Abstract
In both human patients and murine models, the progression from insulitis to diabetes is neither immediate nor inevitable, as illustrated by the innocuous versus destructive infiltrates of BDC2.5 transgenic mice on the nonobese diabetic (NOD) versus C57BL/6.H-2g7 genetic backgrounds. Natural killer (NK)-cell-specific transcripts and the proportion of NK cells were increased in leukocytes from the aggressive BDC2.5/B6.H-2g7 lesions. NK cell participation was also enhanced in the aggressive lesions provoked by CTLA-4 blockade in BDC2.5/NOD mice. In this context, depletion of NK cells significantly inhibited diabetes development. NOD and B6.H-2g7 mice exhibit extensive variation in NK receptor expression, reminiscent of analogous human molecules. NK cells can be important players in type 1 diabetes, a role that was previously underappreciated.
Keywords: type 1 diabetes, mouse model, microarray
Type 1 diabetes (T1D) is an autoimmune disease characterized by immune system attack on the insulin-producing β cells of the pancreatic islets, resulting in the loss of glucose homeostasis and, ultimately, hyperglycemia (1). Somehow breaking through the usual net of immunological self-tolerance, a leukocytic infiltrate, dominated by autoreactive T cells and augmented by bystander populations attracted by the local inflammation, invades the islets. Interestingly, this “insulitis” does not always progress to diabetes. In humans, the presence of anti-β-cell autoantibodies does not always foretell clinical hyperglycemia (2). In nonobese diabetic (NOD) mice, the most commonly used mouse model of T1D, insulitis initiates in all individuals, male and female, but diabetes develops mostly in females, and with variable incidence. In several other murine models, an islet infiltrate is present but does not progress to hyperglycemia (3, 4). Hence, conversion from insulitis to diabetes appears to be a regulated process. Elucidating the elements that control the pathogenicity of an islet infiltrate may be of great therapeutic importance because, given the complications of screening the general population, the best hope of preventing T1D may be to identify individuals with a benign insulitic lesion and block it from becoming pathogenic.
The BDC2.5 T cell receptor (TCR) transgenic (tg) mouse has proven a valuable model for studying the pathogenesis of T1D. The transgene-encoded TCR, derived from a diabetogenic T cell clone isolated from a NOD mouse (5), recognizes a β-cell-derived antigen in the context of the NOD MHC class II molecule Ag7. When the transgenes are carried on the NOD genetic background, massive insulitis initiates in all animals just after 2 weeks of age (6); however, the infiltrate remains quite innocuous and only rarely progresses to diabetes (7), despite the presence of a diabetogenic TCR that is destructive in other circumstances. This state of “respectful” insulitis is histologically quite similar to what has been described for the insulitic lesions of NOD mice protected from diabetes development by strong immunostimulation, i.e., a clear demarcation between the infiltrating leukocytes and residual β cells (8, 9). This state can be disrupted by a number of perturbations. In the early phase of disease, at the time when naïve, potentially diabetogenic T cells are first incited to invade the islets, the blockade of costimulatory molecules, in particular of CTLA-4 (6), prevents the establishment of an innocuous lesion and diabetes develops. Later, after insulitis has been established, artificial induction of pancreatic cell apoptosis via viral infection or administration of the cytotoxic drug cyclophosphamide also rapidly provokes diabetes (10, 11). All of these perturbations result in a more aggressive form of insulitis, wherein the clear border between leukocytes and β cells breaks down, and β cell destruction becomes increasingly evident. A very similar picture is seen when the BDC2.5 transgenes are carried on the C57BL/6-H-2g7 (B6.H-2g7) genetic background: these animals succumb to diabetes within a few weeks of insulitis onset, reflective of histological signs of an aggressive infiltrate. According to genetic analysis, several chromosomal intervals that differ between the NOD and B6.H-2g7 genomes coordinately control this susceptibility to early-onset diabetes (7).
A set of microarray and flow cytometric experiments were performed to further delineate the molecular and cellular mechanisms underlying respectful versus destructive insulitis. We began by comparing the gene expression profiles of the pancreatic infiltrates from BDC2.5/NOD and BDC2.5/B6.H-2g7 mice, leading us to identify some unexpected culprits.
Methods
Mice. The generation and maintenance of BDC2.5/NOD and BDC2.5/B6.H-2g7 TCR tg mice were previously described (6, 12, 13). The congenic NOD.NK1.1 mice (14) were kindly provided by A. Bendelac (Princeton University, Princeton) and typed by PCR on tail DNA (D6Mit52 and D6Mit135). All mice were maintained under barrier conditions in the Joslin Diabetes Center animal facility (protocols 99-19, 99-20).
RNA Preparation, Amplification, and Labeling. Sorted cells were washed and lysed in 500 μl of TRIzol (Invitrogen) for RNA isolation. For microarray analysis, two rounds of amplification were performed by using a MessageAmp aRNA kit (Ambion, Austin, TX); during the second in vitro transcription, biotinylated ribonucleotides were incorporated by using the BioArray HighYield T7 labeling kit (Enzo Life Sciences). Ten to 14 μg of biotinylated amplified RNA was fragmented and hybridized to Affymetrix Mu74Av2 chips. Quantitative real-time PCR assays were performed by using SYBR Green (Applied Biosystems).
Microarray Data Processing. The raw intensity reading of each individual target on the Affymetrix chips (CEL format) was processed and normalized by using the Robust MultiArray transformation (15) run on an S+ ArrayAnalyzer (Insightful, Seattle). To reduce the noise coming from inconsistency among replicates, we ignored the genes for which there was a 10-fold difference in normalized values between any sample and one of its replicates. This excluded 307 genes from the analysis (2.4% of the Mu74Av2 chip). To generate a randomized data set, all replicate intensities for two sample types to be compared were shuffled for each gene separately and assigned randomly to one of the pseudosamples; average values were then calculated as for the experimental analysis.
Diabetes Induction via Anti-CTLA-4 mAb Treatment. Purified anti-CTLA-4 mAb (clone UC10-4F10-11) was purchased from BioExpress (West Lebanon, NH). Diabetes was induced in BDC2.5/NOD mice by injection of 150 μg of antibody on days 10, 12, and 14, and monitoring for hyperglycemia from 16 to 30 days of age (daily urine glucose; positive reads confirmed by blood glucose >250 mg/dl).
Natural Killer (NK) Cell Depletion. Purified anti-mouse NK1.1 mAb (300 μg, clone PK136, BioExpress) was injected three times a week starting at day 10. Anti-Asialo GM1 polyclonal Ab (300 μg, Cedarlane Laboratories) was injected with the same timing; control animals received 300 μg of normal rabbit gamma globulin (Jackson ImmunoResearch).
Results
Differences in Gene Expression Profiles at the Onset of Respectful and Aggressive Insulitis. As described above, an islet infiltrate can follow either of two courses: respectful or aggressive. To elucidate the elements underlying this dichotomous behavior, a microarray analysis of gene expression was performed on early-infiltrating cells in young BDC2.5 TCR tg mice on the NOD and B6.H-2g7 genetic backgrounds, which present with innocuous and destructive insulitis, respectively (7) (Fig. 1 A). The autoimmune T cells themselves were sorted as a CD4+ population expressing high levels of the BDC2.5 clonotype. In addition, a bulk CD45+ population, which includes a variety of hematopoietic cell-types that enter the islets along with the T cells (e.g., B cells, monocytes, and NK cells), was isolated. RNA was prepared from the sorted populations, amplified by using a technique that permits the use of very small cell numbers while still preserving data validity (A. Goldrath, C.B., D.M., unpublished work; see Methods), and converted to labeled probe for hybridization to Affymetrix Mu74Av2 chips. Two to three independent experiments were performed for each cell population. The raw data were processed with the RMA algorithm for probe-level normalization (15), and composite expression values were calculated for each of the genes on the chip, averaging without outlier elimination by using custom-developed S+ scripts for this and subsequent analyses. As indication of statistical significance, a gene-wise P value was derived by Welch's t test. A conservative false-positive rate was estimated from a randomized data set generated by shuffling expression values between categories. Several of the key gene expression differentials were validated by RT-PCR and flow cytometry (Figs. 2 and 5, and data not shown).
Fig. 1.
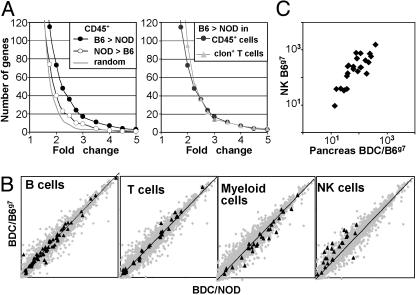
NK-specific genes are up-regulated in BDC2.5/B6.H-2g7 pancreatic infiltrate. (A) Live CD45+ cells and CD4+ BDC2.5-clonotype+ cells were sorted from the pancreas of 23- to 25-day-old BDC2.5/NOD or BDC2.5/B6.H-2g7 mice for microarray analysis. (Left) Cumulative number of up- or down-regulated genes in BDC2.5/B6.H-2g7 versus BDC2.5/NOD pancreatic CD45+ cells for the indicated relative variation. The gray line indicates the level of “background” variation as measured from a randomized data set. (Right) Cumulative number of up-regulated genes in BDC2.5/B6.H-2g7 versus BDC2.5/NOD pancreatic CD45+ cells or CD4+ BDC2.5-clonotype+ cells. (B) Log-scale plot of expression values in pancreatic CD45+ cells with highlights on genes characteristic of the indicated cell type. (C) Microarray expression value of NK-specific genes in B6.H-2g7 purified NK cells versus CD45+ pancreatic infiltrate.
Fig. 2.
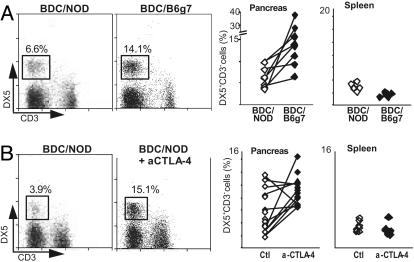
NK frequency correlates with pancreatic infiltrate aggressivity. (A) NK cell were identified as DX5+CD3– by cytofluorimetric analysis; their frequency was measured in the pancreas of 20- to 24-day-old BDC2.5/NOD and BDC2.5/B6.H-2g7 mice. One representative plot is shown as well as the value for all individual mice tested (P < 0.03). Spleen values are shown for control. Gated on live CD45+ cells. (B) NK cell frequency in 16- to 22-day-old anti-CTLA4 treated versus untreated BDC2.5/NOD mice (P < 0.01).
Fig. 5.
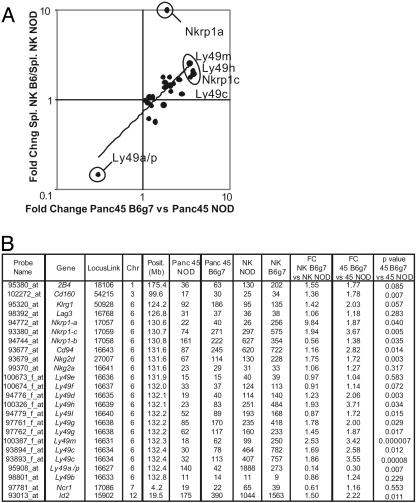
Strain-specific expression of NK-specific genes. (A) Relative variation (fold-change B6.H-2g7/NOD) in expression values of NK-specific genes are plotted for splenic NK cells and CD45+ pancreatic leukocytes. (B) Microarray data for NK-specific gene expression in the pancreatic infiltrate of BDC2.5/NOD and B6.H-2g7, as well as in splenic NOD and B6.H-2g7 NK cells. The genes were sorted by chromosomal position. Fold changes (B6.H-2g7/NOD) are indicated in each tissue. We confirmed the specificity of the probes used in the Affymetrix MuAv2 chip. For some of the Ly49 genes, some cross-reactivity is possible with other Ly49 genes.
We first analyzed the difference between innocuous and destructive insulitis from a global perspective by calculating the number of genes whose expression was increased or decreased in leukocytic infiltrates on the B6.H-2g7 background versus the NOD. For a number of genes, expression values were higher for infiltrating CD45+ cells on the B6.H-2g7 background than on the NOD background. This difference was clearly more than could be attributed to “experimental noise,” as judged by comparison with values issuing from the randomized data set: 73 versus 27 genes at a fold-change cutoff of 2 (Fig. 1A Left; see Table 1, which is published as supporting information on the PNAS web site, for a full tabulation of the differences). Expression values for fewer genes were augmented for the CD45+ cells taken from the innocuous insulitis on the NOD genetic background vis-a-vis the destructive B6.H-2g7 lesion: 40 genes at a fold-change cutoff of 2. A more focused analysis of CD4+ clonotype-positive cells from the infiltrate revealed an induction profile similar to that of unsorted CD45+ cells (Fig. 1A Right). Thus, the B6.H-2g7 versus NOD differential did not pertain just to the autoimmune BDC2.5 T cells, but rather to the infiltrating cells in general. Because clonotype-positive T cells represent only 20–50% of the infiltrate, changes that affected only them would have been diluted out in the CD45+ data sets. This observation suggested that significant differences occurred in the representation or gene expression of cells other than clonotype-positive T cells in the two types of infiltrates.
To explore this possibility, we focused on the behavior of genes representative of the various cell types that make up the CD45+ population (Fig. 1B). Surprisingly, a clear difference was observed in the contribution of gene transcripts characteristic of NK cells, most of which were more abundant on the B6.H-2g7 background. Far fewer changes affected genes characteristic of T, B, or myeloid cells, although there was a trend toward lower representation in the last case. Because several of the genes whose expression was enhanced in B6.H-2g7 leukocytes encoded NK receptors (NKRs) that can also be found on other lymphocytes (reviewed in ref. 16), we wondered whether the overrepresented transcripts included the full complement of genes expressed in NK cells or only a subset. Therefore, we compared the microarray expression values of the NK genes in the CD45+ cells isolated from pancreatic infiltrates and parallel values generated from sorted splenic NK cells (Fig. 1C). The profiles proved very similar, with a good correlation between expression in splenic NK cells and in the mixed pancreatic infiltrate. It was reasonable to hypothesize, then, that this difference reflected an increased frequency of NK cells in the aggressive infiltrate, rather than variation in expression of a subset of NK transcripts expressed in other lymphoid cells.
The Fraction of NK Cells Correlates with Aggressivity. To validate the microarray data, we measured the proportion of NK cells in the insulitic lesions of BDC2.5/NOD and BDC2.5/B6.H-2g7 mice by flow cytometry. NK cells were identified as CD49b(DX5)+CD3– lymphocytes. There were significantly more NK cells in the infiltrates of BDC2.5/B6.H-2g7 than of BDC2.5/NOD animals (P < 0.03) (Fig. 2A Left). No such increase was detected in the splenic NK cell populations of the same animals (Fig. 2A Right), suggesting that the difference observed in the pancreas reflects preferential NK cell recruitment into the aggressive insulitic lesions of BDC2.5/B6.H-2g7 mice.
We wondered whether this observation was also true of other models of rapid-onset diabetes with immediately destructive infiltrates. As discussed above, blockade of the CTLA-4 costimulatory molecule at the initiation of insulitis in BDC2.5/NOD mice provokes diabetes almost immediately, with full hyperglycemia developing between 18 and 30 days of age (6). An increased frequency of NK cells was also observed in this context of aggressive insulitis, again in the absence of a corresponding change in splenic NK cell populations (P < 0.01) (Fig. 2B). Thus, an increased proportion of NK cells might be a general characteristic of aggressive insulitis. Indeed, this result underlines that the destructive influence of NK cells is not just a peculiarity of the B6.H-2g7 context, but can also be induced in NOD mice.
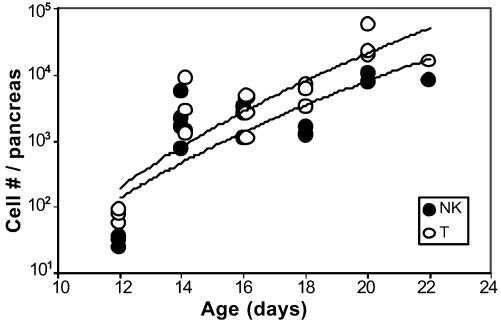
Next, it was necessary to rule out the more trivial possibility that NK cells are recruited to the pancreas only because of a preexisting inflammatory environment, and are merely a marker of an aggressive attack. The kinetics of pancreas infiltration by T and NK cells were followed during the induction of diabetes by anti-CTLA-4 blockade (Fig. 3). In fact, NK cells were a significant component of the infiltrate as soon as it could be detected, from day 14 onward. If anything, the ratio of NK to T cells decreased slightly after this point. Thus, NK cells are very early players in the insulitis of BDC2.5 mice, not late-comers to an already established inflammatory lesion.
Fig. 3.
NK cells are early participants of an aggressive pancreatic lesion. BDC2.5/NOD mice were treated with anti-CTLA4 Ab at days 10, 12, and 14. The number of NK cells and T cells was measured in the pancreas of each mouse from day 12 to day 22. For comparison, the average number of NK cells (and T cells, respectively) found in the insulitis-free pancreas of a young NOD mice (likely arising from blood contamination) is 16 (50) at day 12, 38 (267) at day 15, and 165 (1,120) at day 18.
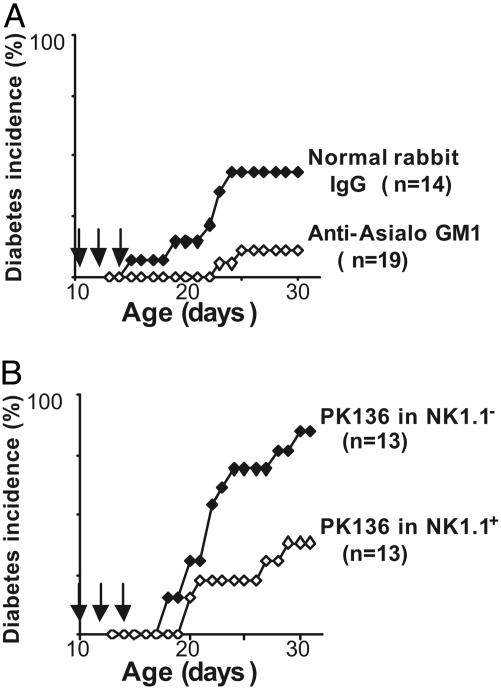
NK Cells Are Important for Islet Cell Destruction. To directly test the significance of NK cells in diabetes progression, we attempted to prevent NK cell activity during the induction of diabetes in BDC2.5/NOD mice by CTLA-4 blockade. An anti-Asialo GM1 polyclonal antibody was first used, a reagent classically used to deplete NK cells, administered at the same time as the anti-CTLA-4 mAb. This antibody led to a significant decrease (P < 0.03, Cox proportional hazards) in diabetes incidence versus mice treated with a polyclonal rabbit IgG (Fig. 4A). Cytofluorimetric analysis confirmed that activated T cells infiltrating the pancreas were not affected by the treatment, whereas >85% of NK cells were eliminated, on average. Yet, because there has been some debate as to the specificity of this reagent (discussed in ref. 17), a second set of experiments was performed by using the anti-NK1.1 mAb PK136, which has been shown to deplete mouse NK cells (18). A genetic control was used in this instance. Many of the NK-cell-specific receptors are encoded in a large multigene complex, the NK gene complex (NKC). The NKC haplotype of NOD mice carries an inactive Nkrpc1 gene and so their NK cells lack the NK1.1 epitope. Thus, we used an intercross/backcross between BDC2.5/NOD mice and the NOD.NK1.1 congenic line, which carries the NKC from B6 origin (14). Initial tests showed that the BDC2.5/NOD.NK1.1 mice were fully susceptible to diabetes induced by CTLA-4 blockade (data not shown). All of the intercross mice were treated with an NK-depleting anti-NK1.1 mAb PK136 administered at the same time as the diabetes-inducing anti-CTLA-4 mAb, monitored for development of diabetes; it was later determined which mice carried the relevant Nkrp1-c gene. A clear reduction in the induction of diabetes was seen in those animals that expressed the NK1.1 epitope compared with NK1.1-negative controls (P < 0.03) (Fig. 4B). The pancreas of animals that proved resistant to diabetes was examined by histology and typical pictures of respectful insulitis were seen (data not shown). In short, the proportion of NK cells in an insulitic lesion correlates with the aggressivity of the autoimmune attack, and their blockade/depletion significantly inhibits a destructive outcome.
Fig. 4.
NK depletion impairs α-CTLA4 Ab-induced diabetes development. Arrows indicate injections with αCTLA-4 Ab. (A) BDC2.5/NOD mice treated with anti-Asialo GM1 or rabbit IgG as control. (B) BDC2.5/NOD or BDC2.5/NOD.NK1.1 were treated with PK136 (300 μg three times a week).
Why and How Are NK Cells Overrepresented in Aggressive Insulitis? These results suggested that the differential installation of a respectful versus aggressive insulitic lesion, and the eventual destruction of β cells in the latter case, may be linked to the activity of NK cells at the onset of insulitis. This begs the questions of why and how NK cell activities differ on the two genetic backgrounds. That NOD-background mice show a difference in NK cell activity has ample precedent: a number of studies have reported that NOD mice have low NK cell activity in general, a characteristic that makes them of great value for xenogeneic transplants (14, 19–21); most recently, Ogasawara et al. (22) described how the NKG2D receptor is dysfunctional in NOD NK cells due to overexpression of its Rae-1 ligand in NOD mice.
We investigated the range of genetically encoded divergences between NOD and B6.H-2g7 mice that might account for the differential aggressivity of their insulitic lesions. First, the microarray analysis was extended by examining gene expression profiles of purified splenic NK cells from juvenile NOD and B6.H-2g7 mice. A comparison of these data sets with those derived from CD45+ participants in the pancreatic infiltrate was expected to reveal which of the differences were intrinsic to NK cells, and thus possibly causative, and which might represent differential expression provoked by the aggressive insulitis. The comparison of NOD versus B6.H-2g7 fold changes indicated that many of the differences found in the pancreatic infiltrate were also present in splenic NK cells (Fig. 5A). Expression of several of the genes was significantly increased on the B6.H-2g7 background in both cases: Ly49m, h, and c, Nkrp1a, and Nkrp1c. Conversely, Ly49a/p expression was augmented in both the normal splenic and pathological pancreatic tissue of NOD animals. Thus, beyond the differential representation of NK cells in their insulitic lesions, as illustrated above, the NOD and B6.H-2g7 genetic backgrounds also seem to encode a range of variations in the expression of NK-cell-specific genes.
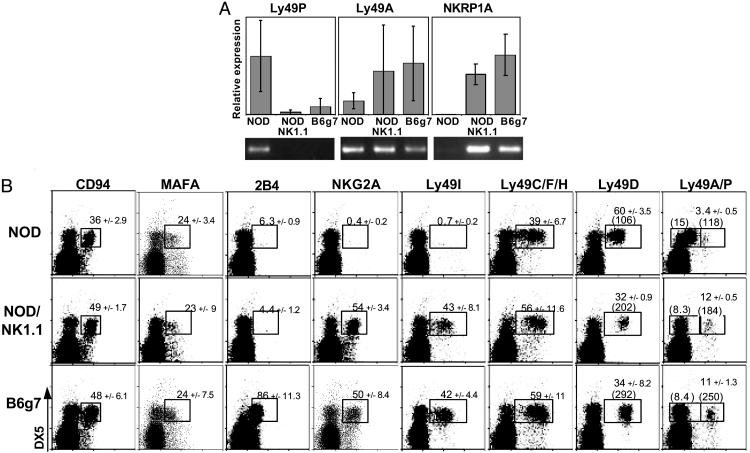
To validate these microarray observations, and because there is some degree of uncertainty as to the precise genes assayed on the array when dealing with multigene families, we measured the expression of NK-cell-specific genes by RT-PCR and flow cytometry. As many of these are encoded in the NKC on chr6, RNA or cells from the NOD, NOD.NK1.1, and B6.H-2g7 lines were analyzed to determine whether the differences were a primary manifestation of the genes themselves or a secondary effect specified by genetic loci located elsewhere. As the MHC is of the same haplotype in all three lines, an indirect influence of MHC ligands could already be ruled out. Real-time quantitative PCR analysis (Fig. 6A) confirmed the microarray results, showing higher levels of Ly49p transcripts in NOD than in B6.H-2g7 mice, and the inverse for Ly49a and Nkrp1-a transcripts (indicating, incidentally, that the chip feature predominantly represents Ly49p expression). Differences clearly tracked with the NKC on chr6.
Fig. 6.
Wide differences in NK surface receptor expression between NOD and B6.H-2g7 mice. (A) Real-time PCR analysis of Ly49a, Ly49p, and Nkrp1-a in sorted splenic NK cells from 4- to 5-week-old NOD, NOD.NK1.1, and B6.H-2g7 mice. (B) Cytofluorimetric analysis of NK surface receptors on NOD, NOD.NK1.1, and B6.H-2g7 splenocytes. Bold numbers indicate the average percentage of NK cells (±SD) positive for the indicated surface molecule (when gated on DX5+CD3–). Numbers in parenthesis indicate the mean fluorescence intensity on the x axis of the designated population. The mAb used to stain Ly49AB6 (clone A1) has been showed to cross-react with Ly49ANOD and partially to Ly49PNOD (40). The preferential expression of Ly49P by NOD NK cells might be reflected by the slight but clear staining observed on Ly49A– NOD NK cells versus Ly49A– B6.H-2g7 NK cells.
Similarly, flow cytometric analyses of cell-surface protein expression (Fig. 6B) confirmed and extended the microarray and PCR data on RNA levels. Of the eight surface receptors analyzed, two did not vary between the strains but six showed clear differences in the number of positive cells, staining intensity, or both: Ly49A/P, Ly49C, Ly49D, Ly49I/F/H, NKG2A, and 2B4 (and patterns were remarkably consistent in independent experiments). Three of the receptors (Ly49I/F/H, NKG2A, and 2B4) exhibited all-or-none staining variations, perhaps denoting missing genes or polymorphic alleles that affected antibody binding. For the other three (Ly49A/P, Ly49C, and Ly49D), the differences influenced the proportion of cells stained and the staining intensity. The surface staining for Ly49A/P confirmed the microarray and RT-PCR data (Fig. 6 and legend), Ly49P being a rare gene up-regulated in NOD relative to B6.H-2g7 NK cells. The staining patterns also indicated that the polymorphism in NKR expression correlated with the NKC, rather than with the rest of the genome. Expression of 2B4 was an exception in that staining levels on NOD and NOD.NK1.1 splenic cells were concordant, which is expected because 2B4 is the only gene in the panel encoded outside the NKC.
In short, the NOD NKC encodes a set of NKR genes with expression patterns very different from those of the B6.H-2g7 NKC, a variation far more extensive than previously recognized. This variability is on par with the extensive polymorphism of the human NKC (23).
Discussion
These data highlight an important role for NK cells in determining the outcome of an autoimmune attack on the pancreatic islets. First, an extraordinary degree of genetic variation in the expression and epitope profile of NK-cell-specific genes was evident when diabetes-resistant and -susceptible mouse strains were compared. Second, the proportion and numbers of NK cells in the islet infiltrate, and the timing of their entry, correlated with the aggressivity of the lesion. Third, depletion/blockade of NK cell function significantly reduced the speed and penetrance of diabetes onset. It seems very unlikely that the outcome of the blocking experiments reflected effects on other NK1.1-positive cells, in particular CD4+ NK-T cells. This T cell subset has been associated with diabetes protection rather than exacerbation in both the NOD and BDC2.5 models of diabetes (14, 24–26), and we observed the same degree of diabetes reduction with the anti-Asialo GM1 reagent, which does not deplete NK-T cells (27), as with the anti-NK1.1 mAb.
An important role for NK cells in setting the tone of insulitis comes as a surprise because their impact on autoimmune diabetes seems to have been little studied, there being only scant mention in the literature (17). NK cells have been observed in the islet infiltrate of NOD mice (28), and an effect of anti-Asialo-GM1 on cyclophosphamide-induced diabetes has been reported (29, 30). Work in the BB rat model had suggested a role for NK cells (31), but this was later questioned on the basis of the specificity of the depleting antibody (17). Furthermore, NOD mice have been reported to exhibit low NK cell activity (14, 19–21), such that a disease-promoting role has seemed counterintuitive. However, it is important to keep in mind that, as a consequence of the selection regime that led to its isolation, which favored late-onset diabetes compatible with reproduction, the NOD genome will harbor alleles of genes disposing to lapses in lymphocyte tolerance pathways but should be impoverished in alleles that promote rapid progression to clinical disease. By delaying the onset of diabetes beyond the peak reproductive age, a relatively low level of NK cell activity may be precisely what permitted derivation of the NOD strain in the first place. The introduction of foreign genetic material might unleash more aggressive pathogenic pathways that are usually not in play in straight NOD mice. Indeed, Brodnicki et al. (32) have nicely illustrated this by crossing the B6 idd14 locus onto the NOD background and thereby provoking a significantly more robust disease. Interestingly, the idd6 and idd19 regions that condition diabetes susceptibility encompass the NKC (33).
How might NK cells impact on diabetes progression? Their best known functions are the direct cytotoxic lysis of virus-infected, tumor, or MHC-deficient cells and the secretion of inflammatory cytokines, such as IFN-γ. Either or both of these could be involved in β cell destruction. Indeed, there is increasing evidence that NK cells can “help” both CD4+ and CD8+ T cell responses in infectious contexts (34, 35). It may be instructive to compare the variations in gene expression profiles observed here with those seen in a kinetic analysis of cyclophosphamide-induced diabetes in BDC2.5/NOD mice (M. Matos, D.M., and C.B., unpublished work). In that case, no changes in NK-cell-specific transcripts were detected. Rather, the gene expression changes in the islets were dominated by a loss of B lymphocyte transcripts, and the induction of IFN-γ and of a number of IFN-γ-controlled genes. Differential expression of IFN-γ was also observed here, as were changes in levels of several of the downstream transcripts (Table 1). Thus, an IFN-γ-dominated response may be a commonality of aggressive end-stages, but the means of arriving to it may differ, sometimes promoted by NK cells, sometimes by cytotoxic agents. Interestingly, in a murine model of myasthenia gravis, the T cell response was critically dependent on IFN-γ production by NK cells, suggesting that NK cells might have a previously underappreciated general function in promoting autoimmune diseases, quite opposed to the protective role previously emphasized (36).
In two quite distinct settings, genetic variation or costimulatory blockade, the stable insulitis of BDC2.5/NOD mice was perturbed, resulting in NK cell-promoted destruction. How can this convergence be explained? Although it is reasonable to suggest that genes of the B6.H-2g7 background control NK cell activity, the link between CTLA-4 and NK cells on the NOD background is less obvious. CTLA-4 has been strongly implicated in the generation and action of T cells with regulatory potential, and we have found that an equilibrium between regulatory and effector T cells is also important in maintaining the stable insulitis of BDC2.5/NOD mice (37). Thus, it is conceivable that a regulatory T cell imbalance resulting from CTLA-4 blockade might promote an early recruitment of NK cells. Alternatively, the anti-CTLA-4 mAb might affect the NK cells themselves. Indeed, although always implicitly assumed, it has never been formally demonstrated that the induction of diabetes by anti-CTLA-4 mAb is via an effect on T cells. It remains possible that, in addition or instead, CTLA-4 has a dampening influence on NK cells, and that the diabetogenic effect of blocking CTLA-4 reflects release from that inhibition.
The NKR repertoire expressed by NK cells is highly variegated, each cell expressing a different subset of NKRs, likely resulting from stochastic regulatory processes (38). Here, many differences were found in the expression of NKC genes when splenic NK cells of BDC2.5/NOD mice were compared with those of BDC2.5/B6.H-2g7 animals. This observation is consistent with a very recent report that NOD mice show a deficiency in multiple pathways of NK cell activation (39). The molecular divergences were not secondary to MHC gene variation, which can strongly influence NKR expression, because the MHC is identical in these two strains. Nor are they determined by differential expression of ligands encoded in other chromosomal regions, as shown for NKG2D (22), because the NOD.NK1.1 congenic strain showed the same expression patterns as B6.H-2g7 for all receptors encoded in the NKC. However, there is reason to believe that other loci located outside the NKC may also play an important role in the differential recruitment and activation of NK cells, not least because NOD.NK1.1 and standard NOD mice develop diabetes with a comparable frequency (14, 21). An earlier genetic analysis of the loci impinging on early-versus late-onset diabetes in BDC2.5 mice on the B6.H-2g7 versus NOD background had implicated four intervals distributed through the genome (7). The most influential of these regions was located proximally on chromosome 7. Several immunologically relevant genes map to this region, some of which are of particular interest because they encode proteins of critical importance in NK cell biology (DAP12, DAP10, and Flt3L). Suggestively, the microarray data revealed that both DAP12 and Flt3l were differentially expressed in B6.H-2g7 and NOD splenic NK cells or T cells (Table 1). It will be of great interest to directly test whether these genes play a part in the differential activity of NK cells in these two strains.
In conclusion, this study highlights an important role for NK cells in unleashing an autoimmune lesion's potential for destruction. It also reveals an extraordinary variance in the NOD and B6 genomes in the expression of NKRs. By extension, one might propose that the very rapid progression to diabetes in the subset of pediatric human patients that presents with severe hyperglycemia in the first years of age might also reflect an influence of NK cells and of the equally extensive NKR polymorphisms in humans.
Supplementary Material
Acknowledgments
We thank Elzbieta Hyatt, Grigoriy Losyev, Rob Saccone, Jennifer Johnson, and Richard Park for assistance with mice, cell sorting, and microarray processing. This work was supported by National Institutes of Health Grants PO1 AI39671-08 and PO1 AI54904-01 and by Joslin Diabetes and Endocrinology Research Center cores (2 P30 DK36836-17).
Abbreviations: NK, natural killer; NKC, NK gene complex; NKR, NK receptor; NOD, nonobese diabetic; TCR, T cell receptor.
References
- 1.Tisch, R. & McDevitt, H. (1996) Cell 85, 291–297. [DOI] [PubMed] [Google Scholar]
- 2.Verge, C. F., Gianani, R., Yu, L., Pietropaolo, M., Smith, T., Jackson, R. A., Soeldner, J. S. & Eisenbarth, G. S. (1995) Diabetes 44, 1176–1179. [DOI] [PubMed] [Google Scholar]
- 3.Anastasi, E., Campese, A. F., Bellavia, D., Bulotta, A., Balestri, A., Pascucci, M., Checquolo, S., Gradini, R., Lendahl, U., Frati, L., et al. (2003) J. Immunol. 171, 4504–4511. [DOI] [PubMed] [Google Scholar]
- 4.Robles, D. T., Eisenbarth, G. S., Dailey, N. J., Peterson, L. B. & Wicker, L. S. (2003) Diabetes 52, 882–886. [DOI] [PubMed] [Google Scholar]
- 5.Haskins, K., Portas, M., Bradley, B., Wegmann, D. & Lafferty, K. J. (1988) Diabetes 37, 1444–1448. [DOI] [PubMed] [Google Scholar]
- 6.Luhder, F., Höglund, P., Allison, J. P., Benoist, C. & Mathis, D. (1998) J. Exp. Med. 187, 427–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gonzalez, A., Katz, J. D., Mattei, M. G., Kikutani, H., Benoist, C. & Mathis, D. (1997) Immunity 7, 873–883. [DOI] [PubMed] [Google Scholar]
- 8.Chatenoud, L., Thervet, E., Primo, J. & Bach, J. F. (1994) Proc. Natl. Acad. Sci. USA 91, 123–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chatenoud, L., Primo, J. & Bach, J. F. (1997) J. Immunol. 158, 2947–2954. [PubMed] [Google Scholar]
- 10.Horwitz, M. S., Bradley, L. M., Harbertson, J., Krahl, T., Lee, J. & Sarvetnick, N. (1998) Nat. Med. 4, 781–785. [DOI] [PubMed] [Google Scholar]
- 11.André-Schmutz, I., Hindelang, C., Benoist, C. & Mathis, D. (1999) Eur. J. Immunol. 29, 245–255. [DOI] [PubMed] [Google Scholar]
- 12.Katz, J. D., Wang, B., Haskins, K., Benoist, C. & Mathis, D. (1993) Cell 74, 1089–1100. [DOI] [PubMed] [Google Scholar]
- 13.Luhder, F., Katz, J., Benoist, C. & Mathis, D. (1998) J. Exp. Med. 187, 379–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carnaud, C., Gombert, J., Donnars, O., Garchon, H. & Herbelin, A. (2001) J. Immunol. 166, 2404–2411. [DOI] [PubMed] [Google Scholar]
- 15.Irizarry, R. A., Hobbs, B., Collin, F., Beazer-Barclay, Y. D., Antonellis, K. J., Scherf, U. & Speed, T. P. (2003) Biostatistics 4, 249–264. [DOI] [PubMed] [Google Scholar]
- 16.McMahon, C. W. & Raulet, D. H. (2001) Curr. Opin. Immunol. 13, 465–470. [DOI] [PubMed] [Google Scholar]
- 17.Ellerman, K., Wrobleski, M., Rabinovitch, A. & Like, A. (1993) Diabetologia 36, 596–601. [DOI] [PubMed] [Google Scholar]
- 18.Seaman, W. E., Sleisenger, M., Eriksson, E. & Koo, G. C. (1987) J. Immunol. 138, 4539–4544. [PubMed] [Google Scholar]
- 19.Kataoka, S., Satoh, J., Fujiya, H., Toyota, T., Suzuki, R., Itoh, K. & Kumagai, K. (1983) Diabetes 32, 247–253. [DOI] [PubMed] [Google Scholar]
- 20.Shultz, L. D., Schweitzer, P. A., Christianson, S. W., Gott, B., Schweitzer, I. B., Tennent, B., McKenna, S., Mobraaten, L., Rajan, T. V., Greiner, D. L., et al. (1995) J. Immunol. 154, 180–191. [PubMed] [Google Scholar]
- 21.Poulton, L. D., Smyth, M. J., Hawke, C. G., Silveira, P., Shepherd, D., Naidenko, D. I. & Baxter, A. G. (2001) Int. Immunol. 13, 887–896. [DOI] [PubMed] [Google Scholar]
- 22.Ogasawara, K., Hamerman, J. A., Hsin, H., Chikuma, S., Bour-Jordan, H., Chen, T., Pertel, T., Carnaud, C., Bluestone, J. A. & Lanier, L. L. (2003) Immunity 18, 41–51. [DOI] [PubMed] [Google Scholar]
- 23.McQueen, K. L. & Parham, P. (2002) Curr. Opin. Immunol. 14, 615–621. [DOI] [PubMed] [Google Scholar]
- 24.Beaudoin, L., Laloux, V., Novak, J., Lucas, B. & Lehuen, A. (2002) Immunity 17, 725–736. [DOI] [PubMed] [Google Scholar]
- 25.Lee, P., Putnam, A., Benlagha, K., Teyton, L., Gottlieb, P. & Bendelac, A. (2002) J. Clin. Invest. 110, 793–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hong, S., Wilson, M. T., Serizawa, I., Wu, L., Singh, N., Naidenko, O. V., Miura, T., Haba, T., Scherer, D. C., Wei, J., et al. (2001) Nat. Med. 7, 1052–1056. [DOI] [PubMed] [Google Scholar]
- 27.Nakagawa, R., Nagafune, I., Tazunoki, Y., Ehara, H., Tomura, H., Iijima, R., Motoki, K., Kamishohara, M. & Seki, S. (2001) J. Immunol. 166, 6578–6584. [DOI] [PubMed] [Google Scholar]
- 28.Miyazaki, A., Hanafusa, T., Yamada, K., Miyagawa, J., Fujino-Kurihara, H., Nakajima, H., Nonaka, K. & Tarui, S. (1985) Clin. Exp. Immunol. 60, 622–630. [PMC free article] [PubMed] [Google Scholar]
- 29.Maruyama, T., Watanabe, K., Takei, I., Kasuga, A., Shimada, A., Yanagawa, T., Kasatani, T., Suzuki, Y., Kataoka, K., Saruta, T., et al. (1991) Diabetes Res. 17, 37–41. [PubMed] [Google Scholar]
- 30.Maruyama, T., Watanabe, K., Yanagawa, T., Kasatani, T., Kasuga, A., Shimada, A., Takei, I., Suzuki, Y., Kataoka, K. & Saruta, T. (1991) Diabetes Res. 16, 171–175. [PubMed] [Google Scholar]
- 31.Jacobson, J. D., Markmann, J. F., Brayman, K. L., Barker, C. F. & Naji, A. (1988) Diabetes 37, 838–841. [DOI] [PubMed] [Google Scholar]
- 32.Brodnicki, T. C., Quirk, F. & Morahan, G. (2003) Diabetes 52, 218–222. [DOI] [PubMed] [Google Scholar]
- 33.Rogner, U. C., Boitard, C., Morin, J., Melanitou, E. & Avner, P. (2001) Genomics 74, 163–171. [DOI] [PubMed] [Google Scholar]
- 34.Mailliard, R. B., Son, Y. I., Redlinger, R., Coates, P. T., Giermasz, A., Morel, P. A., Storkus, W. J. & Kalinski, P. (2003) J. Immunol. 171, 2366–2373. [DOI] [PubMed] [Google Scholar]
- 35.Vankayalapati, R., Klucar, P., Wizel, B., Weis, S. E., Samten, B., Safi, H., Shams, H. & Barnes, P. F. (2004) J. Immunol. 172, 130–137. [DOI] [PubMed] [Google Scholar]
- 36.Baxter, A. G. & Smyth, M. J. (2002) Autoimmunity 35, 1–14. [DOI] [PubMed] [Google Scholar]
- 37.Herman, A. E., Freeman, G. J., Mathis, D. & Benoist, C. (2003) J. Exp. Med., in press. [DOI] [PMC free article] [PubMed]
- 38.Raulet, D. H., Vance, R. E. & McMahon, C. W. (2001) Annu. Rev. Immunol. 19, 291–330. [DOI] [PubMed] [Google Scholar]
- 39.Johansson, S. E., Hall, H., Bjorklund, J. & Hoglund, P. (2004) Int. Immunol 16, 1–11. [DOI] [PubMed] [Google Scholar]
- 40.Silver, E. T., Gong, D. E., Chang, C. S., Amrani, A., Santamaria, P. & Kane, K. P. (2000) J. Immunol. 165, 1771–1781. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.