Abstract
A functional cDNA cloning system was developed by using a retrovirus library encoding CD8-chimeric proteins and a nuclear factor of activated T cells (NFAT)-GFP reporter cell line to identify molecules inducing NFAT activation. By using this strategy, NFAT activating molecule 1 (NFAM1) was cloned as an immunoreceptor tyrosine-based activation motif (ITAM)-bearing cell surface molecule belonging to the Ig superfamily and is predominantly expressed in spleen B and T cells. NFAM1 crosslinking induced ITAM phosphorylation, ZAP-70/Syk recruitment, NFAT activation, and cytokine production. In vivo overexpression of NFAM1 in bone marrow chimeras and transgenic mice induced severe impairment of early B cell development in an ITAM-dependent manner. In NFAM1-expressing B cells, B cell antigen receptor stimulation induced NFAM1 translocation to lipid raft, and NFAM1 co-crosslinking augmented B cell antigen receptor signaling. The results suggest that NFAM1 modulates B cell signaling through its ITAM, which regulates B cell development.
The direction and magnitude of an immune response are determined by the behavior of individual lymphocyte clones bearing distinct Ag receptors and their mutual interactions. Ag recognition signals by Ag receptors on B and T cells, are converted through receptor-associated ITAM (immunoreceptor tyrosine-based activation motif)-bearing molecules into the intracellular signaling cascade. ITAMs are present in the cytoplasmic domains of several molecules that serve as signal transducers for immunoreceptors (1–5), including CD3 (γ, δ, ε, and ζ) for TCR, Igα/β for B cell antigen receptor (BCR), FcRγ for Fc receptors, and DAP12 for natural killer (NK) cell receptors. The mechanism underlying ITAM-dependent signal transduction for lymphocyte activation is well established. Two tyrosines within ITAM are phosphorylated by Src-family kinases upon receptor engagement. Then, ZAP-70/Syk is recruited for binding to the phospho-ITAM and activated (6–8), which transduces a downstream signaling cascade by phosphorylating various adaptor/effector molecules. Thus, ITAM represents a hallmark of signal modules that convert an Ag recognition event to initiate a signal cascade.
The activation of ZAP-70/Syk induces the activation of calcineurin and nuclear factor of activated T cells (NFAT). NFAT plays important roles in the activation of B, T, and NK cells (9–12). NFAT is rapidly dephosphorylated and translocated into the nucleus upon stimulation, and induces the transcription of various cytokine genes. Therefore, NFAT can be used as a standard readout for lymphocyte activation.
We have developed a functional cDNA cloning system [NFAT-activation molecule cloning system (NACS)] for identifying molecules that induce NFAT activation by using a retrovirus cDNA library encoding CD8-chimeric molecules and reporter cells expressing NFAT-GFP, and have succeeded in identifying various cDNA clones that induce NFAT activation, including known surface molecules, kinases, adaptors, and unknown molecules. In particular, we have cloned an ITAM+ molecule, NFAM1 (NFAT activating molecule 1). We describe herein that NFAM1 crosslinking induces ITAM phosphorylation, Ca2+ influx, and NFAT activation. NFAM1 co-crosslinking in B cells enhances BCR signaling, and its in vivo overexpression results in severe impairment of B cell development.
The results suggest that NFAM1 belongs to a category of ITAM+ molecules that regulate B cell signaling and development. The successful cloning of various molecules by NACS indicates its applicability to the systematic cloning of functional molecules involved in a defined signaling pathway.
Materials and Methods
Mice. C57BL/6 mice were purchased from Nihon SLC (Hamamatsu, Japan). NFAM1-transgenic (Tg) mice were generated by injecting the insert of the pHSE3′ expression vector (provided by H. Pircher, University of Freiburg, Freiburg, Germany) (13) containing the H-2Kb promoter and the IgH enhancer in which mouse NFAM1 cDNA was subcloned.
NFAT-GFP Reporter Cell Line. NFAT-GFP construct was prepared by fusing three tandem NFAT-binding sites with enhanced GFP cDNA. T hybridoma 2B4 (14) was transfected with NFAT-GFP and stimulated with immobilized anti-TCRβ (H57.597, 1 μg/ml), and GFP+ cells were collected by FACStarplus (Becton Dickinson). After two cycles of stimulation and sorting, GFP+ single-cell clones were cloned and a representative clone, 43–1, was used.
CD8-Chimeric Retrovirus cDNA Library. Extracellular and transmembrane domains of mouse CD8α cDNA (residues 1–217) lacking the intrinsic EcoRI site were cloned into the pMx vector (15) and a stop codon was created to prepare the pMx-CD8 vector. Poly(A)+ RNA was purified from splenocytes or NK cells by using an mRNA purification kit (Amersham Pharmacia Biotech). cDNAs were made by the Superscript Choice system (GIBCO/BRL), ligated into pMX-CD8, and transduced into ElectroMAX DH5α (GIBCO/BRL). The complexity of the library was 6 × 106.
Screening of cDNA Library. cDNA libraries were transfected into Phoenix-E cells by using Lipofectamine Plus (GIBCO/BRL), and viral supernatants were collected 2 days later. NFAT-GFP-expressing 43–1 cells were infected with the viral supernatants in the presence of 10 μg/ml N-[1-(2,3-dioleoyloxy)propyl]-N,N,N-trimethylammonium methylsulfate (Roche, Basel). CD8+ cells were stained with FITC-anti-CD8 and anti-FITC-conjugated microbeads, purified by magnetic cell sorter (Miltenyi Biotec), and stimulated with immobilized anti-CD8 (53.6.7, 5 μg/ml), and then GFP+ cells were collected by FACStarplus. Restimulation and sorting were repeated, and GFP– cells were sorted without stimulation to eliminate cells expressing GFP constitutively. Finally, GFP+ clone upon anti-CD8 stimulation was isolated, and DNA was extracted. The cDNA was amplified by PCR using primers specific for pMx-CD8 vector (5′ primer, GGATTGGACTTCGCCTGTGA; 3′ primer, CCCTTTTTCTGGAGACAAAT).
Cloning of Full-Length NFAM1. Mouse full-length NFAM1 was cloned by 5′ RACE as described (16). Human full-length NFAM1 was cloned from T cell cDNAs based on a partial homologous sequence from the National Center for Biotechnology Information database.
Quantitative Real-Time PCR. Quantitative real-time PCR was performed on a Bio-Rad iCycler by using SYBR green PCR core reagents (Applied Biosystems). The values were normalized to that of β-actin by dividing the average copy number of the respective transcript sample.
Ca2+ Flux. Cells were loaded with 5 μM fura-2/AM (Dojin) at 37°C for 30 min and Ca2+ flux was measured in a buffer [140 mM NaCl/5 mM KCl,/1 mM MgSO4/5.5 mM glucose (pH 7.4)] by a fluorescence spectrophotometer with excitations at 340 and 380 nm, and emission at 500 nm.
Biochemical Analysis of NFAM1. Cell surface biotinylation, immunoprecipitation, SDS/PAGE and Western blotting were performed as described (17). The Abs used were anti-Flag M2 (Sigma), anti-CD8 (53.6.7), 4G10 (Upstate Biotechnology), and anti-ZAP70 (Transduction Laboratories). Anti-NFAM1 Ab was prepared by immunizing a rabbit with an NFAM1 peptide (amino acids 198–213) coupled with keyhole limpet hemocyanin, and purified by affinity column. For deglycosylation, immunoprecipitates were digested with N-glycosidase F by using a modified manufacturer's protocol (Prozyme). Sucrose density gradient centrifugation for separating lipid raft fractions was performed as described (18).
Transfection and Stimulation of Cell Lines. Retrovirus-mediated gene transfer was performed by using Phoenix E-packaging cells and pMx-IRES-GFP vectors as described (19). Transfectants were collected for GFP expression by using FACStarplus. For biochemical analysis, CD8-NFAM1 transfectants were stimulated with anti-CD8 and goat anti-rat IgG (100 μg/ml, Cappel) for 2 min. For co-crosslinking, Flag-NFAM1-transfected WEHI-231 cells were stimulated with anti-Flag-biotin and anti-IgM-biotin, followed by streptavidin for 2 min.
ITAM Peptide-Binding Assay. Twenty-mer human and mouse peptides corresponding to NFAM1-ITAM and mouse CD3-ζ-ITAM peptides of which the N terminus was biotinylated and two tyrosines were phosphorylated, were synthesized (Funakoshi, Tokyo). Avidin-conjugated Sepharose (Amersham Pharmacia) was mixed with those peptides, and spleen cell lysate was added to the mixtures. After 2 h incubation, the precipitated samples were analyzed by Western blotting.
Generation of Bone Marrow (BM) Chimera. Ly5.1+ C57BL/6 mice (Sankyo) were injected i.p. with 5-fluorouracil (150 mg/kg). Four days later, BM cells were stained with anti-ScaI and sorted by magnetic cell sorter. The cells were cultured for 3 days in a medium containing a cytokine mixture [20 ng/ml murine IL-3, 100 ng/ml murine SCF (Genzyme), 100 ng/ml human IL-6], together with viral supernatant and 10 μg/ml polybrene (Sigma). A total of 5 × 104 GFP+ cells sorted by FACStarplus were injected into lethally irradiated recipient mice.
Results
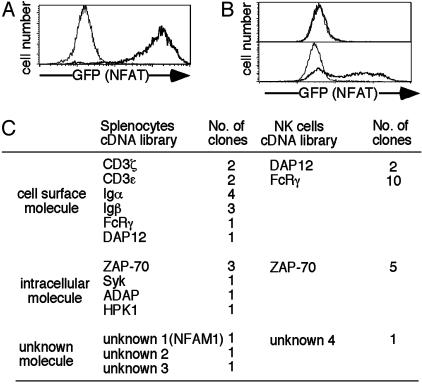
Establishment of NACS. A retrovirus vector for generating CD8-chimeric molecules (pMx-CD8) was constructed and cDNA libraries prepared from splenocytes or NK cells were cloned into this vector to generate retrovirus CD8-chimeric cDNA libraries. A mouse cell line was transfected with an NFAT-GFP reporter gene possessing three tandem NFAT-binding sites. A clone, 43–1, which became almost GFP+ upon TCR stimulation and had no background of spontaneous GFP expression without stimulation, was isolated and used as the recipient cell line for cDNA transfection (Fig. 1A). Clone 43–1 was transfected with the CD8-chimeric retrovirus cDNA library. CD8+ cells were separated by magnetic cell sorter and stimulated with immobilized anti-CD8, and GFP+ cells were sorted by FACS. This stimulation sorting procedure was repeated three or four times. Approximately half of the cells expressed GFP upon CD8 crosslinking after three rounds of enrichment (Fig. 1B). Finally, single-cell clones expressing GFP upon stimulation were isolated and cDNAs were recovered by PCR.
Fig. 1.
Strategy of NACS and cloned genes. (A) Recipient cell line for NACS. The NFAT-GFP reporter cell line, 43–1, was stimulated by immobilized anti-TCR mAb (thick line) or left unstimulated (thin line), and it was analyzed for GFP expression by FACS. (B) Representative flow cytometric profiles during NACS screening. Clone 43–1 cells were transfected with a CD8-chimeric library, stimulated by crosslinking with anti-CD8, and analyzed for GFP expression before purification (Upper) and after three cycles of purification (Lower) by flow cytometry. (C) The number of cloned genes from the cDNA library of splenocytes and NK cells. Each number represents distinct clones. The number of analyzed clones was >10-fold of these numbers due to multiple identical clones.
The cDNA clones obtained by NACS are categorized into four groups, as shown in Fig. 1C: (i) Ag-receptor-associated ITAM+ surface molecules—CD3ε/ζ, Igα/β, DAP12, and FcRγ; (ii) ITAM-associated tyrosine kinases—ZAP-70 and Syk; (iii) intracellular signaling molecules—HPK1, ADAP, and Clast6; and (iv) unknown molecules. Whereas all cDNAs encoding CD3ε/ζ, Igα/β, and DAP12 were cloned as cytoplasmic domains, eight of 10 FcRγ clones contained full-length FcRγ. As with ZAP-70, some contained the kinase domain, whereas most clones consisted mainly of the SH2 domain without the kinase domain. Retransfection of all these clones into 43–1 cells confirmed the induction of NFAT activation upon CD8 crosslinking (data not shown).
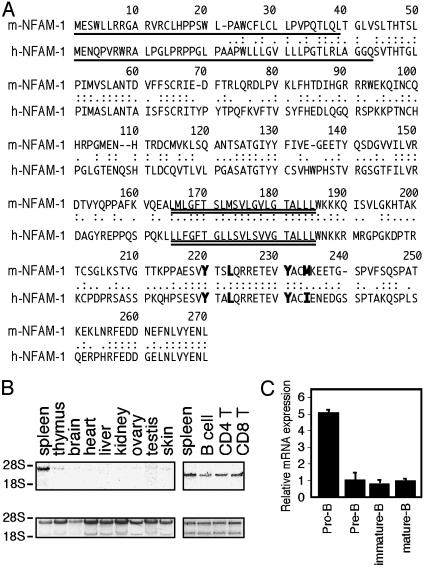
NFAM1 Is an ITAM-Bearing Cell-Surface Molecule. One of the genes inducing NFAT activation was named NFAM1 (GenBank accession no. AF361364). Full-length mouse and human cDNAs of NFAM1 were cloned (GenBank accession no. AY121370) and their amino acid sequences are shown in Fig. 2A. NFAM1 contained a signal peptide and a transmembrane region. The cloned region by NACS was a part of the cytoplasmic domain containing a YxxLx (7)YxxM sequence in mouse and a YxxLx (7)YxxI sequence in human, which is similar to the consensus sequence of ITAM (1, 20). The extracellular domain contained an Ig-like domain. NFAM1 was expressed predominantly in spleen and at very low levels, if any, in other tissues. In spleen, both B and T cells (both CD4+ and CD8+ T cells) as well as non-T and non-B cells, including macrophages and neutrophils, expressed NFAM1 (Fig. 2B).
Fig. 2.
Structure and expression of NFAM1. (A) Amino acid sequences of mouse and human NFAM1. Single and double underlines indicate signal peptide and transmembrane regions, respectively. Bold letters indicate ITAM. The cloned region of mouse NFAM1 by NACS is residues 207–3′UT. (B) Northern blot analysis of NFAM1 transcripts in various tissues. A total of 10 μg of total RNA was analyzed with an NFAM1 C-terminal probe (369 bp, residues 160–3′UT). (C) Quantitative real-time PCR analysis of NFAM1 mRNA in B cell subpopulations. B cell subpopulations in BM and spleen are defined as follows: pro-B, B220+ CD43+; pre-B, B220+ CD25+; splenic immature-B, B220+ IgMhigh IgDlow; splenic mature-B, B220+ IgMlow IgDhigh. Relative mRNA expression of NFAM1 in each population is shown as the ratio to the expression in splenic mature B cells. Data represent means of triplicate experiments.
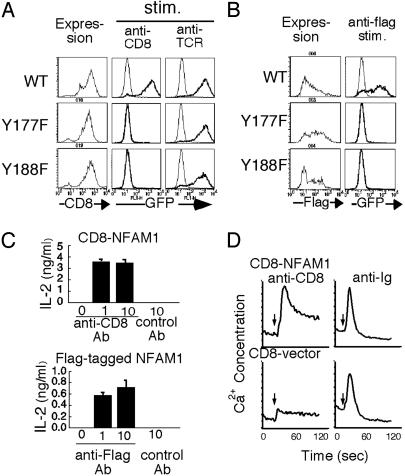
Induction of Activation Signals Through ITAM of NFAM1. To investigate the role of ITAM of NFAM1 in NFAT activation, either of the tyrosines within the ITAM of CD8-NFAM1 was replaced with phenylalanines (Y177F and Y188F). Transfectants expressing either of the two mutations failed to induce GFP expression upon CD8 crosslinking, whereas both cells induced GFP expression upon TCR stimulation (Fig. 3A). To analyze the function of full-length NFAM1, N-terminal Flag-tagged NFAM1 was transfected into a T cell line. NFAM1 was expressed on the cell surface and stained with anti-Flag (Fig. 3B). Similar to CD8-NFAM1, Flag-NFAM1 could induce NFAT activation upon crosslinking with anti-Flag, and either of the tyrosine mutations (Y177F and Y188F) of ITAM abrogated NFAT activation (Fig. 3B). As a functional consequence, crosslinking of transfectants expressing CD8-NFAM1 and Flag-NFAM1 with immobilized anti-CD8 and anti-Flag, respectively, induced IL-2 production, whereas the ITAM mutations of both NFAM1 failed to stimulate production (Fig. 3C).
Fig. 3.
Cell activation through NFAM1 and the requirement of ITAM for NFAT activation. (A) Clone 43–1 cells expressing CD8-NFAM1 (wild type, WT; Y177 mutant, Y177F; Y188 mutant, Y188F) were stimulated with immobilized anti-CD8 (5 μg/ml) or anti-TCR (1 μg/ml) for 12 h and analyzed for GFP expression. (B) Clone 43–1 transfectants expressing Flag-NFAM1 (WT, Y177F, and Y188F) were stained with anti-Flag (Left). These cells were stimulated with immobilized anti-Flag (10 μg/ml; thick line) for 12 h, and GFP expression was analyzed (Right). (C) IL-2 production in transfectants expressing CD8-NFAM1 (Upper) or Flag-NFAM1 (Lower). Transfectants were stimulated with control Ab or immobilized anti-CD8 or anti-Flag for 24 h. (D) Ca2+ mobilization in CD8-NFAM1-transfected B cells by crosslinking with anti-CD8. IIA1.6 B cells lacking FcγR expression were transfected with CD8-NFAM1 or CD8 vector and Ca2+ flux was measured upon stimulation with anti-CD8 or anti-Ig Ab. Arrows indicate time of Ab addition.
Furthermore, anti-CD8 stimulation of a B cell line, IIA1.6, and a T cell line (data not shown) expressing CD8-NFAM1, induced Ca2+ influx (Fig. 3D). NFAT activation and IL-2 production by NFAM1 crosslinking were completely blocked by FK506 (data not shown).
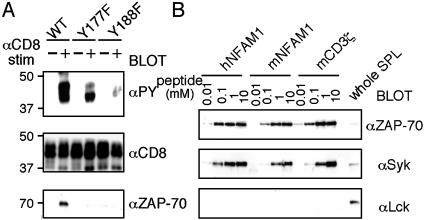
Biochemical Analysis of NFAM1 and Its ITAM. We then analyzed whether NFAM1-ITAM recruits ZAP-70/Syk upon activation. When CD8-NFAM1-expressing T cells were crosslinked with anti-CD8 plus anti-Ig Ab, phosphorylation of NFAM1 and the association with ZAP-70 were observed (Fig. 4A). On the other hand, tyrosine mutants of NFAM1-ITAM reduced NFAM1 phosphorylation and failed to recruit ZAP-70. These results prove that NFAM1-ITAM functions as a conventional ITAM.
Fig. 4.
ITAM-dependent association of ZAP-70/Syk with NFAM1. (A) NFAM1 phosphorylation and ZAP-70 recruitment upon CD8-NFAM1 crosslinking. T cells expressing CD8-NFAM1 (WT, Y177F, and Y188F) were stimulated (+) with anti-CD8 plus anti-mouse Ig Ab for 2 min or were left unstimulated (–). Cell lysates were immunoprecipitated with anti-CD8 and were blotted with the indicated Abs. (B) Binding analysis of tyrosine-phosphorylated ITAM peptides of human (h) and mouse (m) NFAM1 and CD3ζ to ZAP 70 and Syk. Total cell lysate of splenocytes was mixed with each phosphorylated ITAM peptide of which the N terminus was biotinylated at indicated concentrations. The phosphorylated peptides were isolated with avidin-Sepharose and the protein–peptide complexes were electrophoresed and blotted with each Ab. Whole spleen cell lysate (SPL) was used as control.
To confirm the direct association of mouse and human NFAM1 with ZAP-70/Syk, and to analyze their binding affinity compared with that of other known ITAMs, 20-aa peptides containing the tyrosine-phosphorylated ITAM of mouse and human NFAM1 as well as CD3ζ as control were analyzed for binding to ZAP-70/Syk. Both ZAP-70 and Syk associated with the phosphorylated ITAM peptides of human and mouse NFAM1 with similar affinity to the phosphorylated ITAM peptide of CD3ζ (Fig. 4B). No binding to Lck demonstrated specific binding to ZAP-70/Syk. Although mouse NFAM1 contains an unusual YxxM sequence instead of the consensus sequence YxxL/I, this sequence was proven to function as an ITAM with similar affinity.
To analyze NFAM1 on the cell surface, transfectants expressing C-terminal Flag-NFAM1 were surface-biotinylated and immunoprecipitated with anti-Flag (Fig. 5A). Biotinylated NFAM1 was detected as an ≈30-kDa protein. Reduction of its molecular weight by N-glycosidase F digestion revealed an N-linked glycoprotein. Endogenous NFAM1 was also analyzed by surface biotinylation of splenocytes. Similar to the transfectant, biotinylated NFAM1 was immunoprecipitated as an ≈30-kDa molecule (Fig. 5B).
Fig. 5.
Biochemical analysis of cell-surface NFAM1 and augmentation of BCR signaling by NFAM1. (A) Analysis of cell-surface NFAM1 on transfectants. T cell hybridoma expressing C-terminal Flag-NFAM1 was surface-biotinylated and cell lysates were immunoprecipitated (IP) with anti-Flag. Immunoprecipitates were treated (+) or not treated (–) with N-glycosidase F and analyzed under reducing conditions (Upper). The same membrane was reblotted with anti-Flag M2 mAb (Lower). Arrowheads indicate NFAM1. (B) Analysis of endogenous NFAM1 on the cell surface of splenocytes. Splenocytes were analyzed similar to A (Upper). The same membrane was reblotted with anti-NFAM1 Ab (Lower). (C) Co-crosslinking of NFAM1 and BCR enhances tyrosine phosphorylation. Flag-NFAM1- or mock-transfected WEHI-231 cells were stimulated with anti-IgM-biotin and anti-Flag-biotin followed by streptavidin. Total cell lysates were analyzed by Western blotting with anti-PY Ab. Arrowheads indicate particularly augmented phosphorylated proteins. (D) Translocation of NFAM1 to lipid raft upon BCR stimulation. Triton X-100 cell lysates of NFAM1-transfected B cells with (stim.) or without (unstim.) BCR stimulation. BCR stimulation was fractionated on sucrose density gradient centrifugation. Each fraction was analyzed by blotting with anti-NFAM1 or anti-Lyn.
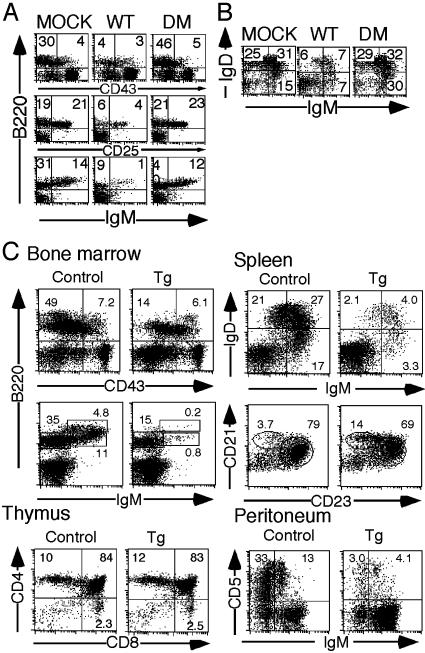
In Vivo Role of NFAM1 in Lymphocyte Development. To investigate the role of NFAM1 in lymphocyte development, we generated BM chimera by transferring BM progenitor cells transfected with NFAM1 in a form of IRES-GFP construct. Enriched Sca-1+ BM stem cells from Ly5.1 C57BL/6 mice were infected with wild-type (WT) and ITAM-mutant (DM) NFAM1 as well as the vector (MOCK) and were transferred into lethally irradiated Ly5.2 mice. Four to six weeks later, BM, spleen, and thymus were analyzed (Fig. 6 A and B). Upon analysis of BM cells, cell numbers in the B220+ CD43– (pre-B and immature B), B220+ CD25+ (pre-B), and B220+ sIgM+ populations (immature and recirculating B) in NFAM1WT BM chimera were notably reduced compared with those in MOCK BM chimera. Similarly, the mature B cell number in spleen was markedly reduced in NFAM1WT BM chimera. Importantly, NFAM1DM BM chimera exhibited no impairment of B cell development in both BM and spleen. As WT- and DM-NFAM1 were expressed at the same level according to GFP expression in Ly5.2+ cells, the results suggest that NFAM1-ITAM is critically involved in the impairment of B cell development. In contrast, T cell development in thymus appeared to be normal. The results suggest that NFAM1overexpression in BM progenitor cells results in the impairment of B cell development in an ITAM-dependent manner.
Fig. 6.
Impaired B cell development in BM chimera and NFAM1-Tg mice. (A and B) Analysis of NFAM1-expressing BM chimera. BM cells from Ly5.1 mice were infected with retrovirus vector (mock), wild-type (WT), or Y177 and Y188 double-mutated NFAM1 (DM), and were transferred into lethally irradiated host mice. BM cells (A) and splenocytes (B) were stained with anti-Ly5.1 and indicated mAbs. All data gated for Ly5.1+ and GFP+ cells are shown. (C) Impairment of B cell development in NFAM1-Tg mice. BM, spleen, thymus, and peritoneum from NFAM1-Tg mice were analyzed by FACS. Percentages of cells in each population are shown. Data from only one Tg line (Tg.10) are shown and similar data were obtained with another line (Tg.7).
The role of NFAM1 in B cell development was further examined in NFAM1-Tg mice under the control of the H-2Kb promoter/IgH enhancer (Eμ). Two lines of NFAM1-Tg mice were established and analysis of Tg.10 expressing higher level of NFAM1 in BM and spleen (Fig. 6C) was conducted. In BM, the proportions of both B220+ CD43– and B220+ sIgM+ populations but not that of B220+CD43+ pro-B cells were significantly reduced. Cellularity in spleen was reduced to approximately half in Tg mice. The number of B220+ cells in spleen was markedly reduced, whereas the maturation of B cells appeared to be normal, as indicated by the IgM/IgD staining profile. Compared with the marked developmental impairment of follicular B cells (CD21int CD23+), the decrease in the number of marginal zone B cells was minimal (approximately half that in WT) in NFAM1-Tg mice, considering the reduction in the number of B220+ cells. Moreover, the number of B1 cells in the peritoneal cavity of these Tg mice was also severely diminished (Fig. 6C). In contrast, T cell development in thymus and spleen of NFAM1-Tg mice was normal.
NFAM1 Expression Decreases with B Cell Differentiation. To understand the basis of the impairment of B cell development in NFAM1-Tg mice, the expression of endogenous NFAM1 during B cell development was analyzed. We found that NFAM1 expression is the highest in pro-B cells and decreases with B cell differentiation (Fig. 2C), suggesting that NFAM1 overexpression may interfere with the normal decrease in NFAM1 expression with B cell development.
NFAM1 Augments BCR Signaling Upon Co-Crosslinking in B Cell Lines. We next analyzed whether NFAM1 modulates BCR signaling. Because NFAM1-Tg B cells are developmentally impaired, we used the Flag-NFAM1-expressing immature B cell line, WEHI-231. Upon co-crosslinking with anti-IgM and anti-Flag, NFAM1, but not ITAM-mutant (DM), transfectants exhibited enhanced tyrosine phosphorylation of several proteins (Fig. 5C), suggesting that NFAM1 enhances BCR-mediated signal transduction by co-crosslinking in B cells in an ITAM-dependent manner.
The enhancement of BCR signaling by NFAM1 might be induced by the functional association of NFAM1 with the BCR signaling complex as the direct association between BCR and NFAM1 was not observed (data not shown). Because the BCR signaling complex is known to be recruited to lipid raft upon stimulation, we analyzed the possible translocation of NFAM1 to lipid raft. Indeed, a significant proportion of NFAM1 was recruited to lipid raft upon BCR stimulation (Fig. 5D).
Discussion
We have developed a cDNA cloning system, NACS, by using retrovirus cDNA libraries and a reporter cell line expressing NFAT-GFP, and cloned a number of signaling molecules inducing NFAT activation, including such ITAM+ surface molecules as CD3ε/ζ, Igα/β, FcRγ, and DAP12, which assemble with Ag receptors of T, B, or NK cells. ITAM-associated tyrosine kinases, ZAP-70/Syk and other intracellular signaling molecules such as an adaptor ADAP and a kinase HPK1 were also cloned. These findings imply that NACS can identify signaling molecules at various signal transduction levels and can potentially identify all signaling molecules involved in NFAT activation. Moreover, NACS can be used in general to clone a wide variety of molecules in a signaling pathway to induce a certain transcription factor (instead of NFAT) by using a reporter cell line expressing GFP fusion protein with the transcription factor.
However, our results suggest that the present experiments were not necessarily performed under optimal conditions because not only CD3ε and ζ but also CD3γ and δ should have been cloned, for instance. Our library (6 × 106 complexity) may not be sufficient for cloning less abundant molecules. Three procedures are vital for the success of the system: the establishment of a good recipient cell line with no background of constitutive GFP expression and that is able to induce all GFP+ cells upon stimulation; the use of an appropriate retrovirus titer for infection as too high a titer results in multiple integrations of cDNA into each clone; and negative cell sorting to avoid cells expressing GFP constitutively.
Although the YxxM sequence of mouse NFAM1-ITAM is not the consensus ITAM of YxxL/I (1, 20), we provided the following evidence that it is indeed an ITAM. First, human NFAM1 contains the consensus ITAM sequence, YxxI. Second, both tyrosines within the ITAM are required for NFAM1 signaling (Fig. 3). Third, NFAM1-ITAM recruits ZAP-70 upon stimulation and binds to ZAP-70/Syk with affinity similar to that of CD3ζ. Finally, NFAM1-ITAM has the same genomic structure as other ITAMs, in which the first YxxL/I is commonly separated from the second one by an intron.
NFAM1 contains an Ig domain in the extracellular region, resembling CD3 and Igα/β molecules. However, unlike other ITAM-bearing molecules possessing negatively charged amino acids within the transmembrane region that are involved in the association with cell-surface receptors bearing positively charged amino acids in the transmembrane region (21), NFAM1 does not have such charged residues within the transmembrane region. Thus, NFAM1 may not be associated with such receptor molecules. CD66d/CEACAM3 is a representative ITAM+ surface molecule, having no transmembrane charged residues and exhibit function as adhesion/signaling and phagocytic receptor in innate immunity (22). In this regard, NFAM1 may belong to another category of ITAM-bearing molecules with distinct functions.
One clue to understanding NFAM1 function is the translocation of NFAM1 to lipid raft upon BCR stimulation. Although NFAM1 does not appear to associate directly with BCR, this finding suggests a functional association between NFAM1 and the BCR signaling complex, and NFAM1 augments BCR signaling upon co-crosslinking in an ITAM-dependent manner. NFAM1 may associate with other molecule(s) susceptible to lipid raft localization. The enhancement of BCR signaling by NFAM1 may be mediated by the increased ITAM expression in the BCR signaling complex or the assembly with a specific signal molecule. NFAM1 may represent another class of ITAM-bearing cell-surface molecules exhibiting costimulatory-like function. Similar to CD66d, NFAM1 may also be activated in vivo upon recognition of a ligand that binds directly or indirectly to it through association with other molecules.
The in vivo overexpression of NFAM1 in lymphoid lineage resulted in severe, although incomplete, blockade of B cell development at the early differentiation stage of pre-B/immature B cells. A similar phenotype was observed in CD19 (23, 24), LMP-2A (25), and BtkE41K (Btk active form)-Tg mice (26). These molecules are known to activate/augment B cell signaling. In the case of BtkE41K-Tg mice, the BCR signaling enhancement results in apoptosis in early B cell development. BCR engagement by self-antigen in early ontogeny also induces apoptosis of immature B cells (27, 28). Considering that NFAM1 co-crosslinking enhances BCR signaling and that NFAM1 expression is the highest in pro-B cells and decreases with B cell maturation, the augmentation of pre-BCR/BCR signaling in early B cell differentiation may inhibit the development of B cells in NFAM1-Tg mice and BM chimeras. Thus, NFAM1 may play a role in modulating B cell development by mediating a kind of costimulatory function. Despite the similarity in NFAM1 expression between T and B cells, NFAM1 overexpression has no obvious effect on T cell development, and there may be some redundancy in T cells. Further analysis by using NFAM1-deficient mice may uncover the physiological role of NFAM1.
Acknowledgments
We thank K. Takase and S. Taki for discussion, M. Sakuma for technical assistance, and H. Yamaguchi and Y. Kurihara for secretarial assistance.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: ITAM, immunoreceptor tyrosine-based activation motif; NFAT, nuclear factor of activated T cells; NFAM1, NFAT activating molecule 1; NACS, NFAT-activation molecule cloning system; BCR, B cell antigen receptor; NK, natural killer; Tg, transgenic; BM, bone marrow.
References
- 1.Irving, B. A., Chan, A. C. & Weiss, A. (1993) J. Exp. Med. 177, 1093–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sanchez, M., Misulovin, Z., Burkhardt, A. L., Mahajan, S., Costa, T., Franke, R., Bolen, J. B. & Nussenzweig, M. (1993) J. Exp. Med. 178, 1049–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Letourneur, F. & Klausner, R. D. (1992) Science 255, 79–82. [DOI] [PubMed] [Google Scholar]
- 4.Reth, M. (1989) Nature 338, 383–384.2927501 [Google Scholar]
- 5.Lanier, L. L., Corliss, B. C., Wu, J., Leong, C. & Phillips, J. H. (1998) Nature 391, 703–707. [DOI] [PubMed] [Google Scholar]
- 6.Iwashima, M., Irving, B. A., van Oers, N. S., Chan, A. C. & Weiss, A. (1994) Science 263, 1136–1139. [DOI] [PubMed] [Google Scholar]
- 7.DeFranco, A. L. (1995) Curr. Opin. Cell Biol. 7, 163–175. [DOI] [PubMed] [Google Scholar]
- 8.Gauen, L. K., Zhu, Y., Letourneur, F., Hu, Q., Bolen, J. B., Matis, L. A., Klausner, R. D. & Shaw, A. S. (1994) Mol. Cell. Biol. 14, 3729–3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peng, S. L., Gerth, A. J., Ranger, A. M. & Glimcher, L. H. (2001) Immunity 14, 13–20. [DOI] [PubMed] [Google Scholar]
- 10.Rao, A., Luo, C. & Hogan, P. G. (1997) Annu. Rev. Immunol. 15, 707–747. [DOI] [PubMed] [Google Scholar]
- 11.Jain, J., McCaffrey, P. G., Miner, Z., Kerppola, T. K., Lambert, J. N., Verdine, G. L., Curran, T. & Rao, A. (1993) Nature 365, 352–355. [DOI] [PubMed] [Google Scholar]
- 12.Crabtree, G. R. (1999) Cell 96, 611–614. [DOI] [PubMed] [Google Scholar]
- 13.Pircher, H., Mak, T. W., Lang, R., Ballhausen, W., Ruedi, E., Hengartner, H., Zinkernagel, R. M. & Burki, K. (1989) EMBO J. 8, 719–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chien, Y. H., Gascoigne, N. R., Kavaler, J., Lee, N. E. & Davis, M. M. (1984) Nature 309, 322–326. [DOI] [PubMed] [Google Scholar]
- 15.Kitamura, T., Onishi, M., Kinoshita, S., Shibuya, A., Miyajima, A. & Nolan, G. P. (1995) Proc. Natl. Acad. Sci. USA 92, 9146–9150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cong, M., Thompson, V. F., Goll, D. E. & Antin, P. B. (1998) J. Biol. Chem. 273, 660–666. [DOI] [PubMed] [Google Scholar]
- 17.Okazaki, Y., Ohno, H., Takase, K., Ochiai, T. & Saito, T. (2000) J. Biol. Chem. 275, 35751–35758. [DOI] [PubMed] [Google Scholar]
- 18.Itoh, K., Sakakibara, M., Yamasaki, S., Takeuchi, A., Arase, H., Miyazaki, M., Nakajima, N., Okada, M. & Saito, T. (2002) J. Immunol. 168, 541–544. [DOI] [PubMed] [Google Scholar]
- 19.Yokosuka, T., Takase, K., Suzuki, M., Nakagawa, Y., Taki, S., Takahashi, H., Fujisawa, T., Arase, H. & Saito, T. (2002) J. Exp. Med. 195, 991–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Romeo, C., Amiot, M. & Seed, B. (1992) Cell 68, 889–897. [DOI] [PubMed] [Google Scholar]
- 21.Ashwell, J. D. & Klusner, R. D. (1990) Annu. Rev. Immunol. 8, 139–167. [DOI] [PubMed] [Google Scholar]
- 22.Schmitter, T., Agerer, F., Peterson, L., Munzner, P. & Hauck, C. R. (2004) J. Exp. Med. 199, 35–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Engel, P., Zhou, L. J., Ord, D. C., Sato, S., Koller, B. & Tedder, T. F. (1995) Immunity 3, 39–50. [DOI] [PubMed] [Google Scholar]
- 24.Zhou, L. J., Smith, H. M., Waldschmidt, T. J., Schwarting, R., Daley, J. & Tedder, T. F. (1994) Mol. Cell. Biol. 14, 3884–3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Caldwell, R. G., Wilson, J. B., Anderson, S. J. & Longnecker, R. (1998) Immunity 9, 405–411. [DOI] [PubMed] [Google Scholar]
- 26.Maas, A., Dingjan, G. M., Grosveld, F. & Hendriks, R. W. (1999) J. Immunol. 162, 6526–6533. [PubMed] [Google Scholar]
- 27.Hardy, R. R. & Hayakawa, K. (2001) Annu. Rev. Immunol. 19, 595–621. [DOI] [PubMed] [Google Scholar]
- 28.Melamed, D., Benschop, R. J., Cambier, J. C. & Nemazee, D. (1998) Cell 92, 173–182. [DOI] [PubMed] [Google Scholar]