Abstract
Recent molecular phylogenetic studies revealed the extraordinary diversity of single-celled eukaryotes. However, the proper assessment of this diversity and accurate reconstruction of the eukaryote phylogeny are still impeded by the lack of molecular data for some major groups of easily identifiable and cultivable protists. Among them, amoeboid eukaryotes have been notably absent from molecular phylogenies, despite their diversity, complexity, and abundance. To partly fill this phylogenetic gap, we present here combined small-subunit ribosomal RNA and actin sequence data for the three main groups of “Heliozoa” (Actinophryida, Centrohelida, and Desmothoracida), the heliozoan-like Sticholonche, and the radiolarian group Polycystinea. Phylogenetic analyses of our sequences demonstrate the polyphyly of heliozoans, which branch either as an independent eukaryotic lineage (Centrohelida), within stramenopiles (Actinophryida), or among cercozoans (Desmothoracida), in broad agreement with previous ultrastructure-based studies. Our data also provide solid evidence for the existence of the Rhizaria, an emerging supergroup of mainly amoeboid eukaryotes that includes desmothoracid heliozoans, all radiolarians, Sticholonche, and foraminiferans, as well as various filose and reticulose amoebae and some flagellates.
Molecular phylogenetic studies have demonstrated the existence of an extraordinary diversity of unicellular eukaryotes, which form up to eight major groups in the eukaryotic tree of life (1, 2). This diversity might even be higher, according to recent environmental DNA studies that revealed a number of extremely small undescribed taxa among these major groups, as well as some candidate phylotypes representing new higher-level diversity among eukaryotes (3, 4). However, the proper assessment of this diversity is impeded by the fact that there are still numerous major groups of easily identifiable and cultivable protists for which only little or no molecular data exist (2). DNA sequences are particularly scarce for amoeboid eukaryotes. Only recently did sequence data for a broad taxonomic sampling of lobose amoebae become available (5–7). Here, we present combined small-subunit ribosomal RNA (SSU rRNA) and actin genes sequence data for the main groups of heliozoans and radiolarians, which together form the bulk of axopodia-bearing protists (the Actinopoda), the last group of amoeboid protists remaining largely unexplored at the molecular level.
Traditionally, all free-living heterotrophic protists characterized by long radial axopodia supported by a bundle of microtubules were grouped into the superclass Actinopoda (8). This group included the marine, usually planktonic, radiolarians and the primarily freshwater heliozoans, also called sun-animalcules. Originally, Radiolaria were classified by Ernst Haeckel (9) into three groups: Acantharea, Phaeodarea, and Polycystinea, considered later as three independent classes based on differences in the composition of the skeleton and the structure of the central capsule (10). The Heliozoa, another group described by Haeckel (11), included initially only two freshwater actinophryids, Actinophrys and Actinosphaerium. Later, several other axopodia-bearing protists were added to this taxon, forming a large heterogeneous class, which comprised from five (12) to eight (13) orders. However, based on differences in the patterns of ultrastructural organization, it has also been proposed that Heliozoa are composed of several evolutionarily unrelated groups (14, 15). In a recent classification of protists, four monophyletic heliozoan orders have been distinguished (Actinophryida, Centrohelida, Desmothoracida, and Gymnosphaerida), whereas the rest of heliozoan-like taxa, including Sticholonche zanclea, the only member of the order Taxopodida, was classified as “other Heliozoa” (16).
The advent of molecular phylogenies did little to resolve the position of Actinopoda in the tree of life. The first analysis of SSU rRNA gene sequences of the radiolarian Acantharea and Polycystinea suggested an independent origin for these groups (17), yet more recent analyses challenged this result using environmental SSU rRNA gene sequences (18). The first SSU rRNA sequences of the heliozoan order Centrohelida and of a Dimorpha-like strain were only recently published (19, 20). Finally, we obtained very recently the first SSU rRNA data on the third group of radiolarians, the Phaeodarea, revealing that this taxon is not related to acanthareans and polycystines (21). However, SSU rRNA gene sequence data are still lacking for most heliozoans, and there are no protein data available yet for any member of the Actinopoda.
To fill this gap and to unravel the origins of the different axopodia-bearing protists, we obtained and analyzed sequence data for most groups classically belonging to Actinopoda. Our study includes previously undescribed SSU rRNA and actin gene sequences for representatives of the three main orders of Heliozoa (Actinophryida, Centrohelida, and Desmothoracida), the SSU rRNA gene sequence of the taxopodid S. zanclea, as well as actin gene sequences for two species of Polycystinea and the filose amoebae Gromia oviformis (Cercozoa: Gromiidae), Lecythium sp. (Cercozoa: Chlamydophryidae), and Nuclearia simplex (Opisthokonts: Nucleariidae).
Materials and Methods
Cell Cultures, DNA, and RNA Extractions. Cultures of the desmothoracids Clathrulina elegans and Hedriocystis cf. spinifera, the freshwater centrohelids Chlamydaster sterni and Pterocystis erinaceoides, the marine centrohelid Heterophrys marina, and the filose amoeba Lecythium sp. were taken from the culture collection of the Institute for the Biology of Inland Waters of the Russian Academy of Sciences (IBIW RAS). Cultures of the centrohelid Raphidiophrys ambigua; the actinophryids Actinosphaerium eichhornii, Actinosphaerium nucleofilum, and Actinophrys sol; and the filose amoeba N. simplex were taken from the Culture Collection of Algae and Protozoa (CCAP) culture collection. Freshwater cultures were maintained on artificial Pratt medium (KNO3 0.1‰/K2HPO4 × 3H2O 0.01‰/MgSO4 × 7H2O 0.01‰/FeCl3 × 6H2O 0.001‰, pH 6.5–7.5). Marine cultures were maintained on artificial Shmaltz–Pratt medium (NaCl 16.07‰/KCl 0.38‰/MgCl2 × 6H2O 3.15‰/MgSO4 × 7H2O 3.95‰/CaCl2 × H2O 0.83‰/KNO3 0.06‰/K2HPO4 × 3H2O 0.006‰, pH 6.5–7.5). Marine heliozoans from the IBIW RAS culture collection were fed with Procryptobia sorokini, and marine heliozoans from the CCAP culture collection were fed with Tetrahymena sp. The freshwater heliozoans were fed with Bodo saltans. All food sources were cultivated separately from their predator and fed with Aerobacter aerogenes. The taxopodid S. zanclea and the polycystines Collozoum inerme and Thalassicolla pellucida were collected in the Mediterranean Sea (Ville-franche-sur-Mer, France). These species were processed directly after isolation; the DNA extracts contained from 9 to 20 specimens in the case of S. zanclea, two specimens in the case of T. pellucida, and one colony per extract in the case of C. inerme. DNA was extracted by using the DNeasy Plant Minikit (Qiagen, Basel). RNA extractions were performed with the Cells-to-cDNA II RNA extraction kit (Ambion, Austin, TX).
DNA Amplification, Cloning, and Sequencing. All SSU rRNA gene sequences were amplified from DNA in one or several unambiguously overlapping fragment(s) by using universal and specific external or internal primers, except the sequence of C. elegans, which was obtained from RNA. Actin genes of the desmothoracid H. cf. spinifera, the centrohelids P. erinaceoides and R. ambigua, all actinophryids, both polycystines, and the three filose amoebae were obtained from DNA. Actin genes of the desmothoracid C. elegans and the centrohelids C. sterni and H. marina were obtained from RNA. A list of all primers used in our study is given in Table 1, which is published as supporting information on the PNAS web site. PCR amplifications, cloning, and sequencing were done as described (6). The 25 previously undescribed sequences reported in this paper were deposited in the GenBank/European Molecular Biology Laboratory database (see Table 2, which is published as supporting information on the PNAS web site, for the species names, taxonomic position, and accession numbers of all SSU rRNA and actin gene sequences used in our analyses). Sequences alignments are available upon request from J.P.
Phylogenetic Analyses. The SSU rRNA gene sequences were manually aligned by using the genetic data environment software (gde), Version 2.2 (22), following the secondary structure model proposed by Wuyts et al. (23). Sequences from public databases were selected so that (i) all available amoeboid organisms and major taxonomic groups of eukaryotes were present, and (ii) the taxon sampling matched the organisms for which actin gene sequences are available; only highly diverging lineages such as Microsporidia and Diplomonadida were omitted. One hundred and eleven sequences were included, and a total of 1,164 unambiguously aligned positions were used in the phylogenetic analyses. A Bayesian analysis of the data was performed with mrbayes, Version 3.0b4 (24), by using the general time reversible (GTR) model of substitution (25, 26) and taking into account a γ-shaped distribution of the rates of substitution among sites, with eight rate categories. Five million generations were run, and 50,000 trees were sampled, 10,000 of which were discarded as burn-in. Parameters estimated from the 40,000 remaining trees were then used to infer an evolutionary tree with the maximum likelihood (ML) method (27) by using paup*, Version 4.0b10 (28). A distance tree was obtained by using the bionj (29) option in paup* with ML-corrected distances and was used as a starting tree for the ML search, then swapped with the tree-bisection-reconnection algorithm. The reliability of internal branches was assessed by using the posterior probabilities (PP) calculated with mrbayes. Additionally, the bootstrap method (30) was used with 1,000 replicates for distance analyses, performed with paup* as described above.
The actin protein sequences were manually aligned by using gde. Sequences from public databases were selected so that all available taxonomic groups of eukaryotes were represented, but the highly diverging actin sequences of ciliates, microsporidians, diplomonads, and trichomonads were discarded. Sixtyeight sequences were included, and a total of 241 amino acid positions were used in the phylogenetic analyses. A Bayesian analysis of the data was performed with mrbayes by using the Whelan and Goldman (WAG) substitution matrix (31) and taking into account a proportion of invariable sites and a γ-shaped distribution of the rates of substitution among variable sites, with eight rate categories. Two million seven hundred fifty thousand generations were run, and 27,500 trees were sampled, 2,500 of which were discarded as burn-in. An evolutionary tree was inferred with the ML method by using the same evolutionary model. All necessary parameters were estimated from the data by using tree-puzzle, Version 5.0 (32), and the tree topology was constructed with the proml program of the phylip package, Version 3.6a3 (33), using the R option with 10 input order jumbles and global rearrangements. Again, the reliability of internal branches was assessed by using the PP calculated with mrbayes. Additionally, the bootstrap method was used with 500 replicates, based on distance analysis using the program neighbor of phylip. For each data resampling, WAG + G + I corrected distances were calculated by tree-puzzle with the utility puzzleboot (shell script available from www.tree-puzzle.de), using the parameters previously estimated.
A supertree was reconstructed from the PP consensus trees of the Bayesian analyses of 111 SSU rRNA sequences and 68 actin sequences, using the program radcon, Version 1.1.5 (34).
Results
We obtained SSU rRNA sequences from four centrohelids, two desmothoracids, one actinophryid, and the taxopodid S. zanclea (see Table 3, which is published as supporting information on the PNAS web site, for a description of the length and GC content of these sequences). Actin sequences were obtained from three centrohelids, two desmothoracids, three actinophryids, and two polycystines. Two distinct gene sequences were found for A. sol and C. inerme, and three for A. nucleofilum, but only one for all other species. Because actin sequences are available only for a limited taxon sampling of amoeboid organisms, we also obtained an actin sequence from the filose amoebae G. oviformis, Lecythium sp., and N. simplex.
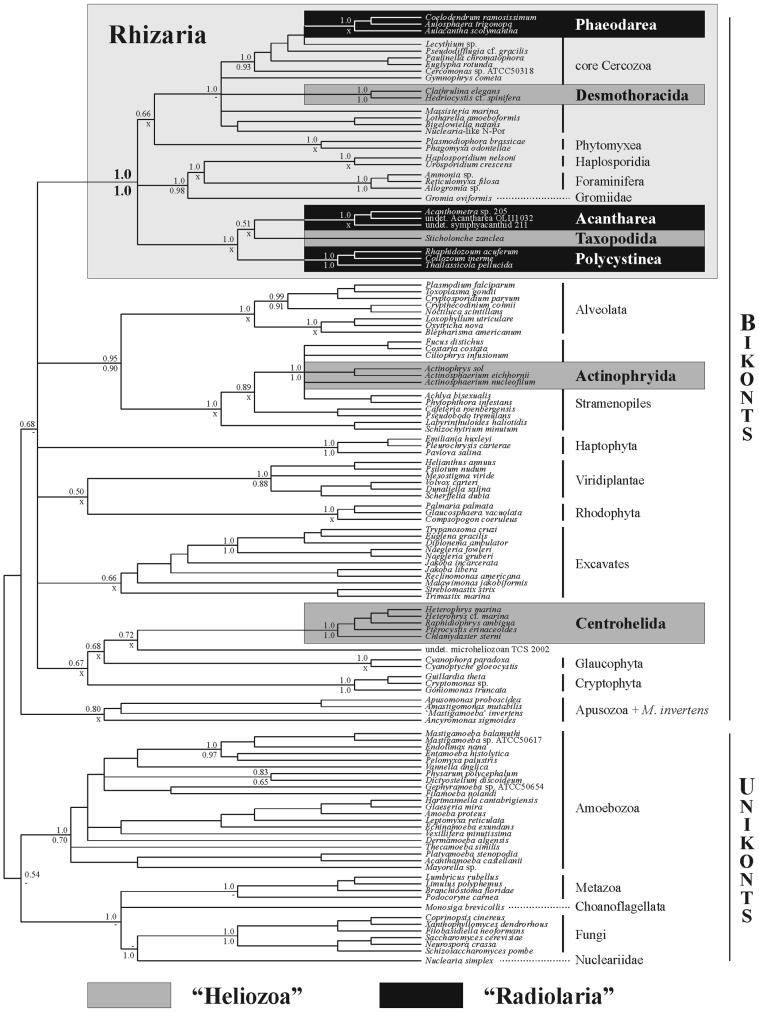
The results of our analyses are presented in the form of a supertree reconstructed from Bayesian PP consensus trees of SSU rRNA and actin genes (Fig. 1). Separate analyses of both genes are in Figs. 2 and 3, which are published as supporting information on the PNAS web site. The tree shown in Fig. 1 is presented with a basal dichotomy between unikonts (opisthokonts + Amoebozoa) and bikonts, following a recent hypothesis (35). The phylogenies of eukaryotes inferred from SSU rRNA and actin genes using a Bayesian method are remarkably congruent, and all major groups of protists are recovered. Provided that the rooting of the trees is correct, Bayesian analyses of SSU rRNA and actin genes strongly support the relationship between heteroloboseans and euglenozoans (PP of 1.0 for both genes) and quite robustly support a sister-group relation between alveolates and stramenopiles (PP of 0.95 and 0.90), as well as the monophyly of Amoebozoa, including mycetozoans and archamoebae (PP of 1.0 and 0.70). Furthermore, the SSU rRNA Bayesian analysis suggests a grouping of apusomonads, Ancyromonas, and the mysterious “Mastigamoeba” invertens (PP of 0.80), which together form the most basal branch of the bikont radiation, albeit with weak support (PP of 0.68). In addition, oxymonads, Trimastix, Malawimonas, jakobids, and discicristates (Heterolobosea + Euglenozoa) cluster weakly (PP of 0.66) in what corresponds to the recently proposed supergroup of excavates (36, 37).
Fig. 1.
Phylogenetic relationships among 121 eukaryotes, emphasizing the polyphyly of heliozoans and radiolarians (gray and black boxes, respectively) and the emergence of the Rhizaria, a previously undescribed supergroup of mainly amoeboid protists (light gray box). The topology shown corresponds to a supertree reconstructed from the consensus trees of Bayesian analyses of 111 SSU rRNA gene sequences and 68 actin gene sequences (see text). Numbers at nodes represent PP of the SSU rRNA (upper) and actin (lower) Bayesian analyses. Dashes indicate support values <0.5, and an X indicates that the considered node is absent in one of the trees due to the differences in available taxa sampling between the two genes. Support values within the lineages were omitted for clarity. The separate ML trees of SSU rRNA and actin analyses are in Figs. 2 and 3.
The phylogenetic position of the three major heliozoan groups is congruent in analyses of both SSU rRNA and actin genes. The Actinophryida branch within stramenopiles, with PP of 1.0 for both genes and bootstrap support values (BV) of 71% and 98%. Desmothoracida form a monophyletic lineage among core Cercozoa (PP of 1.0 and BV of 94% for SSU rRNA analyses; the group is not supported in actin analyses), and Centrohelida form an independent lineage of eukaryotes. The four SSU rRNA sequences of centrohelids that we obtained clearly branch with those published by Cavalier-Smith and Chao (19) and, because sequences from same strains (C. sterni and R. ambigua) were identical, only one of each was kept in our analyses. The monophyly of centrohelids is supported by PP of 1.0 in analyses of both genes, but their relations to other eukaryotes are not identical. In the SSU rRNA tree, centrohelids branch as a sister group to an unidentified microheliozoan, and together they form a weakly supported clade with Glaucophyta and Cryptophyta (PP of 0.67 and BV <50%). In the actin tree, Centrohelida, Cryptophyta, and Haptophyta form a series of independent lineages among the bikonts, whereas glaucophytes branch as the sister group to Viridiplantae. In addition, SSU rRNA analyses reveal that S. zanclea branches between the radiolarian classes Polycystinea and Acantharea (PP of 1.0 and BV of 80% for SSU rRNA analyses), whereas the third radiolarian class, the Phaeodarea, branches within core Cercozoa (PP of 1.0 and BV of 94% for SSU rRNA analyses). Thus, the actinopods as a whole form at least five evolutionarily independent lineages among eukaryotes. Three of these lineages, Phaeodarea, Desmothoracida, and the clade Acantharea + Polycystinea + Taxopodida, cluster together in the Rhizaria, a strongly supported clade (PP of 1.0 and BV of 72% for SSU rRNA analyses, PP of 1.0 and BV of 59% for actin analyses), which also comprises core cercozoans, the parasitic Phytomyxea and Haplosporidia, Gromia, and foraminiferans.
Discussion
Polyphyly of “Heliozoa.” The independent branching of Actinophryida, Centrohelida, and Desmothoracida in SSU rRNA and actin trees provides molecular evidence that “Heliozoa” represent an artificial assemblage, in agreement with previous ultrastructure-based studies (15, 38–40). This finding confirms that heliozoan axopodia are not homologous structures but convergent adaptations, probably for passive predation, the principal mode of feeding of most heliozoans (41). The congruence between molecular and ultrastructural data observed for some heliozoan taxa is remarkable and emphasizes the importance of ultrastructure for protistan phylogeny (42).
The position of actinophryids within stramenopiles is congruent with their close relationship to pedinellid helioflagellates, as suggested by numerous ultrastructure studies (38, 39, 43). Actinophryids and pedinellids resemble each other in the nuclear termination of their axonemes, possessing tubular mitochondrial cristae, and having a particular type of simple extrusomes (39), which differ from the complex kinetocysts of centrohelids, desmothoracids, gymnosphaerids, and dimorphid helioflagellates (41, 44, 45). According to both morphological and molecular data, the pedinellids clearly belong to stramenopiles (46, 47), but for the present, their relation to actinophryids remains unresolved. SSU rRNA analyses performed on stramenopiles indicate only that A. eichhornii branches within the terminal radiation of mainly autotrophic heterokont algae, which also includes the pedinellids. However, due to an unusually rapid rate of substitution in the SSU rRNA gene of A. eichhornii, its exact phylogenetic position cannot be established (data not shown). On the other hand, the lack of monophyly of actinophryids in the actin tree (see Fig. 3) suggests that this marker is not ideal for resolving the phylogeny of stramenopiles.
Ultrastructural and molecular data are also in agreement concerning the placement of Desmothoracida among Cercozoa, both groups having tubular mitochondrial cristae and a similar type of extrusomes (44, 45, 48, 49). In the SSU rRNA tree (Fig. 2), C. elegans and H. cf. spinifera branch between Gymnophrys cometa and Massisteria marina, two amoeboid flagellates that possess complex life cycles, including both a biflagellate and an amoeboid stage similar to that of Desmothoracida (48, 50–52). Actin analyses support the inclusion of desmothoracids within Cercozoa (Fig. 3) and, given the available taxon sampling, suggest a relationship with the chlorarachniophytes, another lineage at the base of core Cercozoa in SSU rRNA trees (Fig. 2). Interestingly, a recent sequence from a Dimorpha-like strain also branches among core cercozoans (20). However, SSU rRNA analyses performed on cercozoans only do not allow convincing testing of the hypothesis of a relation between dimorphids and desmothoracids, as suggested by some authors (38, 41), because of the lack of resolution at the base of core cercozoans (data not shown).
On the other hand, neither ultrastructure nor molecular data help to resolve the position of Centrohelida. Centrohelids have been proposed to be related to dimorphids (53, 54) or to gymnophryids (52), but the first analyses of centrohelid SSU rRNA gene sequences showed that they form an independent protozoan lineage (19). In our analyses, the Centrohelida branch either independently (actin) or within the clade of Cryptophyta + Glaucophyta (SSU rRNA), although the support for the latter relation is weak (PP of 0.67 and BV <50%). Centrohelida and Cryptophyta share the absence of cell walls and the presence of flattened mitochondrial cristae, and some species of Cryptophyta possess siliceous scales, a typical feature of all Centrohelida (16, 40, 55). Thus, a phylogenetic relationship between these taxa cannot be completely discarded and should be explored with additional protein-coding genes.
Polyphyly of “Radiolaria.” Our study confirms the polyphyletic origin of Radiolaria sensu Haeckel. The Bayesian analysis of SSU rRNA gene sequences (Fig. 1; see also Fig. 2) strongly supports the position of Phaeodarea among core Cercozoa, separately from Acantharea and Polycystinea, in agreement with Polet et al. (21). This position is congruent with the fact that phaeodareans display both biflagellate and amoeboid stages in their life cycle (56), as do most cecozoans. The Phaeodarea differ from other radiolarians by (i) the presence of central capsule with only three apertures, (ii) the incapacity to secrete strontium sulfate, (iii) the lack of symbionts, and (iv) the absence of cross bridges between the microtubules of their axopodia (57–59).
Our analyses also support the common origin of Acantharea and Polycystinea, as previously demonstrated (18). In view of our data, however, the Acantharea + Polycystinea clade also includes S. zanclea, the only member of the order Taxopodida. Taxopodida was first described as a separate order of Rhizopoda (60). Some authors have placed it among Radiolaria based on morphological organization (61), but later S. zanclea was moved to Heliozoa based on ultrastructural observations (62), a position retained in most classifications (10, 63). The placement of Taxopodida in the Acantharea + Polycystinea clade is in agreement with the fact that, like these radiolarians, S. zanclea does not have kinetocysts and uses its axopodia for floating rather than for predation, contrary to heliozoans (13).
Interestingly, one of the environmental sequences (DH145-KW16) used by López-García et al. (18) to demonstrate the monophyly of Acantharea and Polycystinea is very close to our sequence of S. zanclea. These two sequences and another partial environmental sequence (CS_E043), recently published by Edgcomb et al. (64), form a robust clade in analyses based on partial SSU rRNA sequences (Fig. 4, which is published as supporting information on the PNAS web site). This suggests that S. zanclea might not be the only member of Taxopodida or might represent a species complex. Four additional environmental clones (DH145-HA2, C1_E045, CS_E020, and AT4–94), the latter published by López-García et al. (4), form four independent lineages branching among Acantharea, the spumellarid Polycystinea, and Taxopodida, suggesting the existence of other groups of eukaryotes closely related to them (Fig. 4). One of these phylotypes might represent the Nassellarida, the yet-unsequenced second order of Polycystinea. These observations stress the importance of a comprehensive molecular taxonomic sampling of identifiable and/or cultivable protists for an accurate interpretation of environmental DNA surveys of eukaryotic diversity.
The Rhizaria, an Emerging Supergroup of Mainly Amoeboid Eukaryotes. The early molecular phylogenies of eukaryotes showed the different lineages of amoeboid protists to be widely dispersed in SSU rRNA trees (17, 65). However, with increasing gene and taxon sampling and improved methods of phylogenetic analysis, it is now becoming evident that at least some lineages of amoeboid eukaryotes are closely related (1, 2). Recent multigene (66) and broad taxon sampling (5, 6) studies confirmed the existence of the phylum Amoebozoa, which comprises all lobose amoebae sequenced to date, as well as the pelobionts, the entamoebids, and the slime molds (see also Fig. 1).
The present study demonstrates that another supergroup of mainly amoeboid eukaryotes is emerging in molecular phylogenies. This group appeared first as the union of the euglyphid filose amoebae with the chlorarachniophytes (67). Cavalier-Smith and Chao (68) showed these taxa to be related to the heterotrophic cercomonad and thaumatomonad flagellates and the plasmodiophorid plant pathogens, and the new phylum Cercozoa was erected to accommodate this assemblage (69). Further SSU rRNA studies confirmed the apparent heterogeneity of this phylum, which also includes the haplosporidian and paramyxean parasites, the Phaeodarea, and other amoeboid protists like Gromia and Gymnophrys (21, 70–72). Protein data indicate that Foraminifera are related to Cercozoa (73–75). An evolutionary link between Cercozoa and Acantharea + Polycystinea was also suggested by some SSU rRNA trees (70, 71). Here, we show that Bayesian analyses based on a broad taxon sampling of SSU rRNA gene sequences lead to a very strong support (PP of 1.0) for the relationship among Acantharea, Polycystinea, Foraminifera, and all Cercozoa, including Phaeodarea (Fig. 1; see also Fig. 2). Besides, Bayesian analyses of actin gene sequences, including the first radiolarian protein data, bring strong independent evidence (PP of 1.0) for the existence of this supergroup (Fig. 1; see also Fig. 3). By adding Taxopodida and Desmothoracida to it, our study further broadens this diverse assemblage of protists, which, in its present form, includes the majority of filose and reticulose amoebae and most actinopods, plus two parasitic lineages and some flagellates.
To name this supergroup, we adopt here the term Rhizaria, proposed by Cavalier-Smith (36), which refers to the root-like filose or reticulose pseudopodia and/or axopodia characterizing the majority of the taxa included in it. Members of the Rhizaria are ancestrally bikonts with tubular mitochondrial cristae; the centrioles ancestrally possess a single root of a microtubular band or fan, and extrusomes, when present, are generally in the form of kinetocysts (36). Our results (Fig. 1) support the recent redefinition of the Rhizaria at the exclusion of centrohelids and apusozoans (76). Rhizaria could thus be defined as the clade including the last common ancestor of Thalassicola, Gromia, Allogromia, Plasmodiophora, and Chlorarachnion, and all of its descendants. However, at this stage, the relationships among the major groups included in this clade cannot be resolved. In SSU rRNA gene analyses (Fig. 2), Foraminifera are more closely related to Gromia and Haplosporidia than to any other eukaryotic lineage (77), whereas the radiolarian clade (Acantharea + Polycystinea + Sticholonche) is the sister group to other Rhizaria. The basal position of this clade seems to be confirmed by unpublished data on polyubiquitin (D. Moreira and T. Cavalier-Smith, personal communication), which show the lack of the one or two amino acid insertions characteristic of Cercozoa and Foraminifera at the junctions between polyubiquitin monomers (74). Nevertheless, because the existence of a diverging second type of actin in Foraminifera embroils relationships at the base of the Rhizaria (Fig. 3), additional protein data will be needed for a better resolution of the rhizarian phylogeny.
Interestingly, with testate filose amoebae, foraminiferans, and all radiolarians being included in Rhizaria, this supergroup comprises the most ancient and the richest source of known indisputable eukaryotic microfossils (78, 79). Future molecular studies on Rhizaria will provide a unique insight into the dawn of skeletonization and the evolution of pseudopodial movement.
Supplementary Material
Acknowledgments
We thank Colette and Jean Febvre for help in identifying and isolating S. zanclea, C. inerme, and T. pellucida. We thank Sandra Baldauf, Samuel Bowser, Thomas Cavalier-Smith, and three anonymous referees for useful comments on the manuscript; Louisette Zaninetti for helpful discussion; and Jackie Guiard for technical assistance. This work was supported by Russian Foundation for Basic Research Grants 02-04-48265, 02-04-48958, 02-04-49987, and 00-15-97905, and Swiss National Science Foundation Grants 3100-064073.00 and 7SUPJ062343.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: BV, bootstrap support value; ML, maximum likelihood; PP, posterior probability; SSU rRNA, small-subunit ribosomal RNA.
Data deposition: The sequences reported in this paper have been deposited in the GenBank/EMBL database (accession nos. AY268041–AY26843, AY268045, AY283744–AY283746, AY283754–AY283762, AY305008–AY305013, and AY507123–AY507125).
References
- 1.Simpson, A. G. B. & Roger, A. J. (2002) Curr. Biol. 12, 691–693. [DOI] [PubMed] [Google Scholar]
- 2.Baldauf, S. L. (2003) Science 300, 1703–1706. [DOI] [PubMed] [Google Scholar]
- 3.Moreira, D. & López-García, P. (2002) Trends Microbiol. 10, 31–38. [DOI] [PubMed] [Google Scholar]
- 4.López-García, P., Philippe, H., Gail, F. & Moreira, D. (2003) Proc. Natl. Acad. Sci. USA 100, 697–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bolivar, I., Fahrni, J. F., Smirnov, A. & Pawlowski, J. (2001) Mol. Biol. Evol. 18, 2306–2314. [DOI] [PubMed] [Google Scholar]
- 6.Fahrni, J. F., Bolivar, I., Berney, C., Nassonova, E., Smirnov, A. & Pawlowski, J. (2003) Mol. Biol. Evol. 20, 1881–1886. [DOI] [PubMed] [Google Scholar]
- 7.Peglar, M. T., Amaral Zettler, L. A., Anderson, O. R., Nerad, T. A., Gillevet, P. M., Mullen, T. E., Frasca, S. J., Silberman, J. D., O'Kelly, C. J. & Sogin, M. (2003) J. Eukaryotic Microbiol. 50, 224–232. [DOI] [PubMed] [Google Scholar]
- 8.Calkins, G. N. (1902) Marine Protozoa from Woods Hole (Government Printing Office, Washington, DC).
- 9.Haeckel, E. (1887) in The Voyage of H. M. S. Challenger, eds. Thompson, C. W. & Murray, J. (Her Majesty's Stationery Office, London), Vol. 18.
- 10.Lee, J. J., Leedale, G. F. & Bradbury, P. (2000) The Illustrated Guide to the Protozoa (Society of Protozoologists, Lawrence, KS), 2nd Ed.
- 11.Haeckel, E. (1866) Generelle Morphologie der Organismen (Georg Reimer, Berlin), Part 2.
- 12.Siemensma, F. J. (1991) in Protozoenfauna: Nackte Rhizopoda und Heliozoea, eds. Page, F. C. & Siemensma, F. J. (Fischer, Stuttgart), Vol. 1, pp. 171–290. [Google Scholar]
- 13.Febvre-Chevalier, C. (1990) in Handbook of Protoctista, eds. Margulis, L., Corliss, J. O., Melkonian, M. & Chapman, D. J. (Jones and Bartlett, Boston), pp. 347–362.
- 14.Patterson, D. J. (1994) in Progress in Protozoology: Proceedings of the Ninth International Congress of Protozoology, eds. Hausmann, K. & Hülsmann, N. (Fischer, Stuttgart), pp. 1–14.
- 15.Patterson, D. J. (1999) Am. Nat. 154, S96–S124. [DOI] [PubMed] [Google Scholar]
- 16.Mikrjukov, K. A., Siemensma, F. J. & Patterson, D. J. (2000) in The Illustrated Guide to the Protozoa, eds. Lee, J. J., Leedale, G. F. & Bradbury, P. (Society of Protozoologists, Lawrence, KS), 2nd Ed., Vol. 2, pp. 860–871. [Google Scholar]
- 17.Amaral Zettler, L. A., Sogin, M. L. & Caron, D. A. (1997) Proc. Natl. Acad. Sci. USA 94, 11411–11416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.López-García, P., Rodriguez-Valera, F. & Moreira, D. (2002) Mol. Biol. Evol. 19, 118–121. [DOI] [PubMed] [Google Scholar]
- 19.Cavalier-Smith, T. & Chao, E. E. (2003a) J. Mol. Evol. 56, 387–396. [DOI] [PubMed] [Google Scholar]
- 20.Cavalier-Smith, T. & Chao, E. E. (2003b) Protist 154, 341–358. [DOI] [PubMed] [Google Scholar]
- 21.Polet, S., Berney, C., Fahrni, J. F. & Pawlowski, J. (2004) Protist 155, 53–63. [DOI] [PubMed] [Google Scholar]
- 22.Larsen, N., Olsen, G. J., Maidak, B. L., McCaughey, M. J., Overbeek, R., Macke, T. J., Marsh, T. L. & Woese, C. R. (1993) Nucleic Acids Res. 21, 3021–3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wuyts, J., De Rijk, P., Van de Peer, Y., Pison, G., Rousseeuw, P. & De Wachter, R. (2000) Nucleic Acids Res. 28, 4698–4708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huelsenbeck, J. P. & Ronquist, F. (2001) Bioinformatics 17, 754–755. [DOI] [PubMed] [Google Scholar]
- 25.Lanave, C., Preparata, G., Saccone, C. & Serio, G. (1984) J. Mol. Evol. 20, 86–93. [DOI] [PubMed] [Google Scholar]
- 26.Rodriguez, F., Oliver, J. L., Marin, A. & Medina, J. R. (1990) J. Theor. Biol. 142, 485–501. [DOI] [PubMed] [Google Scholar]
- 27.Felsenstein, J. (1981) J. Mol. Evol. 17, 368–376. [DOI] [PubMed] [Google Scholar]
- 28.Swofford, D. (1998) paup*: Phylogenetic Analyses Using Parsimony (*and Other Methods) (Sinauer, Sunderland, MA).
- 29.Gascuel, O. (1997) Mol. Biol. Evol. 14, 685–695. [DOI] [PubMed] [Google Scholar]
- 30.Felsenstein, J. (1985) Evolution (Lawrence, Kans.) 39, 783–791. [DOI] [PubMed] [Google Scholar]
- 31.Whelan, S. & Goldman, N. (2001) Mol. Biol. Evol. 18, 691–699. [DOI] [PubMed] [Google Scholar]
- 32.Strimmer, K. & Von Haeseler, A. (1996) Mol. Biol. Evol. 13, 964–969. [Google Scholar]
- 33.Felsenstein, J. (2002) phylip(Phylogeny Inference Package) (Dept. of Genetics, Univ. of Washington, Seattle), Ver. 3.6a3.
- 34.Thorley, J. L. & Page, R. D. (2000) Bioinformatics 16, 486–487. [DOI] [PubMed] [Google Scholar]
- 35.Stechmann, A. & Cavalier-Smith, T. (2003) Curr. Biol. 13, 665–666. [DOI] [PubMed] [Google Scholar]
- 36.Cavalier-Smith, T. (2002) Int. J. Syst. Evol. Microbiol. 52, 297–354. [DOI] [PubMed] [Google Scholar]
- 37.Simpson, A. G. B. (2003) Int. J. Syst. Evol. Microbiol. 53, 1759–1777. [DOI] [PubMed] [Google Scholar]
- 38.Smith, R. M. & Patterson, D. J. (1986) Proc. R. Soc. London Ser. B 227, 325–366. [Google Scholar]
- 39.Mikrjukov, K. A. & Patterson, D. J. (2001) Acta Protozool. 40, 3–25. [Google Scholar]
- 40.Mikrjukov, K. A. (2002) The Centrohelid Heliozoans (Centroheliozoa) (KMK Scientific, Moscow).
- 41.Mikrjukov, K. A. (2000a) Zool. Zhurn. 78, 883–897. [Google Scholar]
- 42.Taylor, F. J. R. (1999) Am. Nat. 154, S125–S136. [DOI] [PubMed] [Google Scholar]
- 43.Davidson, L. A. (1982) J. Protozool. 29, 19–29. [Google Scholar]
- 44.Mikrjukov, K. A. (2000b) Acta Protozool. 39, 81–97. [Google Scholar]
- 45.Mikrjukov, K. A. (2000c) Acta Protozool. 39, 99–115. [Google Scholar]
- 46.Saunders, G. W., Potter, D., Paskind, M. P. & Andersen, R. A. (1995) Proc. Natl. Acad. Sci. USA 92, 244–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sekiguchi, H., Moriya, M., Nakayama, T. & Inouye, I. (2002) Protist 153, 157–167. [DOI] [PubMed] [Google Scholar]
- 48.Brugerolle, G. (1985) Protistologica 21, 259–265. [Google Scholar]
- 49.Mylnikov, A. P. (1988) Tsitologiya 30, 1402–1408. [Google Scholar]
- 50.Bardele, C. F. (1972) Z. Zellforsch. Mikrosk. Anat. 130, 219–242. [DOI] [PubMed] [Google Scholar]
- 51.Patterson, D. J. & Fenchel, T. (1990) Mar. Ecol. Prog. Ser. 62, 11–19. [Google Scholar]
- 52.Mikrjukov, K. A. & Mylnikov, A. P. (1998) Acta Protozool. 37, 179–189. [Google Scholar]
- 53.Brugerolle, G. & Mignot, J. P. (1984) Origins Life 13, 305–314. [Google Scholar]
- 54.Febvre-Chevalier, C. & Febvre, J. (1984) Origins Life 13, 315–338. [Google Scholar]
- 55.Kugrens, P. & Lee, R. E. (1991) in The Biology of Free-Living Heterotrophic Flagellates, eds. Patterson, D. J. & Larsen, J. (Clarendon, Oxford), pp. 219–234.
- 56.Cachon-Enjumet, M. (1964) C. R. Acad. Sci. 259, 2677–2678. [Google Scholar]
- 57.Cachon, J. & Cachon, M. (1973) Arch. Protistenkd. 115, 324–335. [Google Scholar]
- 58.Anderson, O. R. (1983) Radiolaria (Springer, New York).
- 59.Cachon, J., Cachon, M. & Estep, K. W. (1990) in Handbook of Protoctista, eds. Margulis, L., Corliss, J. O., Melkonian, M. & Chapman, D. J. (Jones and Bartlett, Boston), pp. 334–346.
- 60.Fol, H. (1883) Mem. Inst. Nat. Genevois 15, 1–35. [Google Scholar]
- 61.Hollande, A., Cachon, J., Cachon, M. & Valentin, J. (1967) Protistologica 3, 155–166. [Google Scholar]
- 62.Cachon, J. & Cachon, M. (1978) Arch. Protistenkd. 120, 148–168. [Google Scholar]
- 63.Margulis, L., Corliss, J., Melkonian, M. & Chapman, D. (1990) Handbook of Protoctista (Jones and Bartlett, Boston).
- 64.Edgcomb, V. P., Kysela, D. T., Teske, A., de Vera Gomez, A. & Sogin, M. L. (2002) Proc. Natl. Acad. Sci. USA 99, 7658–7662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sogin, M. L. (1991) Curr. Opin. Genet. Dev. 1, 457–463. [DOI] [PubMed] [Google Scholar]
- 66.Bapteste, E., Brinkmann, H., Lee, J. A., Moore, D. V., Sensen, C. W., Gordon, P., Durufle, L., Gaasterland, T., Lopez, P., Muller, M., et al. (2002) Proc. Natl. Acad. Sci. USA 99, 1414–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bhattacharya, D., Helmchen, T. & Melkonian, M. (1995) J. Eukaryotic Microbiol. 42, 65–69. [DOI] [PubMed] [Google Scholar]
- 68.Cavalier-Smith, T. & Chao, E. (1997) Arch. Protistenkd. 147, 227–236. [Google Scholar]
- 69.Cavalier-Smith, T. (1998) Biol. Rev. Camb. Philos. Soc. 73, 203–266. [DOI] [PubMed] [Google Scholar]
- 70.Burki, F., Berney, C. & Pawlowski, J. (2002) Protist 153, 251–260. [DOI] [PubMed] [Google Scholar]
- 71.Cavalier-Smith, T. & Chao, E. E. (2003c) J. Mol. Evol. 56, 540–563. [DOI] [PubMed] [Google Scholar]
- 72.Nikolaev, S. I., Berney, C., Fahrni, J. F., Mylnikov, A. P., Aleshin, V. V., Petrov, N. B. & Pawlowski, J. (2003) Acta Protozool. 42, 183–190. [Google Scholar]
- 73.Keeling, P. J. (2001) Mol. Biol. Evol. 18, 1551–1557. [DOI] [PubMed] [Google Scholar]
- 74.Archibald, J. M., Longet, D., Pawlowski, J. & Keeling, P. J. (2003) Mol. Biol. Evol. 20, 62–66. [DOI] [PubMed] [Google Scholar]
- 75.Longet, D., Archibald, J. M., Keeling, P. J. & Pawlowski, J. (2003) Int. J. Syst. Evol. Microbiol. 53, 1735–1739. [DOI] [PubMed] [Google Scholar]
- 76.Cavalier-Smith, T. (1999) J. Eukaryotic Microbiol. 46, 347–366. [DOI] [PubMed] [Google Scholar]
- 77.Berney, C. & Pawlowski, J. (2003) J. Mol. Evol. 57, S120–S127. [DOI] [PubMed] [Google Scholar]
- 78.Lipps, J. H. (1993) Fossil Prokaryotes and Protists (Blackwell, Boston).
- 79.Porter, S. M. & Knoll, A. H. (2000) Paleobiology 26, 360–385. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.