. No. 11791–020), following the manufacturer's instructions. Agrobacterium tumefaciens C58C1 was transformed by the plasmid and used for subsequent plant transformation. Arabidopsis Col-0 plants were transformed by the floral dip method (Clough and Bent, 1998). T1 transformants were selected on 50 μg mL–1 kanamycin and T2 plants were used for the experiments.
The promoter region of AtSBT3.5, 1560 bp upstream of the start codon, was amplified by PCR from Arabidopsis Col-0 genomic DNA using specific primers (pSBT3.5-F and pSBT3.5-R, Supplementary Data Table S1) and cloned into pCR2.1 TOPO (Invitrogen). After sequence confirmation, the promoter fragment was subcloned into the plant expression vector pGreen 0029 (Hellens et al., 2000) upstream of the coding sequence for a GUS–GFP fusion protein exploiting the NotI and BamHI restriction sites that were included in the PCR primers. The construct was co-transformed with the helper plasmid pSOUP into A. tumefaciens GV3101 and transformed into Arabidopsis Col-0 plants by floral dip (Clough and Bent, 1998). T1 transformants were selected on BASTA and T2 plants were used for the experiments.
GUS assays were performed as described previously (Sessions et al., 1999), with some modifications. Plant samples were harvested and immediately pre-fixed in ice-cold 80 % acetone over 20 min at –20 °C, then washed three times with distilled water. They were vacuum infiltrated twice for 10 min using GUS staining solution [100 mm sodium phosphate buffer, pH7 (Na2HPO4/NaH2PO4), 0·1 % Triton X100, 10 mm EDTA, 0·5 mm potassium ferrocyanide, 0·5 mm potassium ferricyanide and 1 mg mL–1 X-gluc (Duchefa Biochimie, Haarlem, the Netherlands; Cat. No. X1405)) and incubated at 37 °C for different time periods, depending on GUS lines and developmental stages. Samples were destained in 70 % ethanol and images were acquired using a SteREO Discovery V20 stereo microscope (Zeiss, Jena, Germany).
Protein extraction and proteomic analyses by NanoLC-ESI-MS/MS
Cell-wall-enriched proteins from 10-d-old roots were extracted from 50 mg frozen material using 50 mm sodium acetate and 1 m lithium chloride buffer at pH 5, for 1 h at 4 °C under shaking. The extracts were clarified by centrifugation at 20 000 g for 30 min at 4 °C and the supernatants were filtered using an Amicon ultra centrifugal filter 0·5 mL/10 kDa (Millipore, Billerica, MA, USA; Cat. No. UFC5010BK) to remove salts. Protein concentration was determined by the Bradford method (Bradford, 1976) using a protein assay kit (Bio-Rad, Hercules, CA, USA; Cat. No. 500–0006). Equal amounts of proteins (wild-type and mutant) were resolved on SDS-PAGE using Mini-protean® TGXTM gels (Bio-Rad; gradient 4–20 %, Cat. No. 456–1094) at a constant voltage of 200 V for 45 min. Proteins were stained with Coomassie blue (Bio-Rad; Blue G250, Cat. No. 161–0787) and destained with distilled water.
Each SDS–PAGE band was manually excised from the gels to be hydrolysed according to Shevchenko et al. (1996). All digested peptide mixtures were separated online using nanoLC and analysed by nano-electrospray tandem mass spectrometry. The experiments were performed on an Ultimate 3000 RSLC system coupled with an LTQ-Orbitrap XL mass spectrometer (ThermoFisher Scientific). The peptide mixtures were injected onto a nano trap column (Acclaim C18, 100 μm i.d. × 2 cm length) with a flow rate of 5 μL min–1 and subsequently gradient eluted with at a flow rate of 300 nL min–1 from 2–25 % acetonitrile/0·1 % formic acid over 60 min, followed by second linear increase from 25 to 55 % over 20 min.
Xcalibur 2·3 software was used for mass data acquisition. Full MS scans were acquired at high resolution (full width at half maximum, FWHM, 60 000) in an Orbitrap analyser [mass-to-charge ratio (m/z): 400–2000], while collision-induced dissociation (CID) spectra were recorded in centroid mode with low resolution on the ten most intense ions in the linear ion trap. The mass spectrometer was operated in positive mode in a data-dependent mode to automatically switch between orbitrap-MS and linear trap MS/MS (MS2) as previously described (Olsen et al., 2005). CID spectra were recorded in centroid mode at low resolution on the ten most intense ions in the linear ion trap.
For accurate mass measurements the lock mass option was enabled in both MS and MS/MS mode and the polydimethylcyclosiloxane (PCM) ions generated in the electrospray process from ambient air (17) [protonated (Si(CH3)2O))6; m/z = 445·120025] were used for internal recalibration in real time (Schlosser and Volkmer-Engert, 2003).
For protein database searches of MS/MS spectra, data were processed using ProteomeDiscoverer 1·3 (Thermo Fisher Scientific) and Mascot 2·4 (Matrix Science, Boston, MA, USA). Database searches were run against Swissprot from UniProtKB release 2011–09 non-indexed, on any taxonomy, for tryptic peptides with up to two miscleavages, and carbamidomethylation of cysteins (+57·022 uma) and methionin oxidation (+15·995 uma) variable modifications. Protein identifications were validated only if at least two different sequences (in doubly and/or triply charged state) were identified as first candidates in the protein. Mass accuracy tolerance was set to 10 p.p.m. in MS mode and to 0·8 Da in MS/MS mode. The level of confidence for peptide identifications was estimated using the Percolator node with decoy database searching. Strict FDR (false discovery rate) was set to 0·01, relaxed FDR was set to 0·05 and validation was based on the q-value.
PME activity and zymograms
Total PME activity was measured on cell-wall-enriched protein extracts using citrus pectins (DM 85 %, Sigma, St Louis, MO, USA; Cat. No. P9561–2G) and the alcohol oxidase-coupled colorimetric assay adapted from Klavons and Bennett (1986). Data are the means with their SE of three technical and three independent biological replicates. Statistical differences were determined using a non-parametric Mann–Whitney test with the Statistica software (Statistica v9.1, StatSoft).
PME isoforms were separated by isoelectric focusing (IEF) using a 0·5-mm-thick polyacrylamide gel containing a mixture of pharmalytes to form a pH 6–10·5 gradient. The gel was placed horizontally on a cooled plate and anode (25 mm aspartic acid and 25 mm glutamic acid) and cathode (2 m ethylenediamine, 25 mm arginine and 25 mm lysine) strips were positioned at the top and bottom ends of the gel. Pre-focusing was performed (3000 V max., 15 W max. and 15 mA constant, for 20 min) and equal total PME activities, as determined by the colorimetric assay, were loaded into each well. Focusing was realized with the following conditions: 3000 V max., 50 mA max. and 25 W constant for 50 min followed by 5 min at 30 W constant. After IEF, PME activities were visualized following incubation in 1 % citrus pectins (DM 85 %, Sigma; Cat. No. P9561–2G) and subsequent staining in 0·01 % ruthenium red (Sigma; Cat. No. R2751).
Analysis by Fourier transform-infrared (FT-IR) microspectroscopy
Seven-day-old seedlings grown on a plate were collected and incubated in absolute ethanol for 1 week. The samples were subsequently incubated twice in 80 % ethanol for 5 min at 100 °C, twice in absolute acetone for 5 min at 100 °C and stored in water. Seedlings were squashed between two BaF2 windows and thoroughly rinsed in distilled water for 2 min. The samples were then dried on the window at 37 °C for 20 min. For each condition, 10–15 spectra were collected in the root hair area, where PME17 and SBT3.5 are strongly expressed, for individual seedlings, from three independent cultures (five seedlings from each culture), as described by Mouille et al. (2003). An area of 30 × 30 μm was selected for FT-IR microspectroscopy, using a Thermo-Nicolet Nexus iN 10 MX spectrometer equipped with a continuum microscope accessory (Thermo Scientific). Normalization of the data and the discriminant variable selection method were performed as described by Mouille et al. (2003). Various absorbance wavenumbers were assigned to cell-wall polymer bonds according to the literature (Mouille et al., 2003; Pelletier et al., 2010; Guénin et al., 2011; Peaucelle et al., 2011b; Szymanska-Chargot and Zdunek, 2013).
Structural homology modelling
The protein sequence of AtSBT3.5, without signal peptide (SP) and prodomain, was used for homology searches in the protein data bank (PDB). The best template suitable for the protein was selected using different servers: I-tasser (Roy et al., 2010), Sparks-X (Yang et al., 2011), 3D-Jury (Ginalski et al., 2003), HH-Pred (Söding et al., 2005) and FUGUE (Shi et al., 2001). The model of AtSBT3.5 monomer was built using the tomato Solanum lycopersicon SBT, SlSBT3 (PDB code: 3I6S, Ottmann et al., 2009) as a structural template. The tertiary structure was modelled using Modeller 9v11 (Sali and Blundell, 1993), based on the sequence alignment obtained from FUGUE (Shi et al., 2001).
To determine homodimer 3-D structure prediction, two protein sequences of AtSBT3.5 without SP and prodomain were fused and a model was built using SlSBT3 homodimer as template, using similar methods as for the monomer model. Structural models were visualized and labelled in PyMol software (De Lano, 2002). Potentially important amino acid residues were identified according to the literature (Ottmann et al., 2009; Rose et al., 2010).
Root mean-square deviation (RMSD) values and template modelling (TM) score values were determined according to TM-align (Zhang and Skolnick, 2005). A TM-score >0·5 means the structures share the same fold.
Processing analysis by co-expression of PME17 and SBT3.5 in N. benthamiana
The coding sequence of AtPME17, without stop codon, was amplified from clone pda01681 (RIKEN, http://www.brc.riken.jp/lab/epd/catalog/cdnaclone.html), using Phusion®Taq polymerase and specific forward and reverse primers (Supplementary Data Table S1). The Gateway procedure was used for PME17, with the destination vector ImpGWB417 (Nakagawa et al., 2007, 2009). The open reading frame of AtSBT3.5 was amplified by PCR from pUni51 clone (Clone U19516; Arabidopsis Biological Resource Center, https://abrc.osu.edu) with specific primers (Table S1) and cloned into pCR2.1 TOPO-vector (Invitrogen). The sequence was verified and the fragment cloned into the EcoRI sites of pART7, between the CaMV-35S promoter and the terminator sequence. The expression cassette was then subcloned into pART27 (Gleave, 1992).
N. benthamiana plants were grown for 6 weeks in the greenhouse (25 °C, 12 h photoperiod). For transient expression of PME17 and SBT3.5, they were infiltrated with suspensions of A. tumefaciens C58C1 harbouring the expression constructs (PME17–4 × myc in ImpGWB417 and SBT3.5 in pART27) and pART27 as the empty vector control. For enhanced protein expression, the bacteria were always co-infiltrated with another C58C1 strain containing the p19 silencing suppressor. For co-expression of PME17 and SBT3.5, the respective constructs were co-infiltrated at equal optical density, and for the expression of PME17 alone, the PME17 construct was co-infiltrated with bacteria containing the empty vector pART27.
Five days after agro-infiltration, three leaves from 3–4 plants were pooled and vacuum-infiltrated with 50 mm Na-phosphate buffer, pH 7·0, containing 300 mm NaCl. Apoplastic washes were collected by centrifugation at 1000 g at 4 °C for 7 min. Apoplastic proteins were analysed by SDS–PAGE (Laemmli, 1970) and western blot using monoclonal mouse anti-myc (9E10 hybridoma supernatant, 1:20; ATCC number CRL-1729) as the primary antibody, and horseradish-conjugated antimouse IgG (Calbiochem, San Diego, CA, USA; 1:5000) as the secondary antibody. Western blots were developed by enhanced chemiluminescence on X-ray film. For total protein extraction, the leaf material was ground in 1·5 mL extraction buffer (0·5 m Na-acetate, pH 5·2, 15 mm β-mercaptoethanol, 1 % activated charcoal) per gram fresh weight and the extract cleared by centrifuging (15 000 g, 4 °C, 2 min).
To determine the degree of cytoplasmic contamination, α-mannosidase activity was assayed in apoplastic washes and total protein extracts. Ten microlitres of apoplastic and total protein extracts was incubated with 0·5 mg substrate (4-nitrophenyl-α-d-mannopyranoside) in 0·1 m Na-acetate buffer, pH 5·2. After 15 min at 37 °C, the reaction was stopped with 10 % Na-carbonate and absorption was measured at 405 nm. α-Mannosidase activity was calculated as OD405 per gram fresh weight and the contamination of the apoplastic wash was estimated as percentage of the activity in total protein extracts.
RESULTS
PME17 and SBT3.5 genes are co-expressed during Arabidopsis development
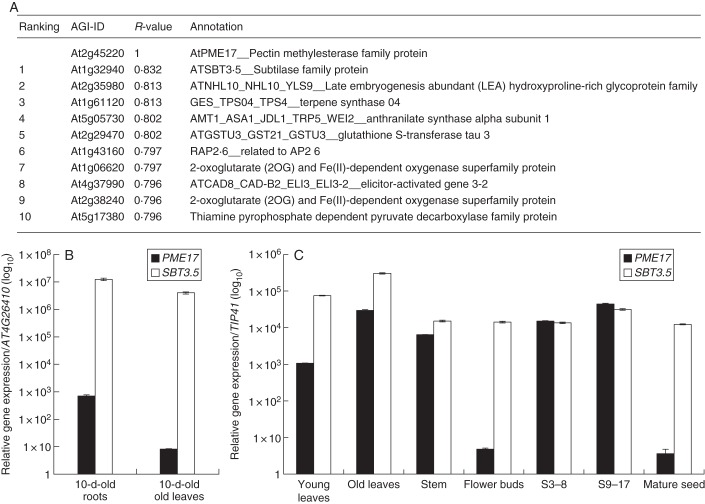
To identify putative PME–SBT pairs, we used the Expression Angler tool of the Bio-Analytic Resource for Plant Biology (BAR, http://bar.utoronto.ca/welcome.htm) and PME17 as the query. Among the top ten genes that were found to be co-expressed with PME17, SBT3.5 ranked number one with an R-value of 0·832 (Fig. 1A). Other genes on this list included amino acids biosynthesis-related (At2g29470, At1g06620, At2g38240) and response to stress-related (At2g35980, At4g37990) genes. Other SBTs (At1g32960) and other cell-wall-related genes were potentially co-expressed with PME17, but with much lower R-value (data not shown). To confirm PME17–SBT3.5 co-expression, we first used RT-qPCR to measure the relative expression of PME17 and SBT3.5 in various organs and developmental stages [mature seeds, siliques (S3–8 DAF, S9–17 DAF), flowers buds, stems, roots and leaves] of Arabidopsis Col-0. As compared with stably expressed reference genes, the relative expression of both genes followed the same trend in all organs and developmental stages tested, except for flower buds and mature seeds, where PME17 was expressed at very low levels, while SBT3.5 was strongly expressed (Fig. 1B, C). Expression of both genes was particularly high in roots of plants grown in vitro.
Fig. 1.
Identification of SBT3.5 as being co-expressed with PME17. (A) Top ten genes co-expressed with AtPME17. Co-expression analysis was performed using the Expression Angler tool of the Bio-Analytic Resource for Plant Biology (BAR, Toufighi et al., 2005). (B) Relative gene expression of PME17 (closed bars) and SBT3.5 (open bars) in Arabidopsis seedlings was measured using stably expressed reference genes (AT4G26410 and PEX4) with similar results. Only results obtained with At4g26410 are shown. (C) Relative gene expression of PME17 (closed bars) and SBT3.5 (open bars) in various organs of Arabidopsis grown on soil was measured using stably expressed reference genes (TIP41 and APT1) with similar results. Only results obtained with TIP41 are shown.
To localize the expression of PME17 and SBT3.5, approx. 1·5 kb of their promoters was PCR amplified and cloned upstream of a GUS coding sequence. Following plant transformation, GUS staining was visualized in light-grown seedlings during development. PME17 and SBT3.5 promoters were particularly active in roots, from 2 d after germination onwards (Fig. 2). Our results show that the activities of the promoters were overlapping, in particular in the root-hair zone, in lateral roots and in the root outer cell layer. While PME17 and SBT3.5 promoter activities were higher in primary roots than lateral roots, no apparent activity was detected in the central cylinder of the roots. Analysis of sequences revealed that specific transcription factor binding sites were conserved when comparing the PME17 and SBT3.5 promoters, including putative DNA binding sites for ARF, BES1/BIM1–3, BLR or LFY transcription factors (Supplementary Data Table S2). These transcription factors are known to regulate the expression of genes involved in control of cell-wall modifications and plant development.
Fig. 2.
Promoter activities of PME17 and SBT3.5. GUS staining of pPME17:GUS (A, C, E, G, I, K) and pSBT3.5:GUS (B, D, F, H, J, L) are shown for seedlings at different age: 1 d (A, B), 2 d (C, D), 3 d (E, F), 4 d (G, H), 7 d (I, J) and 10 d (K, L). Scale bars: 0·2 mm (A, B), 0·5 mm (C–F), 1 mm (G, H), 2 mm (I, J) and 5 mm (K, L).
Processed PME17 and SBT3.5 proteins are identified in cell-wall enriched protein extracts
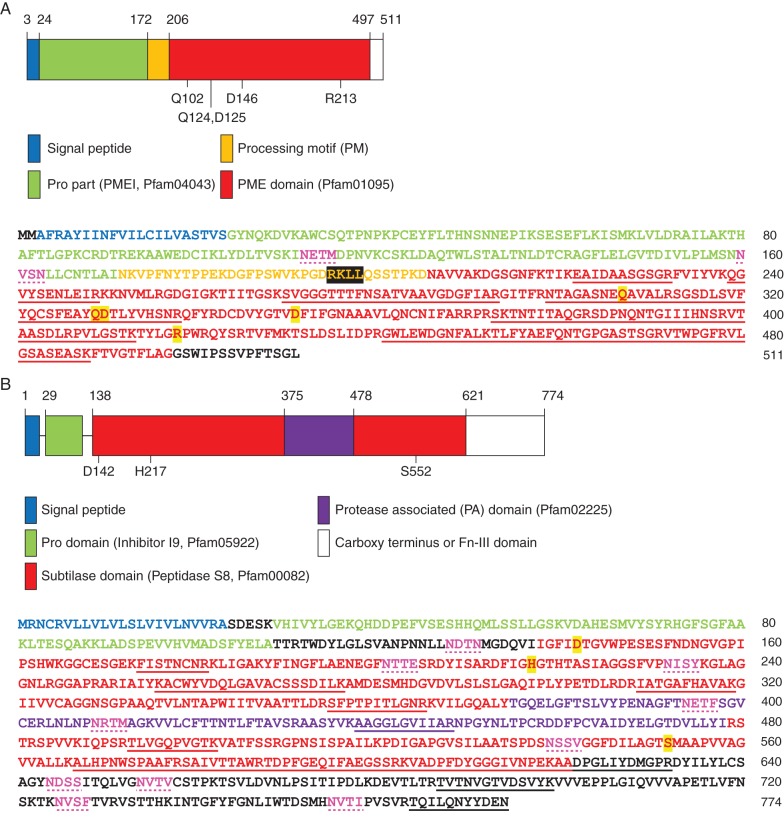
Proteins from 10-d-old roots and cell-wall-enriched extracts isolated from Ws, Col-0, pme17–1 and sbt3.5–1 were resolved by SDS–PAGE and identified using LC-MS Orbitrap analyses. Thirty proteins that are potentially involved in HG modifications were identified in these extracts, including PME17 and SBT3.5 (Table 1). The analysis further revealed 13 specific peptides mapping PME17, ranging from amino acids 222 to 488, resulting in 56 % coverage of the predicted PME domain (Pfam01095, Fig. 3A). In contrast, no peptide mapping the putative PMEI domain (Pfam04043) was detected. Fourteen peptides ranging from amino acids 174 to 774 were identified for SBT3.5, covering 25 % of the sequence of the mature protease lacking SP and prodomain. Peptides were identified within the subtilase domain (Pfam00082), the protease-associated (PA) domain as well as in the fibronectin-III (Fn-III) domain, including the extreme C terminus (Fig. 3B and Supplementary Data Table S3). This suggests that SBT3.5, in contrast to, for example, cucumisin, is only processed at the N terminus of the protein. This is consistent with the reported relevance of the Fn-III domain and the C terminus for secretion and the stability of SBTs (Cedzich et al., 2009; Ottmann et al., 2009) and appears to be a common feature of Arabidopsis SBTs, given that for the majority of SBTs retrieved in our study, peptides mapping the C-terminal Fn-III domain of the protein were identified (Table S3). After sequence comparisons (Supplementary Data Fig. S1), the tomato subtilase (SlSBT3) was used as a template for the structural modelling of the SBT3.5 isoform (Supplementary Data Fig. S2). SBT3.5 showed the same overall structural organization as SlSBT3 with RMSD = 1·36 Å, TM score = 0·95298 for the modelled monomer, and RMSD = 6·73 Å, TM score = 0·60861 for the homodimer, respectively (Ottmann et al., 2009).
Table 1.
Proteomics analysis of 10-d-old root cell-wall-enriched protein extracts from wild-type (WS and Col-0), pme17 and sbt3.5 plants
| Locus | Protein name | WS | pme17–1 | Col-0 | sbt3.5–1 |
|---|---|---|---|---|---|
| Subtilases (SBTs) | |||||
| At1g30600 | AtSBT2.1 | x | x | x | |
| At1g32940 | AtSBT3.5 | x | x | ||
| At2g04160 | AtSBT5.3, AIR3 | x | x | x | x |
| At2g05920 | AtSBT1.8 | x | x | x | x |
| At2g19170 | AtSBT2.5, SLP3 | x | |||
| At3g14067 | AtSBT1.4 | x | x | x | x |
| At4g20430 | AtSBT2.2 | x | x | x | x |
| At4g21650 | AtSBT3.13 | x | x | ||
| At4g30020 | AtSBT2.6 | x | |||
| At4g34980 | AtSBT1.6, SLP2 | x | x | x | x |
| At5g44530 | AtSBT2.3 | x | x | x | x |
| At5g59090 | AtSBT4.12 | x | x | x | x |
| At5g67360 | AtSBT1.7, ARA12, SLP1 | x | x | x | x |
| Pectin methylesterases (PMEs) | |||||
| At1g53830 | AtPME2 | x | x | x | x |
| At2g45220 | AtPME17 | x | x | x | |
| At3g14310 | AtPME3 | x | x | x | x |
| At3g43270 | AtPME32 | x | x | ||
| At4g33220 | AtPME44 | x | x | ||
| At5g04960 | AtPME46 | x | |||
| At5g09760 | AtPME51 | x | x | x | x |
| Pectin acetylesterases (PAEs) | |||||
| At2g46930 | AtPAE | x | x | ||
| At4g19410 | AtPAE | x | x | x | x |
| At5g45280 | AtPAE | x | x | x | x |
| Polygalacturonases (PGs) | |||||
| At3g16850 | AtPG | x | x | x | x |
| At3g62110 | AtPG | x | x | ||
| At4g23500 | AtPG | x | x | x | x |
| At3g57790 | AtPG | x | x | x | x |
| Pectin methylesterase inhibitors (PMEIs) | |||||
| At4g12390 | AtPMEI | x | |||
| At4g25260 | AtPMEI7 | x | x | ||
| At5g62350 | AtPMEI | x | x | ||
Equal amounts of cell-wall-enriched protein extracts from 10-d-old roots of wild-type, pme17–1 and sbt3.5–1 were resolved by SDS-PAGE. Protein bands were dissected, trypsin digested and analysed by LC-MS. The presence of peptides mapping the sequences of SBT, PME, PG, PAE, PMEI is indicated.
Bold indicates the presence/absence of the two proteins of interest: PME17 and SBT3.5.
Fig. 3.
PME17 and SBT3.5 proteins are identified in cell-wall-enriched 10-d-old root protein extracts. Structural domains (top) and amino acid sequences with peptides identified by MS (bottom) are shown for PME17 (A) and SBT3.5 (B). On the structural domains, numbers indicate amino acids of the catalytic site, according to numbering of crystalized models, and domain boundaries. On the amino acid sequences, peptides identified by MS are underlined. Residues involved in catalysis are highlighted in yellow and putative N-glycosylation sites are coloured in pink and dotted underlined. In the PME17 sequence, the putative basic processing motif RKLL is highlighted in black.
pme17 and sbt3.5 mutants display similar phenotypes
Two T-DNA insertion lines were identified for both PME17 and SBT3.5. The insertions were localized in the first exon and in the intron for pme17–1 (FLAG_208G03) and pme17–2 (SALK_059908), respectively. For SBT3.5, the insertions were localized in the first and second intron for sbt3.5–1 (SAIL_400F09) and sbt3.5–2 (GABI_672C08), respectively (Fig. 4A). PCR on 10-d-old root cDNAs confirmed pme17–1, sbt3.5–1 and sbt3.5–2 as true KO lines, while pme17–2 was a knock-down line which displayed, as assessed by qPCR, 100-fold reduction of target gene expression compared with the wild-type (Fig. 4B and data not shown). Levels of PME17 and SBT3.5 transcripts were further measured in the sbt3.5 and pme17 mutant backgrounds showing that SBT3.5 expression was significantly increased in the two pme17 mutant alleles. In parallel, PME17 transcript levels were increased by twofold in sbt3.5 mutants (Fig. 4C). Apparently, the plant compensates for the loss of PME17 function by overexpressing SBT3.5, and vice versa, which will need to be further investigated. pme17–1 and sbt3.5–1 were also confirmed as KO mutants by proteomic analysis, which did not detect any PME17- or SBT3.5-derived peptides in 10-d-old root cell wall-enriched protein extracts in mutants compared with respective wild-types (Table 1). Interestingly, peptides matching the mature part of PME17 were identified in sbt3.5–1, suggesting that other root SBTs (Table 1) could compensate for the lack of SBT3.5 and thus process PME17 into a mature active protein. In addition, peptides mapping to several other cell-wall proteins [SBTs, polygalacturonases (PGs), PMEs, pectin acetylesterases (PAEs)] were identified in roots of wild-type (Ws and Col-0), pme17–1 and sbt3.5–1, and some of these proteins appear to be differentially expressed in wild-type and mutant contexts. This included At3g62110 (PG), for which peptides were identified in sbt3.5–1 but not in the corresponding wild-type roots (Col-0). Peptides mapping At4g30020 (AtSBT2.6), At5g04960 (AtPME46) and At4g12390 (AtPMEI) were identified in pme17–1 but not in the corresponding wild-type (Ws). In contrast, peptides mapping AtSBT2.5 and At3g62110 were identified in Ws but not in pme17–1. These observations indicate that mutations in PME17 and SBT3.5 have consequences that go far beyond the sole extinction of the genes of interest, and these indirect effects may contribute to some of the phenotypes observed in the mutants.
The defects in PME17 and SBT3.5 expression lead to transient delay in germination at 24 h (Supplementary Data Fig. S3), which was unlikely to be related to changes in the release and structure of seed coat mucilage (data not shown), and a small but significant increase in the length of the primary root after 10 d of culture (Fig. 4D). Averages of 6 and 3 % increase in root length over the wild-type were observed for pme17 and sbt3.5 mutants, respectively. The results were similar for both mutant alleles, with a more marked effect for pme17–1 and sbt3.5–1. Thus, we further investigated the consequences of the mutations on PME activity and cell wall structure in these two lines.
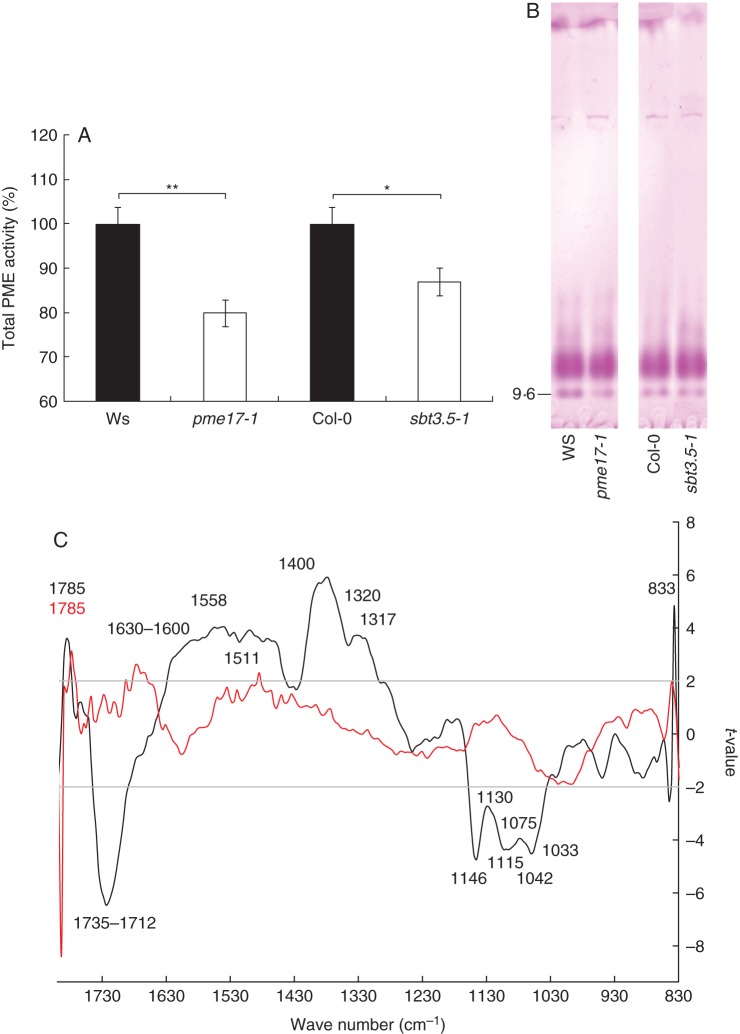
Total PME activity is decreased in pme17 and sbt3.5 mutants, with consequent effects on the DM of pectins
Using similar protein extraction procedures as described for proteomic analysis, we measured total PME activity in pme17–1 and sbt3.5–1 roots. A significant 20 and 13 % decrease in total PME activity was observed for pme17–1 and sbt3.5–1, respectively (Fig. 5A). The loss of SBT3.5 function could thus impair the processing of root-expressed PMEs, with consequent effects on the production of mature active isoforms. The decrease in total PME activity was related, at least for pme17–1, to a decrease in the activity of a PME isoform (pI = 9) revealed by IEF (Fig. 5B). In contrast, no apparent changes in the balance between the activities of PME isoforms could be observed when comparing sbt3.5–1 and wild-type plants. In accordance with proteomic analysis, this showed that PME17 was effectively processed in sbt3.5–1 by root-expressed SBTs, which could potentially compensate for the disappearance of SBT3.5. Together with in silico analysis, these results suggest that PME17 could be part of a pool of basic PME isoforms which also includes the previously identified PME3 (Guénin et al., 2011). To investigate whether the decrease in total PME activity in the pme17–1 mutant could be related to changes in the expression of some other PME and PMEI genes, the expression of PME2, PME3, PME32, PMEI4 and PMEI7 was assessed by RT-qPCR in 10-d-old roots. These five genes were previously reported to be expressed in roots and to play a role in pectin modifications during development (Pelletier et al., 2010; Guénin et al., 2011). Our results showed that the expression of PME3 was significantly down-regulated (<2-fold) and that of PMEI4 up-regulated (>5-fold) in the pme17–1 mutant compared with the wild-type (Supplementary Data Fig. S4).
Fig. 5.
Changes in cell-wall structure are associated with changes in PME activities. (A) Total PME activity in 10-d-old roots of wild-type, pme17–1 and sbt3.5–1 KO mutants. Data represent the means ± SE of three independent experiments. Significant differences were determined with non-parametric Mann–Whitney test (*P < 0·05 and **P < 0·01). (B) Isoelectric focusing (IEF) of cell-wall-enriched protein extracts prepared from 10-d-old roots of wild-type, pme17–1 and sbt3.5–1 KO plants. The same PME activities (15 mU) were loaded for each condition. After IEF, PME activity was detected by incubation in a pectin (DM 85 %) solution, followed by staining with ruthenium red. Similar observations were obtained for three independent experiments. (C) Comparison between FT-IR spectra collected on wild-type and pme17 or sbt3.5 mutant plants. WS versus pme17–1 is represented as a black line. Col-0 versus sbt3.5–1 is represented as a red line. Horizontal lines refer to the P = 0·95 significance threshold (Student's test). Wavenumbers for which significant differences were observed are indicated in black for Ws versus pme17–1 and in red for Col-0 versus sbt3.5–1.
Next we assessed the consequences of the mutations in PME17 and SBT3.5 on root cell-wall structure using FT-IR microspectroscopy at the site of the main promoter activities in the root-hair zone. A strong and highly significant (P < 0·001) increase in absorbance at 1735–1712 cm–1, the wavenumber assigned to one pattern of ester linkages, was observed for pme17–1 compared with the wild-type (Fig. 5C). Similar results were observed for pme17–2 (Supplementary Data Fig. S5). A higher abundance of ester linkages is in accordance with the observed decrease in total PME activity in the mutant and confirms the biochemical activity of PME17. Significant differences in absorbance were also observed for other wavenumbers (Mouille et al., 2003; Pelletier et al., 2010; Szymanska-Chargot and Zdunek, 2013). In particular, a decrease in the absorbance for wavenumbers corresponding to amide bonds (1558 and 1511 cm–1), cellulose (1426, 1370 and 1317 cm–1), xyloglucan (1370 cm–1), pectin (1320 and 833 cm–1) and carboxylate of the pectin ester group (1630–1600 and 1400 cm–1) was observed in pme17–1 compared with the wild-type. In contrast, the absorbance for wavenumbers corresponding to the polysaccharide fingerprint of cellulose (1115 and 1033 cm–1), xyloglucan (1130, 1075 and 1042 cm–1) and pectin glycosidic link (1146 cm–1) were significantly increased in pme17–1 compared with wild-type. This suggests that alteration of PME activity had consequent effects on other cell-wall polymers. Although FT-IR spectra for the sbt3.5 mutants showed no overall drastic changes, a significant decrease (P < 0·01) in the absorbance for wavenumber 1785 cm–1 was observed in the sbt3.5 mutants (Fig. 5C and Supplementary Data Fig. S5). This wavenumber could correspond to a distinct pattern of methylester (for instance in the distribution of methylesters on the HG chain), as chemical environment surrounding methylesters in the cell wall could lead to a shift of absorbances. Although the changes observed between wild-type and mutant for this specific wavenumber were similar for pme17 and sbt3.5, the lack of strong differences in the absorbance for 1735–1712 cm–1 in sbt3.5 suggests potential compensatory effects within the SBT gene family.
PME17 is processed by SBT3.5
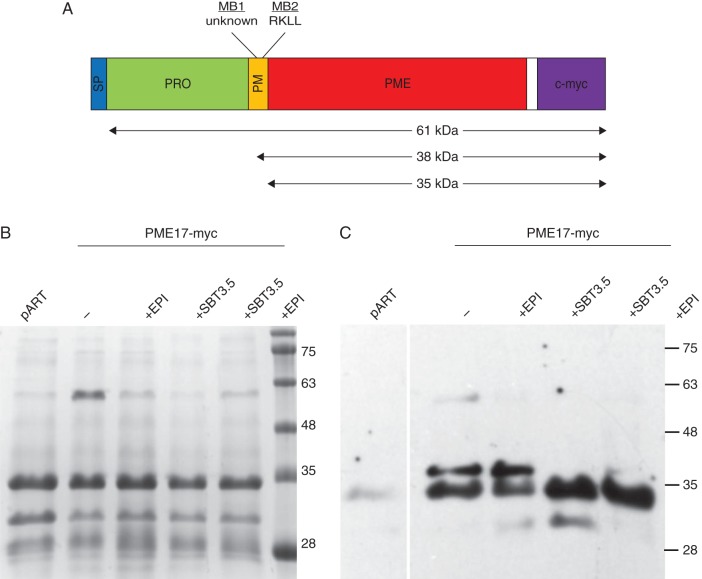
To assess if SBT3.5 can indeed process full-length PME17 and mediate the release of the PME domain into the apoplasm, transient co-expression experiments were performed in N. benthamiana, followed by apoplastic protein extraction and western blotting. For this, expression constructs for a C-terminally myc-tagged version of PME17 were agro-infiltrated in tobacco leaves with SBT3.5 (Fig. 6A) in the presence or absence of EPI1 and EPI10, SBT inhibitors belonging to the Kazal family of serine protease inhibitors (Tian and Kamoun, 2005). Following apoplastic washes, equal amounts of extracted proteins were resolved by SDS–PAGE (Fig. 6B), transferred to nitrocellulose membrane and probed with anti-c-Myc antibodies (Fig. 6C). In the absence of SBT3.5, two bands in a molecular mass range of 35–38 kDa were detected in the apoplasm (Fig. 6C). This suggests that, even though a single RKLL sequence was identified, two processing motifs could be present in the PME17 amino acid sequence, both of which are cleaved by an endogenous tobacco subtilase/protease. An additional band at a molecular mass close to 61 kDa probably represents the non-processed form of PME17. The recovery of this non-processed form in apoplastic washes is likely to be explained by a slight contamination (5 %) with cytosolic content, as measured through an α-mannosidase enzymatic assay (Supplementary Data Table S4). When SBT3.5 was co-infiltrated with PME17, the larger band disappeared, suggesting that PME17 is cleaved by SBT3.5 at at least one of the two processing sites, probably the RKLL motif. An additional lower band was detected that could indicate the presence of N-terminal degradation products of PME17. In the presence of the SBT inhibitor EPI, no difference in the processing of PME17 was revealed. These results indicate that SBT3.5 is able to process PME17 and because both proteins are co-expressed in Arabidopsis roots where they are co-targeted to the secretory pathway and apoplasm, they support a role for SBT3.5 in the maturation and regulation of PME17 in vivo.
Fig. 6.
Processing of proPME17:c-myc by SBT3.5. (A) Schematic representation of the c-Myc tagged version of PME17. Cleavage on a cryptic processing motif (MB1, see below) leads to the production of a 38-kDa protein. Cleavage at the RKLL motif (MB2) leads to the production of a 35-kDa isoform. Non-processed PME17 has an expected molecular mass of 61 kDa. (B) SDS-PAGE of apoplastic washes from N. benthamiana leaves infiltrated with either proPME17:c-myc, or proPME17:c-myc and the SBT inhibitor EPI, proPME17:c-myc and SBT3.5 and the combination of the three. Equal amounts of proteins were loaded. Proteins were stained using Commassie blue. (C) Western blot analysis of apoplastic proteins using a monoclonal antibody against the c-myc epitopes as the primary and horseradish peroxidase-conjugated anti-mouse IgG as the secondary antibodies. Western blots were developed by enhanced chemiluminescence and exposure to X-ray film.
DISCUSSION
To investigate the relevance of the proteolytic processing of group 2 PMEs by SBTs in vivo, we first looked for spatially and temporally co-expressed isoforms during Arabidopsis development. Among the wealth of available data, PME17 and SBT3.5 appeared to be two candidates of interest, being strongly co-expressed in roots. To our knowledge, no such co-expression approach on group 2 PMEs and SBTs has been undertaken so far, despite the fact that this approach has previously revealed relevant candidate genes for the tuning of pectin methylesterification during plant development. For instance, PME1 and PMEI2, which are co-expressed in pollen, were shown to interact during pollen tube elongation (Rôckel et al., 2008). Similarly, PME5 and PMEI3, which are co-expressed at the shoot apical meristem, play a key role in mediating local changes in HG structure with consequences for primordia emergence (Peaucelle et al., 2008). Up to now, although the processing of group 2 PMEs was shown to occur in plants and SBTs have been implicated in the process, the SBTs responsible for PME processing were either not identified, for instance in tobacco (Bosch et al., 2005; Dorokhov et al., 2006), or rather atypical as in the case of AtS1P (Wolf et al., 2009). AtS1P is more similar to mammalian SBTs than to other plant SBTs (Schaller et al., 2012) and in addition, AtS1P is a Golgi-resident protein (Liu and Howell, 2010a, b), while most other SBTs are secreted, or predicted to be secreted, by the cell wall (Von Groll et al., 2002; Hamilton et al., 2003; Rautengarten et al., 2005; Srivastava et al., 2008; Albenne et al., 2013; Ramírez et al., 2013). The relevance of S1P for the processing of PMEs may thus be questioned and while S1P was found to be co-localized with the group 2 PME VGD1, the identification of other co-expressed PME–SBT pairs in specific developmental processes is warranted.
The identification of PME17 and SBT3.5 as a highly co-expressed SBT–PME pair prompted us to develop two distinct approaches to address the potential role of the SBT3.5 protein in the processing of PME17. The first approach used specific Arabidopsis homozygous T-DNA insertion lines to investigate whether PME17 and SBT3.5 are linked functionally in planta. The second approach used N. benthamiana as a heterologous system to determine the ability of SBT3.5 to cleave the PRO domain of PME17.
As PME17 and SBT3.5 are strongly expressed in root epidermis and particularly in the root hair area, the role of the encoded proteins was determined in this organ. Despite this rather specific localization, the expression patterns of the PME and SBT gene families show that potential redundancy of isoforms is likely to occur in roots (Rautengarten et al., 2005; Wang et al., 2013). For instance, AtPME3 and AtSBT4.12 were previously shown to have partially overlapping expression patterns when compared with PME17 and SBT3.5 (Kuroha et al., 2009; Guénin et al., 2011). Interestingly, pme17 and sbt3.5 display similar phenotypes, at the level of both total PME activity and root growth. The decrease in total PME activity measured in the pme17–1 mutant, and its consequent effects on the DM of HG revealed by FT-IR, is similar to what was previously reported for the pme3 mutant (Guénin et al., 2011). In addition, changes in the DM of HG were previously reported to mediate growth phenotypes (Mouille et al., 2003; Hewezi et al., 2008; Pelletier et al., 2010; Guénin et al., 2011).
The activity of the PME17 promoter, being excluded from the root elongation zone, suggested that the observed root elongation phenotype may be an indirect effect of the loss of PME17 function. Indeed, several genes implicated in HG modification were found to be up-regulated in the pme17 mutant. Proteomics analyses of pme17–1 detected peptides mapping one PME (At5g04960) and one PMEI (At4g12390) that were absent in the wild-type. Furthermore, expression analysis of various PME and PMEI genes known to be expressed in roots (Pelletier et al., 2010; Guénin et al., 2011) showed that PME3 was down-regulated and PMEI4 was up-regulated in the pme17 mutant. Both genes are expressed in the root elongation zone and could thus contribute to the overall changes in total PME activity as well as to the increased root length observed in pme17 mutants.
In other studies, using KO for PME genes or overexpressors for PMEI genes, alteration of primary root growth is correlated with a decrease in total PME activity and related increase in DM (Lionetti et al., 2007; Hewezi et al., 2008). Similarly, total PME activity was decreased in the sbt3.5–1 KO as compared with the wild-type, despite increased levels of PME17 transcripts. Considering previous work with S1P (Wolf et al., 2009), one obvious explanation would be that processing of group 2 PMEs, including PME17, may be impaired in the sbt3.5 mutant resulting in the retention of unprocessed, inactive PME isoforms inside the cell. However, for other sbt mutants, different consequences on PME activity were reported. In the atsbt1.7 mutant, for instance, an increase in total PME activity was observed (Rautengarten et al., 2008; Saez-Aguayo et al., 2013). This discrepancy probably reflects the dual, isoform-dependent function of SBTs: in contrast to the processing function we propose here for SBT3.5, SBT1.7 may rather be involved in the proteolytic degradation of extracellular proteins, including the degradation of some PME isoforms (Hamilton et al., 2003; Schaller et al., 2012).
While the similar root elongation phenotypes of the sbt3.5 and pme17 mutants imply a role for SBT3.5 in the regulation of PME activity and the DM, a contribution of other processes cannot be excluded. For instance, root growth defects could be also be explained by impaired proteolytic processing of other cell-wall proteins, including growth factors such as AtPSKs (phytosulfokines) or AtRALFs (rapid alkalinization growth factors) (Srivastava et al., 2008, 2009). Some of the AtPSK and AtRALF precursors may be direct targets of SBT3.5 or, alternatively, may be processed by other SBTs that are up-regulated in compensation for the loss of SBT3.5 function. AtSBT4.12, for instance, is known to be expressed in roots (Kuroha et al., 2009), and peptides mapping its sequence were retrieved in cell-wall-enriched protein fractions of pme17 roots in our study. SBT4.12, as well as other root-expressed SBTs, could target group 2 PMEs identified in our study at the proteome level (i.e. PME3, PME32, PME41 and PME51), all of which show a dibasic motif (RRLL, RKLL, RKLA or RKLK) between the PRO and the mature part of the protein.
The co-expression of PME17 and SBT3.5 in N. bethamiana formally demonstrated the ability of SBT3.5 to cleave the PME17 protein and to release the mature form in the apoplasm. Given that the structural model of SBT3.5 is very similar to that of tomato SlSBT3 previously crystallized (Ottmann et al., 2009), a similar mode of action of the homodimer could be hypothesized (Cedzich et al., 2009). Interestingly, unlike the majority of group 2 PMEs, which show two conserved dibasic processing motifs, most often RRLL or RKLL, a single motif (RKLL) was identified in the PME17 protein sequence upstream of the PME domain. Surprisingly, in the absence of SBT3.5, cleavage of PME17 by endogenous tobacco proteases/subtilases leads to the production of two proteins that were identified by the specific anti-c-myc antibodies. This strongly suggests that, in addition to the RKLL motif, a cryptic processing site is present in the PME17 protein sequence. Although the presence of two processed PME isoforms was previously described for PMEs with two clearly identified dibasic processing motifs (tobacco proPME1, Arabidopsis VGD1 and PME3), their roles remained have remained elusive (Dorokhov et al., 2006; Wolf et al., 2009; Weber et al., 2013). For all of these proteins, a strong preference of processing was found at the RRLL site, regardless of whether it was placed in the first or in second position, compared with RKLK, RKLM and RKLR motifs. When SBT3.5 was co-expressed with PME17, a shift in the equilibrium between the two processed PME17 isoforms was observed. The isoform with the lowest molecular mass, probably the one processed at the RKLL site, was more abundant than the larger one, probably to be processed at a cryptic site upstream of the RKLL motif. Based on these results, we postulate that SBT3.5 has a preference for the RKLL motif, and is able to process PME17 as a possible mechanism to fine tune its activity.
CONCLUSIONS
Following the identification, through data mining, of two co-expressed genes encoding a putative pectin methylesterase (PME) and a subtilisin-type serine protease (SBT), we used RT-qPCR and promoter:GUS fusions to confirm that both genes had overlapping expression patterns during root development. We further identified processed isoforms for both proteins in cell-wall-enriched protein extracts of roots. Using Arabidopsis pme17 and sbt3.5 T-DNA insertion lines we showed that total PME activity in roots was impaired. This notably confirmed the biochemical activity of PME17 and suggested that in a wild-type context, SBT3.5 could target group 2 PMEs, possibly including PME17. Mutations in both genes led to similar root phenotypes. Using biochemical approaches we finally showed that SBT3.5 can indeed process PME17, releasing the mature form into the apoplasm. Our study brings new insights into the complexity of the post-translational regulation of group 2 PMEs, and highlights the need to identify SBT isoforms involved in the process.
SUPPLEMENTARY DATA
ACKNOWLEDGEMENTS
We thank Professor Nakagawa from the Department of Molecular and Functional Genomics Center for Integrated Research in Science of Shimane University (Japan) for the kind gift of ImpGWB417 Gateway vector. This work was supported by a grant from the Agence Nationale de la Recherche (ANR-09-BLANC-0007-01, GROWPEC project), the Conseil Régional de Picardie through a PhD studentship awarded to F.S and by the Trans Channel Wallnet project (INTERREG IVA program France (Channel) - England European cross-border cooperation programme, co-financed by the ERDF). The financial support from the Institut Universitaire de France (IUF) to J.P. is gratefully acknowledged.
LITERATURE CITED
- Albenne C, Canut H, Jamet E. Plant cell wall proteomics: the leadership of Arabidopsis thaliana. Frontiers in Plant Science. 2013;4:1–17. doi: 10.3389/fpls.2013.00111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Qsous S, Carpentier E, Klein-Eude D, et al. Identification and isolation of a pectin methylesterase isoform that could be involved in flax cell wall stiffening. Planta. 2004;219:369–378. doi: 10.1007/s00425-004-1246-1. [DOI] [PubMed] [Google Scholar]
- Beers EP, Jones AM, Dickerman AW. The S8 serine, C1A cysteine and A1 aspartic protease families in Arabidopsis. Phytochemistry. 2004;65:43–58. doi: 10.1016/j.phytochem.2003.09.005. [DOI] [PubMed] [Google Scholar]
- Berger D, Altmann T. A subtilisin-like serine protease involved in the regulation of stomatal density and distribution in Arabidopsis thaliana. Genes & Development. 2000;14:1119–1131. [PMC free article] [PubMed] [Google Scholar]
- Bosch M, Cheung A, Hepler P. Pectin methylesterase, a regulator of pollen tube growth. Plant Physiology. 2005;138:1334–1346. doi: 10.1104/pp.105.059865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudart G, Jamet E, Rossignol M, et al. Cell wall proteins in apoplastic fluids of Arabidopsis thaliana rosettes: identification by mass spectrometry and bioinformatics. Proteomics. 2005;5:212–221. doi: 10.1002/pmic.200400882. [DOI] [PubMed] [Google Scholar]
- Bradford M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical Biochemistry. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Braybrook SA, Peaucelle A. Mechano-chemical aspects of organ formation in Arabidopsis thaliana: the relationship between auxin and pectin. PLoS ONE. 2013;8:e57813. doi: 10.1371/journal.pone.0057813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cedzich A, Huttenlocher F, Kuhn BM, et al. The protease-associated domain and C-terminal extension are required for zymogen processing, sorting within the secretory pathway, and activity of tomato subtilase 3 (SlSBT3) Journal of Biological Chemistry. 2009;284:14068–14078. doi: 10.1074/jbc.M900370200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chichkova NV, Shaw J, Galiullina RA, et al. Phytaspase, a relocalisable cell death promoting plant protease with caspase specificity. The EMBO Journal. 2010;29:1149–1161. doi: 10.1038/emboj.2010.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S, Bent A. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- D'Erfurth I, Signor C, Aubert G, et al. A role for an endosperm-localized subtilase in the control of seed size in legumes. The New Phytologist. 2012;196:738–751. doi: 10.1111/j.1469-8137.2012.04296.x. [DOI] [PubMed] [Google Scholar]
- DeLano . PyMOL: An open-sources molecular graphics tool. San Carlos, CA: 2002. http://www.pymol.org/ [Google Scholar]
- Derbyshire P, McCann MC, Roberts K. Restricted cell elongation in Arabidopsis hypocotyls is associated with a reduced average pectin esterification level. BMC Plant Biology. 2007;7:1–12. doi: 10.1186/1471-2229-7-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorokhov YL, Skurat EV, Frolova OY, et al. Role of the leader sequence in tobacco pectin methylesterase secretion. FEBS Letters. 2006;580:3329–3334. doi: 10.1016/j.febslet.2006.04.090. [DOI] [PubMed] [Google Scholar]
- Feiz L, Irshad M, Pont-Lezica RF, Canut H, Jamet E. Evaluation of cell wall preparations for proteomics: a new procedure for purifying cell walls from Arabidopsis hypocotyls. Plant Methods. 2006;2:1–13. doi: 10.1186/1746-4811-2-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis KE, Lam SY, Copenhaver GP. Separation of Arabidopsis pollen tetrads is regulated by QUARTET1, a pectin methylesterase gene. Plant Physiology. 2006;142:1004–1013. doi: 10.1104/pp.106.085274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginalski K, Elofsson A, Fischer D, Rychlewski L. 3D-Jury: a simple approach to improve protein structure predictions. Bioinformatics. 2003;19:1015–1018. doi: 10.1093/bioinformatics/btg124. [DOI] [PubMed] [Google Scholar]
- Gleave A. A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Molecular Biology. 1992;20:1203–1207. doi: 10.1007/BF00028910. [DOI] [PubMed] [Google Scholar]
- Gutierrez L, Bussell JD, Pacurar DI, Schwambach J, Pacurar M, Bellini C. Phenotypic plasticity of adventitious rooting in Arabidopsis is controlled by complex regulation of AUXIN RESPONSE FACTOR transcripts and microRNA abundance. The Plant Cell. 2009;21:3119–3132. doi: 10.1105/tpc.108.064758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guénin S, Mareck A, Rayon C, et al. Identification of pectin methylesterase 3 as a basic pectin methylesterase isoform involved in adventitious rooting in Arabidopsis thaliana. New Phytologist. 2011;192:114–126. doi: 10.1111/j.1469-8137.2011.03797.x. [DOI] [PubMed] [Google Scholar]
- Hamilton JM, Simpson DJ, Hyman SC, Ndimba BK, Slabas AR. Ara12 subtilisin-like protease from Arabidopsis thaliana: purification, substrate specificity and tissue localization. The Biochemical Journal. 2003;370:57–67. doi: 10.1042/BJ20021125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens R, Edwards E, Leyland N, Bean S, Mullineaux P. pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Molecular Biology. 2000;42:819–832. doi: 10.1023/a:1006496308160. [DOI] [PubMed] [Google Scholar]
- Hewezi T, Howe P, Maier TR, et al. Cellulose binding protein from the parasitic nematode Heterodera schachtii interacts with arabidopsis pectin methylesterase: cooperative cell wall modification during parasitism. The Plant Cell. 2008;20:3080–3093. doi: 10.1105/tpc.108.063065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hongo S, Sato K, Yokoyama R, Nishitani K. Demethylesterification of the primary wall by PECTIN METHYLESTERASE35 provides mechanical support to the Arabidopsis stem. The Plant Cell. 2012;24:2624–2634. doi: 10.1105/tpc.112.099325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irshad M, Canut H, Borderies G, Pont-Lezica R, Jamet E. A new picture of cell wall protein dynamics in elongating cells of Arabidopsis thaliana: confirmed actors and newcomers. BMC Plant Biology. 2008;8:1–16. doi: 10.1186/1471-2229-8-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Yang SL, Xie LF, et al. VANGUARD1 encodes a pectin methylesterase that enhances pollen tube growth in the Arabidopsis style and transmitting tract. The Plant Cell. 2005;17:584–596. doi: 10.1105/tpc.104.027631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juge N. Plant protein inhibitors of cell wall degrading enzymes. Trends in Plant Science. 2006;11:359–367. doi: 10.1016/j.tplants.2006.05.006. [DOI] [PubMed] [Google Scholar]
- Klavons J, Bennett R. Determination of methanol using alcohol oxidase and its application to methyl ester content of pectins. Journal of Agricultural and Food Chemistry. 1986;34:597–599. [Google Scholar]
- Kuroha T, Okuda A, Arai M, et al. Identification of Arabidopsis subtilisin-like serine protease specifically expressed in root stele by gene trapping. Physiologia Plantarum. 2009;137:281–288. doi: 10.1111/j.1399-3054.2009.01281.x. [DOI] [PubMed] [Google Scholar]
- Laemmli U. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lionetti V, Raiola A, Camardella L, et al. Overexpression of pectin methylesterase inhibitors in Arabidopsis restricts fungal infection by Botrytis cinerea. Plant Physiology. 2007;143:1871–1880. doi: 10.1104/pp.106.090803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JX, Howell SH. bZIP28 and NF-Y transcription factors are activated by ER stress and assemble into a transcriptional complex to regulate stress response genes in Arabidopsis. The Plant Cell. 2010a;22:782–796. doi: 10.1105/tpc.109.072173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JX, Howell SH. Endoplasmic reticulum protein quality control and its relationship to environmental stress responses in plants. The Plant Cell. 2010b;22:2930–2942. doi: 10.1105/tpc.110.078154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mareck A, Lamour R, Schaumann A, et al. Analysis of LuPME3, a pectin methylesterase from Linum usitatissimum, revealed a variability in PME proteolytic maturation. Plant Signaling & Behavior. 2012;7:59–61. doi: 10.4161/psb.7.1.18632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matos JL, Fiori CS, Silva-Filho MC, Moura DS. A conserved dibasic site is essential for correct processing of the peptide hormone AtRALF1 in Arabidopsis thaliana. FEBS Letters. 2008;582:3343–3347. doi: 10.1016/j.febslet.2008.08.025. [DOI] [PubMed] [Google Scholar]
- Minic Z, Jamet E, San-Clemente H, et al. Transcriptomic analysis of Arabidopsis developing stems: a close-up on cell wall genes. BMC Plant Biology. 2009;9:1–17. doi: 10.1186/1471-2229-9-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouille G, Robin S, Lecomte M, Pagant S, Höfte H. Classification and identification of Arabidopsis cell wall mutants using Fourier-Transform InfraRed (FT-IR) microspectroscopy. The Plant Journal. 2003;35:393–404. doi: 10.1046/j.1365-313x.2003.01807.x. [DOI] [PubMed] [Google Scholar]
- Müller K, Levesque-Tremblay G, Bartels S, et al. Demethylesterification of cell wall pectins in Arabidopsis plays a role in seed germination. Plant Physiology. 2013;161:305–316. doi: 10.1104/pp.112.205724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Suzuki T, Murata S, et al. Improved Gateway binary vectors: high-performance vectors for creation of fusion constructs in transgenic analysis of plants. Bioscience, Biotechnology, and Biochemistry. 2007;71:2095–2100. doi: 10.1271/bbb.70216. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Ishiguro S, Kimura T. Gateway vectors for plant transformation. Plant Biotechnology. 2009;26:275–284. [Google Scholar]
- Olsen JV, De Godoy LM, Li G, et al. Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Molecular & Cellular Proteomics. 2005;4:2010–2021. doi: 10.1074/mcp.T500030-MCP200. [DOI] [PubMed] [Google Scholar]
- Osorio S, Castillejo C, Quesada MA, et al. Partial demethylation of oligogalacturonides by pectin methylesterase 1 is required for eliciting defence responses in wild strawberry (Fragaria vesca) The Plant Journal. 2008;54:43–55. doi: 10.1111/j.1365-313X.2007.03398.x. [DOI] [PubMed] [Google Scholar]
- Ottmann C, Rose R, Huttenlocher F, et al. Structural basis for Ca2+-independence and activation by homodimerization of tomato subtilase 3. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:17223–17228. doi: 10.1073/pnas.0907587106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peaucelle A, Louvet R, Johansen JN, et al. Arabidopsis phyllotaxis is controlled by the methyl-esterification status of cell-wall pectins. Current Biology. 2008;18:1943–1948. doi: 10.1016/j.cub.2008.10.065. [DOI] [PubMed] [Google Scholar]
- Peaucelle A, Braybrook S, Le Guillou L, Bron E, Kuhlemeier C, Höfte H. Pectin-induced changes in cell wall mechanics underlie organ initiation in Arabidopsis. Current Biology. 2011a;21:1720–1726. doi: 10.1016/j.cub.2011.08.057. [DOI] [PubMed] [Google Scholar]
- Peaucelle A, Louvet R, Johansen JN, et al. The transcription factor BELLRINGER modulates phyllotaxis by regulating the expression of a pectin methylesterase in Arabidopsis. Development. 2011b;138:4733–4741. doi: 10.1242/dev.072496. [DOI] [PubMed] [Google Scholar]
- Pelletier S, Van Orden J, Wolf S, et al. A role for pectin de-methylesterification in a developmentally regulated growth acceleration in dark-grown Arabidopsis hypocotyls. New Phytologist. 2010;188:726–739. doi: 10.1111/j.1469-8137.2010.03409.x. [DOI] [PubMed] [Google Scholar]
- Pelloux J, Rusterucci C, Mellerowicz E. New insights into pectin methylesterase structure and function. Trends in Plant Science. 2007;12:267–277. doi: 10.1016/j.tplants.2007.04.001. [DOI] [PubMed] [Google Scholar]
- Raiola A, Lionetti V, Elmaghraby I, et al. Pectin methylesterase is induced in Arabidopsis upon infection and is necessary for a successful colonization by necrotrophic pathogens. Molecular Plant–Microbe Interactions. 2011;24:432–440. doi: 10.1094/MPMI-07-10-0157. [DOI] [PubMed] [Google Scholar]
- Ramírez V, López A, Mauch-Mani B, Gil MJ, Vera P. An extracellular subtilase switch for immune priming in Arabidopsis. PLoS Pathogens. 2013;9:e1003445. doi: 10.1371/journal.ppat.1003445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rautengarten C, Steinhauser D, Büssis D, et al. Inferring hypotheses on functional relationships of genes: analysis of the Arabidopsis thaliana subtilase gene family. PLoS Computational Biology. 2005;1:e40. doi: 10.1371/journal.pcbi.0010040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rautengarten C, Usadel B, Neumetzler L, Hartmann J, Büssis D, Altmann T. A subtilisin-like serine protease essential for mucilage release from Arabidopsis seed coats. The Plant Journal. 2008;54:466–480. doi: 10.1111/j.1365-313X.2008.03437.x. [DOI] [PubMed] [Google Scholar]
- Rôckel N, Wolf S, Kost B, Rausch T, Greiner S. Elaborate spatial patterning of cell-wall PME and PMEI at the pollen tube tip involves PMEI endocytosis, and reflects the distribution of esterified and de-esterified pectins. The Plant Journal. 2008;53:133–143. doi: 10.1111/j.1365-313X.2007.03325.x. [DOI] [PubMed] [Google Scholar]
- Rose R, Schaller A, Ottmann C. Structural features of plant subtilases. Plant Signaling & Behavior. 2010;5:180–183. doi: 10.4161/psb.5.2.11069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nature Protocols. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez-Aguayo S, Ralet MC, Berger A, et al. PECTIN METHYLESTERASE INHIBITOR6 promotes Arabidopsis mucilage release by limiting methylesterification of homogalacturonan in seed coat epidermal cells. The Plant Cell. 2013;25:308–323. doi: 10.1105/tpc.112.106575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sali A, Blundell TL. Comparative protein modeling by satisfaction of spatial constraints. Journal of Molecular Biology. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- Schaller A, Stintzi A, Graff L. Subtilases – versatile tools for protein turnover, plant development, and interactions with the environment. Physiologia Plantarum. 2012;145:52–66. doi: 10.1111/j.1399-3054.2011.01529.x. [DOI] [PubMed] [Google Scholar]
- Schlosser A, Volkmer-Engert R. Volatile polydimethylcyclosiloxanes in the ambient laboratory air identified as source of extreme background signals in nanoelectrospray mass spectrometry. Journal of Mass Spectrometry. 2003;38:523–525. doi: 10.1002/jms.465. [DOI] [PubMed] [Google Scholar]
- Sessions A, Weigel D, Yanofsky M. The Arabidopsis thaliana MERISTEM LAYER 1 promoter specifies epidermal expression in meristems and young primordia. The Plant Journal. 1999;20:259–263. doi: 10.1046/j.1365-313x.1999.00594.x. [DOI] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Analytical Chemistry. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- Shi J, Blundell TL, Mizuguchi K. FUGUE: sequence-structure homology recognition using environment-specific substitution tables and structure-dependent gap penalties. Journal of Molecular Biology. 2001;310:243–257. doi: 10.1006/jmbi.2001.4762. [DOI] [PubMed] [Google Scholar]
- Söding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Research. 2005;33:244–248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava R, Liu JX, Howell SH. Proteolytic processing of a precursor protein for a growth-promoting peptide by a subtilisin serine protease in Arabidopsis. The Plant Journal. 2008;56:219–227. doi: 10.1111/j.1365-313X.2008.03598.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava R, Liu JX, Guo H, Yin Y, Howell SH. Regulation and processing of a plant peptide hormone, AtRALF23, in Arabidopsis. The Plant Journal. 2009;59:930–939. doi: 10.1111/j.1365-313X.2009.03926.x. [DOI] [PubMed] [Google Scholar]
- Szymanska-Chargot M, Zdunek A. Use of FT-IR spectra and PCA to the bulk characterization of cell wall residues of fruits and vegetables along a fraction process. Food Biophysics. 2013;8:29–42. doi: 10.1007/s11483-012-9279-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H, Onouchi H, Kondo M, et al. A subtilisin-like serine protease is required for epidermal surface formation in Arabidopsis embryos and juvenile plants. Development. 2001;128:4681–4689. doi: 10.1242/dev.128.23.4681. [DOI] [PubMed] [Google Scholar]
- Tian M, Kamoun S. A two disulfide bridge Kazal domain from Phytophthora exhibits stable inhibitory activity against serine proteases of the subtilisin family. BMC Biochemistry. 2005;6:1–9. doi: 10.1186/1471-2091-6-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toufighi K, Brady SM, Austin R, Ly E, Provart NJ. The Botany Array Resource: e-northerns, expression angling, and promoter analyses. The Plant Journal. 2005;43:153–163. doi: 10.1111/j.1365-313X.2005.02437.x. [DOI] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biology. 2002;3:1–12. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voiniciuc C, Dean GH, Griffiths JS, et al. FLYING SAUCER1 is a transmembrane RING E3 ubiquitin ligase that regulates the degree of pectin methylesterification in Arabidopsis seed mucilage. The Plant Cell. 2013;25:944–959. doi: 10.1105/tpc.112.107888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Groll U, Berger D, Altmann T. The subtilisin-like serine protease SDD1 mediates cell-to-cell signaling during Arabidopsis stomatal development. The Plant Cell. 2002;14:1527–1539. doi: 10.1105/tpc.001016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Yuan D, Gao W, Li Y, Tan J, Zhang X. A comparative genome analysis of PME and PMEI families reveals the evolution of pectin metabolism in plant cell walls. PLoS ONE. 2013;8:e72082. doi: 10.1371/journal.pone.0072082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M, Deinlein U, Fischer S, et al. A mutation in the Arabidopsis thaliana cell wall biosynthesis gene pectin methylesterase 3 as well as its aberrant expression cause hypersensitivity specifically to Zn. The Plant Journal. 2013;76:151–164. doi: 10.1111/tpj.12279. [DOI] [PubMed] [Google Scholar]
- Wolf S, Rausch T, Greiner S. The N-terminal pro region mediates retention of unprocessed type-I PME in the Golgi apparatus. The Plant Journal. 2009;58:361–375. doi: 10.1111/j.1365-313X.2009.03784.x. [DOI] [PubMed] [Google Scholar]
- Xing Q, Creff A, Waters A, Tanaka H, Goodrich J, Ingram GC. ZHOUPI controls embryonic cuticle formation via a signalling pathway involving the subtilisin protease ABNORMAL LEAF-SHAPE1 and the receptor kinases GASSHO1 and GASSHO2. Development. 2013;140:770–779. doi: 10.1242/dev.088898. [DOI] [PubMed] [Google Scholar]
- Yang Y, Faraggi E, Zhao H, Zhou Y. Improving protein fold recognition and template-based modeling by employing probabilistic-based matching between predicted one-dimensional structural properties of query and corresponding native properties of templates. Bioinformatics. 2011;27:2076–2082. doi: 10.1093/bioinformatics/btr350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Skolnick J. TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Research. 2005;33:2302–2309. doi: 10.1093/nar/gki524. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.