Abstract
Background
The secondary cell wall is a defining feature of xylem cells and allows them to resist both gravitational forces and the tension forces associated with the transpirational pull on their internal columns of water. Secondary walls also constitute the majority of plant biomass. Formation of secondary walls requires co-ordinated transcriptional regulation of the genes involved in the biosynthesis of cellulose, hemicellulose and lignin. This co-ordinated control appears to involve a multifaceted and multilayered transcriptional regulatory programme.
Scope
Transcription factor MYB46 (At5g12870) has been shown to function as a master regulator in secondary wall formation in Arabidopsis thaliana. Recent studies show that MYB46 not only regulates the transcription factors but also the biosynthesis genes for all of the three major components (i.e. cellulose, hemicellulose and lignin) of secondary walls. This review considers our current understanding of the MYB46-mediated transcriptional regulatory network, including upstream regulators, downstream targets and negative regulators of MYB46.
Conclusions and Outlook
MYB46 is a unique transcription factor in that it directly regulates the biosynthesis genes for all of the three major components of the secondary wall as well as the transcription factors in the biosynthesis pathway. As such, MYB46 may offer a useful means for pathway-specific manipulation of secondary wall biosynthesis. However, realization of this potential requires additional information on the ‘MYB46-mediated transcriptional regulatory programme’, such as downstream direct targets, upstream regulators and interacting partners of MYB46.
Keywords: Plant cell wall, secondary wall biosynthesis, MYB46, transcription factor, At5g12870, transcriptional regulation, biomass, Arabidopsis thaliana
INTRODUCTION
Vascular plants have evolved to have secondary cell wall structure between the plasma membrane and the primary cell wall in fibres and tracheid/vessel elements, which provide mechanical support for the growing body and serve as a conduit for long-distance transport of water and solutes, respectively. The secondary walls of these cells allow them to resist gravitational forces and the forces of the tension associated with the transpirational pull on their internal columns of water. Economically, secondary walls constitute the vast majority of plant biomass, which is of primary importance to humans for fibre, pulp and paper manufacture, and as an environmentally cost-effective renewable source of energy. Formation of secondary walls requires co-ordinated transcriptional regulation of the genes involved in the biosynthesis of the major secondary wall components (e.g. cellulose, hemicellulose and lignin). Complex transcriptional networks appear to be involved in the co-ordinated regulation of secondary wall biosynthesis (Demura and Ye, 2010; Ko et al., 2011, 2012; Wang and Dixon, 2011).
Many transcription factors (TFs) have been identified as central regulators of secondary wall biosynthesis (for recent reviews, see Yamaguchi and Demura, 2010; Zhang et al., 2010; Zhong et al., 2010a; Wang and Dixon, 2011; Zhao and Dixon, 2011; Ko et al., 2012; Pimrote et al., 2012; Hussey et al., 2013; Schuetz et al., 2013). Of these, MYB46 (At5g12870) and its paralogue, MYB83 (At3g08500), have been shown to function as a master switch for the secondary wall biosynthetic programme in Arabidopsis thaliana (Zhong et al., 2007a; Ko et al., 2009). Recent studies on this MYB46-mediated regulation have provided novel insights into the transcriptional control of secondary wall biosynthesis (Kim et al., 2013a, b, 2014). This review describes the current understanding of the MYB46-mediated transcriptional regulatory network and its implication on pathway-specific engineering of the properties and quantity of plant biomass.
MYB46-MEDIATED TRANSCRIPTIONAL REGULATION OF SECONDARY WALL BIOSYNTHESIS
The TF MYB46, a central regulator in secondary wall formation (Zhong et al., 2007a; Ko et al., 2009), is specifically expressed in both fibres and xylem cells undergoing secondary wall thickening (Zhong et al., 2007a). Promoter activity of MYB46 was also detected in both protoxylem and metaxylem (Nakano et al., 2010). Constitutive overexpression of either MYB46 or its close homologue MYB83 upregulates the genes involved in secondary wall biosynthesis (e.g. cellulose, hemicellulose and lignin biosynthesis genes), resulting in ectopic deposition of secondary walls even in epidermis, cortex and pith cells that are normally parenchymatous. On the other hand, dominant suppression of MYB46 significantly reduced secondary wall thickening in the fibres and vessels of the transgenic plants (Zhong et al., 2007a; Ko et al., 2009). Indeed, myb46myb83 double mutants show lack of secondary wall formation in both the vessels and fibres, and severe growth arrest in young seedlings followed by wilting and subsequent death (McCarthy et al., 2009). These observations clearly suggest that MYB46/MYB83 function as an essential regulator for secondary cell wall biosynthesis in arabidopsis. However, single myb46 or myb83 loss-of-function mutants do not produce any observable phenotype, while the myb46myb83 double mutant results in a ‘seedling-lethal’ phenotype, suggesting functional redundancy between the two TFs.
Furthermore, several MYB46 orthologues from other plant species have also been shown to function as a master switch for secondary wall biosynthesis, including PtMYB4 from pine, EgMYB2 from eucalyptus, OsMYB46 from rice, PtrMYB2/3/20/21 from poplar and ZmMYB46 from maize (Table 1) (Patzlaff et al., 2003; Goicoechea et al., 2005; Zhong et al., 2011, 2013).
Table 1.
Regulators identified in the transcriptional network of secondary wall biosynthesis in plants
| Protein name | ID | Plant | Function | Regulation | References |
|---|---|---|---|---|---|
| MYB family transcription factors | |||||
| MYB46 | At5g12870 | Arabidopsis thaliana | A direct target of SND1 and regulates secondary wall biosynthesis | Activator | Zhong et al. (2007a); Ko et al. (2009) |
| MYB83 | At3g08500 | Arabidopsis thaliana | Act redundantly with MYB46 | Activator | McCarthy et al. (2009) |
| MYB26/MS35 | At3g13890 | Arabidopsis thaliana | Regulates secondary wall thickening in the endothecium | Activator | Yang et al. (2007) |
| MYB52 | At1g17950 | Arabidopsis thaliana | Regulates secondary wall biosynthesis | Activator | Zhong et al. (2008) |
| MYB54 | At1g73410 | Arabidopsis thaliana | Regulates secondary wall biosynthesis | Activator | Zhong et al. (2008) |
| MYB85 | At4g22680 | Arabidopsis thaliana | Regulates lignin biosynthesis | Activator | Zhong et al. (2008) |
| MYB103 | At1g63910 | Arabidopsis thaliana | Regulates secondary wall biosynthesis | Activator | Zhong et al. (2008) |
| MYB58 | At1g16490 | Arabidopsis thaliana | Activates lignin biosynthetic pathway | Activator | Zhou et al. (2009) |
| MYB63 | At1g79180 | Arabidopsis thaliana | Activates lignin biosynthetic pathway | Activator | Ko et al. (2009); Zhou et al. (2009) |
| MYB75/PAP1 | At1g56650 | Arabidopsis thaliana | Represses lignin biosynthetic pathway | Repressor | Bhargava et al. (2010) |
| MYB32 | At4g34990 | Arabidopsis thaliana | Represses SND1 and lignin biosynthesis | Repressor | Wang et al. (2011) |
| MYB4 | At4g38620 | Arabidopsis thaliana | Probably similar function to MYB32 | Repressor | Wang et al. (2011) |
| MYB7 | At2g16720 | Arabidopsis thaliana | Probably similar function to MYB32 | Repressor | Wang et al. (2011) |
| PtMYB1 | AY356372 | Pinus taeda | Regulates secondary wall biosynthesis | Activator | Bomal et al. (2008) |
| PtMYB4 | AY356371 | Pinus taeda | Regulates lignin biosynthesis | Activator | Patzlaff et al. (2003) |
| PtMYB8 | DQ399057 | Pinus taeda | Regulates secondary wall biosynthesis | Activator | Bomal et al. (2008) |
| PtrMYB2 | Potri.001G258700 | Populus trichocarpa | Regulates secondary wall biosynthesis | Activator | Zhong et al. (2013) |
| PtrMYB3 | Potri.001G267300 | Populus trichocarpa | Regulates secondary wall biosynthesis | Activator | Zhong et al. (2013) |
| PtrMYB20 | Potri.009G061500 | Populus trichocarpa | Regulates secondary wall biosynthesis | Activator | Zhong et al. (2013) |
| PtrMYB21 | Potri.009G053900 | Populus trichocarpa | Regulates secondary wall biosynthesis | Activator | Zhong et al. (2013) |
| PttMYB21a | AJ567345 | P. tremula × tremuloides | Negatively regulates lignin biosynthesis | Repressor | Karpinska et al. (2004) |
| EgMYB1 | AJ576024 | Eucalyptus gunnii | Negatively regulates secondary wall formation | Repressor | Legay et al. (2010) |
| EgMYB2 | AJ576023 | Eucalyptus gunnii | Positvely regulates secondary wall formation | Activator | Goicoechea et al. (2005) |
| OsMYB46 | Os12g0515300 | Oryza sativa | Regulates secondary wall biosynthesis | Activator | Zhong et al. (2011) |
| ZmMYB46 | JN634085 | Zea mays | Regulates secondary wall biosynthesis | Activator | Zhong et al. (2011) |
| ZmMYB31 | NM_001112479 | Zea mays | Directly represses lignin biosynthesis | Repressor | Fornale et al. (2006, 2010) |
| ZmMYB42 | NM_001112539 | Zea mays | Represses lignin biosynthesis | Repressor | Sonbol et al. (2009) |
| TaMYB4 | JF746995 | Triticum aestivum | Negatively regulates lignin biosynthesis | Repressor | Ma et al. (2011) |
| PvMYB4 | JF299185 | Panicum virgatum | Negatively regulates lignin biosynthesis | Repressor | Shen et al. (2012) |
| NAC family transcription factors | |||||
| NST1 | At2g46770 | Arabidopsis thaliana | Regulates secondary wall thickenings and required for anther dehiscence | Activator | Mitsuda et al. (2005) |
| NST2 | At3g61910 | Arabidopsis thaliana | Regulates secondary wall thickenings and required for anther dehiscence | Activator | Mitsuda et al. (2005) |
| NST3/ANAC012/SND1 | At1g32770 | Arabidopsis thaliana | Regulates secondary wall synthesis in fibres | Activator | Zhong et al. (2006); Ko et al. (2007) |
| SND2 | At4g28500 | Arabidopsis thaliana | Regulates secondary wall biosynthesis | Activator | Zhong et al. (2008) |
| SND3 | At1g28470 | Arabidopsis thaliana | Regulates secondary wall biosynthesis | Activator | Zhong et al. (2008) |
| VND6 | At5g62380 | Arabidopsis thaliana | Promotes protoxylem differentiation | Activator | Kubo et al. (2005) |
| VND7 | At1g71930 | Arabidopsis thaliana | Promotes metaxylem differentiation | Activator | Kubo et al. (2005) |
| VNI2 | At5g13180 | Arabidopsis thaliana | Negatively regulates xylem vessel formation | Repressor | Yamaguchi et al. (2010) |
| XND1 | At5g64530 | Arabidopsis thaliana | Negatively regulates lignocellulose synthesis and PCD in xylem. | Repressor | Zhao et al. (2008) |
| MtNST1 | Medicago truncatula | Regulates secondary wall biosynthesis | Activator | Zhao et al. (2010b) | |
| Other transcription factors | |||||
| ASL19/LBD30 | At4g00220 | Arabidopsis thaliana | Positively regulates xylem differentiation | Activator | Soyano et al. (2008) |
| ASL20/LBD18 | At2g45420 | Arabidopsis thaliana | Positively regulates xylem differentiation | Activator | Soyano et al. (2008) |
| AtC3H14 | At1g66810 | Arabidopsis thaliana | Regulates secondary wall biosynthesis | Activator | Ko et al. (2009) |
| KNAT7 | At1g62990 | Arabidopsis thaliana | Negatively regulates secondary wall synthesis | Repressor | Li et al. (2012) |
| OFP4 | At1g06920 | Arabidopsis thaliana | Forms a functional complex with KNAT7 to repress secondary cell wall formation | Repressor | Li et al. (2011) |
| SHN2 | At5g25390 | Arabidopsis thaliana | Co-ordinated activation of cellulose and repression of lignin biosynthesis | Activator | Ambavaram et al. (2011) |
| WRKY12 | At2g44745 | Arabidopsis thaliana | Represses secondary wall formation in pith | Repressor | Wang et al. (2010) |
| MtSTP | HM622067 | Medicago truncatula | Represses secondary wall formation in pith | Repressor | Wang et al. (2010) |
| Ntlim1 | AB079513 | Nicotiana tabacum | Regulates lignin biosynthesis | Activator | Kawaoka et al. (2000) |
Upstream regulators of MYB46/MYB83
Upstream direct regulators of MYB46 are largely unknown. Several studies using inducible expression and/or activation systems coupled with comparative transcriptome analyses demonstrated that secondary wall NAC (NAM, ATAT1/2 and CUC2) TFs, such as NST1, NST2, NST3/ANAC012/SND1, VND6 and VND7, are direct upstream regulators of MYB46/MYB83 (Demura and Ye, 2010; Ohashi-Ito et al., 2010; Yamaguchi et al., 2011). The NAC TF family proteins are characterized by a conserved NAC domain located at the N-terminal region and a highly divergent C-terminal activation domain (Olsen et al., 2005). These TFs are specific to plants and play diverse roles in plant defence, growth and development (Olsen et al., 2005).
Several VASCULAR RELATED NAC DOMAIN (VND1–VND7) TF genes were isolated by whole-genome microarray analysis of transdifferentiating tracheary elements (TEs) in arabidopsis cell culture (Kubo et al., 2005). Expression of these VND genes is spatially and temporally correlated with TE differentiation. Among these, overexpression of VND6 and VND7 induces ectopic metaxylem and protoxylem formation, respectively, even in highly specialized cell types such as epidermis, stomata, trichomes and root hairs of arabidopsis and poplar (Kubo et al., 2005; Yamaguchi et al., 2008, 2010a). On the other hand, dominant suppression of VND6 and VND7 resulted in defective metaxylem and protoxylem formation, respectively, in arabidopsis roots (Kubo et al., 2005). However, single VND loss-of-function mutants are apparently not defective in TE formation, which indicates the functional redundancy with other members of the VND family (Kubo et al., 2005). These results indicate that VND6 and VND7 function as key regulators in xylem vessel differentiation (Fig. 1; Table 1).
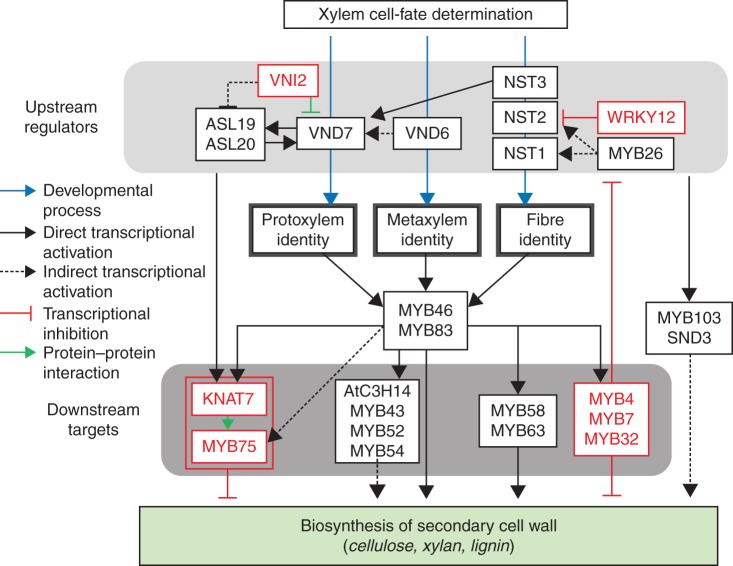
Fig. 1.
MYB46/MYB83 function as a central regulator of secondary wall biosynthesis. Secondary wall biosynthesis is controlled by a complex and multifaceted transcriptional network including positive and negative feedback/forward regulation. Secondary wall NACs are upstream regulators of MYB46/MYB83, while several transcription factors are downstream targets. MYB46/MYB83 regulate the secondary wall biosynthesis genes either directly or co-operatively with downstream target transcription factors. Negative regulators are highlighted in red.
In addition, other NAC family TFs, such as, NAC SECONDARY WALL THICKENING PROMOTING FACTOR1 (NST1), NST2 and NST3/ANAC012/SECONDARY WALL-ASSOCIATED NAC-DOMAIN 1 (SND1), were identified as key regulators of secondary wall biosynthesis in fibre cells (Mitsuda et al., 2005, 2007; Zhong et al., 2006; Ko et al., 2006, 2007; Demura and Ye, 2010). Overexpression of these NAC genes resulted in ectopic deposition of secondary walls in non-vascular cell types, while their suppression reduced secondary wall thickness. For example, the nst1nst3 double knockout showed complete loss of secondary wall thickening in the fibres (Mitsuda et al., 2007; Zhong et al., 2007b). However, their double knockout or simultaneous RNAi (RNA interference) inhibition did not affect secondary wall biosynthesis in the xylem vessels (Mitsuda et al., 2007; Zhong et al., 2007b). NST3/ANAC012/SND1 is specifically expressed in fibres, while VND6 and VND7 are expressed in xylem vessels. Overexpression of any of the NST or VND genes is able to activate the entire secondary wall biosynthetic programme, indicating that NST1 and NST3/ANAC012/SND1 are responsible for secondary wall biosynthesis in fibres, and VND6 and VND7 are responsible in xylem vessels. NST2 was found to be responsible for the secondary wall thickening of endothecium in anther development (Mitsuda et al., 2005).
Members of the ASYMMETRIC LEAVES2-LIKE/LATERAL ORGAN BOUNDARIES DOMAIN (ASL/LBD) protein family were identified as a part of a positive feedback loop regulating VND6 and VND7 in the transcriptional network of secondary wall biosynthesis (Soyano et al., 2008). Overexpression of ASL19/LBD30 and ASL20/LBD18 genes transdifferentiated non-vascular tissues into TE-like cells in arabidopsis, similar to those induced by VND6 or VND7 overexpression, while their dominant suppression caused aberrant TEs (Soyano et al., 2008). Both ASL19 and ASL20 were expressed in immature TEs, and their expression depends on VND6 and VND7 (Soyano et al., 2008) (Table 2). Moreover, ectopic expression of VND7 was detected in plants overexpressing ASL20. Therefore, ASL20/LBD18 and ASL19/LBD30 function in a positive feedback loop that amplifies the expression of VND6 and VND7 (Soyano et al., 2008).
Table 2.
Transcriptional regulators of secondary wall biosynthesis and their downstream target genes
| Protein name | Downstream target genes | Cis-acting element | References |
|---|---|---|---|
| MYB46/83 | MYB4, MYB7, MYB32, MYB52, MYB54, KNAT7, MYB43, MYB58, MYB63, AtC3H14, CesA4, CesA7, CesA8 | M46RE: (T/C)ACC(A/T)A(A/C) (T/C) | Ko et al. (2009) |
| Zhong et al. (2007a) | |||
| PAL, C4H, 4CL, HCT, C3H, CCoAOMT, F5H, CCR, CAD, FRA8, IRX8, IRX9, IRX14 | SMRE: ACC(A/T)A(A/C) (T/C) | Kim et al. (2012) | |
| Zhong and Ye (2012) | |||
| VND6 | MYB46/83, MYB5, MYB63, VND7, MYB103 | SNBE: (T/A)NN(C/T) (T/C/G) TNNNNNNNA(A/C)GN(A/C/T) (A/T) | Demura and Ye (2010) |
| Ohashi-Ito et al. (2010) | |||
| Zhong et al. (2010b) | |||
| VND7 | MYB46/83, MYB58, MYB63, BFN1, XCP1, XCP2, XSP1, LBD18, LBD30, MYB103, ASL19, ASL20, CesA4, CesA8 | SNBE: (T/A)NN(C/T) (T/C/G) TNNNNNNNA(A/C)GN(A/C/T) (A/T) | Yamaguchi et al. (2011) |
| Soyano et al. (2008) | |||
| Zhong et al. (2010b) | |||
| NST1 | MYB46/83, MYB58, MYB63 | SNBE: (T/A)NN(C/T) (T/C/G)TNNNNNNNA(A/C)GN(A/C/T) (A/T) | Mitsuda et al. (2007) |
| Zhong et al. (2010b) | |||
| SND1/NST3/ANAC012 | MYB4, MYB7, MYB20, MYB32, MYB42, MYB43, MYB52, MYB54, MYB58, MYB69, MYB85, MYB46/83, SND2, SND3, MYB103, KNAT7, VND7 | SNBE: (T/A)NN(C/T) (T/C/G) TNNNNNNNA(A/C)GN(A/C/T) (A/T) | Zhong et al. (2006) |
| Zhong et al. (2007a) | |||
| Ko et al. (2007) | |||
| McCarthy et al. (2009) | |||
| Zhong et al. (2010b) | |||
| MYB58/63 | LAC4, PAL1, C4H, 4CL1, HCT, C3H1, CCoAOMT1, CCR1, COMT, CAD6 | AC-I: ACCTACC, AC-II: ACCAACC, AC-III: ACCTAAC | Zhou et al. (2009) |
| Zhong et al. (2008) | |||
| MYB85 | 4CL1 | ND | Zhong et al. (2008) |
| KNAT7 | CesA1, CesA3, CesA6, IRX8, IRX9, IRX10, FRA8, CesA4, CesA7, CesA8, PAL1, C4H, 4CL1, HCT, C3H1, CCoAOMT1, CCR1, F5H1, COMT1, CAD5 | ND | Li et al. (2012) |
| MYB4 | C4H | ND | Jin et al. (2000) |
Bold and underlined genes are suggested as direct targets.
ND, not determined.
Taken together, it appears that the transcriptional network activated by the secondary wall NACs functions through MYB46/MYB83 (Ko et al., 2012; Schuetz et al., 2013) (Fig. 1). Yeast-one hybrid (Y1H) analysis using MYB46 as bait might lead to identification of additional upstream regulators of MYB46.
Downstream targets of MYB46/MYB83
In order to study the downstream transcriptional network leading to secondary wall biosynthesis controlled by MYB46, a comprehensive time-course transcriptome profiling was performed with an inducible secondary wall thickening system in arabidopsis plants by overexpressing MYB46 under the control of a dexamethasone-inducible promoter (Ko et al., 2009). This study identified a total of 42 TFs whose expression either coincides with, or precedes, the induction of secondary wall biosynthetic genes. Subsequent transient transcriptional activation assays confirmed that MYB46 activates the expression of MYB4, MYB7, MYB32, KNAT7, MYB52, MYB54, MYB63 and AtC3H14 (Fig. 1; Tables 1 and 2). Among them, AtC3H14, MYB52 and MYB63 were shown to activate the genes involved in secondary wall biosynthesis (Ko et al., 2009) (Table 2).
Identification of cis-acting regulatory elements (i.e. TF-binding motifs) that are recognized by MYB46 may facilitate the search for direct downstream target genes of MYB46. Independent studies by Kim et al (2012) and Zhong and Ye (2012) identified such motifs. Kim et al. (2012) analysed the promoter region of the TF gene AtC3H14, a known direct target of MYB46 (Ko et al., 2009), and identified an eight-nucleotide core motif ([A/G][G/T]T[A/T]GGT[A/G]) named M46RE (MYB46-responsive cis-regulatory element) that is recognized by MYB46. Various in vitro and in vivo experimental approaches [e.g. electrophoretic mobility shift assay (EMSA), transient transcriptional activation assay and chromatin immunoprecipitation (ChIP) analysis] were used to confirm that M46RE is a ‘necessary and sufficient’ element for MYB46-mediated transcriptional activation of the target genes. Zhong and Ye (2012) also reported a cis-acting motif called SMRE (secondary wall MYB-responsive element, ACC[A/T]A[A/C][T/C]) that is recognized by MYB46. This motif was identified by MYB46 binding assays with serial deletions of the promoter of MYB63, another direct target of MYB46. These two cis-elements (i.e. M46RE and SMRE) are essentially identical except for the eighth nucleotide in M46RE that is absent in SMRE (Table 2).
Several TFs identified as putative direct target of MYB46 have these elements in their promoter region, including AtC3H14, MYB43, MYB58, MYB63 and KNAT7 (Kim et al., 2012; Zhong and Ye, 2012) (Table 2). The TFs MYB58 and MYB63, which are direct targets of MYB46, have been shown to function as direct transcriptional activators of lignin biosynthesis during secondary wall formation in arabidopsis (Ko et al., 2009; Zhou et al., 2009; Demura and Ye, 2010). MYB85 also regulates lignin biosynthesis by activating the lignin biosynthetic genes and causes ectopic lignin deposition when overexpressed (Zhong et al., 2008).
A bioinformatics survey of the arabidopsis genome using the M46RE motif as bait found that many of the cell wall biosynthesis genes (e.g. cellulose, xylan and lignin) or the genes involved in cell wall biosynthesis-related cellular processes (e.g. cytoskeletal organization and signal transduction) have the motif in their promoters (Kim et al., 2012). The list includes three secondary wall-associated cellulose synthase genes (IRX1/CESA8, IRX3/CESA7 and IRX5/CESA4) (Taylor et al., 2003), xylan biosynthesis genes (IRX8, IRX9, IRX14 and IRX15-L) (Peña et al., 2007; Jensen et al., 2010; Wu et al., 2010; Brown et al., 2011), three laccase genes (IRX12/LACCASE4, LACCASE10 and LACCASE11) involved in lignin biosynthesis (Brown et al., 2005), two cytoskeleton-related genes (MYOSIN5 and microtubule-associated protein) (Kaneda et al., 2010; Pesquet et al., 2010) and two DUF579 genes (At1g33800 and At4g09990) (Jensen et al., 2010; Brown et al., 2011). This result suggests that MYB46 may directly regulate not only TFs but also structural genes of secondary wall biosynthesis (Kim et al., 2012).
MYB46 directly regulates secondary wall-associated cellulose synthases
Cellulose, the most abundant biopolymer on Earth, is a central component of plant cell walls and highly abundant (up to 50 %) in the secondary walls (Somerville, 2006). Recently, conversion of cellulose from energy crops into biofuels (e.g. cellulosic ethanol) has attracted global attention as an alternative fuel source. In the secondary cell walls of arabidopsis, three cellulose synthases (CESA4, CESA7 and CESA8) are necessary for cellulose production (Turner and Somerville, 1997; Taylor et al., 1999, 2000, 2003; Doblin et al., 2002; Williamson et al., 2002). However, little was known about the transcriptional regulation of these CESA genes. Interestingly, all three of the secondary wall-associated CESA genes (CESA4, CESA7 and CESA8) have the M46RE motif in their promoters, suggesting that their expression may be directly regulated by MYB46. Kim et al. (2013a) reported several lines of experimental evidence in support of this hypothesis. First, all three of the CESA genes were highly upregulated in both constitutive and inducible overexpression of MYB46 in planta. Secondly, MYB46 directly activates the transcription of the three CESA genes in a steroid receptor-based inducible activation system. Thirdly, MYB46 protein directly binds the promoters of the three CESA genes both in vitro and in vivo, which was confirmed by EMSA and ChIP analysis, respectively. Fourthly, ectopic upregulation of MYB46 resulted in a significant increase of crystalline cellulose content in arabidopsis (Kim et al., 2013a). Taken together, the evidence is quite convincing that MYB46 is a direct regulator of all three secondary wall-associated CESA genes.
Since cellulose biosynthesis in the secondary wall is critical to the plant's survival, it is prudent to speculate that MYB46 is not the only direct regulator of the secondary wall cellulose synthases. Previously, it has been demonstrated that VND6 binds to the TE-specific cis-element TERE (Pyo et al., 2007) in the promoter of CESA4 (Ohashi-Ito et al., 2010). VND7 was also suggested as a direct transcriptional regulator of CESA4 and CESA8 (Yamaguchi et al., 2011) (Table 2). In addition, Y1H screening using the promoter sequences of CESA4, CESA7 and CESA8 as bait identified multiple TFs that bind to the promoter sequences (Kim et al., 2013a). The Y1H identified 13 TFs for the CESA4 promoter and one TF for the CESA7 promoter. However, none of them appears to be involved in the MYB46-mediated regulation pathway because their expression is not altered by MYB46 nor are they co-expressed with MYB46 (Ko et al., 2009; Kim et al., 2012). Thus, the presence of multiple regulators, independent of the MYB46-mediated regulatory pathway, supports the notion that the transcriptional regulation of cellulose biosynthesis is multifaceted and complex.
MYB46 is required for functional expression of the secondary wall cellulose synthases
Considering the elaborate nature of transcriptional control of secondary wall cellulose biosynthesis, one pertinent question is whether MYB46 is the necessary regulator for functional expression of the secondary wall CESA genes. To address this question, Kim et al. (2013b) used a series of genetic complementation experiments using cesa knockout mutants with the CESA coding sequence driven by either the native or the mutated promoter of the genes. The mutant promoters have two-nucleotide point mutations in the M46RE such that MYB46 cannot bind to the promoter, while the binding of other non-MYB-type secondary wall TFs is not affected. The results showed that MYB46 binding to the intact M46RE is essential to restore the normal phenotype of the cesa mutants (Kim et al., 2013b), suggesting that MYB46 is an obligate component of the transcriptional regulatory complex involved in the commitment to secondary wall cellulose synthesis in arabidopsis.
MYB46 directly regulates hemicellulose and lignin biosynthesis genes
The genome-wide survey of promoter sequences in arabidopsis revealed that many genes involved in hemicellulose and lignin biosynthesis have one or more M46RE motifs in their promoter region (Kim et al., 2012), leading to the hypothesis that the expression of these genes may be directly regulated by MYB46. For example, cellulose synthase-like A9 (CSLA9) is responsible for the majority of glucomannan synthesis in both primary and secondary walls of arabidopsis inflorescence stems (Liepman et al., 2005; Goubet et al., 2009). Both in vitro (EMSA) and in vivo (ChIP) binding assays clearly showed that MYB46 binds to the promoter of CSLA9 (Kim et al., 2014). Overexpression of MYB46 resulted in a significant increase in mannan content (Kim et al., 2014). Recently, we obtained experimental evidence for direct regulation of four xylan biosynthesis genes (FRA8, IRX8, IRX9 and IRX14) by MYB46 (W.-C. Kim and K.-H. Han, unpubl. res.). These four genes encode glycosyltransferases that are required for glucuronoxylan synthesis in secondary cell walls (Peña et al., 2007; Keppler and Showalter, 2010). FRA8 and IRX8 are involved in the reducing end synthesis of xylan chains (Scheller and Ulvskov, 2010), while IRX9 and IRX14 are responsible for the xylan backbone synthesis (Lee et al., 2012). Therefore, it appears that MYB46 directly regulates the biosynthesis of the xylan backbone. However, it is notable that the other four known xylan biosynthesis genes (PARVUS, IRX10, IRX15 and IRX15-L) do not seem to be directly regulated by MYB46, indicating the multifaceted nature of the regulation of xylan biosynthesis.
The genes involved in monolignol biosynthesis have been identified (Boerja et al., 2003). Nine out of ten monolignol biosynthesis genes (PAL, C4H, 4CL, HCT, C3H, CCoAOMT, F5H, CCR and CAD) are directly regulated by MYB46 (W.-C. Kim and K.-H. Han, unpubl. res.) (Table 2). Previously, two TFs, MYB58 and MYB63, were identified as master regulators of lignin biosynthesis (Zhou et al., 2009). These TFs directly control the expression of seven monolignol biosynthesis genes (PAL, 4CL, C3H, CCoAOMT, CCR and CAD), but not of F5H, a key gene in syringyl (S) lignin biosynthesis (Raes et al., 2003; Zhou et al., 2009). The TF MYB46 directly regulates F5H as well as MYB58 and MYB63 (W.-C. Kim and K.-H. Han, unpubl. res.). In Medicago truncatula, a secondary wall master switch SND1 directly regulates F5H but not the other monolignol genes (i.e. C4H, COMT, CCoAOMT and 4CL) (Zhao et al., 2010a, b). Whether SND1 regulates F5H in arabidopsis is not known. Recently, Ohman et al. (2013) showed that a loss-of-function mutation of MYB103 substantially reduced F5H expression, resulting in a 70–75 % decrease in S-lignin, while it did not transactivate F5H expression. Taken together, these observations further support the hypothesis that a multifaceted regulatory network exists for the control of lignin biosynthesis, and MYB46 is a key regulator in the network.
Interacting partners of MYB46/MYB83
In eukaryotes, gene expression is frequently controlled by multiprotein complexes. The formation of protein complexes enables the combinatorial action of TFs on the basis of both specific protein–DNA and protein–protein interactions, which facilitate the complex regulatory networks found in higher eukaryotes (Du et al., 2009).
The physical interaction and regulatory synergy between particular sub-classes of MYB and bHLH (basic helix–loop–helix) family TFs is well known in plant gene regulation (Du et al., 2009). In addition, members of the MYB and bHLH families also interact with a number of other regulatory proteins, forming complexes that either activate or repress the expression of sets of target genes (Feller et al., 2011). Examples of such complexes include the PAP1 (MYB)–GL3/EGL3/TT8 (bHLH)–TTG1 (WD40) complex in anthocyanin production, WER (MYB)–GL3/EGL3–TTG1 in root hair development, MYB61–TT8–TTG1 in seed coat mucilage production and GL1 (MYB)–GL3–TTG1 in trichome development (Petroni and Tonelli, 2011). To date, no interacting partners of MYB46 have been identified. However, it is probable that MYB46 can interact with bHLH family TFs. To test this hypothesis, we carried out yeast two-hybrid (Y2H) screening using MYB46 as bait and the bHLH TF library as prey. Our preliminary results indicate that two bHLH TFs strongly interact with MYB46 in yeast (J.-H. Ko and K.-H. Han, unpubl. res.). In planta interaction with MYB46 and the functional significance of the bHLH TFs remain to be elucidated.
Negative regulators of secondary wall biosynthesis
In terms of adaptation to the changing environmental and developmental contexts, negative regulation of secondary wall biosynthesis may be required for tissue-type fine-tuning of secondary wall deposition. Two NAC TFs, VNI2 and XND1, were identified as negative regulators of xylem formation in arabidopsis (Zhao et al., 2008; Yamaguchi et al., 2010b). VND-INTERACTING2 (VNI2) can bind to VND proteins and has been shown to function as a transcriptional repressor of VND7-mediated gene transcription (Yamaguchi et al., 2010b). During xylem differentiation, VNI2 protein is targeted for degradation to unleash VND7 that is required for xylem differentiation, while VNI2 expression precedes that of VND7 in procambial cells (Yamaguchi et al., 2010b). However, VNI2 expression persists in neighbouring xylary parenchyma cells, suggesting that VNI2 functions as a negative regulator of xylem differentiation (Yamaguchi et al., 2010b). Overexpression of XYLEM NAC DOMAIN1 (XND1) causes the complete suppression of xylem vessel secondary wall biosynthesis and programmed cell death (PCD), but not phloem marker gene expression, suggesting that XND1 negatively regulates xylem differentiation (Zhao et al., 2008). Interestingly, both VNI2 and XND1 appear to be targeted for proteasomal degradation by the 20S proteasome (20SP) (Zhao et al., 2008; Yamaguchi et al., 2010b; Han et al., 2012). The 20SP is thought to be a part of the ubiquitin–26SP proteolytic system, possessing caspase-3-like activity, which is a characteristic of animal cell apoptosis (Han et al., 2012). This fact suggests that 20SP may degrade VNI2 and XND1 to induce xylem differentiation in arabidopsis and Populus (Han et al., 2012).
Mutation of the arabidopsis WRKY12 gene caused secondary cell wall thickening in pith cells associated with ectopic deposition of lignin, xylan and cellulose by upregulation of downstream genes encoding NST2 and AtC3H14 TFs that activate secondary wall synthesis (Wang et al., 2010). Direct binding of WRKY12 to the NST2 gene promoter and repression of NST2 and AtC3H14 were confirmed by in vitro assays and in planta transgenic experiments (Wang et al., 2010). The WRKY12 gene is expressed in both pith and cortex that do not have secondary wall thickening. These results suggest that WRKY12 controls the parenchymatous nature of pith cells by acting as a negative regulator of secondary wall NACs.
Recently, a homeodomain TF KNAT7 was described as a negative regulator of secondary wall formation despite being a direct downstream target of both MYB46 and NST3/ANAC012/SND1 (Zhong et al., 2008; Ko et al., 2009; Li et al., 2012). KNAT7 is specifically expressed in vascular tissues and shown to function as a transcriptional repressor. In a transient activation assay using protoplasts, the expression of secondary wall biosynthetic genes was increased in the absence of KNAT7 function (Li et al., 2012). Overexpression of KNAT7 resulted in thinner secondary cell walls in interfascicular fibres, while its loss-of-function mutant knat7 forms thicker secondary cell walls (Li et al., 2011, 2012).
MYB4, MYB7 and MYB32, further downstream targets of both SND1 and MYB46/MYB83, were identified as negative regulators in secondary wall biosynthesis (Ko et al., 2009; Zhong et al., 2010b; Wang et al., 2011). As potent transcriptional repressors, these MYBs can reduce both their target gene expression and the expression of the SND1 upstream regulator (Ko et al., 2009; Zhong et al., 2010b; Wang et al., 2011) (Fig. 1). MYB4 and MYB32 were shown to regulate general phenylpropanoid biosynthesis genes negatively (Preston et al., 2004). MYB4 specifically suppresses the expression of the C4H gene, which encodes the first committed step in the phenylpropanoid pathway (Jin et al., 2000).
Taken together, negative regulators identified so far may function to fine-tune the expression of other TFs or genes involved in secondary wall biosynthesis. This mechanism provides a potential negative feedback regulation that may be a critical homeostatic control within the MYB46-mediated transcriptional regulation network (Fig. 1; Table 1).
Functional homology between MYB46 and its homologues in other plant species
The poplar genome has four close homologues of arabidopsis MYB46: PtrMYB2, PtrMYB3, PtrMYB20 and PtrMYB21 (McCarthy et al., 2010). Both PtrMYB3 and PtrMYB20 have been shown to function as a master switch for secondary wall biosynthesis in poplar (McCarthy et al., 2010). These MYB TFs are directly activated by PtrWND2, an activator of secondary wall biosynthesis in the wood tissue of poplar (Zhong et al., 2010a). Based on these findings and the high degree of collinearity between arabidopsis and Populus, it is hypothesized that the MYB46-mediated transcriptional regulatory programme may control the biosynthesis of secondary walls in poplars. This hypothesis was supported by the results from our EMSA analyses, clearly showing that arabidopsis MYB46 binds to the promoters of Populus secondary wall CESA genes and PtrMYB21 to the promoters of both arabidopsis and poplar secondary wall CESA genes (W.-C. Kim, J.-Y. Kim and K.-H. Han, unpubl. res.). In addition, rice and maize MYB TFs, OsMYB46 and ZmMYB46, were suggested as functional orthologues of MYB46 (Zhong et al., 2011). Both OsMYB46 and ZmMYB46 were directly regulated by the secondary wall NAC TFs OsSWNs and ZmSWNs, respectively, and were able to activate the secondary wall biosynthetic programme when they were overexpressed in arabidopsis (Zhong et al., 2011). These observations, along with the reciprocal binding of arabidopsis MYB46 and PtrMYB21 to the promoters of secondary wall CESA genes, suggest that the MYB46-mediated regulation of secondary wall biosynthesis may be functionally conserved among plant species.
PERSPECTIVES
Multiple observations in the literature indicate that a complex transcriptional regulatory programme appears to be involved in the control of secondary wall biosynthesis. MYB46 and its functional paralogue MYB83 play a central role in the regulatory programme, evidenced by the fact that myb46myb83 double knockout mutants show severe growth arrest in the early seedling stage. Recent studies have shown that MYB46 regulates not only the TFs in the secondary wall biosynthesis pathway but also the biosynthesis genes for all three of the major components (i.e. cellulose, hemicellulose and lignin) of secondary walls. Having the ability to regulate directly the biosynthesis genes for the major components, MYB46 may be useful in pathway-specific manipulation of secondary wall biosynthesis. For example, upregulation of MYB46 can increase the biosynthesis of cellulose and hemicellulose, while lignin biosynthesis is reduced. In order to realize this potential fully, additional information on the upstream regulators, downstream targets and interacting partners of MYB46 is critical. Considering the high degree of functional homology between arabidopsis MYB46 and its homologues in other plant species, the knowledge gained from the model arabidopsis plant on MYB46-mediated transcriptional regulation can be applicable in economically important crop species for production of biofuel and bioproducts.
ACKNOWLEDGEMENTS
This work was funded by the DOE Great Lakes Bioenergy Research Center (DOE Office of Science BER DR-FC02-07ER64494), in part by a grant to J.-H.K. from the Basic Science Research Program through the National Research Foundation of Korea (NRF) (2011-0008840) and a grant to J.-H.K. from the Korea Forest Service (S111213L080110).
LITERATURE CITED
- Ambavaram MM, Krishnan A, Trijatmiko KR, Pereira A. Coordinated activation of cellulose and repression of lignin biosynthesis pathways in rice. Plant Physiology. 2011;155:916–31. doi: 10.1104/pp.110.168641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhargava A, Mansfield SD, Hall HC, Douglas CJ, Ellis BE. MYB75 functions in regulation of secondary cell wall formation in the Arabidopsis inflorescence stem. Plant Physiology. 2010;154:1428–1438. doi: 10.1104/pp.110.162735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhargava A, Ahad A, Wang S, et al. The interacting MYB75 and KNAT7 transcription factors modulate secondary cell wall deposition both in stems and seed coat in Arabidopsis. Planta. 2013;237:1199–1211. doi: 10.1007/s00425-012-1821-9. [DOI] [PubMed] [Google Scholar]
- Bomal C, Bedon F, Caron S, et al. Involvement of Pinus taeda MYB1 and MYB8 in phenylpropanoid metabolism and secondary cell wall biogenesis: a comparative in planta analysis. Journal of Experimental Botany. 2008;59:3925–3939. doi: 10.1093/jxb/ern234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerjan WW, Ralph JJ, Baucher MM. Lignin biosynthesis. Annual Review of Plant Biology. 2003;54:519–546. doi: 10.1146/annurev.arplant.54.031902.134938. [DOI] [PubMed] [Google Scholar]
- Brown DM, Zeef LA, Ellis J, Goodacre R, Turner SR. Identification of novel genes in Arabidopsis involved in secondary cell wall formation using expression profiling and reverse genetics. The Plant Cell. 2005;17:2281–2295. doi: 10.1105/tpc.105.031542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown D, Wightman R, Zhang Z, et al. Arabidopsis genes IRREGULAR XYLEM (IRX15) and IRX15L encode DUF579-containing proteins that are essential for normal xylan deposition in the secondary cell wall. The Plant Journal. 2011;66:401–413. doi: 10.1111/j.1365-313X.2011.04501.x. [DOI] [PubMed] [Google Scholar]
- Demura T, Ye ZH. Regulation of plant biomass production. Current Opinion in Plant Biology. 2010;13:299–304. doi: 10.1016/j.pbi.2010.03.002. [DOI] [PubMed] [Google Scholar]
- Doblin MS, Kurek I, Jacob-Wilk D, Delmer DP. Cellulose biosynthesis in plants: from genes to rosettes. Plant and Cell Physiology. 2002;43:1407–1420. doi: 10.1093/pcp/pcf164. [DOI] [PubMed] [Google Scholar]
- Du H, Zhang L, Liu L, et al. Biochemical and molecular characterization of plant MYB transcription factor family. Biochemistry. 2009;74:1–11. doi: 10.1134/s0006297909010015. [DOI] [PubMed] [Google Scholar]
- Feller A, Machemer K, Braun EL, Grotewold E. Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. The Plant Journal. 2011;66:94–116. doi: 10.1111/j.1365-313X.2010.04459.x. [DOI] [PubMed] [Google Scholar]
- Fornale S, Sonbol FM, Maes T, et al. Down-regulation of the maize and Arabidopsis thaliana caffeic acid O-methyl-transferase genes by two new maize R2R3-MYB transcription factors. Plant Molecular Biology. 2006;62:809–823. doi: 10.1007/s11103-006-9058-2. [DOI] [PubMed] [Google Scholar]
- Fornale S, Shi X, Chai C, et al. ZmMYB31 directly represses maize lignin genes and redirects the phenylpropanoid metabolic flux. The Plant Journal. 2010;64:633–644. doi: 10.1111/j.1365-313X.2010.04363.x. [DOI] [PubMed] [Google Scholar]
- Goicoechea M, Lacombe E, Legay S, et al. EgMYB2, a new transcriptional activator from Eucalyptus xylem, regulates secondary cell wall formation and lignin biosynthesis. The Plant Journal. 2005;43:553–567. doi: 10.1111/j.1365-313X.2005.02480.x. [DOI] [PubMed] [Google Scholar]
- Goubet F, Barton CJ, Mortimer JC, et al. Cell wall glucomannan in Arabidopsis is synthesised by CSLA glycosyltransferases, and influences the progression of embryogenesis. The Plant Journal. 2009;60:527–538. doi: 10.1111/j.1365-313X.2009.03977.x. [DOI] [PubMed] [Google Scholar]
- Han JJ, Lin W, Oda Y, Cui KM, Fukuda H, He XQ. The proteasome is responsible for caspase-3-like activity during xylem development. The Plant Journal. 2012;72:129–141. doi: 10.1111/j.1365-313X.2012.05070.x. [DOI] [PubMed] [Google Scholar]
- Hussey SG, Mizrachi E, Creux NM, Myburg AA. Navigating the transcriptional roadmap regulating plant secondary cell wall deposition. Frontiers in Plant Science. 2013;4:325. doi: 10.3389/fpls.2013.00325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen JK, Kim H, Cocuron JC, Orler R, Ralph J, Wilkerson CG. The DUF579 domain containing proteins IRX15 and IRX15-L affect xylan synthesis in Arabidopsis. The Plant Journal. 2010;66:387–400. doi: 10.1111/j.1365-313X.2010.04475.x. [DOI] [PubMed] [Google Scholar]
- Jin H, Cominelli E, Bailey P, et al. Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO Journal. 2000;19:6150–6161. doi: 10.1093/emboj/19.22.6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneda M, Rensing K, Samuels L. Secondary cell wall deposition in developing secondary xylem of poplar. Journal of Integrative Plant Biology. 2010;52:234–243. doi: 10.1111/j.1744-7909.2010.00925.x. [DOI] [PubMed] [Google Scholar]
- Karpinska B, Karlsson M, Srivastava M, et al. MYB transcription factors are differentially expressed and regulated during secondary vascular tissue development in hybrid aspen. Plant Molecular Biology. 2004;56:255–270. doi: 10.1007/s11103-004-3354-5. [DOI] [PubMed] [Google Scholar]
- Kawaoka A, Kaothien P, Yoshida K, Endo S, Yamada K, Ebinuma H. Functional analysis of tobacco LIM protein Ntlim1 involved in lignin biosynthesis. The Plant Journal. 2000;22:289–301. doi: 10.1046/j.1365-313x.2000.00737.x. [DOI] [PubMed] [Google Scholar]
- Keppler B, Showalter AM. IRX14 and IRX14-LIKE, two glycosyl transferases involved in glucuronoxylan biosynthesis and drought tolerance in Arabidopsis. Molecular Plant. 2010;3:834–841. doi: 10.1093/mp/ssq028. [DOI] [PubMed] [Google Scholar]
- Kim WC, Ko JH, Han K-H. Identification of a cis-acting regulatory motif recognized by MYB46, a master transcriptional regulator of secondary wall biosynthesis. Plant Molecular Biology. 2012;78:489–501. doi: 10.1007/s11103-012-9880-7. [DOI] [PubMed] [Google Scholar]
- Kim WC, Ko JH, Kim JY, Kim JM, Bae HJ, Han K-H. MYB46 directly regulates the gene expression of secondary wall-associated cellulose synthases in Arabidopsis. The Plant Journal. 2013a;73:26–36. doi: 10.1111/j.1365-313x.2012.05124.x. [DOI] [PubMed] [Google Scholar]
- Kim WC, Kim JY, Ko JH, Kim J, Han K-H. Transcription factor MYB46 is an obligate component of the transcriptional regulatory complex for functional expression of secondary wall-associated cellulose synthases in Arabidopsis thaliana. Journal of Plant Physiology. 2013b;170:1374–1378. doi: 10.1016/j.jplph.2013.04.012. [DOI] [PubMed] [Google Scholar]
- Kim WC, Reca I-B, Kim YS, et al. Transcription factors that directly regulate the expression of CSLA9 encoding mannan synthase in Arabidopsis thaliana. Plant Molecular Biology. 2014;84:577–587. doi: 10.1007/s11103-013-0154-9. [DOI] [PubMed] [Google Scholar]
- Ko JH, Beers EP, Han K-H. Global comparative transcriptome analysis identifies gene network regulating secondary xylem development in Arabidopsis thaliana. Molecular Genetics and Genomics. 2006;276:517–531. doi: 10.1007/s00438-006-0157-1. [DOI] [PubMed] [Google Scholar]
- Ko JH, Yang SH, Park AH, Lerouxel O, Han K-H. ANAC012, a member of the plant-specific NAC transcription factor family, negatively regulates xylary fiber development in Arabidopsis thaliana. The Plant Journal. 2007;50:1035–1048. doi: 10.1111/j.1365-313X.2007.03109.x. [DOI] [PubMed] [Google Scholar]
- Ko JH, Kim WC, Han K-H. Ectopic expression of MYB46 identifies transcriptional regulatory genes involved in secondary wall biosynthesis in Arabidopsis. The Plant Journal. 2009;60:649–665. doi: 10.1111/j.1365-313X.2009.03989.x. [DOI] [PubMed] [Google Scholar]
- Ko JH, Kim HT, Han K-H. Biotechnological improvement of lignocellulosic feedstock for enhanced biofuel productivity and processing. Plant Biotechnology Reports. 2011;5:1–7. [Google Scholar]
- Ko JH, Kim WC, Kim JY, Ahn SJ, Han K-H. MYB46-mediated transcriptional regulation of secondary wall biosynthesis. Molecular Plant. 2012;5:961–963. doi: 10.1093/mp/sss076. [DOI] [PubMed] [Google Scholar]
- Kubo M, Udagawa M, Nishikubo N, et al. Transcription switches for protoxylem and metaxylem vessel formation. Genes and Development. 2005;19:1855–60. doi: 10.1101/gad.1331305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Zhong R, Ye ZH. Arabidopsis family GT43 members are xylan xylosyltransferases required for the elongation of the xylan backbone. Plant and Cell Physiology. 2012;53:135–143. doi: 10.1093/pcp/pcr158. [DOI] [PubMed] [Google Scholar]
- Legay S, Sivadon P, Blervacq AS, et al. EgMYB1, an R2R3 MYB transcription factor from eucalyptus negatively regulates secondary cell wall formation in Arabidopsis and poplar. New Phytologist. 2010;188:774–786. doi: 10.1111/j.1469-8137.2010.03432.x. [DOI] [PubMed] [Google Scholar]
- Li E, Wang S, Liu Y, Chen JG, Douglas CJ. OVATE FAMILY PROTEIN4 (OFP4) interaction with KNAT7 regulates secondary cell wall formation in Arabidopsis thaliana. The Plant Journal. 2011;67:328–341. doi: 10.1111/j.1365-313X.2011.04595.x. [DOI] [PubMed] [Google Scholar]
- Li E, Bhargava A, Qiang W, et al. The Class II KNOX gene KNAT7 negatively regulates secondary wall formation in Arabidopsis and is functionally conserved in Populus. New Phytologist. 2012;194:102–115. doi: 10.1111/j.1469-8137.2011.04016.x. [DOI] [PubMed] [Google Scholar]
- Liepman AH, Wilkerson CG, Keegstra K. Expression of cellulose synthase-like (Csl) genes in insect cells reveals that CslA family members encode mannan synthases; Proceedings of the National Academy of Sciences; USA. 2005. pp. 2221–2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma QH, Wang C, Zhu HH. TaMYB4 cloned from wheat regulates lignin biosynthesis through negatively controlling the transcripts of both cinnamyl alcohol dehydrogenase and cinnamoyl-CoA reductase genes. Biochimie. 2011;93:1179–1186. doi: 10.1016/j.biochi.2011.04.012. [DOI] [PubMed] [Google Scholar]
- McCarthy RL, Zhong R, Ye ZH. MYB83 is a direct target of SND1 and acts redundantly with MYB46 in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant and Cell Physiology. 2009;50:1950–1964. doi: 10.1093/pcp/pcp139. [DOI] [PubMed] [Google Scholar]
- McCarthy RL, Zhong R, Fowler S, et al. The poplar MYB transcription factors, PtrMYB3 and PtrMYB20, are involved in the regulation of secondary wall biosynthesis. Plant and Cell Physiology. 2010;51:1084–1090. doi: 10.1093/pcp/pcq064. [DOI] [PubMed] [Google Scholar]
- Mitsuda N, Seki M, Shinozaki K, Ohme-Takagi M. The NAC transcription factors NST1 and NST2 of Arabidopsis regulate secondary wall thickenings and are required for anther dehiscence. The Plant Cell. 2005;17:2993–3006. doi: 10.1105/tpc.105.036004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuda N, Iwase A, Yamamoto H, et al. NAC transcription factors, NST1 and NST3, are key regulators of the formation of secondary walls in woody tissues of Arabidopsis. The Plant Cell. 2007;19:270–280. doi: 10.1105/tpc.106.047043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano Y, Nishikubo N, Goue N, et al. MYB transcription factors orchestrating the developmental program of xylem vessels in Arabidopsis roots. Plant Biotechnology. 2010;27:267–272. [Google Scholar]
- Ohashi-Ito K, Oda Y, Fukuda H. Arabidopsis VASCULAR-RELATED NAC-DOMAIN6 directly regulates the genes that govern programmed cell death and secondary wall formation during xylem differentiation. The Plant Cell. 2010;22:3461–3473. doi: 10.1105/tpc.110.075036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohman D, Demedts B, Kumar M, et al. MYB103 is required for FERULATE-5-HYDROXYLASE expression and syringyl lignin biosynthesis in Arabidopsis stems. The Plant Journal. 2013;73:63–76. doi: 10.1111/tpj.12018. [DOI] [PubMed] [Google Scholar]
- Olsen AN, Ernst HA, Leggio LL, Skriver K. NAC transcription factors: structurally distinct, functionally diverse. Trends in Plant Science. 2005;10:79–87. doi: 10.1016/j.tplants.2004.12.010. [DOI] [PubMed] [Google Scholar]
- Patzlaff A, McInnis S, Courtenay A, et al. Characterisation of a pine MYB that regulates lignification. The Plant Journal. 2003;36:743–754. doi: 10.1046/j.1365-313x.2003.01916.x. [DOI] [PubMed] [Google Scholar]
- Peña MJ, Zhong R, Zhou GK, et al. Arabidopsis irregular xylem8 and irregular xylem9: implications for the complexity of glucuronoxylan biosynthesis. The Plant Cell. 2007;19:549–563. doi: 10.1105/tpc.106.049320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesquet E, Korolev AV, Calder G, Lloyd CW. The microtubule-associated protein AtMAP70-5 regulates secondary wall patterning in Arabidopsis wood cells. Current Biology. 2010;20:744–749. doi: 10.1016/j.cub.2010.02.057. [DOI] [PubMed] [Google Scholar]
- Petroni K, Tonelli C. Recent advances on the regulation of anthocyanin synthesis in reproductive organs. Plant Science. 2011;181:219–229. doi: 10.1016/j.plantsci.2011.05.009. [DOI] [PubMed] [Google Scholar]
- Pimrote K, Tian Y, Lu X. Transcriptional regulatory network controlling secondary cell wall biosynthesis and biomass production in vascular plants. African Journal of Biotechnology. 2012;11:13928–13937. [Google Scholar]
- Preston J, Wheeler J, Heazlewood J, Li SF, Parish RW. AtMYB32 is required for normal pollen development in Arabidopsis thaliana. The Plant Journal. 2004;40:979–995. doi: 10.1111/j.1365-313X.2004.02280.x. [DOI] [PubMed] [Google Scholar]
- Pyo H, Demura T, Fukuda H. TERE; a novel cis-element responsible for a coordinated expression of genes related to programmed cell death and secondary wall formation during differentiation of tracheary elements. The Plant Journal. 2007;51:955–965. doi: 10.1111/j.1365-313X.2007.03180.x. [DOI] [PubMed] [Google Scholar]
- Raes J, Rohde A, Christensen JH, Van de Peer Y, Boerjan W. Genome-wide characterization of the lignification toolbox in Arabidopsis. Plant Physiology. 2003;133:1051–1071. doi: 10.1104/pp.103.026484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheller HV, Ulvskov P. Hemicelluloses. Annual Review of Plant Biology. 2010;61:263–289. doi: 10.1146/annurev-arplant-042809-112315. [DOI] [PubMed] [Google Scholar]
- Schuetz M, Smith R, Ellis B. Xylem tissue specification, patterning, and differentiation mechanisms. Journal of Experimental Botany. 2013;64:11–31. doi: 10.1093/jxb/ers287. [DOI] [PubMed] [Google Scholar]
- Shen H, He X, Poovaiah CR, et al. Functional characterization of the switchgrass (Panicum virgatum) R2R3-MYB transcription factor PvMYB4 for improvement of lignocellulosic feedstocks. New Phytologist. 2012;193:121–136. doi: 10.1111/j.1469-8137.2011.03922.x. [DOI] [PubMed] [Google Scholar]
- Somerville C. Cellulose synthesis in higher plants. Annual Review of Cell and Developmental Biology. 2006;22:53–78. doi: 10.1146/annurev.cellbio.22.022206.160206. [DOI] [PubMed] [Google Scholar]
- Sonbol FM, Fornale S, Capellades M, et al. The maize ZmMYB42 represses the phenylpropanoid pathway and affects the cell wall structure, composition and degradability in Arabidopsis thaliana. Plant Molecular Biology. 2009;70:283–296. doi: 10.1007/s11103-009-9473-2. [DOI] [PubMed] [Google Scholar]
- Soyano T, Thitamadee S, Machida Y, Chua NH. ASYMMETRIC LEAVES2-LIKE19/LATERAL ORGAN BOUNDARIES DOMAIN30 and ASL20/LBD18 regulate tracheary element differentiation in Arabidopsis. The Plant Cell. 2008;20:3359–3373. doi: 10.1105/tpc.108.061796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor NG, Scheible WR, Cutler S, Somerville CR, Turner SR. The irregular xylem3 locus of Arabidopsis encodes a cellulose synthase required for secondary cell wall synthesis. The Plant Cell. 1999;11:769–780. doi: 10.1105/tpc.11.5.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor NG, Laurie S, Turner SR. Multiple cellulose synthase catalytic subunits are required for cellulose synthesis in Arabidopsis. The Plant Cell. 2000;12:2529–2540. doi: 10.1105/tpc.12.12.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor NG, Howells RM, Huttly AK, Vickers K, Turner SR. Interactions among three distinct CesA proteins essential for cellulose synthesis; Proceedings of the National Academy of Sciences; USA. 2003. pp. 1450–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner SR, Somerville CR. Collapsed xylem phenotype of Arabidopsis identifies mutants deficient in cellulose deposition in the secondary cell wall. The Plant Cell. 1997;9:689–701. doi: 10.1105/tpc.9.5.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HZ, Dixon RA. On–off switches for secondary cell wall biosynthesis. Molecular Plant. 2011;5:297–303. doi: 10.1093/mp/ssr098. [DOI] [PubMed] [Google Scholar]
- Wang H, Avci U, Nakashima J, Hahn MG, Chen F, Dixon RA. Mutation of WRKY transcription factors initiates pith secondary wall formation and increases stem biomass in dicotyledonous plants; Proceedings of the National Academy of Sciences; USA. 2010. pp. 22338–22343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Zhao Q, Chen F, Wang M, Dixon RA. NAC domain function and transcriptional control of a secondary cell wall master switch. The Plant Journal. 2011;68:1104–1114. doi: 10.1111/j.1365-313X.2011.04764.x. [DOI] [PubMed] [Google Scholar]
- Williamson RE, Burn JE, Hocart CH. Towards the mechanism of cellulose synthesis. Trends in Plant Science. 2002;7:461–467. doi: 10.1016/s1360-1385(02)02335-x. [DOI] [PubMed] [Google Scholar]
- Wu AM, Hornblad E, Voxeur A, et al. Analysis of the Arabidopsis IRX9/IRX9-L and IRX14/IRX14-L pairs of glycosyltransferase genes reveals critical contributions to biosynthesis of the hemicellulose glucuronoxylan. Plant Physiology. 2010;153:542–554. doi: 10.1104/pp.110.154971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi M, Demura T. Transcriptional regulation of secondary wall formation controlled by NAC domain proteins. Plant Biotechnology. 2010;27:237–242. [Google Scholar]
- Yamaguchi M, Kubo M, Fukuda H, Demura T. Vascular-related NAC-DOMAIN7 is involved in the differentiation of all types of xylem vessels in Arabidopsis roots and shoots. The Plant Journal. 2008;55:652–664. doi: 10.1111/j.1365-313X.2008.03533.x. [DOI] [PubMed] [Google Scholar]
- Yamaguchi M, Goue N, Igarashi H, et al. VASCULAR-RELATED NAC-DOMAIN6 and VASCULAR-RELATED NAC-DOMAIN7 effectively induce transdifferentiation into xylem vessel elements under control of an induction system. Plant Physiology. 2010a;153:906–914. doi: 10.1104/pp.110.154013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi M, Ohtani M, Mitsuda N, et al. VND-INTERACTING2, a NAC domain transcription factor, negatively regulates xylem vessel formation in Arabidopsis. The Plant Cell. 2010b;22:1249–1263. doi: 10.1105/tpc.108.064048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi M, Mitsuda N, Ohtani M, Ohme-Takagi M, Kato K, Demura T. VASCULAR-RELATED NAC-DOMAIN7 directly regulates the expression of a broad range of genes for xylem vessel formation. The Plant Journal. 2011;66:579–590. doi: 10.1111/j.1365-313X.2011.04514.x. [DOI] [PubMed] [Google Scholar]
- Yang C, Xu Z, Song J, Conner K, Vizcay Barrena G, Wilson ZA. Arabidopsis MYB26/MALE STERILE35 regulates secondary thickening in the endothecium and is essential for anther dehiscence. The Plant Cell. 2007;19:534–548. doi: 10.1105/tpc.106.046391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Elo A, Helariutta Y. Arabidopsis as a model for wood formation. Current Opinion in Biotechnology. 2010;22:293–299. doi: 10.1016/j.copbio.2010.11.008. [DOI] [PubMed] [Google Scholar]
- Zhao C, Avci U, Grant EH, Haigler CH, Beers EP. XND1, a member of the NAC domain family in Arabidopsis thaliana, negatively regulates lignocellulose synthesis and programmed cell death in xylem. The Plant Journal. 2008;53:425–436. doi: 10.1111/j.1365-313X.2007.03350.x. [DOI] [PubMed] [Google Scholar]
- Zhao Q, Dixon RA. Transcriptional networks for lignin biosynthesis: more complex than we thought? Trends in Plant Science. 2011;16:227–233. doi: 10.1016/j.tplants.2010.12.005. [DOI] [PubMed] [Google Scholar]
- Zhao Q, Wang H, Yin Y, Xu Y, Chen F, Dixon RA. Syringyl lignin biosynthesis is directly regulated by a secondary cell wall master switch; Proceedings of the National Academy of Sciences; USA. 2010a. pp. 14496–14501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Gallego-Giraldo L, Wang H, et al. An NAC transcription factor orchestrates multiple features of cell wall development in Medicago truncatula. The Plant Journal. 2010b;63:100–114. doi: 10.1111/j.1365-313X.2010.04223.x. [DOI] [PubMed] [Google Scholar]
- Zhong R, Demura T, Ye ZH. SND1, a NAC domain transcription factor, is a key regulator of secondary wall synthesis in fibers of Arabidopsis. The Plant Cell. 2006;18:3158–3270. doi: 10.1105/tpc.106.047399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Ye ZH. MYB46 and MYB83 bind to the SMRE sites and directly activate a suite of transcription factors and secondary wall biosynthetic genes. Plant and Cell Physiology. 2012;53:368–380. doi: 10.1093/pcp/pcr185. [DOI] [PubMed] [Google Scholar]
- Zhong R, Richardson EA, Ye ZH. The MYB46 transcription factor is a direct target of SND1 and regulates secondary wall biosynthesis in Arabidopsis. The Plant Cell. 2007a;19:2776–2792. doi: 10.1105/tpc.107.053678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Richardson EA, Ye ZH. Two NAC domain transcription factors, SND1 and NST1, function redundantly in regulation of secondary wall synthesis in fibers of Arabidopsis. Planta. 2007b;225:1603–1611. doi: 10.1007/s00425-007-0498-y. [DOI] [PubMed] [Google Scholar]
- Zhong R, Lee C, Zhou J, McCarthy RL, Ye ZH. A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. The Plant Cell. 2008;20:2763–2782. doi: 10.1105/tpc.108.061325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Lee C, Ye ZH. Evolutionary conservation of the transcriptional network regulating secondary cell wall biosynthesis. Trends in Plant Science. 2010a;15:625–632. doi: 10.1016/j.tplants.2010.08.007. [DOI] [PubMed] [Google Scholar]
- Zhong R, Lee C, Ye ZH. Global analysis of direct targets of secondary wall NAC master switches in Arabidopsis. Molecular Plant. 2010b;3:1087–1103. doi: 10.1093/mp/ssq062. [DOI] [PubMed] [Google Scholar]
- Zhong R, Lee C, McCarthy RL, Reeves CK, Jones EG, Ye ZH. Transcriptional activation of secondary wall biosynthesis by rice and maize NAC and MYB transcription factors. Plant and Cell Physiology. 2011;52:1856–1871. doi: 10.1093/pcp/pcr123. [DOI] [PubMed] [Google Scholar]
- Zhong R, McCarthy RL, Haghighat M, Ye ZH. The poplar MYB master switches bind to the SMRE site and activate the secondary wall biosynthetic program during wood formation. PLoS One. 2013;8:e69219. doi: 10.1371/journal.pone.0069219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Lee C, Zhong R, Ye ZH. MYB58 and MYB63 are transcriptional activators of the lignin biosynthetic pathway during secondary cell wall formation in Arabidopsis. The Plant Cell. 2009;21:248–266. doi: 10.1105/tpc.108.063321. [DOI] [PMC free article] [PubMed] [Google Scholar]