Abstract
The evolution of ecological specialization is expected to carry a cost, due to either antagonistic pleiotropy or mutation accumulation. In general, it has been difficult to distinguish between these two possibilities. Here, we demonstrate that the experimental evolution of niche-specialist genotypes of the bacterium Pseudomonas fluorescens that colonize the air–broth interface of spatially structured microcosms is accompanied by pleiotropic fitness costs in terms of reduced carbon catabolism. Prolonged selection in spatially structured microcosms caused the cost of specialization to decline without loss of the benefits associated with specialization. The decline in the cost of specialization can be explained by either compensatory adaptation within specialist lineages or clonal competition among specialist lineages. These results provide a possible explanation of conflicting accounts for the cost of specialization.
The evolution of a diverse community in a heterogeneous environment requires that no single type, whether it be a sexual species or an asexual genotype, be able to simultaneously outcompete all other types in all patches of the environment (1–3). According to the pleiotropy hypothesis of niche evolution, the evolution of such a universally superior type is unlikely, because alleles that increase fitness in one patch will inevitably cause a regress of fitness in other patches (2, 4–7). In contrast, recent theoretical work has demonstrated that pleiotropic fitness costs do not need to be invoked to explain the evolution of a diverse community of specialists. The evolution of a universally superior type could be prevented by demographic constraints on the ability of broadly distributed types to rapidly adapt to a variety of patches or conditions (8). In the absence of pleiotropy, a cost of specialization is likely to evolve as conditionally deleterious mutations that reduce fitness in rarely encountered or unproductive patches stochastically accumulate in specialist populations (9–11). In summary, both the antagonistic pleiotropy and mutation accumulation theories predict the evolution of tradeoffs in fitness across patches of a heterogeneous environment; they also predict that specialization will be accompanied by a cost, but they provide very different accounts of the genetic basis of this cost. According to the pleiotropy theory, the same mutations that create a specialist phenotype create a cost of specialization. The mutation accumulation theory assumes that mutations that create specialist phenotypes are not directly associated with a cost, but that specialist populations will stochastically accumulate mutations at other loci that carry a fitness cost.
As a result of the relative ease with which organisms and environments can be manipulated in the laboratory, selection experiments have become a powerful tool for testing alternative hypotheses concerning ecological and evolutionary causes and consequences of specialization (12–15). The general protocol for these experiments is as follows: replicate selection lines founded from a common ancestral genotype are allowed to adapt to different environments that represent different conditions for growth and reproduction. After some period of selection, lines adapted to different environments are competed against their common ancestor in a range of environments to assay for the benefits and costs of adaptation. Although a number of experiments of this kind have been able to detect the existence of costs of adaptation, only a few studies have been able to attribute this cost convincingly to either antagonistic pleiotropy (16–19) or mutation accumulation (20–22).
When inoculated into a spatially structured microcosm containing a nutrient-rich culture medium, a broth-colonizing genotype of the bacterium Pseudomonas fluorescens rapidly divides, reaching a density of ≈109 cells per ml–1 after 2 days of incubation. After 3 days of incubation, mutant genotypes descended from the ancestral genotype begin to appear. One of the most ecologically successful derived types is the “wrinkly spreader” (WS), which colonizes the air–liquid interface of the microcosm and forms colonies on agar plates that can be easily distinguished from the broth-colonizing ancestral genotype “smooth” (SM). Both the frequency and density of WS morphs rapidly increases, such that both SM and WS morphs are present at a density of ≈109 cells per ml–1 after 7 days of incubation (15, 23). In undisturbed microcosms, SM and WS morphs can stably coexist at similar frequencies for hundreds of generations, because the fitness of each morph is negative frequency-dependent. As such, this simple population provides a tractable model for studying the evolutionary and ecological causes and consequences of specialization. Here, we investigate costs associated with the evolution of the WS morph by using phenotypic microarrays (24); we examine the evolution of metabolic costs associated with the WS morph, determine whether the costs are due to mutation accumulation or antagonistic pleiotropy, and examine changes in the magnitude of tradeoffs due to compensatory mutation.
Materials and Methods
Diversification Experiment. We used clonal isolates of Pseudomonas fluorescens strain SBW25 or SBW25ΔpanB, an isogenic strain of SBW25 containing a deletion of the panB gene, to inoculate 20 [12 ΔpanB (pan–) 8 SBW25 (pan+)] microcosms containing 5 ml of M9KB plus pantothenate [glycerol, 10 g·l–1; proteose peptone 3, 20 g·l–1; Na2HPO4, 6g·l–1; KH2PO4, 3g·l–1; NH4Cl, 1 g·l–1; NaCl, 0.5 g·l–1; and pantothenate, 0.0024% (wt/vol)]. These microcosms were incubated at 28°C without shaking. After 7 days of selection, 12 microcosms (six pan+ and six pan–) were destructively harvested by vortexing (≈45 sec) and diluted samples were spread on KB plus pantothenate agar plates. One randomly chosen WS colony was sampled from each replicate population and stored in 50% (wt/vol) glycerol at –80°C. Picking either the last isolated colony on a streaked plate, or the colony closest to an arbitrarily chosen point on a spread plate, ensured random colony selection. We checked the correspondence between marker and WS genotype by streaking on indicator plates (glycerol, 5 g·l–1; casamino acids, 10 g·l–1; Na2HPO4, 6g·l–1; KH2PO4, 3g·l–1; NH4Cl, 1g·l–1; and NaCl, 0.5 g·l–1) containing a low concentration of pantothenate (2.4 × 10–6 %) that allows pan– and pan+ colonies to be easily distinguished. The niche occupancy of each colony was determined by reinoculating the colony into a M9KB plus pantothenate microcosm and recording the region of the microcosm that the genotype colonized. In every case, WS genotypes had the same marker (pan+ or pan–) as their ancestor (SBW25 or SBW25ΔpanB, respectively) and colonized the air–broth interface of the microcosm.
Continued Selection Experiments. The remaining eight genotypically diverse microcosms were thoroughly vortexed and serially diluted by 1,000-fold into a fresh microcosm containing 5 ml of M9KB plus pantothenate. After 2 days of selection, the populations were once again serially transferred to fresh medium. After 10 transfers, representing ≈100 generations of continued selection, a single WS genotype was randomly sampled from each of the lines, as described above.
WS Selection Experiments. We used WS genotypes that evolved during the diversification experiment to found two subsequent sets of experimental populations. The first set of populations was founded by inoculating six randomly chosen WS genotypes into M9KB plus pantothenate microcosms. After 1 week of incubation without physical disturbance, microcosms were destructively harvested and plated on M9KB plus pantothenate agar plates. A single SM colony (derived-SM) was randomly chosen from each selected line and frozen at –80°C. In every case, the marker (pan– or pan+) of each derived-SM genotype was the same as that of its WS ancestor, and all derived-SM genotypes colonized the broth phase of an undisturbed microcosm.
In the second experiment, 10 WS genotypes and their reciprocally marked SM ancestor were together used to found two replicate selection lines. Microcosms were incubated without disturbance at 28°C and were transferred by a 1,000-fold serial dilution into 5 ml of fresh M9KB plus pantothenate every 2 days. After 10 transfers, one WS colony was randomly sampled from each replicate of each line and the niche preference and marker (pan+/pan–) of each genotype were measured and recorded. In every case, evolved WS genotypes were descended from a WS ancestor; all WS genotypes colonized the air–broth interface of unshaken microcosms.
Competitive Fitness Assays. To estimate the competitive fitness of different SM and WS genotypes, we carried out a series of competition assays against marked strains. In every case, a competitor genotype was matched against a tester genotype carrying the opposing pantothenate marker so that the frequency of the two types could be readily determined on competition plates. In all fitness assays, single colonies of all tester and competitor genotypes were first inoculated into microcosms containing 5 ml of M9KB plus pantothenate and were incubated overnight at 28°C with vigorous shaking (200 rpm). This procedure ensured that the populations had comparable initial physiological states and cell densities. To initiate a competitive fitness assay, tester and competitor genotypes were mixed together to form a common pool that was thoroughly mixed before inoculation into three replicate competition microcosms containing 5 ml of KB plus pantothenate. Common pools were then diluted and spread on indicator plates to determine the initial ratio of the two competing genotypes. Competition microcosms were incubated at 28°C for either 1 or 2 days (depending on the experiment) without physical disturbance before being vortexed, diluted, and spread on indicator plates. In all competition experiments, the number of colonies counted from the common pool and in the replicated fitness assays was large (>100 colonies). The fitness of a competitor genotype, relative to its tester, was estimated as log w, the change in the ratio of log [(Ncompetitor)/(Ntester)] over the course of the competition (25), where equal fitness is defined as log w = 0.
Biolog Fitness Assays. To assay for pleiotropic changes in the physiology and fitness of Pseudomonas as a result the evolution of specialization, we assayed evolved and ancestral genotypes on commercially available Biolog GN2 microplates (Biolog, Hayward CA). Biolog plates are 96-well microtiter plates in which all wells contain an indicator dye of carbon metabolism as well as a simple growth medium. Ninety-five wells contain a unique carbon compound that acts as the sole available carbon source in the well and the remaining well is a carbon-free control. To assay a given genotype on a Biolog plate, we first grew a single colony overnight in M9KB plus pantothenate (28°C at 200 rpm). A total of 20 μl of this culture was then diluted in 20 ml of M9 salts supplemented with excess pantothenate [Na2HPO4, 6g·l–1; KH2PO4, 3 g·l–1; NH4Cl, 1 g·l–1; and NaCl, 0.5 g·l–1 pantothenate 0.0024% (wt/vol)] and incubated for ≈2hat28°Ctostarve the cells. A total of 150 μl of starved cells was then added to each well on the microplate, which was read at 660 nm by using an automated Bio-Tek Elx800-NB microplate reader after 48 h of incubation ± 5 min. We calculated a quantitative score for each well (OD660 corr) as the observed absorbance minus the blank control. Under these conditions, OD660 corr values are well correlated with cell densities (r2 = 0.76) and no growth occurs in the carbon-free control well. Quantitative absorbance data were used to generate qualitative growth scores for each substrate on a three point scale: 0, or no detectable growth, 1/2, which corresponded to weak growth, and 1, which corresponded to strong growth. The cut-off points (OD660corr <0.1 = 0; 0.1< OD660corr <.15 = 1/2; and OD660 corr>.15 = 1) for these binary measures was chosen on the basis of the distribution of OD660 corr values and on visual inspection of Biolog plates. In addition, the data from these experiments corresponded closely with the results obtained in numerous preceding experiments with Biolog plates (20).
Cluster Analysis. We used clustering techniques to visualize differences in the Biolog profile among genotypes. The phenotypic distance between all possible pairs of genotypes was calculated as the percentage of substrates where two genotypes produced different qualitative growth scores. Only substrates where the ancestral clone was consistently able to grow were used. Clustering was performed by applying a upgma algorithm to the distance matrix.
Results and Discussion
Fitness Benefits of the WS Phenotype. To determine the competitive fitness advantage associated with each WS genotype, we assayed the ability of independently evolved WS genotypes to invade a population that consisted of 99% of the ancestral SM broth-colonizing genotype. In agreement with previous results (23), we found that WS genotypes that evolve after 1 week of selection have a large competitive advantage in an SM-dominated population (mean log w = 3.65, SD = 1.19, two-tailed t11 = 10.55, P < 0.001), although this advantage varies widely among genotypes (F11,22 = 15.9, P < 0.0001; range = 1.25–5.04; coefficient of variation = 32%). Note that a value of log w of 0 indicates equal competitive ability, and that extensive preliminary results have demonstrated that the pantothenate marker that we used in competition assays does not confer a detectable fitness effect.
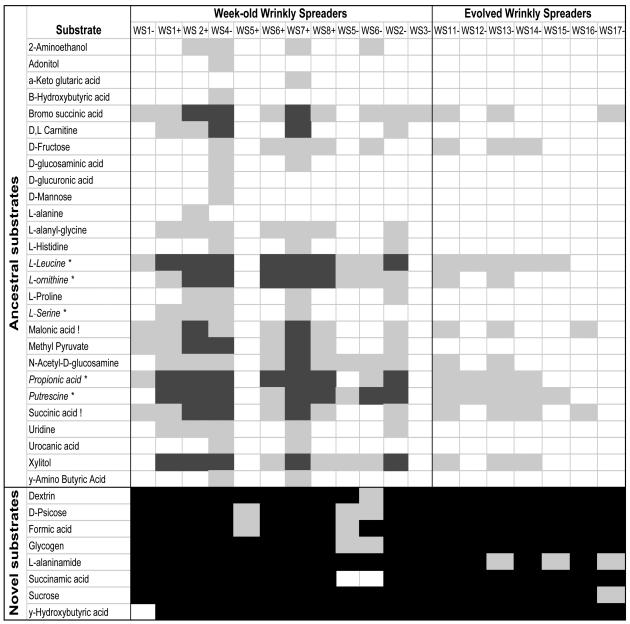
Metabolic Costs Associated with the WS Phenotype. To assay for costs associated with the evolution of the WS morph, we measured the stationary density of independently evolved WS genotypes and the ancestral SM genotype in the 95 environments provided by Biolog microtiter plates. Most (18 of 19) WS genotypes express catabolic defects, in that they produce marginal or undetectable growth on substrates that always support strong growth of numerous replicates of the ancestral SM genotype (Fig. 1). A comparison with the Poisson distribution reveals that the number of catabolic defects per substrate is not randomly distributed; rather, losses of growth tend to occur on a narrow range of substrates (Log-likelihood = 221, df = 12, P < 0.0001), most of which are tricarboxylic acid cycle intermediates (Fig. 1, substrates marked with exclamation points) or amino acids that are ultimately fed into the tricarboxylic acid cycle (Fig. 1, substrates marked with asterisks) without first passing through glycolysis (Fig. 4, which is published as supporting information on the PNAS web site). It is also interesting to note that WS genotypes occasionally gain the ability to metabolize carbon compounds (Fig. 1).
Fig. 1.
Biolog profile of WS genotypes. The substrates shown here either always support the growth of the ancestral SM genotype (ancestral substrates) or never support the growth of this genotype (novel substrates). Blank boxes, substrates that support substantial growth of WS genotypes; gray boxes, substrates that support weak growth of WS genotypes; black boxes, substrates that do not support visible growth of WS genotypes.
Fitness and Biolog Changes Are the Results of Mutations at the Same Locus. To test the hypothesis that the differences between WS genotypes and the ancestral SM clone in Biolog substrate utilization are a pleiotropic effect of the same mutation that causes the WS phenotype, we reinoculated six WS genotypes that express varying degrees of catabolic impairment (mean catabolic defects = 16, SD = 7.9, range = 6–25) into undisturbed microcosms. Derived SM genotypes directly descended from WS evolved in every reverse-selection line, presumably because of the fitness benefits associated with colonizing the uninhabited broth phase of a WS-dominated microcosm (23, 26). Four derived SM genotypes completely recover the full spectrum of ancestral catabolic function. The remaining two derived-SM genotypes express a catabolic defect, in terms of marginal growth on a single substrate (Fig. 5, which is published as supporting information on the PNAS web site). These results directly demonstrate that the same mutations that generate a WS morph from an SM ancestor are responsible for the catabolic defects associated with the WS morph.
If the transition from WS to derived-SM is caused by a reversion of the original mutation that generates WS from ancestral SM, we would expect that the fitness of derived-SM genotypes should equal that of ancestral SM. To test this hypothesis, we directly competed derived-SM genotypes against ancestral SM. Although the average fitness of derived-SM (mean log w = 0.37, SD = 0.41) does not differ from ancestral SM (two-tailed t5 = 2.26, P = 0.07), significant genetic variance in fitness exists among derived SM genotypes (F5,12 = 29.1, P < 0.001), suggesting that derived SM genotypes are generated by second-site mutations in WS genotypes, and not by reversions.
The Evolution of WS Metabolic Profile. Biolog assays document extensive variation in catabolic performance among WS genotypes (Fig. 1). The metabolic phenotype of WS genotypes from continued selection lines can be distinguished from that of WS genotypes that evolve after 1 week in two ways: (i) WS genotypes from continued selection lines are always able to produce detectable growth on all of the substrates that support the growth of ancestral SM (Fig. 1), and (ii) the average number of catabolic defects associated with the WS phenotype declines from 11.5 (SE = 1.94) to 4.8 (SE = 2.5) with subsequent adaptation (two-tailed t17 = 2.1, P = 0.05).
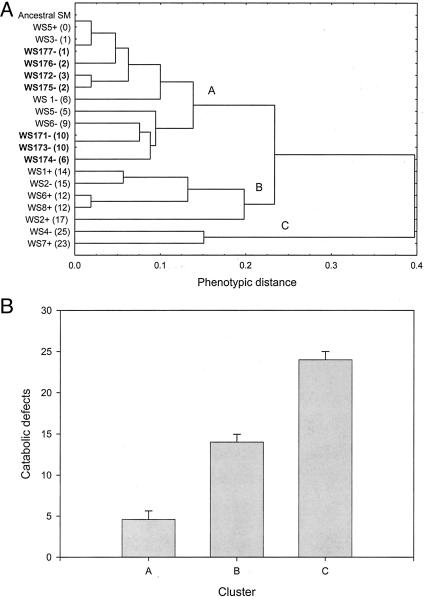
Clustering methods provide a complementary technique to analyze the evolution of the WS metabolic profile. WS genotypes that evolve in the short and long term can be classified into three principle clusters (Fig. 2A) that are well differentiated with respect to catabolic performance (Fig. 2B). The distribution of genotypes among these three clusters changes significantly after ≈100 generations of selection, according to the null hypothesis that the fraction of genotypes in each cluster is constant through time (χ2 = 9.33, df = 2, P < 0.01). This shift is attributable to the fact that WS genotypes that evolve after 1 week of selection fall into all three clusters, whereas WS genotypes that are allowed to evolve for a further 100 generations fall into a single cluster (Fig. 2 A). This cluster is characterized by minor catabolic defects when compared with the remaining clusters (Fig. 2B).
Fig. 2.
The evolution of the WS metabolic profile. (A) Phenotypic clustering of WS genotypes and ancestral SM. Each tip corresponds to a single genotype. WS genotypes from continued selection lines are in bold. Numbers in brackets denote the number of catabolic defects expressed by each genotype, such that the value of ancestral SM is 0 by definition. Letters (A, B, and C) denote the three principle phenotypic clusters. (B) Mean (± SE) number catabolic defects of WS genotypes belonging to the three principle clusters identified in A.
The Fitness Cost of Impaired Metabolism. The most straightforward hypothesis to explain why the catabolic performance of WS genotypes improves after continued selection is that impaired catabolism represents a general fitness cost to WS genotypes that evolve after a week of selection. Our results strongly support this possibility; catabolic performance is positively correlated with competitive fitness against the ancestral SM genotype (Fig. 3A: F1,11 = 9.6, P = 0.011, r2 = 0.49) or a randomly chosen WS genotype (Fig. 3B: F1,6 = 8.35, P = 0.03, r2 = 0.62). These two measures of WS competitive performance are independent, because measures of competitive fitness against these two tester genotypes are not correlated with each other (r = –0.41, F1,9 = 1.8, P = 0.2).
Fig. 3.
Competitive fitness and catabolic performance. Plotted points show the competitive fitness of WS genotypes that evolve after 1 week of selection and catabolic defects, measured as the number of ancestral Biolog substrates that fail to support vigorous growth of WS genotypes. Dashed lines show linear regressions of fitness on catabolic performance. (A) Fitness was assayed as the ability of a WS genotype to invade a population of the ancestral broth-colonizing genotype from an initial frequency of ≈1% over a 2-day competition period. (B) Fitness was assayed as the competitive ability of a WS genotype against a randomly chosen WS tester genotype at an initial frequency of 1:1 over a 1-day competition period.
Compensatory Adaptation. One hypothesis to explain how the metabolic cost associated with the WS phenotype declines after continued selection is that second-site compensatory mutations ameliorate the biochemical defects associated with mutations that generate the WS phenotype from an SM ancestor. To investigate the plausibility of this mechanism, we selected two replicates of 10 WS genotypes from week-old selection lines for an additional ≈100 generations. Our statistical test for compensatory adaptation used phenotypic clustering techniques based on Biolog data. We define compensatory adaptation as a shift from one of the principle clusters of WS catabolic phenotype to a cluster characterized by improved catabolic performance. Increased fitness costs of the WS phenotype after prolonged selection would result in a downward shift to a cluster characterized by inferior catabolic performance (Fig. 2). Of the 20 possible opportunities for cluster shifts, we observed shifts in only five cases (Table 1). In every case, these shifts represented compensatory adaptation. Given that the maximum possible number of upward and downward shifts are 14 and 16, respectively, the likelihood of observing this outcome under the null hypothesis of an equal probability of upward and downward shifts is low [14/(14 + 16)]5 = 0.022. Note that compensatory adaptation resulted in shifts from cluster B to A and from cluster C to B. We conclude that there is a tendency toward compensatory adaptation in evolving WS lineages, although the overall frequency of compensatory adaptation over is modest (5 shifts/14 opportunities) = 35.7%.
Table 1. Compensatory adaptation in WS lineages.
| Genotype | Ancestral WS cluster | Replicate line 1 cluster | Replicate line 2 cluster |
|---|---|---|---|
| WS1- | A | A | A |
| WS3- | A | A | A |
| WS5- | A | A | A |
| WS1+ | B | B | B |
| WS2- | B | B | A |
| WS6+ | B | B | A |
| WS8+ | B | B | B |
| WS2+ | B | B | B |
| WS4- | C | B | B |
| WS7+ | C | B | C |
Conclusion
A central concept in evolutionary ecology is that the evolution of ecological specialization is expected to be accompanied by both benefits and costs. Previous studies (23) have established that WS morph genotypes are able to invade SM-dominated microcosms as a result of the ability to colonize the oxygen-rich surface of the microcosm. Many of the WS morphs that initially colonize the air–broth interface express extensive catabolic defects (Figs. 1 and 2). Impaired catabolism represents a general fitness cost to WS in that it results in reduced competitive ability against both SM and WS competitors (Fig. 3). The fitness cost associated with impaired catabolism provides a convincing explanation for why WS catabolic performance improves with continued selection (Figs. 1 and 2).
Two explanations can be put forward to explain how WS metabolic performance improves after continued selection. In the first place, it is possible that catabolic performance improves as a result of clonal competition among WS genotypes derived from ancestral SM (27). According to this hypothesis, the air–broth interface of microcosms is first colonized by a number of WS genotypes with various degrees of catabolic impairment. Competition among WS genotypes at the air–broth interface then leads to the competitive exclusion of WS genotypes with impaired metabolism. The results of our competitive fitness assays are consistent with this hypothesis: all WS genotypes, including those with severely impaired catabolism, enjoy a large competitive advantage in an SM-dominated population (Fig. 3A), and WS genotypes with impaired catabolism perform poorly in competition with a rival WS genotype at the air–broth interface (Fig. 3B). Alternatively, it is possible that WS catabolic performance improves as a result of compensatory adaptation. According to this hypothesis, all WS genotypes directly descended from ancestral SM express severe catabolic defects. Second-site compensatory mutations lead to the appearance of WS genotypes with restored catabolic function that then exclude WS competitors that lack compensatory mutations. In support of this hypothesis, we were able to directly detect compensatory adaptation in individual WS lineages (Table 1). Our results do not allow us to determine whether clonal interference or compensatory adaptation was responsible for the evolution of improved WS catabolic performance in selection lines founded by ancestral SM. The critical distinction between these two hypotheses is that the clonal interference hypothesis predicts that WS genotypes with unimpaired catabolism are directly descended from ancestral SM, whereas the compensatory adaptation hypothesis predicts that WS genotypes with unimpaired catabolism are descended from WS genotypes. A detailed knowledge of the phylogeny of evolving Pseudomonas populations would, therefore, allow us to discriminate between these hypotheses. At present, the phylogenies of microcosm populations that adapt on a short time scale (i.e., many thousands of generations) can only be reconstructed for viruses, where it is possible to sequence entire genomes.
In a recent study, Cooper and Lenski (17) found that specialization on a glucose-limited culture environment is accompanied by pleiotropic fitness costs that do not decline, even after 20,000 generations of selection. How can these findings be reconciled with the present study? We argue that two fundamentally different classes of pleiotropic costs exist: those that are a direct cause of increased fitness and those that are an indirect consequence of increased fitness. For example, Cooper et al. (28) found that insertion mutations in the ribose operon become fixed in populations of Escherichia coli evolving in a glucose-limited environment because they directly cause an increase in fitness when ribose is not present in the culture medium. Costs such as this are stable, because reversion of the mutation causing the cost will cause a decline in fitness. On the other hand, it is possible that beneficial mutations will be associated with deleterious side effects that do not directly cause an increase in fitness. The best-studied example of this class of cost is the evolution of antibiotic and resistance. Chromosomal mutations that confer antibiotic resistance often alter the structure of the proteins that antibiotics target, resulting in both increased resistance to antibiotic and decreased function of the target protein, which is often associated with a considerable fitness cost (29). This cost declines with subsequent selection in the presence of antibiotic as a result of second site compensatory mutations that diminish the cost of resistance, without increasing sensitivity to the antibiotic (30–32). We argue that the evolution of the WS morph is also an excellent example of this second class of cost. The WS phenotype is generated by mutations in the wsp operon (33), which encodes a chemosensory pathway (E. Bantinaki, A. J. Spiers, and P.B.R., unpublished work), that regulates the expression of an acetylated cellulose polymer (33, 34). Although impaired catabolic function is not required for the expression of the WS phenotype (Fig. 1), the indirect consequence of mutations at this locus is that central pathways of carbon catabolism become impaired, which imposes a fitness cost that is expressed in competition with a range of ecologically different competitors (Fig. 3). Costs such as these are expected to decline during the evolution of specialization because reversion of the cost will be associated with an increase in fitness.
Providing a rigorous account of the consequences of ecological specialization is critical to our understanding of the evolution and subsequent maintenance of ecological and genetic diversity. Models of the evolution of specialization assume either that antagonistic pleiotropy is ubiquitous, or that it is absent. In this system, neither of these assumptions is upheld: the evolution of a simple specialist morph of bacteria is accompanied by a large pleiotropic cost, but this cost rapidly diminishes as a result of compensatory mutation. Given that there is no obvious link between acetylated cellulose biosynthesis and the regulation of central metabolism, we can only speculate as to why mutations in wsp are associated with such dramatic defects. One possibility is that the overproduction of cellulose directly impairs central metabolism by shunting carbon into cellulose production. In this scenario, pleiotropy results from conflicts between energetic allocation to different organismal functions. A second plausible explanation is that mutations at this locus perturb cell-wide signaling networks that regulate metabolism. This scenario invokes the disruption of a well integrated genome as the cause of antagonistic pleiotropy. In future experiments, we will attempt to discriminate between these possibilities.
Supplementary Material
Acknowledgments
We thank A. J. Spiers, C. Knight, R. Kassen, A. Gonzalez, and two anonymous reviewers for comments and insightful discussion. This work was supported by grants from the Natural Sciences and Engineering Research Council of Canada (to G.B. and R.C.M.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: WS, wrinkly spreader; SM, smooth.
References
- 1.Levene, H. (1953) Am. Nat. 88, 690–692. [Google Scholar]
- 2.Levins, R. (1968) Evolution in Changing Environments (Princeton Univ. Press, Princeton).
- 3.Chesson, P. (2000) Annu. Rev. Ecol. Syst. 31, 343–366. [Google Scholar]
- 4.Lynch, M. & Gabriel, W. (1987) Am. Nat. 129, 282–303. [Google Scholar]
- 5.Roughgarden, J. (1972) Am. Nat. 106, 683–718. [Google Scholar]
- 6.Wilson, D. S. & Yoshimura, J. (1994) Am. Nat. 144, 692–707. [Google Scholar]
- 7.Futuyma, D. J. & Moreno, G. (1988) Annu. Rev. Ecol. Syst. 19, 207–233. [Google Scholar]
- 8.Whitlock, M. C. (1996) Am. Nat. 148, S65–S77. [Google Scholar]
- 9.Fry, J. D. (1996) Am. Nat. 148, S84–S107. [Google Scholar]
- 10.Kawecki, T. J. (1994) Am. Nat. 144, 833–838. [Google Scholar]
- 11.Kawecki, T. J., Barton, N. H. & Fry, J. D. (1997) J. Evol. Biol. 10, 407–429. [Google Scholar]
- 12.Elena, S. F. & Lenski, R. E. (2003) Nat. Rev. Genet. 4, 457–469. [DOI] [PubMed] [Google Scholar]
- 13.Ebert, D. (1998) Science 282, 1432–1435. [DOI] [PubMed] [Google Scholar]
- 14.Kassen, R. (2002) J. Evol. Biol. 15, 173–190. [Google Scholar]
- 15.Travisano, M. & Rainey, P. B. (2000) Am. Nat. 156, S35–S44. [DOI] [PubMed] [Google Scholar]
- 16.Crill, W. D., Wichman, H. A. & Bull, J. J. (2000) Genetics 154, 27–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cooper, V. S. & Lenski, R. E. (2000) Nature 407, 736–739. [DOI] [PubMed] [Google Scholar]
- 18.Cooper, V. S., Bennett, A. F. & Lenski, R. E. (2001) Evolution (Lawrence, Kans.) 55, 889–896. [DOI] [PubMed] [Google Scholar]
- 19.Bull, J. J., Badgett, M. R. & Wichman, H. A. (2000) Mol. Biol. Evol. 17, 942–950. [DOI] [PubMed] [Google Scholar]
- 20.MacLean, R. C. & Bell, G. (2002) Am. Nat. 160, 569–581. [DOI] [PubMed] [Google Scholar]
- 21.Reboud, X. & Bell, G. (1997) Heredity 78, 507–514. [Google Scholar]
- 22.Giraud, A., Matic, I., Tenaillon, O., Clara, A., Radman, M., Fons, M. & Taddei, F. (2001) Science 291, 2606–2608. [DOI] [PubMed] [Google Scholar]
- 23.Rainey, P. B. & Travisano, M. (1998) Nature 394, 69–72. [DOI] [PubMed] [Google Scholar]
- 24.Bochner, B. R. (2003) Nat. Rev. Genet. 4, 309–314. [DOI] [PubMed] [Google Scholar]
- 25.Hartl, D. L. & Clark, A. G. (1997) Principles of Population Genetics (Sinauer, Sunderland, MA).
- 26.Rainey, P. B. & Rainey, K. (2003) Nature 425, 72–74. [DOI] [PubMed] [Google Scholar]
- 27.Gerrish, P. J. & Lenski, R. E. (1998) Genetica (The Hague) 103, 127–144. [PubMed] [Google Scholar]
- 28.Cooper, V. S., Schneider, D., Blot, M. & Lenski, R. E. (2001) J. Bacteriol. 183, 2834–2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Andersson, D. I. & Levin, B. R. (1999) Curr. Opin. Microbiol. 2, 489–493. [DOI] [PubMed] [Google Scholar]
- 30.Maisnier-Patin, S., Berg, O. G., Liljas, L. & Andersson, D. I. (2002) Mol. Microbiol. 46, 355–366. [DOI] [PubMed] [Google Scholar]
- 31.Cohan, F. M., King, E. C. & Zawadzki, P. (1994) Evolution (Lawrence, Kans.) 48, 81–95. [DOI] [PubMed] [Google Scholar]
- 32.Shrag, S. J., Perrot, V. & Levin, B. R. (1997) Proc. R. Soc. London Ser. B 264, 1287–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spiers, A. J., Kahn, S. G., Bohannon, J., Travisano, M. & Rainey, P. B. (2002) Genetics 161, 33–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spiers, A. J., Bohannon, J., Gehrig, S. M. & Rainey, P. B. (2003) Mol. Microbiol. 50, 15–27. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.