Abstract
Cancer cells undergo distinct metabolic changes to cope with their hypoxic environment. These changes are achieved at least partly by the action of transcriptional factors called hypoxia-inducible factors (HIFs). We investigated gene expression in cultured human colon cancer cells induced by hypoxic conditions with special reference to cell-adhesion molecules and carbohydrate determinants having cell-adhesive activity by using DNA-microarray and RT-PCR techniques. Hypoxic culture of colon cancer cells induced a marked increase in expression of selectin ligands, the sialyl Lewis x and sialyl Lewis a determinants at the cell surface, which led to a definite increase in cancer cell adhesion to endothelial E-selectin. The transcription of genes for fucosyltransferase VII (FUT7), sialyltransferase ST3Gal-I (ST3O), and UDP-galactose transporter-1 (UGT1), which are all known to be involved in the synthesis of the carbohydrate ligands for E-selectin, was significantly induced in cancer cells by hypoxic culture. In addition, a remarkable induction was detected in the genes for syndecan-4 (SDC4) and α5-integrin (ITGA5), the cell-adhesion molecules involved in the enhanced adhesion of cancer cells to fibronectin. The transcriptional induction by hypoxia was reproduced in the luciferase-reporter assays for these genes, which were significantly suppressed by the co-transfection of a dominant-negative form of HIF. These results indicate that the metabolic shifts of cancer cells partly mediated by HIFs significantly enhance their adhesion to vascular endothelial cells, through both selectin- and integrin-mediated pathways, and suggest that this enhancement further facilitates hematogenous metastasis of cancers and tumor angiogenesis.
Adhesion of cancer cells to endothelial cells is known to be involved in the hematogenous metastasis of cancer (1, 2). This adhesion is mediated by the interaction of selectins on endothelial cells and their specific carbohydrate ligands, sialyl Lewis x and sialyl Lewis a, on cancer cells, and this is followed by integrin-mediated cell adhesion (3–5). Selectin- and integrin-mediated interaction of cancer cells with endothelial cells also is known to be implicated in tumor angiogenesis (6).
Expression of sialyl Lewis x and sialyl Lewis a is known to be increased significantly in cancers, but the mechanism for its induction has remained unclear. Specific alteration of transcription of genes for several glycosyltransferases and sugar transporters in cancer has been suggested to be involved in the induction of the selectin ligands on cancer cells. We previously proposed that an increased expression of a sialyltransferase gene (ST3O) in cancer is closely associated with sialyl Lewis a expression (7). Increased expression of UDP-galactose transporter gene (UGT1) in cancers was also suggested to be involved in the induction of these selectin ligands, because the introduction of the gene for UDP-galactose transporter conferred the induction of sialyl Lewis a and sialyl Lewis x expression on cultured cancer cells (8). Other investigators have suggested a role for fucosyltransferase VII (9–11) and even the involvement of another sugar transporter, GLUT1, for induction of sialyl Lewis x in cancers (12). However, the exact mechanism underlying this transcriptional induction has remained uncharacterized.
In this study we will show that a transcriptional factor, hypoxia-inducible factor (HIF), known to be increased in cancers, induces transcription of all these four genes and leads to the significant increase of selectin-mediated cancer cell adhesion to endothelial cells. We also show that HIF induces transcription of syndecan-4 and α5-integrin, both of which significantly enhance integrin-mediated adhesion of cancer cells to endothelial fibronectin.
Materials and Methods
Cells, Hypoxic Culture, Antibodies, and Flow-Cytometric Analysis. Human colon cancer cell lines, SW480, C-1, and Colo201, were routinely maintained in Dulbecco's modified MEM (high glucose) supplemented with 10% FCS. For hypoxia experiments, the cells were cultured at 37°C with 5% CO2/94% N2/1% O2 in a multigas incubator (Juji Field, Tokyo). Desferrioxamine (Sigma) was added to the culture medium at a final concentration of 100 μM. Monoclonal anti-sialyl Lewis x (SNH-3, murine IgM) and anti-sialyl Lewis a (2D3, murine IgM) antibodies were prepared as described (13, 14), and anti-α5-integrin antibody (P1D6, murine IgG3) was obtained from Life Technologies (Gaithersburg, MD). Anti-syndecan-4 ectodomain antibody (rabbit polyclonal) was prepared as described (15). For flow-cytometric analysis, cells were harvested at a semiconfluent stage and stained with the respective antibody by using purified antibody at 1 μg/ml or culture supernatant at a dilution of 1:5. Cells then were stained with a 1:200 dilution of FITC-conjugated second antibody and analyzed with FACScalibur (Becton Dickinson).
Generation of Dominant-Negative HIF Transfectant Cells. Expression vector pcDNA3.1+ (Invitrogen) containing HIF-1α cDNA lacking DNA-binding domain was prepared as described (16) and transfected to SW480 cells (1 × 106) by the lipofection method using Lipofectamine Plus reagent (Invitrogen) according to manufacturer protocol. Cells were allowed to grow for 2 days before being subjected to selection for the ability to grow in medium containing 600 μg/ml geneticin (G418-sulfate, Invitrogen).
Cell-Adhesion Assay to E-Selectin and Fibronectin. Cell-adhesion experiments were performed as described (17–19). SW480 cells were cultured in monolayer in 24-well plates for 7 days under hypoxic or normoxic conditions. 2′7′-bis-(Carboxyethyl)-5-(and-6)-carboxyf luorescein acetoxymethyl ester (BCECF-AM)-labeled 300.19/E-selectin cells (1 × 106 cells per 0.5 ml per well, murine B lymphoma 300.19 cells transfected with human E-selectin cDNA, kindly supplied by Geoffrey S. Kansas, Department of Microbiology-Immunology, Northwestern University Medical School, Chicago) (20) were added to each well, and the plate was incubated on a rotating platform for incubation under shear (90 rpm with a rotary shaker R-20mini, Taitek, Tokyo) for 20 min at room temperature. Adhesion to fibronectin was assayed by using 24-well plates coated with 30 μg/ml human fibronectin (Seikagaku Kogyo, Tokyo) at 4°C for overnight. SW480 cells (5 × 105), cultured under hypoxic or normoxic conditions, were added in a volume of 0.5 ml per well after being labeled with BCECF-AM and incubated for 1 h at 37°C without rotation. After nonadherent cells were washed out 3 times with PBS, the cells were lysed with 0.5% Nonidet P-40, and the attached cells were counted by measuring fluorescence intensity with an Arvo 1420 multilabel counter (Wallac, Gaithersburg, MD).
DNA-Microarray and RT-PCR Analyses. Total RNA was isolated from SW480 and C-1 cells (1 × 106 cells), which had been incubated for 24 h under hypoxic and normoxic conditions, by using mTRAP mRNA isolation kit (Active Motif, Carlsbad, CA). Cy3-dUTP (normoxia) or Cy5-dUTP (hypoxia) was incorporated into cDNA by using CyScribe First-Strand cDNA-labeling kit (Amersham Pharmacia Biotech). Two cDNA samples, labeled with either Cy3 or Cy5 fluorescent dyes, were applied competitively to the microarray, which contained >1,000 human cDNAs mapped in the UniGene database related with complex carbohydrate synthesis and metabolism and adhesion molecules. A fluorescence image of the microarray was obtained by using an Affymetrix 428 array scanner (Affymetrix, Santa Clara, CA). Data obtained from the scanner were analyzed further by imagene (BioDiscovery, Marina del Rey, CA). The genes expressed at >2-fold higher levels under hypoxic conditions than under normoxic conditions were defined as possible hypoxia-inducible genes. Additional conventional RT-PCR was performed when the transcription levels of genes related to the complex carbohydrate metabolism were below the detection limit for DNA microarray. Semiquantitative conventional RT-PCR was performed as described (7, 8). The primers used for conventional RT-PCR analysis in this study are provided in Table 1, which is published as supporting information on the PNAS web site.
Luciferase-Reporter Assays. The SW480 cells cultured in 24-well plates were cotransfected with firefly luciferase-reporter plasmids (1.0 μg/ml) for each gene mentioned below and pRL Renilla luciferase plasmids (0.1 μg/ml) by using Lipofectamine reagent (Life Technologies) with or without the expression vector for the dominant-negative form of HIF (1.0 μg/ml). After culturing the cells under either hypoxic (1% O2) or normoxic (20% O2) conditions for the indicated number of hours in each experiment, the cell lysates were obtained by using 100 μl per well of cell lysis buffer (Promega). Luciferase activity was measured by using 20 μl of cell lysate per assay tube in a single-photon channel of a scintillation counter (Beckman). Luciferase transfection efficiency was normalized by dualluciferase analysis using pRL Renilla luciferase control (dualluciferase reporter assay system, Promega) (21).
Reporter constructs, pGL2/pΔFT7pro(–791) containing from –791 to +104, equivalent to just 5′ of the ATG codon of the human fucosyltransferase VII gene (FUT7) (22), pGL3–1009pI for sialyltransferase ST3O (21), and pGL36hRG-781 for syndecan-4 (SDC4) (23) have been described already. The reporter construct for UDP-galactose transporter-1 (UGT1) (pGL3/UGT1-1042), which contains –1,042 to +1 base from the putative transcription start site, was prepared by PCR amplification of the genomic clone containing exons for human UDP-galactose transporter that had been obtained by screening of a λDASH (Stratagene) phage library constructed from Sau3AI partial digest of human genome DNA. Details of the construct will be published elsewhere.
Clinical Samples and Real-Time RT-PCR Analysis. Surgical specimens were obtained from 20 patients with primary colorectal cancer at surgical operation and processed as described (7, 8). The median age of patients was 59.8 years. Malignant and nonmalignant portions of each specimen were used for RNA extraction. Nonmalignant mucosa was scraped off by using slide glasses, and tissue specimens of cancer were excised carefully so as to eliminate noncancerous tissue components. Real-time quantitative RT-PCR analysis was performed by using TaqMan PCR detector Prism7000 (Perkin–Elmer Applied Biosystems) with TaqMan probes: Hs00161688_m1 (for ST3O), Hs00194644_m1 (for UGT1), Hs00197884_ml (for GLUT1), Hs00233743_m1 (for α5-integrin, ITGA5), and Hs00161617_m1 (for syndecan-4, SDC4) as provided by the manufacturer. The probe and primers for FUT7 were designed by using primer express software (probe, 5′-CTGGCATGAATGAGAGCCGATACCAAC-3′; forward primer, 5′-AGAGCTGGCGGCTTTCCT-3′; reverse primer, 5′-CTTCTTTGCCTGGCGTGAC-3′).
Results
Induction of Sialyl Lewis a and Sialyl Lewis x Determinants by Hypoxic Culture of Cancer Cells. A hypoxic culture of colon cancer cells under 1% O2 resulted in a significant induction of sialyl Lewis x and sialyl Lewis a determinants, which was evident after 3 days of hypoxic culture. A representative result on a cultured colon cancer cell line, SW480, is shown in Fig. 1A, and similar results were obtained with other cultured colon cancer cell lines including C-1 and Colo201 (see Fig. 6, which is published as supporting information on the PNAS web site). A similar induction of sialyl Lewis a and sialyl Lewis x was observed when the cells were cultured in the presence of hypoxia mimetics such as desferrioxamine, an iron-chelating agent known to activate HIF-1 (Fig. 1B). The induction was reversible; when the cells were cultured under normoxic conditions for an additional 7 days, the expression of sialyl Lewis a and x returned nearly to the starting level. Involvement of the HIF in the induction of the carbohydrate determinants was evident, because a prior transfection of the cells with dominant-negative form of HIF almost completely abrogated the induction (Fig. 1 A). The dominant-negative form of HIF was effective for both HIF-1 and HIF-2, but the major HIF species in these cells was found to be HIF-1, as ascertained by Western blotting analysis (see Fig. 7, which is published as supporting information on the PNAS web site). The induction of sialyl Lewis a/x expression resulted in the significant enhancement of cancer cell adhesion to E-selectin (Fig. 1C).
Fig. 1.
Hypoxia-induced sialyl Lewis x (sialyl Lex) and sialyl Lewis a (sialyl Lea) expression and E-selectin-mediated adhesion of colon cancer cells. (A) A human colon cancer cell line, SW480, was cultured under 1% O2 for 7 days (hypoxia) and subsequently under 20% O2 for additional 7 days (reoxygenation) and analyzed for sialyl Lewis x/a expression by flow cytometry. Results on the SW480 cells transfected with dominant-negative HIF are shown also. (B) SW480 cells were cultured in the presence of 100 μM desferrioxamine for 6 days before analyses of carbohydrate determinants. (C) Hypoxia-induced enhancement of SW480 cell adhesion to 300.19/E-selectin cells. Filled bars, adhesion of 300.19/E-selectin cells; open bars, adhesion of parental 300.19 cells (negative control); N.S., not significant.
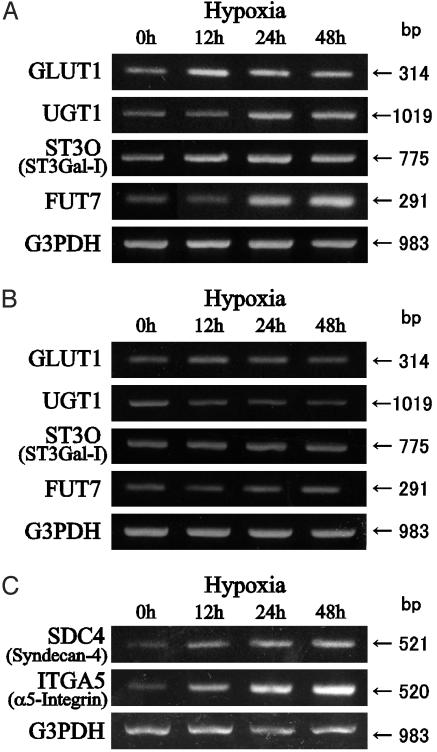
Transcriptional Induction of Carbohydrate-Related Genes by Hypoxic Culture. We used DNA microarray to determine which gene expression was induced by hypoxic culture. The DNA microarray covered >1,000 genes including all known human glycosyltransferases, sugar transporters, and other genes related to carbohydrate metabolism as well as core proteins for important cell-adhesive glycoproteins. Two colon cancer cell lines, SW480 and C-1, were analyzed, and the genes showing a significant change in their transcription in both cell lines were chosen. Because the transcription levels of 40 important genes for the complex carbohydrate metabolism were below the detection limit of the DNA-microarray technique, additional RT-PCR was used for such genes. From these analyses, four genes that have a possible involvement in the enhanced synthesis of sialyl Lewis a and x determinants were picked up: GLUT1, fucosyltransferase VII (FUT7), sialyltransferase ST3O (ST3Gal-I), and UDP-galactose transporter-1 (UGT1). Transcription of genes for other fucosyltransferases (fucosyltransferases III–VI), sialyltransferases (ST3Gal-II–ST3Gal-IV), other known glycosyltransferases, and sugar transporters showed no appreciable changes by hypoxic culture (see Fig. 8, which is published as supporting information on the PNAS web site). Unexpectedly, a marked increase in the transcription of the gene for core proteins of syndecan-4 (SDC4) and α5-integrin (ITGA5) was also detected after hypoxic culture. The increase of transcription of these genes was ascertained by RT-PCR as shown in Fig. 2, thus excluding possible experimental error associated with the DNA-microarray technique.
Fig. 2.
RT-PCR analyses of the genes up-regulated in SW480 cells cultured under hypoxic conditions. Hypoxia-induced up-regulation of carbohydrate-related genes in parental SW480 cells (A) and dominant-negative HIF transfectant cells (B) and hypoxia-induced up-regulation of genes for syndecan-4 and α5-integrin in parental SW480 cells (C) are shown. For other genes, see Fig. 8.
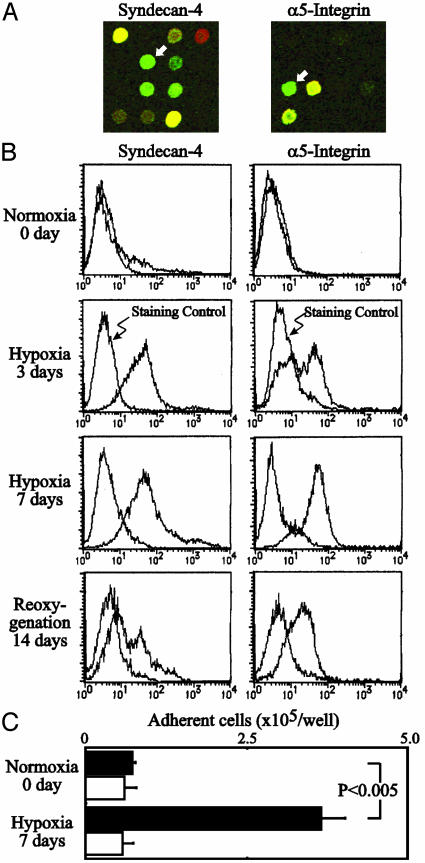
Enhanced Expression of Syndecan-4 and α5-Integrin by Hypoxic Culture. The unexpected induction of genes for syndecan-4 and α5-integrin core protein (Fig. 3A) prompted us to study the expression of these cell-adhesion molecules on the surface of cells after hypoxic culture. As shown in Fig. 3B, hypoxic culture of human colon cancer SW480 cells resulted in markedly enhanced expression of these proteins on the cell surface. Syndecan-4 and α5-integrin are known to be involved in the cell adhesion to fibronectin; syndecan-4 is known to bind to the so-called heparan-sulfate binding site of fibronectin, and α5-integrin is widely acknowledged to be a typical ligand for the integrin-binding site of fibronectin. Both ligands are reportedly necessary for focal adhesion formation and cell migration (24, 25). Expression of the gene for αv-integrin, another integrin that is implicated in tumor angiogenesis, was found to be markedly down-regulated under hypoxic conditions in the present DNA microarray. A remarkable enhancement of adhesion was observed when the cancer cells cultured under hypoxic conditions were assayed for adhesion to immobilized human fibronectin (Fig. 3C).
Fig. 3.
Hypoxia-induced syndecan-4 and α5-integrin expression and adhesion to fibronectin of colon cancer cells cultured under hypoxic conditions. (A) Signals of syndecan-4 and α5-integrin in the DNA microarray (white arrows). Cy3-dUTP (normoxia: red) or Cy5-dUTP (hypoxia, 1% O2 for 24 h: green) was incorporated into cDNA prepared from a human colon cancer cell line SW480. (B) SW480 cells were cultured under 1% O2 for 7 days and subsequently under 20% O2 for an additional 14 days and analyzed for syndecan-4 and α5-integrin expression by flow cytometry. (C) Hypoxia-induced enhancement of SW480 cell adhesion to immobilized fibronectin. Filled bars, adhesion to immobilized fibronectin; open bars, adhesion to BSA (negative control).
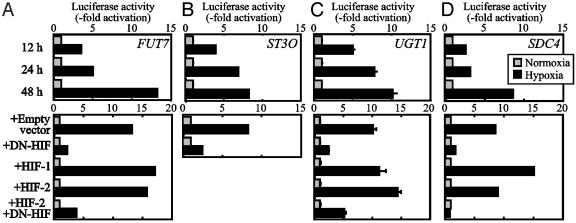
Luciferase-Reporter Assays for Transcriptional Induction of Genes Related to Cancer Cell Adhesion by Hypoxia. Induction of the genes for GLUT1, SDC4, and ITGA5 by hypoxia is already documented (26–29), whereas significant induction of the other genes was noted in the present study. To further ascertain the effect of HIFs on the induction of these genes, luciferase-reporter assays were performed by using the reporter constructs containing 5′-regulatory regions of these genes. As shown in Fig. 4, the construct for FUT7, ST3O, UGT1, and SDC4 showed remarkable transcriptional induction by hypoxic culture. Again, cotransfection of the dominant-negative form of HIF abrogated the transcriptional induction, indicating the involvement of HIF in the transcriptional induction.
Fig. 4.
Effect of hypoxia on FUT7 (A), ST3O (B), UGT1 (C), and SDC4 (D) promoter activity. The reporter construct of each gene was transfected to SW480 cells and cultured for 12, 24, or 48 h under hypoxic (1% O2) or normoxic (20% O2) conditions. (Lower) Effect of cotransfection of the dominant-negative form of HIF (DN-HIF). The same amount of empty vector served as negative control, and nonmutated HIF-1 and HIF-2 constructs in the same vector served as positive controls.
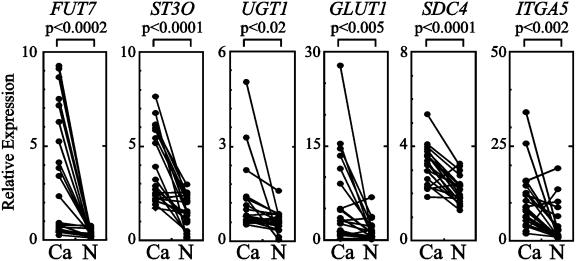
Clinical Relevance of Transcriptional Induction of Genes Related to Cancer Cell Adhesion. To know the clinical relevance of transcriptional induction of these genes in patients with cancers, we studied expression of these genes in the cancer tissues prepared from 20 patients with colon cancers and compared the results with the expression levels of the genes in nonmalignant epithelial cells prepared from the same patients. As shown in Fig. 5, transcription of all six genes was increased significantly in cancer cells compared with nonmalignant epithelial cells. The increase in transcription was more prominent in Dukes C and D patients than in Dukes A and B patients (data not shown), suggesting that the transcription of these genes increases along with the progression of colon cancers.
Fig. 5.
Real-time RT-PCR analyses of expression of genes for fucosyltransferase VII (FUT7), sialyltransferase ST3Gal-I (ST3O), UDP-galactose transporter-1 (UGT1), GLUT1, syndecan-4 (SDC4), and α5-integrin (ITGA5) in cancer tissues prepared from the patients with colon cancer. Ca, cancer tissues; N, nonmalignant mucosa prepared from the same patient. A paired t test was performed to ascertain statistical significance between the amount in cancer tissue and in nonmalignant mucosa.
Discussion
Cancer cells are known to exhibit a metabolic shift from oxidative to elevated anaerobic glycolysis (the Warburg effect) to cope with hypoxic environments (30), which is achieved by the alteration of sugar transporters and enzymes related in carbohydrate metabolism (31, 32). Activation of various oncogenes including v-src, K-ras, fps, and c-myc and SV40 or loss of antioncogenes such as VHL is known to facilitate this metabolic shift. More recently a transcription factor called HIF, the expression of which is shown to be increased in various human cancers (33–40), is known to be involved in the transcriptional induction of the genes related to sugar transporters and glycolytic enzymes (41–43). The present study indicated that the increased sialyl Lewis x/a expression in cancers is clearly a link in the chains of these events, because our results demonstrated that HIFs are involved, directly or indirectly, in the induction of sialyl Lewis x/a expression and enhanced adhesion of cancer cells to endothelial E-selectin under hypoxic culture. Moreover, our results also demonstrated that HIFs are involved in the induction of syndecan-4 and α5-integrin expression, which mediates adhesion of cancer cells to fibronectin. Vascular endothelial cells are known to significantly express fibronectin as well as E-selectin (44–46).
This promotion of cancer cell adhesion is accompanied by the transcriptional induction of a set of genes closely related to sialyl Lewis x/a expression. Among the genes shown to be increased by HIFs in this study, fucosyltransferase VII and sialyltransferase ST3O have an ability to directly contribute to enhanced synthesis of sialyl Lewis x or sialyl Lewis a, respectively, because they can synthesize these carbohydrate determinants, which carry fucose and sialic acid residues. The contribution of UDP-galactose transporter-1 would be more indirect compared with them, because UDP-galactose transporter-1 is primarily related to the galactose metabolism, and is involved in the synthesis of precursors for sialyl Lewis x/a synthesis. However, our previous results indicated that introduction of the UDP-galactose transporter gene to cells led to a prominent increase in the sialyl Lewis x/a expression (8), suggesting that the galactose metabolism may have a strong effect on the overall expression of these carbohydrate ligands for selectins. This result is supported also by a recent finding that disruption of the gene for a major galactosyltransferase results in a remarkable reduction of selectin-ligand expression in mice (47). Although the exact mechanism of the relationship between GLUT1 induction and sialyl Lewis x/a is not clear, the good correlation of GLUT1 expression and sialyl Lewis x expression in clinical samples is known (12). Interestingly, GLUT1 encodes the major cellular transporter for not only glucose but also galactose. The transcription levels of these six genes are all significantly increased in the clinical tumor specimens prepared from patients with cancers.
Expression of sialyl Lewis x/a determinants is known to be markedly enhanced in cancer cells, but the molecular mechanism that leads to cancer-associated expression of sialyl Lewis x/a has not been well understood. In general, two principal mechanisms have been proposed for the cancer-associated change of cell-surface carbohydrates: “incomplete synthesis,” the inhibition of synthesis of normal carbohydrate determinants resulting in the precursor accumulation, and “neosynthesis,” the de novo induction of carbohydrate determinant expression caused by cancer-associated transcriptional anomaly (48, 49). The HIF-mediated increment of sialyl Lewis x/a expression can be regarded as an example of the latter mechanism.
Cancer-cell adhesion to endothelial cells mediated by selectins and their carbohydrate ligands has been proposed to figure heavily in cancer metastasis. Our recent results indicated that this adhesion is involved also in the vasculogenesis associated with tumor progression (6). HIFs are known to be intimately involved in the promotion of tumor angiogenesis by triggering the transcription of vascular endothelial growth factor in cancer cells to overcome their hypoxic environments (41–43, 50, 51). Simultaneous induction of cell-adhesion molecules, which mediate cancer cell adhesion to vascular endothelial cells by HIFs, also would facilitate growth of tumors.
Supplementary Material
Acknowledgments
This work was supported in part by Ministry of Education, Culture, Sports, Science, and Technology (Japan) Grants-in-Aid 15590263 and On Priority Areas 14030092, grants-in-aid for the Second Term Comprehensive Ten-Year Strategy for Cancer Control from the Ministry of Health, Labor, and Welfare (Japan), and a grant from the Core Research for Evolution of Science and Technology program of the Japan Science and Technology Agency.
Abbreviation: HIF, hypoxia-inducible factor.
References
- 1.Hakomori, S. (2002) Proc. Natl. Acad. Sci. USA 99, 10231–10233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hakomori, S. (1996) Cancer Res. 56, 5309–5318. [PubMed] [Google Scholar]
- 3.Kannagi, R. (2004) Glycoconj. J. 20, 353–364. [DOI] [PubMed] [Google Scholar]
- 4.Kannagi, R. (2002) Curr. Opin. Struct. Biol. 12, 599–608. [DOI] [PubMed] [Google Scholar]
- 5.Kannagi, R. (1997) Glycoconj. J. 14, 577–584. [DOI] [PubMed] [Google Scholar]
- 6.Tei, K., Kawakami-Kimura, N., Taguchi, O., Kumamoto, K., Higashiyama, S., Taniguchi, N., Toda, K., Kawata, R., Hisa, Y. & Kannagi, R. (2002) Cancer Res. 62, 6289–6296. [PubMed] [Google Scholar]
- 7.Ito, H., Hiraiwa, N., Sawada-Kasugai, M., Akamatsu, S., Tachikawa, T., Kasai, Y., Akiyama, S., Ito, K., Takagi, H. & Kannagi, R. (1997) Int. J. Cancer 71, 556–564. [DOI] [PubMed] [Google Scholar]
- 8.Kumamoto, K., Goto, Y., Sekikawa, K., Takenoshita, S., Ishida, N., Kawakita, M. & Kannagi, R. (2001) Cancer Res. 61, 4620–4627. [PubMed] [Google Scholar]
- 9.Martín-Satué, M., Marrugat, R., Cancelas, J. A. & Blanco, J. (1998) Cancer Res. 58, 1544–1550. [PubMed] [Google Scholar]
- 10.Ogawa, J., Inoue, H. & Koide, S. (1996) Cancer Res. 56, 325–329. [PubMed] [Google Scholar]
- 11.Liu, F., Qi, H. L., Zhang, Y., Zhang, X. Y. & Chen, H. L. (2001) Eur. J. Biochem. 268, 3501–3512. [DOI] [PubMed] [Google Scholar]
- 12.Ogawa, J., Inoue, H. & Koide, S. (1997) Int. J. Cancer 74, 189–192. [DOI] [PubMed] [Google Scholar]
- 13.Phillips, M. L., Nudelman, E., Gaeta, F. C. A., Perez, M., Singhal, A. K., Hakomori, S. & Paulson, J. C. (1990) Science 250, 1130–1132. [DOI] [PubMed] [Google Scholar]
- 14.Takada, A., Ohmori, K., Yoneda, T., Tsuyuoka, K., Hasegawa, A., Kiso, M. & Kannagi, R. (1993) Cancer Res. 53, 354–361. [PubMed] [Google Scholar]
- 15.Ishiguro, K., Kojima, T., Taguchi, O., Saito, H., Muramatsu, T. & Kadomatsu, K. (1999) Histochem. Cell Biol. 112, 25–33. [DOI] [PubMed] [Google Scholar]
- 16.Chen, J., Zhao, S., Nakada, K., Kuge, Y., Tamaki, N., Okada, F., Wang, J., Shindo, M., Higashino, F., Takeda, K., et al. (2003) Am. J. Pathol. 162, 1283–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kawakami-Kimura, N., Narita, T., Ohmori, K., Yoneda, T., Matsumoto, K., Nakamura, T. & Kannagi, R. (1997) Br. J. Cancer 75, 47–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kimura, N., Mitsuoka, C., Kanamori, A., Hiraiwa, N., Uchimura, K., Muramatsu, T., Tamatani, T., Kansas, G. S. & Kannagi, R. (1999) Proc. Natl. Acad. Sci. USA 96, 4530–4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mitsuoka, C., Ohmori, K., Kimura, N., Kanamori, A., Komba, S., Ishida, H., Kiso, M. & Kannagi, R. (1999) Proc. Natl. Acad. Sci. USA 96, 1597–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kansas, G. S., Ley, K., Munro, J. M. & Tedder, T. F. (1993) J. Exp. Med. 177, 833–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taniguchi, A., Yoshikawa, I. & Matsumoto, K. (2001) Glycobiology 11, 241–247. [DOI] [PubMed] [Google Scholar]
- 22.Hiraiwa, N., Yabuta, T., Yoritomi, K., Hiraiwa, M., Tanaka, Y., Suzuki, T., Yoshida, M. & Kannagi, R. (2003) Blood 101, 3615–3621. [DOI] [PubMed] [Google Scholar]
- 23.Takagi, A., Kojima, T., Tsuzuki, S., Katsumi, A., Yamazaki, T., Sugiura, I., Hamaguchi, M. & Saito, H. (1996) J. Biochem. (Tokyo) 119, 979–984. [DOI] [PubMed] [Google Scholar]
- 24.Mostafavi-Pour, Z., Askari, J. A., Parkinson, S. J., Parker, P. J., Ng, T. T. & Humphries, M. J. (2003) J. Cell Biol. 161, 155–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Woods, A. & Couchman, J. R. (2001) Curr. Opin. Cell Biol. 13, 578–583. [DOI] [PubMed] [Google Scholar]
- 26.Ebert, B. L., Firth, J. D. & Ratcliffe, P. J. (1995) J. Biol. Chem. 270, 29083–29089. [DOI] [PubMed] [Google Scholar]
- 27.Kojima, T., Takagi, A., Maeda, M., Segawa, T., Shimizu, A., Yamamoto, K., Matsushita, T. & Saito, H. (2001) Thromb. Haemost. 85, 793–799. [PubMed] [Google Scholar]
- 28.Lal, A., Peters, H., St Croix, B., Haroon, Z. A., Dewhirst, M. W., Strausberg, R. L., Kaanders, J. H., van der Kogel, A. J. & Riggins, G. J. (2001) J. Natl. Cancer Inst. 93, 1337–1343. [DOI] [PubMed] [Google Scholar]
- 29.Denko, N. C., Fontana, L. A., Hudson, K. M., Sutphin, P. D., Raychaudhuri, S., Altman, R. & Giaccia, A. J. (2003) Oncogene 22, 5907–5914. [DOI] [PubMed] [Google Scholar]
- 30.Warburg, O. (1956) Science 123, 309–314. [DOI] [PubMed] [Google Scholar]
- 31.Kalckar, H. M. (1965) Science 150, 305–313. [DOI] [PubMed] [Google Scholar]
- 32.Hatanaka, M. (1973) Proc. Natl. Acad. Sci. USA 70, 1364–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhong, H., De Marzo, A. M., Laughner, E., Lim, M., Hilton, D. A., Zagzag, D., Buechler, P., Isaacs, W. B., Semenza, G. L. & Simons, J. W. (1999) Cancer Res. 59, 5830–5835. [PubMed] [Google Scholar]
- 34.Talks, K. L., Turley, H., Gatter, K. C., Maxwell, P. H., Pugh, C. W., Ratcliffe, P. J. & Harris, A. L. (2000) Am. J. Pathol. 157, 411–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Birner, P., Schindl, M., Obermair, A., Plank, C., Breitenecker, G. & Oberhuber, G. (2000) Cancer Res. 60, 4693–4696. [PubMed] [Google Scholar]
- 36.Giatromanolaki, A., Koukourakis, M. I., Sivridis, E., Turley, H., Talks, K., Pezzella, F., Gatter, K. C. & Harris, A. L. (2001) Br. J. Cancer 85, 881–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bos, R., Zhong, H., Hanrahan, C. F., Mommers, E. C., Semenza, G. L., Pinedo, H. M., Abeloff, M. D., Simons, J. W., Van Diest, P. J. & van Der Wall, E. (2001) J. Natl. Cancer Inst. 93, 309–314. [DOI] [PubMed] [Google Scholar]
- 38.Beasley, N. J., Leek, R., Alam, M., Turley, H., Cox, G. J., Gatter, K., Millard, P., Fuggle, S. & Harris, A. L. (2002) Cancer Res. 62, 2493–2497. [PubMed] [Google Scholar]
- 39.Kuwai, T., Kitadai, Y., Tanaka, S., Onogawa, S., Matsutani, N., Kaio, E., Ito, M. & Chayama, K. (2003) Int. J. Cancer 105, 176–181. [DOI] [PubMed] [Google Scholar]
- 40.Kurokawa, T., Miyamoto, M., Kato, K., Cho, Y., Kawarada, Y., Hida, Y., Shinohara, T., Itoh, T., Okushiba, S., Kondo, S. & Katoh, H. (2003) Br. J. Cancer 89, 1042–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Semenza, G. L. (2003) Nat. Rev. Cancer 3, 721–732. [DOI] [PubMed] [Google Scholar]
- 42.Semenza, G. L. (2001) Curr. Opin. Cell Biol. 13, 167–171. [DOI] [PubMed] [Google Scholar]
- 43.Semenza, G. L. (2001) Cell 107, 1–3. [DOI] [PubMed] [Google Scholar]
- 44.Carreiras, F., Thiebot, B., Leroy-Dudal, J., Maubant, S., Breton, M. F. & Darbeida, H. (2002) Int. J. Cancer 99, 800–808. [DOI] [PubMed] [Google Scholar]
- 45.Boyle, D. L., Shi, Y., Gay, S. & Firestein, G. S. (2000) Cell Immunol. 200, 1–7. [DOI] [PubMed] [Google Scholar]
- 46.Peters, J. H., Sporn, L. A., Ginsberg, M. H. & Wagner, D. D. (1990) Blood 75, 1801–1808. [PubMed] [Google Scholar]
- 47.Asano, M., Nakae, S., Kotani, N., Shirafuji, N., Nambu, A., Hashimoto, N., Kawashima, H., Hirose, M., Miyasaka, M., Takasaki, S. & Iwakura, Y. (2003) Blood 102, 1678–1685. [DOI] [PubMed] [Google Scholar]
- 48.Hakomori, S. & Kannagi, R. (1983) J. Natl. Cancer Inst. 71, 231–251. [PubMed] [Google Scholar]
- 49.Hakomori, S. (1985) Cancer Res. 45, 2405–2414. [PubMed] [Google Scholar]
- 50.Semenza, G. L. (2001) J. Clin. Invest. 108, 39–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Harris, A. L. (2002) Nat. Rev. Cancer 2, 38–47. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.