Abstract
Aminoacyl-tRNA synthetases (ARSs) are in charge of cellular protein synthesis and have additional domains that function in a versatile manner beyond translation. Eight core ARSs (EPRS, MRS, QRS, RRS, IRS, LRS, KRS, DRS) combined with three nonenzymatic components form a complex known as multisynthetase complex (MSC).We hypothesize that the single-nucleotide polymorphisms (SNPs) of the eight core ARS coding genes might influence the susceptibility of sporadic congenital heart disease (CHD). Thus, we conducted a case-control study of 984 CHD cases and 2953 non-CHD controls in the Chinese Han population to evaluate the associations of 16 potentially functional SNPs within the eight ARS coding genes with the risk of CHD. We observed significant associations with the risk of CHD for rs1061248 [G/A; odds ratio (OR) = 0.90, 95% confidence interval (CI) = 0.81–0.99; P = 3.81×10−2], rs2230301 [A/C; OR = 0.73, 95%CI = 0.60–0.90, P = 3.81×10−2], rs1061160 [G/A; OR = 1.18, 95%CI = 1.06–1.31; P = 3.53×10−3] and rs5030754 [G/A; OR = 1.39, 95%CI = 1.11–1.75; P = 4.47×10−3] of EPRS gene. After multiple comparisons, rs1061248 conferred no predisposition to CHD. Additionally, a combined analysis showed a significant dosage-response effect of CHD risk among individuals carrying the different number of risk alleles (P trend = 5.00×10−4). Compared with individuals with “0–2” risk allele, those carrying “3”, “4” or “5 or more” risk alleles had a 0.97-, 1.25- or 1.38-fold increased risk of CHD, respectively. These findings indicate that genetic variants of the EPRS gene may influence the individual susceptibility to CHD in the Chinese Han population.
Introduction
Congenital heart disease(CHD) is the most common human birth defect and the leading cause of perinatal mortality, with an incidence of approximately 6–8 per 1000 live births or even higher [1], [2], [3]. With the advances in surgical techniques, the prognosis of children with complicated and uncomplicated CHDs continues to improve, but the reported incidence remains unchanged [4]. The etiology of CHD is complex and possibly includes the interaction of inherited factors and environmental exposures [5], [6], [7]. A multitude of research studies have identified both chromosomal abnormality and gene mutations as causation for the syndromic heart malfunction [8]. However, the origin of non-syndromic CHD, which accounts for most of all congenital cardiac abnormalities, is waiting to be uncovered further.
Over the past decades, plenty of genes have been identified as candidates to be responsible for CHD [9], [10], [11]. However, aminoacyl-tRNA synthetases (ARSs) that seemed to be in charge of only cellular protein synthesis were overlooked. ARSs catalyze the attachment of amino acids to their cognate tRNAs with high fidelity [12], [13]. Recent research has shown that eukaryote ARSs, distinguished from their prokaryotic counterparts, have additional domains and motifs such as glutathione S-transferase (GST), WHEP domains, leucine zipper domains, and α-helicalappendices that function beyond translation [14] and may link with a variety of human diseases, such as cancer, neuronal pathologies, autoimmune disorders, and disrupted metabolic conditions [13], [15]. Recently, the nontranslational functions of vertebrate ARSs have been associated with cytoplasmic forms and nuclear and secreted extracellular forms that impact cardiovascular development pathways [16].
Eight core aminoacyl-tRNA synthetases (ARSs), bifunctional glutamyl-prolyl-tRNA synthetase (EPRS), isoleucyl-tRNA synthetase (IRS), leucyl-tRNA synthetase (LRS), methionyl-tRNA synthetase (MRS), glutaminyl-tRNA synthetase (QRS), lysyl-tRNA synthetase (KRS), aspartyl-tRNA synthetase (DRS), and arginyl-tRNA synthetase (RRS), form a macromolecular protein complex with three auxiliary factors, designated ARS-interacting multifunctional protein 1 (AIMP1), AIMP2 and AIMP3. This complex is known as the multisynthetase complex (MSC). The MSC may act as a depot for ARSs, which could be subsequently released from the macromolecular complexes to participate in auxiliary tasks beyond translation [17], generate a channel for the delivery of tRNAs [18], [19] and help the proofreading of newly synthesized nuclear tRNAs in the nucleus [20].
According to the expressed sequence tags (EST) profile in the public database UniGene (http://www.ncbi.nlm.nih.gov/UniGene), all eight of the core ARS coding genes were expressed in human heart tissues, with transcripts ranging from 44 to 502 per million (Figure S1). Thus, it is plausible that changes in the core ARSs may affect heart development and are related to the occurrence of CHD. However, to date, no research has reported a relation between the genetic variants of the core ARS genes and CHD susceptibility.
To determine the effect of genetic variants in the core ARS genes on CHD development, we conducted a case-control study by investigating the genotype frequency distribution of the 16 potential functional polymorphisms in the eight members of the MSC.
Materials and Methods
Ethics Statement
This study was approved by the institutional review board of Nanjing Medical University and adhered to the tenets of the Declaration of Helsinki. The design and performance of the current study involving human subjects were clearly described in a research protocol. All participants and/or their parents were voluntary and completed the informed consent in writing before taking part in this research.
Study populations
The case-control analysis included 984 affected children with sporadic CHD and 2953 unrelated non-CHD controls. All subjects were genetically unrelated ethnic Han Chinese. Subjects for the study were consecutively recruited from the Affiliated Nanjing Children’s Hospital of Nanjing Medical University and the First Affiliated Hospital of Nanjing Medical University, Nanjing, China, from March 2009 to December 2011. All CHD patients were diagnosed based on echocardiography, with some diagnoses further confirmed by cardiac catheterization and/or surgery. Potential study subjects were initially surveyed with a brief questionnaire at clinics to determine whether they were willing to participate in a research study; we then conducted a face-to-face interview to obtain demographic information. Cases that had clinical features of developmental syndromes, multiple major developmental anomalies or known chromosomal abnormalities were excluded. The exclusion criteria also included a positive family history of CHD in a first-degree relative (parents, siblings and children), maternal diabetes mellitus, phenylketonuria, maternal teratogen exposure (e.g., pesticides and organic solvents), and maternal therapeutic drug exposure during the intrauterine period. Controls were non-CHD outpatients from the same geographic areas. They were recruited from the hospitals listed above during the same time period. Controls with congenital anomalies or cardiac disease were excluded. For each participant, approximately 2 ml of whole blood was obtained to extract genomic DNA for genotyping analysis.
SNP selection and genotyping
Eight ARSs (EPRS, MRS, QRS, RRS, IRS, LRS, KRS, DRS) that formed MSC were selected. For each ARS-coding gene, we first used the public HapMap single nucleotide polymorphism (SNP) database (phase II+ III Feb 09, on NCBI B36 assembly and dbSNP b126) to search for SNPs that localized within gene regions, with MAF≥0.05, in the Chinese Han population. Then, a web-based analysis tool was used to predict the function of these SNPs (http://snpinfo.niehs.nih.gov/snpinfo/snpfunc.htm). Finally, a total of 27 potentially functional SNPs were selected in 8 ARS-coding genes. We next conducted linkage disequilibrium (LD) analysis by the Haploview 4.2 software, and only one SNP was selected in the case of multiple SNPs in the same haplotype block (r2>0.8). Eighteen (rs1061160, rs1061248, rs2230301 and rs5030754 in EPRS; rs508904 in MRS; rs193466, rs2305737 and rs244903 in RRS; rs1058751, rs10820966 and rs556155 in IRS; rs10988 in LRS; rs2233805 and rs3784929 in KRS; rs2164331, rs309142, rs309143 and rs6738266 in DRS) of 27 SNPs remained. Two SNPs (rs6738266 and rs2164331) were excluded due to primer design failure.
Genomic DNA was isolated from leukocyte pellets of venous blood by proteinase K digestion, followed by phenol-chloroform extraction and ethanol precipitation. Nanodrop and DNA electrophoresis were used to check the quality and quantity of DNA samples before genotyping. The genotyping was performed by Illumina Infinium BeadChip (Illumina, Inc.). All SNPs were successfully genotyped with call rates >95% ( Table 1 ).
Table 1. Primary information for 16 functional SNPs in ARS-coding genes.
| Gene | ARS | Chr. (cytoband) | SNP | Position (bp)a | Location | Predicted functionb | MAFc | Allelec | HWEd | Genotyping call rate (%) |
| EPRS | Glutamyl-prolyl-tRNA synthetase | 1q41 | rs1061248 | 219968681 | 3′UTR | miRNAe | 0.378 | G/A | 0.63 | 99.5 |
| rs1061160 | 219981426 | exon | Splice sitesf | 0.366 | G/A | 0.76 | 99.8 | |||
| rs5030754 | 219983387 | exon | Splice sites | 0.012 | G/A | 0.82 | 99.9 | |||
| rs2230301 | 220024283 | exon | Splice sites, nsSNPg | 0.073 | A/C | 0.90 | 99.9 | |||
| DRS | Aspartyl-tRNA synthetase | 2q21.3 | rs309143 | 135956608 | intron | TFBSh | 0.207 | A/G | 0.40 | 99.8 |
| rs309142 | 1395957754 | intron | TFBS | 0.488 | A/G | 0.29 | 99.8 | |||
| LRS | Leucyl-tRNA synthetase | 5q32 | rs10988 | 146120433 | exon | nsSNP | 0.280 | G/A | 0.04 | 99.9 |
| RRS | Arginyl-tRNA synthetase | 5q34 | rs244903 | 168486505 | exon | Splice sites, nsSNP | 0.146 | A/G | 0.35 | 99.7 |
| rs193466 | 1684865598 | intron | TFBS | 0.305 | A/G | 0.58 | 99.7 | |||
| rs2305737 | 168519256 | 3′UTR | Splice sites, miRNA | 0.146 | C/A | 0.71 | 99.8 | |||
| IRS | Isoleucyl-tRNA synthetase | 9q22.31 | rs1058751 | 92210634 | 3′UTR | miRNA | 0.122 | A/T | 0.22 | 99.5 |
| rs556155 | 92223355 | exon | nsSNP | 0.195 | A/G | 0.25 | 99.9 | |||
| rs10820966 | 92293147 | intron | TFBS | 0.078 | A/T | 0.16 | 99.9 | |||
| MRS | Methionyl-tRNA synthetase | 12q13.3 | rs508904 | 57488311 | intron | TFBS | 0.289 | A/G | 0.70 | 99.8 |
| KRS | Lysyl-tRNA synthetase | 16q23.1 | rs3784929 | 75643129 | intron | TFBS | 0.159 | G/A | 0.66 | 99.8 |
| rs2233805 | 75647244 | intron | TFBS | 0.049 | A/G | 1.00 | 99.9 |
Derived from the UCSC Genome Browser on Human Feb. 2009 (GRCh37/hg19) Assembly (http://genome.ucsc.edu/);
Derived from an online tool-SNPinfo (http://snpinfo.niehs.nih.gov/snpfunc.htm);
Major/minor allele;
Hardy-Weinberg equilibrium test among controls;
miRNA: microRNA
Splice sites: Exonic splicing enhancer (ESE) or exonic splicing silencer (ESS) binding sites;
nsSNP: non-synonymous polymorphisms.
TFBS: Transcription factor binding sites;
Statistical analyses
The differences between the CHD patients and control subjects were evaluated in the distributions of demographic characteristics, selected variables, and frequencies of genotypes of the 16 polymorphisms using Student’s t-test (for continuous variables) or the χ2 test (for categorical variables). The χ2 test determined the Hardy-Weinberg equilibrium of the genotype distribution of polymorphisms in the control group. LD between SNPs was evaluated using Haploview 4.2.Odds ratios (ORs) and 95% confidence intervals (CIs) were estimated by logistic regression analyses in the additive model to estimate the associations between the variants genotypes and risk of CHD. Chi-square-based Q-test was applied to test the heterogeneity of associations between subgroups, and the heterogeneity was considered significant when P<0.05. All statistical analyses were performed using the Statistical Analysis System software (v.9.1.3; SAS Institute, Cary, NC, USA). All tests were two-sided, and P<0.05 was considered significant.
Results
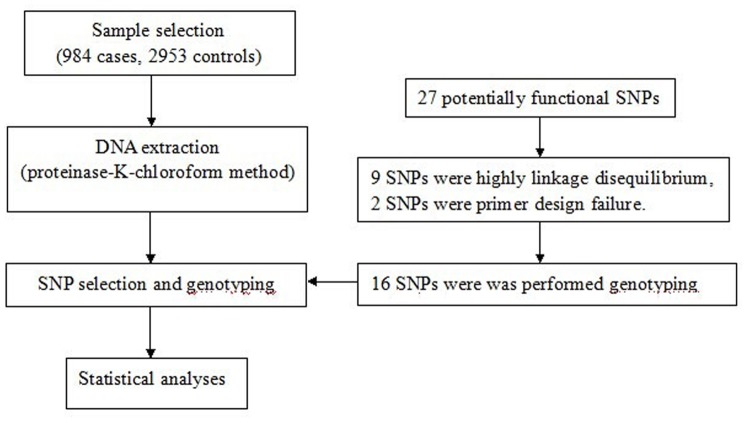
An overview of the study design using a flowchart was performed as shown in Figure 1 . We systematically investigated the association of potentially functional SNPs with CHD susceptibility in 984 cases and 2953 controls in a Chinese population. There were no statistically significant differences for the distributions of age and gender between cases and controls (P = 0.261 and P = 0.832, respectively). Among the 984 CHD patients, 312 had atrial septal defect (ASD), 585 were diagnosed with ventricular septal defect (VSD), and 87 were diagnosed with ASD combined with VSD.
Figure 1. Study design procedures for association of ARSs gene polymorphisms with the risk of congenital heart disease in the Chinese Han population.
The genotype distributions of the 16 SNPs and the associations with CHD risk are summarized in Table 2 . The observed genotype frequencies of these SNPs were in agreement with Hardy-Weinberg equilibrium in the controls (P value from 0.16 to 1.00) except rs10988 (P = 0.04). Among the 16 SNPs, significant associations were observed between 4 SNPs (rs1061248, rs1061160, rs5030754 and rs2230301) and CHD risk following a logistic regression analysis in the additive model. All four SNPs were in the ERPS gene. The G allele of rs1061248 and the A allele of rs2230301 were associated with a decreased risk of CHD [additive model: odds ratio (OR) = 0.90, 95% confidence interval (CI) = 0.81–0.99, P = 3.81×10−2; and OR = 0.73, 95%CI = 0.60–0.90, P = 3.53×10−3, respectively]; however, the G allele of rs1061160 and the G allele of rs5030754 were associated with an increased risk of CHD (OR = 1.18, 95%CI = 1.06–1.31, P = 1.28×10−3; and OR = 1.39, 95%CI = 1.11–1.75, P = 4.47×10−3, respectively). We further calculated P values for the false discovery rate to perform multiple comparisons. After comparisons, we found that rs2230301, rs5030754 and rs1061160 correlated with CHD risk, whereas rs1061248 lost its significant association with the risk of CHD. In contrast, no obvious evidence of a significant association between the other 12 SNPs and CHD risk was found.
Table 2. Summary of associations between 16 SNPs of MSC genes with congenital heart disease.
| Chr. | Gene | SNP | Allelea | Caseb (N = 984) | Controlb (N = 2953) | MAFc | HWEd | Additive model | P FDR e | ||
| (cytoband) | Cases | Controls | OR(95%CI) | P | |||||||
| 1q41 | EPRS | rs1061248 | G/A | 217/486/271 | 741/1459/744 | 0.47 | 0.50 | 0.63 | 0.90 (0.81–0.99) | 3.81×10 −2 | 0.152 |
| rs1061160 | G/A | 184/486/311 | 465/1402/1083 | 0.44 | 0.40 | 0.76 | 1.18 (1.06–1.31) | 1.82×10 −3 | 0.029 | ||
| rs5030754 | G/A | 1/113/869 | 6/241/2704 | 0.06 | 0.04 | 0.82 | 1.39 (1.11–1.75) | 4.47×10 −3 | 0.023 | ||
| rs2230301 | A/C | 3/114/865 | 20/440/2490 | 0.06 | 0.08 | 0.90 | 0.73 (0.60–0.90) | 3.53×10 −3 | 0.028 | ||
| 2q21.3 | DRS | rs309143 | A/G | 38/308/634 | 100/927/1921 | 0.20 | 0.19 | 0.40 | 1.03 (0.91–1.17) | 6.40×10−1 | 1.024 |
| rs309142 | A/G | 172/499/310 | 526/1472/949 | 0.43 | 0.43 | 0.29 | 1.01 (0.91–1.12) | 9.11×10−1 | 0.911 | ||
| 5q32 | LRS | rs10988 | G/A | 47/340/596 | 120/1041/1789 | 0.22 | 0.22 | 0.04 | 1.02 (0.90–1.16) | 7.32×10−1 | 0.901 |
| 5q34 | RRS | rs244903 | A/G | 15/213/754 | 50/622/2270 | 0.12 | 0.12 | 0.35 | 1.01 (0.87–1.18) | 9.06×10−1 | 0.966 |
| rs193466 | A/G | 74/399/509 | 229/1162/1553 | 0.28 | 0.28 | 0.58 | 1.02 (0.91–1.14) | 7.72×10−1 | 0.505 | ||
| rs2305737 | C/A | 20/270/691 | 60/744/2145 | 0.16 | 0.15 | 0.71 | 1.10 (0.95–1.26) | 2.12×10−1 | 0.565 | ||
| 9q22.31 | IRS | rs1058751 | A/T | 0/150/828 | 11/412/2516 | 0.08 | 0.07 | 0.22 | 1.04 (0.86–1.27) | 6.71×10−1 | 0.976 |
| rs556155 | A/G | 16/206/760 | 34/627/2291 | 0.12 | 0.12 | 0.25 | 1.03 (0.88–1.21) | 6.79×10−1 | 0.905 | ||
| rs10820966 | A/T | 10/213/760 | 40/681/2230 | 0.12 | 0.13 | 0.16 | 0.91 (0.77–1.06) | 2.21×10−1 | 0.505 | ||
| 12q13.3 | MRS | rs508904 | A/G | 105/442/433 | 301/1268/1380 | 0.33 | 0.32 | 0.70 | 1.07 (0.96–1.20) | 2.01×10−1 | 0.643 |
| 16q23.1 | KRS | rs3784929 | G/A | 33/302/644 | 95/849/2005 | 0.19 | 0.18 | 0.66 | 1.08 (0.95–1.23) | 2.40×10−1 | 0.480 |
| rs2233805 | A/G | 0/31/951 | 1/106/2844 | 0.02 | 0.02 | 1.00 | 0.86 (0.57–1.29) | 4.63×10−1 | 0.823 | ||
Major/minor allele;
Variant homozygote/Heterozygote/Wild type homozygote;
Minor allele frequency among cases/controls;
Hardy-Weinberg equilibrium test among controls.
Multiple comparisons P values for false discovery rate.
We have listed the results of the genotypic association analysis in Table 3 . In dominant genetic model, for rs1061160 and rs5030754 polymorphisms, AG+AA and AG+GG genotypes were associated with an increased risk of CHD compared with the GG genotype, respectively(OR = 1.25, 95%CI = 1.07–1.46; OR = 1.44, 95%CI = 1.14–1.82). For rs2230301 polymorphism, AC+CC genotypes were associated with a decreased risk of CHD compared with the AA genotype(OR = 0.73, 95% CI = 0.59–0.91).
Table 3. Summary of genotypic association analysis of four SNPs of the EPRS gene with congenital heart disease.
| SNP | Genotype | Cases | Controls | OR (95%CI) | P |
| rs1061248 | GG | 271 | 744 | 1.00 | – |
| AG | 486 | 1459 | 0.91 (0.77–1.09) | 3.11×10−1 | |
| AA | 217 | 741 | 0.80 (0.65–0.99) | 3.74×10 −2 | |
| AG+AA | 703 | 2200 | 0.88 (0.75–1.03) | 1.15×10−1 | |
| rs1061160 | GG | 311 | 1083 | 1.00 | – |
| AG | 486 | 1402 | 1.21 (1.03–1.42) | 2.35×10 −2 | |
| AA | 164 | 465 | 1.38 (1.11–1.70) | 3.07×10 −3 | |
| AG+AA | 650 | 1867 | 1.25 (1.07–1.46) | 4.54×10 −3 | |
| rs5030754 | GG | 869 | 2704 | 1.00 | – |
| AG | 113 | 241 | 1.46 (1.15–1.85) | 1.72×10 −3 | |
| AA | 1 | 6 | 0.52 (0.06–4.31) | 5.44×10−1 | |
| AG+GG | 114 | 247 | 1.44 (1.14–1.82) | 2.51×10 −3 | |
| rs2230301 | AA | 865 | 2490 | 1.00 | – |
| AC | 114 | 440 | 0.75 (0.60–0.93) | 8.99×10 −3 | |
| CC | 3 | 20 | 0.43 (0.13–1.46) | 1.76×10−1 | |
| AC+CC | 117 | 460 | 0.73 (0.59–0.91) | 4.90×10 −3 |
Additionally, we performed haplotype analysis ( Table 4 ). As shown, the haplotype “GAAA” (combination of risk alleles of the four SNPs) was associated with an increased risk of CHD, whereas the protective allele combination “AGGC” was associated with a decreased risk of CHD. In the stratification analysis, we further evaluated the associations of the four SNPs in EPRS with CHD risk in subgroups stratified by gender and specific CHD phenotypes. As shown in Table 5 , similar effects were observed among the subgroups.
Table 4. The haplotypic association of the four SNPs of the EPRS gene with congenital heart disease.
| Haplotypea | case (%) | control (%) | OR (95%CI) | P |
| AGGA | 905 (45.99) | 2835 (48.0) | 1.00 (referent) | |
| GAGA | 740 (37.60) | 2075 (35.13) | 1.12 (0.99–1.25) | 5.34×10−2 |
| GGGC | 100 (5.08) | 370 (6.26) | 0.85 (0.67–1.07) | 1.62×10−1 |
| GGGA | 84 (4.27) | 259 (4.39) | 1.02 (0.79–1.31) | 9.04×10−1 |
| GAAA | 113 (5.74) | 253 (4.28) | 1.40 (1.11–1.77) | 4.91×10 −3 |
| AGGC | 20 (1.02) | 107 (1.81) | 0.59 (0.36–0.95) | 3.00×10 −2 |
| Others | 6 (0.30) | 7 (0.12) | 2.69 (0.90–8.01) | 7.65×10−2 |
SNP order: rs1061248, rs1061160, rs5030754 and rs2230301.
Table 5. Stratified analysis on the associations between four SNPs in EPRS with congenital heart disease.
| Characteristics | rs1061248 | rs1061160 | ||||||||
| Case a | Control a | OR (95%CI) b | P b | P c | Case a | Control a | OR (95%CI) b | P b | P c | |
| Gender | ||||||||||
| male | 105/241/138 | 472/856/464 | 0.87 (0.76–1.00) | 4.90×10−2 | 0.527 | 91/237/160 | 286/844/667 | 1.16 (1.00–1.33) | 4.79×10−2 | 0.751 |
| female | 112/245/133 | 269/603/280 | 0.93 (0.80–1.09) | 3.74×10−1 | 93/249/151 | 179/558/416 | 1.20 (1.03–1.40) | 1.83×10−2 | ||
| Diagnostic groups | ||||||||||
| ASD | 71/139/98 | 741/1459/744 | 0.84 (0.71–0.99) | 4.27×10−2 | 0.483 | 68/146/98 | 465/1402/1083 | 1.26 (1.07–1.49) | 6.44×10−3 | 0.489 |
| VSD | 126/299/154 | 741/1459/744 | 0.91 (0.80–1.03) | 1.41×10−1 | 104/294/184 | 465/1402/1083 | 1.16 (1.02–1.32) | 2.21×10−2 | ||
| ASD/VSD | 20/48/19 | 741/1459/744 | 1.03 (0.76–1.39) | 8.71×10−1 | 12/46/29 | 465/1402/1083 | 1.03 (0.76–1.40) | 8.52×10−1 | ||
| Characteristics | rs5030754 | rs2230301 | ||||||||
| Case a | Control a | OR(95%CI) b | P b | P c | Case a | Control a | OR(95%CI) b | P b | P c | |
| Gender | ||||||||||
| male | 1/58/429 | 4/135/1659 | 1.58 (1.16–2.15) | 3.90×10−3 | 0.211 | 3/64/421 | 13/280/1504 | 0.83 (0.63–1.09) | 1.77×10−1 | 0.260 |
| female | 0/55/440 | 2/106/1045 | 1.18 (0.84–1.65) | 3.37×10−1 | 0/50/444 | 7/160/986 | 0.65 (0.47–0.90) | 9.74×10−3 | ||
| Diagnostic groups | ||||||||||
| ASD | 0/42/270 | 6/241/2704 | 1.62 (1.15–2.27) | 5.54×10−3 | 0.119 | 0/31/280 | 20/440/2490 | 0.59 (0.41–0.86) | 5.93×10−3 | 0.281 |
| VSD | 1/67/516 | 6/241/2704 | 1.40 (1.07–1.85) | 1.56×10−2 | 2/76/506 | 20/440/2490 | 0.83 (0.65–1.06) | 1.37×10−1 | ||
| ASD/VSD | 0/4/83 | 6/241/2704 | 0.53 (0.19–1.43) | 2.08×10−1 | 1/7/79 | 20/440/2490 | 0.62 (0.31–1.21) | 1.62×10−1 | ||
Major/minor allele;
Calculated by additive model;
P for heterogeneity.
We also conducted a combined analysis of the four promising SNPs to test their joint effects on CHD risk. There was a significant dosage-response effect among individuals carrying the different number of risk alleles and CHD risk (P trend = 5.00×10−4). Compared with individuals with “0–2” risk allele, those carrying “3”, “4” or “5 or more” risk alleles had a 0.97- (95% CI = 0.69–1.37), 1.25- (95% CI = 1.05–1.50) or 1.38-fold (95% CI = 1.14–1.68) increased risk of CHD, respectively ( Table 6 ).
Table 6. Combined effects of rs1061248, rs1061160, rs1061160, and rs2230301 on CHD.
| Number of risk allelesa | Case (%) | Control (%) | OR (95% CI)b | P b |
| 0–2 | 262 (26.95) | 939 (31.96) | 1.00 | |
| 3 | 50 (5.14) | 184 (6.26) | 0.97 (0.69–1.37) | 8.79×10−1 |
| 4 | 384 (39.51) | 1098 (37.37) | 1.25 (1.05–1.50) | 1.37×10−2 |
| ≥5 | 276 (28.40) | 717 (24.40) | 1.38 (1.14–1.68) | 1.20×10−3 |
| Trend | 5.00×10−4 |
rs1061248 G, rs1061160 A, rs5030754 A, and rs2230301 A were assumed as risk alleles;
Calculated by additive model.
Discussion
In this study, we systematically investigated the association of potentially functional SNPs in ARS-coding genes of the MSC with CHD susceptibility in 984 cases and 2953 controls in a Chinese population. We observed significant association of four SNPs (rs1061248, rs1061160, rs5030754 and rs2230301) in the EPRS gene with the risk of CHD, and the risk remarkably accelerated in the individuals who carried more risk alleles. Although ASD and VSD represent the most common congenital heart malfunctions, the accurate pathogenesis is poorly understood. Based on previous research, the ARS-coding genes of MSC take part in diverse functional activities, and some of them have been proven to be crucial for heart development and proper functioning. Few studies have linked the variants of MSC genes to congenital heart disease. To our knowledge, we provide the first evidence that SNPs in EPRS, one of the core coding genes in MSC, may modulate the process of CHD.
Some ARSs in MSC have been demonstrated to have a close correlation with cardiovascular development. Glutamyl-prolyl-tRNA synthetase (EPRS) is a bifunctional enzyme that could translationally suppress vascular endothelial growth factor-A (VEGF-A) to regulate angiogenesis [21] and seems to act as a key gatekeeper of inflammatory gene translation [22]. Lysyl-tRNA synthetase (KRS) is secreted to trigger pro-inflammatory response [23] and plays a key role via Ap4A as an important signaling molecule in the transcriptional activity of microphthalmia transcription factor(MITF) [24], which has been demonstrated to be necessary in heart growth [25]. Glutaminyl-tRNA synthetase (QRS) can bind and inhibit the apoptotic activity of apoptosis signal-regulating kinase 1 (ASK1) [26], which has been demonstrated to be a new intracellular regulator of p38 MAPK activation in cardiac myogenic differentiation [27]. Han and colleagues [28] reported that Leucyl-tRNA synthetase (LRS) acts as a vital mediator for amino acid signaling to mTORC1, and the latter has been found to be related to the normal development of cardiovascular tissue [29].
Human EPRS, the largest polypeptide from the complex, is a bifunctional enzyme in which the two domains exhibiting each catalytic activity are linked by three tandem WHEP motifs [30]. EPRS contains 29 exons and 28 introns. In response to interferon-γ (IFNγ), EPRS is phosphorylated and released from its residence in the MSC. MSC then forms another multi-component complex, known as IFN-γ–activated inhibitor of translation (GAIT), with other regulatory proteins at a 3′UTR region that is involved in the translational silencing of target transcripts, such as VEGF-A [31], [32], [33]. As documented in many studies, VEGF-A shares a close relationship with CHD, and both the increased and decreased expression of VEGF-A during heart development can result in various CHD [34], [35], [36]. The SNP rs2230301, a missense SNP located at the 23rd exon of the EPRS gene, may act as a part of the exonic splicing enhancer based on the online tool SNPinfo [37]. The missense mutation would change the sequence of EPRS and may lead to protein misfolding and malfunction. We used a web-based analysis tool to predict the potential function of the SNPs, and rs2230301 was predicted to be a missense variant that may result in an amino acid alteration from aspartic acid (Asp) to glutamic acid (Glu) (http://snpinfo.niehs.nih.gov/snpinfo/snpfunc.htm). The NCBI database confirmed the results (http://www.ncbi.nlm.nih.gov/). However, the predicted results differed from the in-silico analysis. To further validate the function of this variant, some functional studies should be performed in some follow-up studies. The SNP rs1061248 is located at the 3′ regulatory region of the EPRS gene with a predicted function as a MicroRNA-binding site. Considering its potentially functional role, it is likely that this polymorphism might alter miRNA binding, thereby modulating the biological function of EPRS. The two synonymous SNPs rs1061160 and rs5030754 were localized on the seventh exon and the eleventh exon, respectively. Recently, a synonymous SNP was reported to alter the function of the protein in certain circumstances [38].
Several limitations of the present study need to be addressed. First, we did not replicate the results in additional individuals; this may contribute to potential false positive errors. The present analysis was restricted to individuals of Chinese Han descent, and therefore, the findings may not hold true for individuals of other races and ethnicities. Additionally, the limited sample size may contribute to the failed validation in the stratified analysis concerning the association between the SNPs and CHD. We performed the statistic power analysis of the significant SNPs in the studied population. The powers of three SNPs (rs1061248, rs5030754, and rs2230301) are lower than 0.6 because the sample size of our study is relatively small (984 CHD cases and 2953 non-CHD controls) and the effects of our target common SNPs are weak. Further replication of the association signal in an independent cohort for the four SNPs would support the conclusions. Therefore, the results are required to be further replicated by well-designed studies in additional large-scale Chinese Han populations.
In conclusion, we conducted a case-control study to investigate the role of genetic variants in ARS-coding genes of MSC in the development of CHD in a Chinese population. We observed that four SNPs (rs1061248, rs1061160, rs5030754, and rs2230301) in the EPRS gene may confer susceptibility to sporadic CHD and that the risk significantly increased with the number of risk alleles. However, further studies with functional evaluations are warranted to elucidate the potentially biological mechanisms of these polymorphisms in the development of CHD.
Supporting Information
Expressed sequence tags (EST) profile of the 8 core ARSs coding genes.
(DOC)
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported in part by National Natural Science Foundation of China (grant numbers 81370277 and 81300128); Jiangsu Provincial Special Program of Medical Science (grant number BL2013013); Ph.D. Programs Foundation of Ministry of Education of China (grant number 20123234120015); and Jiangsu Natural Science Foundation (grant number BK20131025). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Hoffman JI, Kaplan S (2002) The incidence of congenital heart disease. J Am Coll Cardiol 39: 1890–1900. [DOI] [PubMed] [Google Scholar]
- 2. Botto LD, Correa A, Erickson JD (2001) Racial and temporal variations in the prevalence of heart defects. Pediatrics 107: E32. [DOI] [PubMed] [Google Scholar]
- 3. Rosamond W, Flegal K, Friday G, Furie K, Go A, et al. (2007) Heart disease and stroke statistics–2007 update: a report from the American Heart Association Statistics Committee and Stroke Statistics Subcommittee. Circulation 115: e69–171. [DOI] [PubMed] [Google Scholar]
- 4. Gatzoulis MA (2004) Adult congenital heart disease: a cardiovascular area of growth in urgent need of additional resource allocation. Int J Cardiol 97 Suppl 1 1–2. [DOI] [PubMed] [Google Scholar]
- 5. van der Bom T, Zomer AC, Zwinderman AH, Meijboom FJ, Bouma BJ, et al. (2011) The changing epidemiology of congenital heart disease. Nat Rev Cardiol 8: 50–60. [DOI] [PubMed] [Google Scholar]
- 6. Jenkins KJ, Correa A, Feinstein JA, Botto L, Britt AE, et al. (2007) Noninherited risk factors and congenital cardiovascular defects: current knowledge: a scientific statement from the American Heart Association Council on Cardiovascular Disease in the Young: endorsed by the American Academy of Pediatrics. Circulation 115: 2995–3014. [DOI] [PubMed] [Google Scholar]
- 7. Huhta J, Linask KK (2013) Environmental origins of congenital heart disease: the heart-placenta connection. Semin Fetal Neonatal Med 18: 245–250. [DOI] [PubMed] [Google Scholar]
- 8. Pierpont ME, Basson CT, Benson DW Jr, Gelb BD, Giglia TM, et al. (2007) Genetic basis for congenital heart defects: current knowledge: a scientific statement from the American Heart Association Congenital Cardiac Defects Committee, Council on Cardiovascular Disease in the Young: endorsed by the American Academy of Pediatrics. Circulation 115: 3015–3038. [DOI] [PubMed] [Google Scholar]
- 9. Wessels MW, Willems PJ (2010) Genetic factors in non-syndromic congenital heart malformations. Clin Genet 78: 103–123. [DOI] [PubMed] [Google Scholar]
- 10. Wolf M, Basson CT (2010) The molecular genetics of congenital heart disease: a review of recent developments. Curr Opin Cardiol 25: 192–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Garg V (2006) Insights into the genetic basis of congenital heart disease. Cell Mol Life Sci 63: 1141–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schimmel P (1987) Aminoacyl tRNA synthetases: general scheme of structure-function relationships in the polypeptides and recognition of transfer RNAs. Annu Rev Biochem 56: 125–158. [DOI] [PubMed] [Google Scholar]
- 13. Park SG, Ewalt KL, Kim S (2005) Functional expansion of aminoacyl-tRNA synthetases and their interacting factors: new perspectives on housekeepers. Trends Biochem Sci 30: 569–574. [DOI] [PubMed] [Google Scholar]
- 14. Guo M, Yang XL, Schimmel P (2010) New functions of aminoacyl-tRNA synthetases beyond translation. Nat Rev Mol Cell Biol 11: 668–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Park SG, Schimmel P, Kim S (2008) Aminoacyl tRNA synthetases and their connections to disease. Proc Natl Acad Sci U S A 105: 11043–11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Guo M, Schimmel P (2013) Essential nontranslational functions of tRNA synthetases. Nat Chem Biol 9: 145–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ray PS, Arif A, Fox PL (2007) Macromolecular complexes as depots for releasable regulatory proteins. Trends Biochem Sci 32: 158–164. [DOI] [PubMed] [Google Scholar]
- 18. Simos G, Grosshans H, Hurt E (2002) Nuclear export of tRNA. Results Probl Cell Differ 35: 115–131. [DOI] [PubMed] [Google Scholar]
- 19. Hopper AK, Phizicky EM (2003) tRNA transfers to the limelight. Genes Dev 17: 162–180. [DOI] [PubMed] [Google Scholar]
- 20. Nathanson L, Deutscher MP (2000) Active aminoacyl-tRNA synthetases are present in nuclei as a high molecular weight multienzyme complex. J Biol Chem 275: 31559–31562. [DOI] [PubMed] [Google Scholar]
- 21. Ray PS, Fox PL (2007) A post-transcriptional pathway represses monocyte VEGF-A expression and angiogenic activity. EMBO J. 26: 3360–3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mukhopadhyay R, Jia J, Arif A, Ray PS, Fox PL (2009) The GAIT system: a gatekeeper of inflammatory gene expression. Trends Biochem Sci 34: 324–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Park SG, Kim HJ, Min YH, Choi EC, Shin YK, et al. (2005) Human lysyl-tRNA synthetase is secreted to trigger proinflammatory response. Proc Natl Acad Sci U S A 102: 6356–6361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lee YN, Nechushtan H, Figov N, Razin E (2004) The function of lysyl-tRNA synthetase and Ap4A as signaling regulators of MITF activity in FcepsilonRI-activated mast cells. Immunity 20: 145–151. [DOI] [PubMed] [Google Scholar]
- 25. Tshori S, Gilon D, Beeri R, Nechushtan H, Kaluzhny D, et al. (2006) Transcription factor MITF regulates cardiac growth and hypertrophy. J Clin Invest 116: 2673–2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Aizenshtat LI, Rozenblyum YZ (1975) The OPD-1 trial frame for children. Biomed Eng (NY) 8: 177–178. [DOI] [PubMed] [Google Scholar]
- 27. Choi TG, Lee J, Ha J, Kim SS (2011) Apoptosis signal-regulating kinase 1 is an intracellular inducer of p38 MAPK-mediated myogenic signalling in cardiac myoblasts. Biochim Biophys Acta 1813: 1412–1421. [DOI] [PubMed] [Google Scholar]
- 28. Han JM, Jeong SJ, Park MC, Kim G, Kwon NH, et al. (2012) Leucyl-tRNA synthetase is an intracellular leucine sensor for the mTORC1-signaling pathway. Cell 149: 410–424. [DOI] [PubMed] [Google Scholar]
- 29. Malhowski AJ, Hira H, Bashiruddin S, Warburton R, Goto J, et al. (2011) Smooth muscle protein-22-mediated deletion of Tsc1 results in cardiac hypertrophy that is mTORC1-mediated and reversed by rapamycin. Hum Mol Genet 20: 1290–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rho SB, Lee JS, Jeong EJ, Kim KS, Kim YG, et al. (1998) A multifunctional repeated motif is present in human bifunctional tRNA synthetase. J Biol Chem 273: 11267–11273. [DOI] [PubMed] [Google Scholar]
- 31. Arif A, Jia J, Moodt RA, DiCorleto PE, Fox PL (2011) Phosphorylation of glutamyl-prolyl tRNA synthetase by cyclin-dependent kinase 5 dictates transcript-selective translational control. Proc Natl Acad Sci U S A 108: 1415–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sampath P, Mazumder B, Seshadri V, Gerber CA, Chavatte L, et al. (2004) Noncanonical function of glutamyl-prolyl-tRNA synthetase: gene-specific silencing of translation. Cell 119: 195–208. [DOI] [PubMed] [Google Scholar]
- 33. Ray PS, Jia J, Yao P, Majumder M, Hatzoglou M, et al. (2009) A stress-responsive RNA switch regulates VEGFA expression. Nature 457: 915–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhao W, Wang J, Shen J, Sun K, Zhu J, et al. (2010) Mutations in VEGFA are associated with congenital left ventricular outflow tract obstruction. Biochem Biophys Res Commun 396: 483–488. [DOI] [PubMed] [Google Scholar]
- 35. Ackerman C, Locke AE, Feingold E, Reshey B, Espana K, et al. (2012) An excess of deleterious variants in VEGF-A pathway genes in Down-syndrome-associated atrioventricular septal defects. Am J Hum Genet. 91: 646–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Vannay A, Vasarhelyi B, Kornyei M, Treszl A, Kozma G, et al. (2006) Single-nucleotide polymorphisms of VEGF gene are associated with risk of congenital valvuloseptal heart defects. Am Heart J 151: 878–881. [DOI] [PubMed] [Google Scholar]
- 37. Xu Z, Taylor JA (2009) SNPinfo: integrating GWAS and candidate gene information into functional SNP selection for genetic association studies. Nucleic Acids Res 37: W600–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kimchi-Sarfaty C, Oh JM, Kim IW, Sauna ZE, Calcagno AM, et al. (2007) A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science 315: 525–528. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expressed sequence tags (EST) profile of the 8 core ARSs coding genes.
(DOC)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.