Abstract
As mediators of adhesion, autoaggregation and bacteria-induced plasma membrane reorganization, type IV pili are at the heart of Neisseria meningitidis infection. Previous studies have proposed that two minor pilins, PilV and PilX, are displayed along the pilus structure and play a direct role in mediating these effects. In contrast with this hypothesis, combining imaging and biochemical approaches we found that PilV and PilX are located in the bacterial periplasm rather than along pilus fibers. Furthermore, preventing exit of these proteins from the periplasm by fusing them to the mCherry protein did not alter their function. Deletion of the pilV and pilX genes led to a decrease in the number, but not length, of pili displayed on the bacterial surface indicating a role in the initiation of pilus biogenesis. By finely regulating the expression of a central component of the piliation machinery, we show that the modest reductions in the number of pili are sufficient to recapitulate the phenotypes of the pilV and pilX mutants. We further show that specific type IV pili-dependent functions require different ranges of pili numbers.
Keywords: adhesion, infectious disease, meningitis, septicemia, type IV pili
See also: V Karuppiah & JP Derrick (August 2014)
Introduction
Type IV pili are filamentous organelles found on the surface of a large number of bacterial species including several pathogens (Pelicic, 2008). They can be found in different groups of proteobacteria in species such as Vibrio cholerae, enteropathogenic Escherichia coli, Pseudomonas aeruginosa, and Neisseria spp. More recently, they were also found in firmicutes such as Clostridium perfringens, illustrating the wide distribution of these structures (Varga et al, 2006). Type IV pili are part of a larger group of bacterial machineries. It is becoming increasingly clear that type II secretion systems share several features with type IV pili (Hobbs & Mattick, 1993; Giltner et al, 2012). Competence systems and flagella in archaea also appear to function through common mechanisms (Peabody et al, 2003). Type IV pili and related structures are thus one of the building blocks of the archeal and bacterial world.
The diversity and importance of functions carried out by type IV pili are illustrated by the case of Neisseria meningitidis. In 5–30% of the total human population, N. meningitidis thrives in the nasopharynx without triggering any damage (Caugant et al, 1994). Bacteria proliferate in bacterial aggregates, referred to as microcolonies, in tight association with the epithelial surface (Stephens et al, 1983). In vitro studies indicate that both adhesion to epithelial cells and auto aggregation are mostly mediated by type IV pili (Carbonnelle et al, 2006). In addition, DNA exchange between different N. meningitidis strains occurs via the natural transformation properties mediated by type IV pili (Weyand et al, 2013). From the throat, bacteria can occasionally access the bloodstream and trigger septic shocks and meningitis. In the circulation, bacteria bind to the endothelium and proliferate, often filling the vessel lumen (Guarner et al, 2004; Mairey et al, 2006). A recent study involving a humanized model of infection points to the importance of type IV pili in the ability of the bacteria to adhere to human vessels and to trigger vascular damage (Melican et al, 2013). Following adhesion to endothelial cells an intense crosstalk is initiated by type IV pili that leads to the reshaping of the host cell plasma membrane with the formation of filopodia-like protrusions (Eugene et al, 2002; Soyer et al, 2014). Type IV pili are therefore involved in numerous steps of the N. meningitidis infection process, in particular those involving interaction with the host cells.
Type IV pili are composed of one main component, the major pilin, PilE in Neisseria spp. This protein is composed of two domains, a hydrophobic alpha helix and a beta sheet head that contains a carboxy-terminal disulfide bond. The major pilin assembles into a helical fiber with the alpha helix buried inside the center of the structure (Parge et al, 1995; Craig et al, 2006). Determining how this fibrous organelle carries out such diverse functions remains a central challenge and requires a better understanding of pilus biogenesis. Genetic screens have revealed that building a fully functional type IV pilus requires over 20 proteins (Carbonnelle et al, 2006; Brown et al, 2010) although the function of most of these proteins remains unknown. After translocation to the periplasm via the general secretion pathway, pilins are cleaved by the PilD peptidase (Lory & Strom, 1997). Once cleaved, the major pilin, assembled in fibers in the periplasm, exits the outer membrane through a pore composed of the PilQ secretin (Wolfgang et al, 2000). The system is powered by several ATPases, which provide the energy for the extension or retraction of the pili fibers.
A subclass of proteins referred to as ‘minor pilins’ or ‘pilin-like proteins’ share structural features with the major pilin. These proteins contain a hydrophobic amino-terminal alpha helix and a carboxy-terminal disulfide bond (Winther-Larsen et al, 2005). Although a clear homology can be detected in the first 20–30 amino acids of the minor pilins, the rest of the primary sequence bears no homology, either among them or with the major pilin. Among this subclass, a triad of proteins, PilV, PilX, and ComP, displays the unique feature of conferring specific functions to pili rather than promoting pilus biogenesis. The corresponding mutants lose specific functions but still display type IV pili on their surface and maintain other functions. These three proteins and the major pilin also have in common a similar molecular weight range (15–20 kDa). The pilV, pilX, and comP genes are located in unrelated regions of the bacterial chromosome, however. Because type IV pili are still expressed by the corresponding mutants, these genes were qualified as ‘accessory’ to pilus biogenesis (Brown et al, 2010). In the absence of PilX, bacteria fail to aggregate, adhere to host cells, and trigger plasma membrane reorganization although they maintain competence (Helaine et al, 2005; Brissac et al, 2012). In the absence of the ComP minor pilin, competence is lost but pili are otherwise functional (Wolfgang et al, 1999). Finally, the pilV mutant adheres to host cells and is competent but fails to trigger plasma membrane reshaping upon bacterial adhesion (Mikaty et al, 2009). An attractive hypothesis is that these minor pilins are localized in the pilus fiber to carry out their effect (Helaine et al, 2007). In this view, the minor pilins are of central importance in pilus biology as they are the ‘effectors’ of this complex organelle. A number of reports have provided arguments in favor of this hypothesis. In particular, PilX was detected by immuno-electron microscopy associated with type IV pili (Helaine et al, 2007). The crystallographic structure of PilX showed the presence of a hook that could explain its role in aggregation by a homodimerization process (Helaine et al, 2007). In addition, the purified globular domain of PilV coated on the surface of beads was sufficient to lead to the accumulation of the β2-adrenergic receptor under the beads suggesting a direct role in triggering intracellular signals (Coureuil et al, 2010). In the case of ComP, the purified globular domain was recently shown to directly interact with DNA in a sequence-specific manner (Cehovin et al, 2013).
Although the genetic evidence is compelling, questions remain concerning the mode of action of these proteins. First, in terms of localization, pilus association was addressed by electron microscopy in the case of PilX but not for PilV or ComP. In addition, localization of a proportion of the protein at a given site does not necessarily reflect the actual site of action. Furthermore, recent evidence suggests that PilX has a global effect on the conformation of pili implying a possible indirect impact on pilus function (Brissac et al, 2012).
Because PilV and PilX are key players in type IV pili biology, the objective of this study was to determine how these minor pilins exert their functions. We provide evidence that these two minor pilins are required for efficient initiation of pilus biogenesis and that this effect is sufficient to explain the phenotype of the corresponding mutants.
Results
The majority of PilV and PilX proteins is localized in the periplasm rather than associated with pilus fibers
The low amounts of protein expression, together with structural similarities among the minor pilins and the major pilin, make determination of the localization of the minor pilins difficult. To address this technical difficulty, minor pilins were tagged with the Flag epitope (Munro & Pelham, 1984). This strategy offers high sensitivity and specificity due to the affinity of available antibodies directed against this peptide.
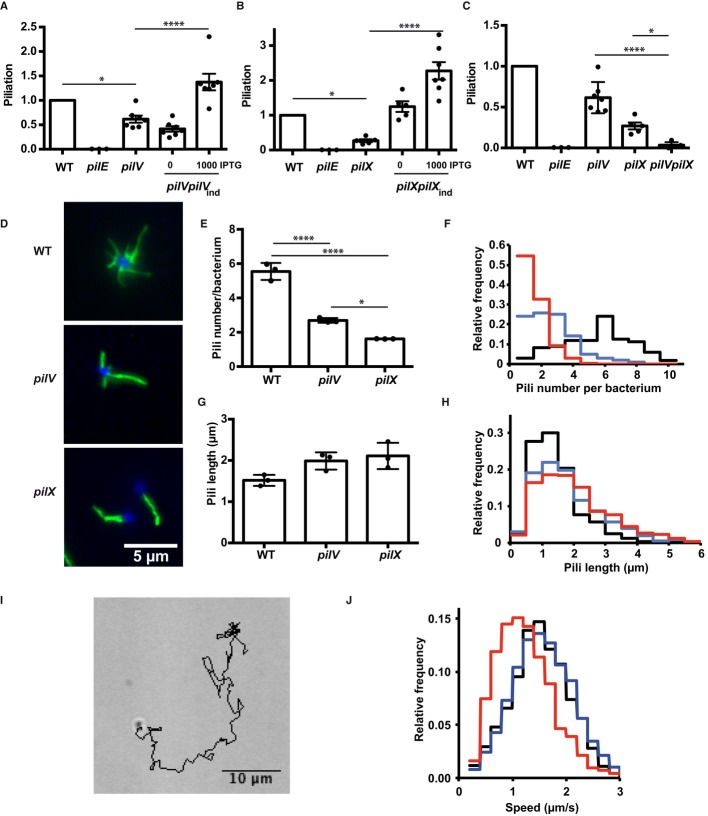
To determine the localization of PilV and PilX, strains expressing the minor pilins with a Flag tag placed in the loop formed by the disulfide bond (D-region) were generated. Flagged constructs were placed under the control of the lac promoter in the mutant background (pilVpilVFlagind and pilXpilXFlagind strains). Bacteria were cultured in the presence of 25 μM IPTG to mimic endogenous expression (Supplementary Fig S1). As a positive control, a Flag tag was inserted in the same site in the major pilin, PilE, and expressed under the control of the lac promoter (pilEpilEFlagind strain). Staining of the pilEpilEFlagind strain immobilized on coverslips with the anti-Flag monoclonal antibody showed fibers typical of type IV pili (Fig1A, left panel). In contrast, in the pilVpilVFlagind and the pilXpilXFlagind strain, Flag staining could not be detected despite repeated attempts using a highly sensitive back-illuminated camera (Evolve, Photometrics). Occasionally, a single bacterium on these samples showed a strong staining around the bacterial body, and we speculated that these bacteria may have ruptured outer membranes. Accordingly, bacteria with experimentally lyzed outer membranes showed intense staining around the bacterial body, indicative of a periplasmic localization of both of the tagged proteins (Fig1A, spheroplasts). To demonstrate the sensitivity of the technique, we co-expressed the pilEFlag construct under the control of the lac promoter with the endogenous pilE gene (pilEFlagind strain). In the presence of 10 μM of IPTG, reflecting a lower expression level than pilX or pilV genes, the tagged major pilin could be readily detected along type IV pili in the form of aligned dots (Fig1B). Importantly, in both cases, for pilV and pilX, complementation indicated that the Flag-tagged proteins were fully functional including at expression levels mimicking the endogenous expression. The Flag-tagged PilV was able to restore bacteria-induced plasma membrane reorganization upon adhesion to human umbilical endothelial cells (HUVECs, Fig1C), and the Flag-tagged PilX construct provided normal adhesion levels on the same cells (Fig1D). These results indicate that a large amount of these proteins are localized in the periplasm rather than inside the pilus fiber.
Figure 1. Periplasmic localization of the minor pilins PilV and PilX.
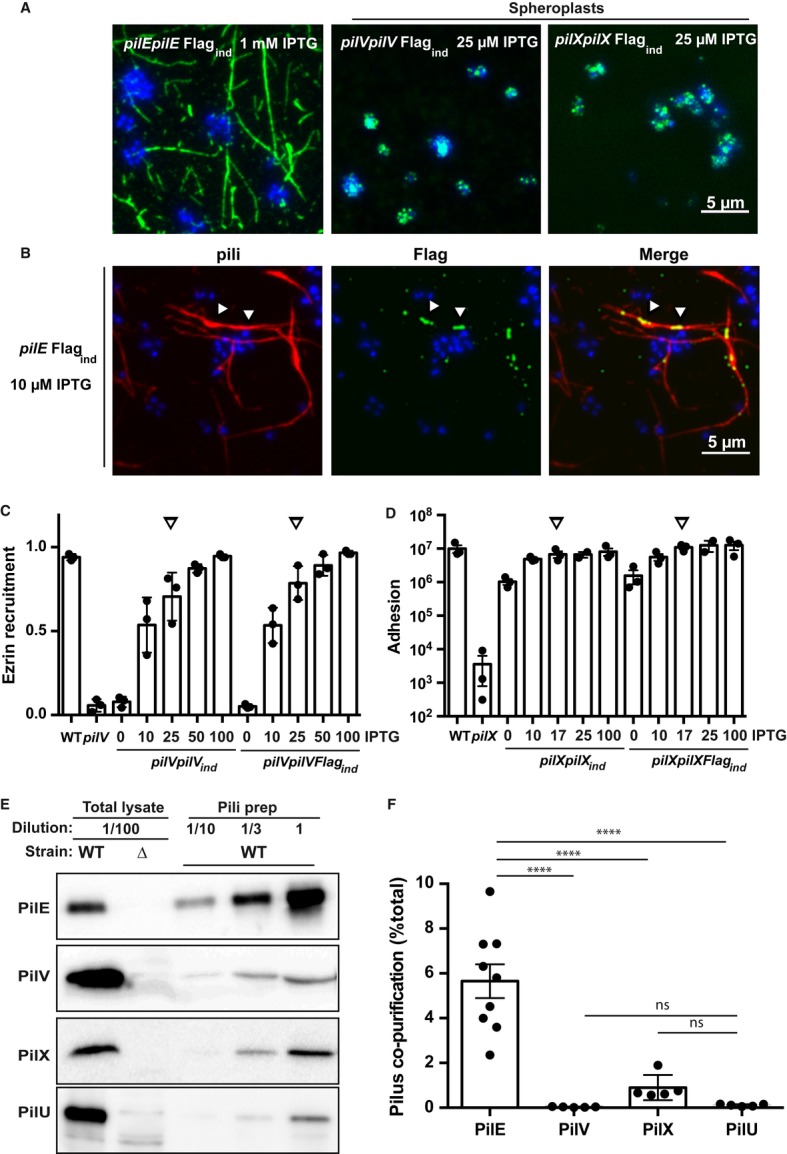
- Immunofluorescence detection of Flag-tagged PilE (left), PilV (middle), and PilX (right). Bacteria were visualized by staining their DNA with DAPI (blue), and anti-Flag detection appears in green. In the case of PilV and PilX, the outer membrane had to be removed thus generating spheroplasts to obtain a signal.
- Demonstration of the sensitivity of the immunofluorescence detection. Flag-tagged PilE was co-expressed with the endogenous PilE although at a low level. Detection was done in the presence of 10 μM IPTG generating a lower level of expression for the Flag-tagged protein than endogenous PilX or PilV levels. Bacteria are visualized in blue with DAPI, pili in red using the 20D9 monoclonal antibody, and the PilEFlag in green with the anti-Flag polyclonal antibody. Arrowheads indicate examples of co-localization of the PilEFlag signal and type IV pili.
- Ability of the Flag-tagged PilV (pilVpilVFlagind strain) to promote plasma membrane reshaping. Bacteria were incubated with endothelial cells for a period of 2 h, and the fraction of microcolonies with efficient plasma membrane reorganization was determined using ezrin as a marker. The amount of added inducer (IPTG, μM) is indicated. Arrows indicate conditions mimicking wild-type levels of expression. As in all the figures, each dot represents an independent experiment.
- Ability of the Flag-tagged PilX (pilXpilXFlagind strain) to promote adhesion. Bacteria were incubated with endothelial cells for a period of 4 h, and the number of cell-associated bacteria was determined.
- Pili from the wild-type strain were purified by mechanical shearing and ammonium sulfate precipitation, and the amounts of the proteins of interest were analyzed by Western blot. A total lysate diluted 100-fold from the starting bacterial suspension assesses the total amount of proteins, inside and outside of the bacteria. A total lysate from the corresponding mutant indicates the specificity of the detection. Different dilutions of the result of the pilus preparation were loaded (tenfold, 1/10; threefold 1/3, and undiluted, 1).
- Quantitative analysis of pilus association. Intensity of the bands on the immunoblots generated by pilus purification was analyzed, and the proportion of pilus-associated protein was plotted relative to the total. Each dot represents one experiment.
Data information: Data represent mean ± SEM; ****P ≤ 0.0001; ns, not significant.
In order to determine the proportion of periplasmic versus pili-associated minor pilins, we took advantage of a well-established pilus purification approach based on mechanical shearing and ammonium sulfate precipitation (Wolfgang et al, 1998). A number of reports describe the co-purification of a certain amount of PilV and PilX with type IV pili but the proportion of co-purified minor pilins relative to the total amount of these proteins is unknown (Winther-Larsen et al, 2001; Helaine et al, 2005). Pili were purified from wild-type bacteria, and the amount of PilV, PilX and the major pilin PilE were determined in the pilus fraction as well as in the initial bacterial suspension (Fig1E). Quantitative analysis of Western blot signals determined that 5.6 ± 0.7% of the total major pilin could be purified in these conditions reflecting the purification efficiency and the balance between periplasmic and pili-associated PilE (Fig1F). In contrast, in the case of PilV and PilX, the proportion of pilus-associated protein was much lower with 0.03 ± 0.01 and 0.9 ± 0.3%, respectively. These results were not significantly different from those obtained with PilU, a cytoplasmic protein not expected to associate with pili. These results confirm that, at best, only a minute proportion of minor pilins are associated with pili.
PilV and PilX are necessary for optimal initiation of pilus biogenesis
The presence of the minor pilins in the periplasm suggests a role in the pilus machinery and pilus biogenesis. In addition, given their common pilin-like structural features, we reasoned that PilV and PilX could exert redundant functions in pilus biogenesis. To evaluate this possibility, we reassessed the piliation levels of the pilV and pilX mutants and the double mutant by ELISA on whole bacteria, and the morphology of pili was observed using immunofluorescence.
Inactivation of pilV led to a minor but reproducible defect in piliation with 61 ± 7% of piliation relative to the wild-type strain (FigA). PilV overexpression not only rescued the amount of pili but increased pili levels above wild-type levels. Similar results were obtained for pilX with the mutant displaying a more severe phenotype with only 27 ± 4% of piliation and the complemented strain showing higher piliation than the wild type (Fig2B). In addition, the pilV and pilX mutations had a synergistic effect. The double mutant could not be distinguished from the pilE deficient strain by ELISA (Fig2C). These results were confirmed by quantitative biochemical pilus preparations (Supplementary Fig S2).
Figure 2. Role of PilV and PilX in pilus biogenesis.
- ELISA analysis of surface-exposed pilin in the pilV mutant and complemented strains. Results are presented relative to the wild-type strain, given the value of 1.
- ELISA analysis of surface-exposed pilin in the pilX mutant.
- ELISA analysis of surface-exposed pilin in the pilVpilX double mutant.
- Immunofluorescence analysis of pili on the surface of the indicated bacterial strains. DAPI is in blue and pilus staining in green (20D9).
- Quantitative determination of the number of pili detected per individual diplococcus. The results of 3 independent experiments each with over 50 bacteria are included.
- Frequency distribution of the number of pili per diplococcus expressed by the wild-type strain (black line), the pilV strain (blue line), and the pilX strain (red line).
- Pili length measurement (μm). The results of 3 independent experiments each with over 50 bacteria are included.
- Frequency distribution of the length of pili expressed by the wild-type, pilV and the pilX strains (same color code as in F).
- A typical track representing bacterial movement resulting from twitching motility over a 2-min period.
- Frequency representation of the different speeds of bacteria for the wild-type, pilV and pilX strains (same color code as in F).
Data information: Data represent mean ± SEM; *P ≤ 0.05; ****P ≤ 0.001.
Decreased steady state amount of pili can be explained by a decreased initiation of biogenesis, slower extension, or faster retraction. In the case of slower extension or faster retraction, pili should be shorter but with the same number per bacterium. In contrast, lower initiation rate would be predicted to produce a lower number of pili with the same length. To further characterize these piliation defects, pili were visualized on individual bacteria by immunofluorescence and their number and length determined (Fig2D). While wild-type bacteria displayed an average of 5.5 ± 0.3 pili on their surface, pilV and pilX strains displayed 2.7 ± 0.08 and 1.6 ± 0.003, respectively (Fig2E and F). Pili lengths, however, were similar with a trend toward longer pili in the pilV and pilX strains (Fig2G and H). These results indicate that PilV and PilX exert their effect at the initiation of pilus biogenesis rather than extension or retraction.
Since a role for PilV and PilX in opposing PilT-dependent pilus retraction was previously proposed (Carbonnelle et al, 2006), we sought an additional experimental approach to confirm the above result. If the effect of PilV and PilX was to slow down retraction, a prediction would be that twitching motility, a process powered by retraction, would be higher in the pilV and pilX mutants. Motility assays indicate that the twitching motility of the pilV mutant is indistinguishable from that of the wild-type strain (Fig2I and J). The pilX mutant has a slower motility, probably due to the decreased number of pili expressed by this strain. These results confirm that PilV and PilX do not participate in the control of pilus retraction but rather play a role in the initiation phase of pilus biogenesis.
PilV and PilX exert their functions from the periplasm
As shown above, the PilV and PilX minor pilins are mainly localized in the periplasm. This does not however rule out the possibility that a small proportion of the proteins located in the pilus could carry out their functional roles, particularly in terms of interaction with host cells. To address this point, we designed a strategy to block these proteins in the periplasm and assessed their function under these conditions. Based on available structures of the secretin family of proteins (Korotkov et al, 2012), we reasoned that fusing the bulky mCherry protein to the carboxy-terminus of the minor pilins would prevent their exit from the periplasm through the PilQ outer-membrane pore.
The mCherry fusion constructs with PilE, PilV, and PilX all localized in the bacterial periplasm and, as expected, no evidence of organization as pilus fibers could be seen (FigA). To confirm that the PilE-mCherry fusion was retained in the periplasm, the amount of pili expressed at the surface of this strain was determined by ELISA on whole bacteria (Fig3B). As previously described, the lac promoter was insufficiently strong to provide full complementation of pilE (Long et al, 2003). Nevertheless, expression of the unmodified pilE gene reached a piliation value of 0.21 ± 0.04 relative to wild-type level (value of 1). Validating our experimental strategy, expression of the pilEmCherry fusion did not lead to any detectable pili. In striking contrast, fusion of the mCherry protein to PilV and PilX did not have any obvious impact on the capacity of these proteins to initiate pilus biogenesis (Fig3C). The functionality of the PilV-mCherry fusion protein was also tested for its ability to provide efficient crosstalk with host cells. At the concentration of inducer mimicking the wild-type level of PilV (25 μM, Supplementary Fig S1), the pilVmCherry construct fully complemented the deletion strain (FigD). Similar results were obtained with the pilXmCherry construct, and full complementation was observed for adhesion and aggregation with 17 μM of IPTG (Fig3E and Supplementary Figs S1 and S3). These results show that preventing the exit from the outer membrane of the minor pilins PilV and PilX does not affect their function. This demonstrates that PilV and PilX exert their functions, both in terms of pilus biogenesis and interaction with host cells, inside the periplasm and not from the bacterial surface.
Figure 3. Fusion of PilV or PilX to mCherry does not prevent PilV or PilX function.
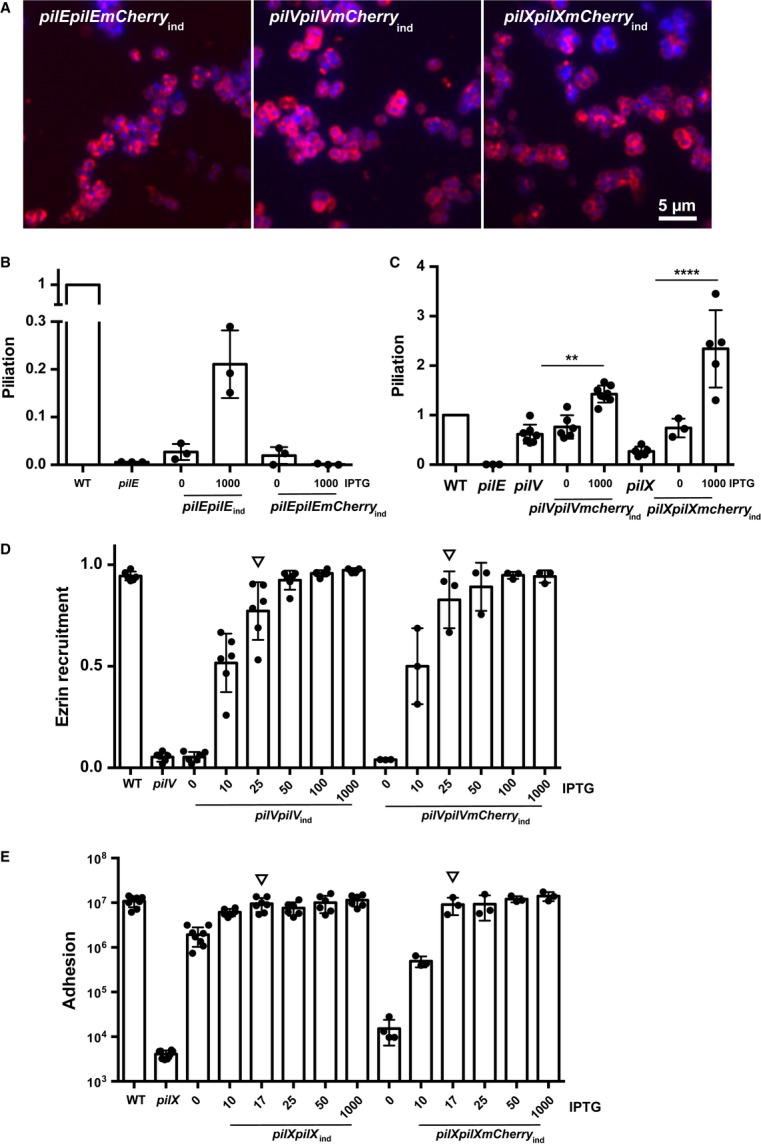
- Fluorescence analysis of the localization of the mCherry fusions (mCherry is in red, DAPI is in blue), pilEmCherry (left), pilVmCherry (middle) and pilXmCherry (right).
- Impact of the mCherry fusion on the ability of PilE to form pilus fibers. The amount of surface-exposed pili was determined by ELISA on whole bacteria for the indicated strains. The amount of added IPTG to induce the Lac promoter is indicated (μM). Each dot represents one experiment.
- Impact of the mCherry fusion on the ability of PilV and PilX to promote pilus biogenesis.
- Ability of the PilV-mCherry fusion to promote plasma membrane reorganization following adhesion was determined using ezrin as a marker. Bacteria were incubated with endothelial cells for 2 h, and the proportion of microcolonies efficiently triggering plasma membrane reorganization was determined. Arrows point to the conditions mimicking wild-type levels of PilV.
- Ability of the PilX-mCherry fusion to promote adhesion. Bacteria were incubated with endothelial cells for a period of 4 h, and the number of cell-associated bacteria was determined. The amount of added inducer is indicated. Arrows point to the conditions mimicking wild-type levels of PilX.
Data information: Data represent mean ± SEM; **P ≤ 0.01; ****P ≤ 0.0001. In (B–E) each dot represents one experiment.
PilV and PilX do not require assembly into a fiber to exert their functions
To further characterize the mechanism of action of PilV and PilX, we investigated whether assembly into a fiber was necessary to exert their functions. Mutation of glutamate at position 5 into an alanine (E5A) is a well-established way to block pilin-like monomer assembly into fibers in different piliation systems (Pasloske & Paranchych, 1988; Aas et al, 2007; Campos et al, 2010). We therefore generated E5A point mutations in pilV and pilX and evaluated the functionality of these mutants. While the pilEE5A mutant did not show any pili on its surface, the pilVE5A and pilXE5A constructs fully restored piliation (FigA). Furthermore, PilVE5A was functional in mediating plasma membrane reshaping under microcolonies in conditions mimicking the endogenous expression level (Fig4B). Similarly, PilXE5A restored bacterial adhesion to host cells and aggregation (Fig4C and Supplementary Fig S3). These results show that PilV and PilX do not require the glutamate residue in position 5 to exert their functions. Since this residue is typically necessary for assembly into fibers, it is likely that PilV and PilX do not need to assemble into fibers to modulate piliation and to mediate interaction with host cells.
Figure 4. The E5A mutation does not affect PilV or PilX function.
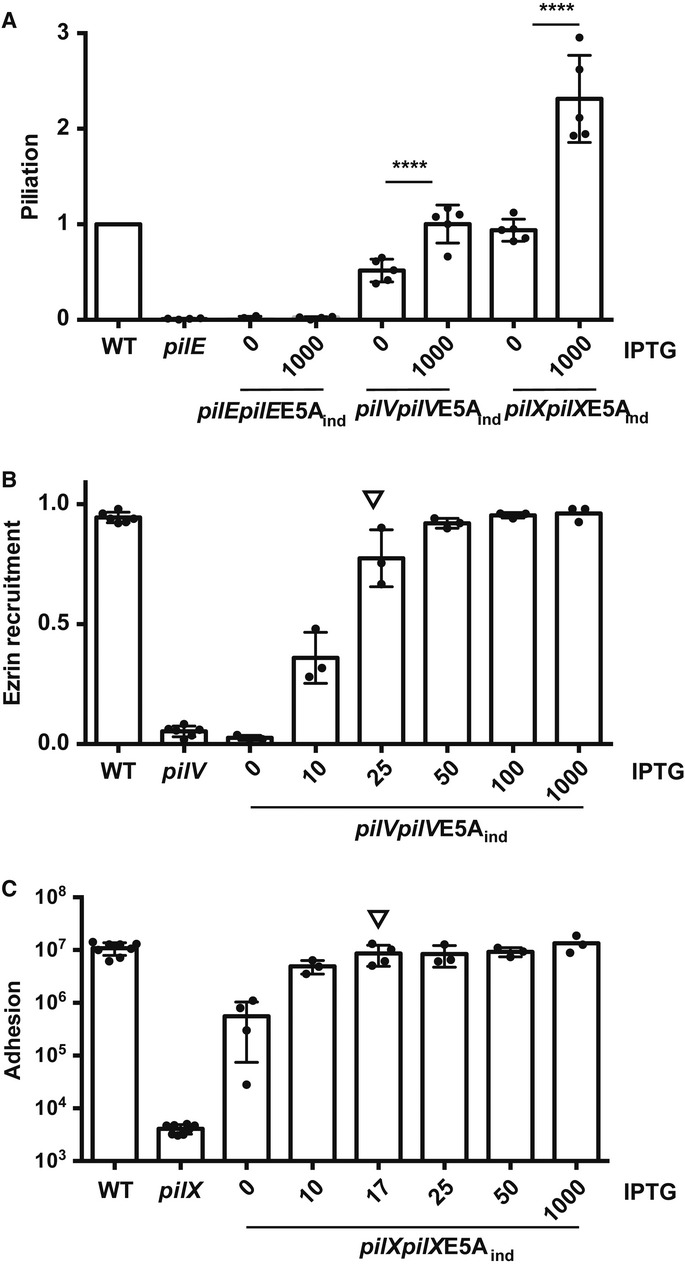
- Effect of the E5A mutation in pilE, pilV, and pilX on pilus biogenesis determined by ELISA.
- Effect of the E5A mutation in PilV on its ability to promote plasma membrane reshaping. The amount of inducer is indicated (IPTG, μM). The arrow indicates the amount of inducer mimicking the wild-type level of expression.
- Effect of the E5A mutation on the ability of PilX to promote bacterial adhesion. The arrow indicates the amount of inducer mimicking the wild-type level of expression.
Data information: Data represent mean ± SEM; ****P ≤ 0.0001.
PilV and PilX require cleavage by the PilD peptidase to be functional
Considering the evidence described above pointing to a possible site of action of the minor pilins PilV and PilX in the periplasm, we considered whether cleavage by the PilD peptidase was necessary. Point mutations in the PilD cleavage site, glycine in position −1 to asparagine (G-1N), were thus generated. First, the impact of the mutation on protein cleavage was checked (Fig5A). Importantly, the G-1N mutations only affected the cleavage of the mutated protein but not the other minor or major pilins. As expected, the major pilin carrying the G-1N mutation was unable to form pilus fibers (Fig5B). When pilVG-1N and pilXG-1N were expressed, not only were those constructs unable to complement the mutation but they appeared to block pilus formation. To confirm this observation, the pilVG-1N and pilXG-1N constructs were expressed in the wild-type background. Expression of the PilD-resistant PilV and PilX proteins restricted pilus formation thus exhibiting a dominant-negative effect (Fig5C). It can be concluded that cleavage by PilD is necessary for proper function of PilV and PilX and that, surprisingly, the uncleaved forms are able to interact functionally with the piliation machinery and block biogenesis. These results highlight the tight functional association of PilV and PilX with the piliation machinery.
Figure 5. Absence of PilD cleavage in G-1N mutants blocks the function of PilV and PilX.
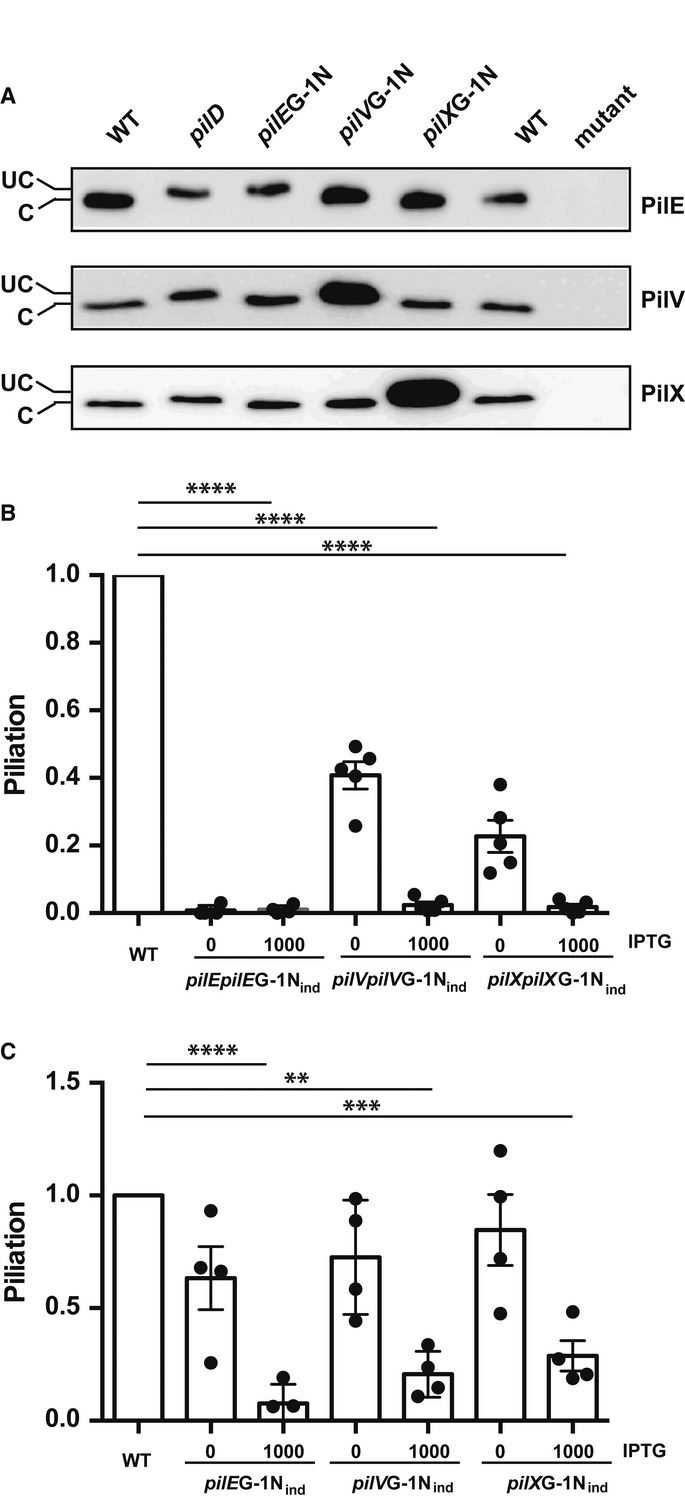
- Mobility shift due to the absence of cleavage by PilD of mutated PilE, PilV, and PilX. Western blots with polyclonal antibodies directed against PilE (top), PilV (middle), and PilX (bottom) are shown. Uncleaved proteins (UC) migrate slower than cleave proteins (C).
- Piliation levels determined by ELISA in the G-1N mutants.
- Dominant-negative effect of the G-1N mutation. Mutant proteins were expressed with intact endogenous protein and piliation levels were determined by ELISA.
Data information: Data represent mean ± SEM; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001.
The number of pili displayed on the bacterial surface tightly regulates function
The results described above can be grouped into two lines of evidence: (i) PilV and PilX participate in the pilus machinery necessary for the initiation of pilus biogenesis; (ii) PilV and PilX exert all their functions involving interaction with host cells such as adhesion and plasma membrane reshaping from the periplasm rather than from outside of the bacteria in association with pili. A straightforward hypothesis that arises from the above results is that the phenotypes displayed by the individual mutants are due to their effect on piliation. To test this hypothesis, we designed a genetic system that allows fine regulation of the amount of pili on the bacterial surface. The PilF ATPase that provides energy for pilus biogenesis was expressed under the control of the lac promoter in a pilF background. When expressed relative to the wild-type level of piliation, the system allows to finely regulate the amount of pili between 3% in the absence of inducer up to 95% in the presence of 100 μM IPTG (Fig6A). Intermediate values follow a sigmoidal curve (R2 = 0.89). Notably, 10 μM of inducer corresponds to the amount of pili expressed by pilX, 20 μM to pilV, and 100 μM to the wild-type strain. Beyond the amount of pili displayed on the surface, these concentrations of inducer reproduced the average number (Fig6B) and length of pili expressed by individual bacteria (Fig6C). Using this system, the different properties mediated by type IV pili were evaluated with variable amounts of pili: competence, adhesion, aggregation, and bacteria-induced plasma membrane reorganization.
Figure 6. Impact of the number of pili on pilus-dependent functions.
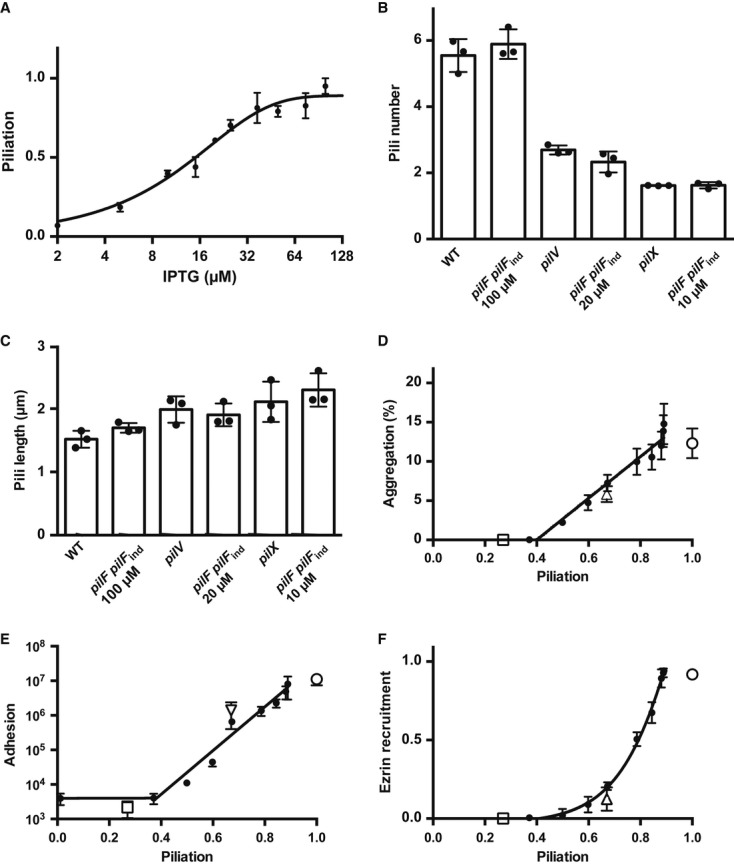
- ELISA analysis of the piliation levels of the pilFpilFind strain. The level of piliation relative to the wild-type strain was plotted as a function of the amount of inducer (IPTG). A sigmoidal curve fit was included.
- Number of pili per bacterium expressed by the pilFpilFind strain in conditions mimicking the amount of pili present in the pilV (20 μM IPTG) and pilX (10 μM IPTG) mutants. A total of 150 bacteria were analyzed in three independent experiments.
- Length of pili expressed by the pilFpilFind strain in conditions mimicking the amount of pili present in the pilV and pilX mutants.
- Ability of the pilFpilFind strain to aggregate in the presence of different amounts of inducer. Aggregation is indicated as the percentage of bacteria involved in aggregates relative to the total amount of bacteria. The X-axis is the amount of pili in each condition determined by ELISA.
- Ability of the pilFpilFind strain to adhere to HUVEC cells in the presence of different amounts of inducer.
- Ability of the pilFpilFind strain to trigger plasma membrane reorganization upon adhesion to host cells in the presence of different amounts of inducer.
Data information: In (D–F) open squares, triangles and circles represent the values for the pilX, pilV, and wild-type strains respectively.
To measure aggregation, bacteria grown in the presence of different amounts of IPTG were incubated for a period of 1 h, aggregates were visualized under a microscope, and images were analyzed automatically with the ImageJ software package to determine the ratio of bacteria involved in aggregates relative to the total amount of bacteria (Chamot-Rooke et al, 2011; Schneider et al, 2012). Results are presented as a function of the level of piliation as indicated in FigA. No aggregation could be detected with < 40% piliation (Fig6D). Beyond this value, the level of aggregation increased in a linear fashion with piliation. For adhesion, results were similar to aggregation, with up to about 40% of piliation resulting in a low level of adhesion corresponding to a non-piliated strain (Fig6E). In the range of 40–95% of piliation, adhesion increased linearly when the log of bacterial number was plotted. The ability of the bacteria to induce plasma membrane reorganization under microcolonies was measured in different conditions of induction. This response could only be evaluated in conditions where adhesion was observed, that is, above 40% of piliation. The efficiency of the cellular response was low when piliation was lower than 70% and then increased sharply with higher levels of piliation (Fig6F). In the case of competence, small amounts of piliation were sufficient and a plateau value was reached with as low as 20% of piliation (Supplementary Fig S4). Optimal levels of type IV pili are thus necessary to foster the biologic effects carried out by this bacterial organelle.
The level of piliation thus tightly regulates the ability of the bacteria to form aggregates, to adhere to host cells, and to induce a host cell response. The pilV and pilX mutants were also included in these different assays. As expected, the pilX mutant was unable to adhere, aggregate, or trigger a cellular response (open square, Fig6D, E and F). Plotting the amount of pili of the pilX mutant and its performance in the different assays showed a striking association. The pilV mutant displayed a mild decrease in aggregation and adhesion and was nearly completely defective in its ability to trigger a cellular response (open triangle, Fig6D, E and F). Taken together, these results show that the mild effects of the minor pilins PilV and PilX on type IV pilus biogenesis are sufficient to explain the phenotype of the corresponding mutants.
Discussion
An important result of our study is that the minor pilins PilV and PilX participate in pilus biogenesis. More specifically, they participate in the initiation phase of pilus biogenesis. A first argument that supports this idea is that deletion of these genes leads to a reduction in the number of pili displayed by the bacteria and overexpression of the genes leads to a higher amount of pili. The expression level of PilV and PilX thus tightly correlates with the levels of piliation. Three independent experimental approaches were used to demonstrate this: (i) surface-exposed piliation was determined by an ELISA approach; (ii) pili were counted on individual diplococci by immunofluorescence; (iii) the amount of pilin that could be precipitated by ammonium sulfate was determined. A modest reduction in the amount of piliation (76% of piliation compared to the wild type) was previously described for the pilX mutant in N. meningitidis (Brown et al, 2010). In N. gonorrheae, a more robust reduction in piliation was described although not precisely quantified for the pilL mutant, which is the pilX ortholog (Winther-Larsen et al, 2005). Although the exact value of the reduction might vary depending on the precise experimental conditions, there is good agreement on the reduction itself. For the pilV mutant, the subtle reduction in the amount of pili has gone unnoticed (Winther-Larsen et al, 2001; Mikaty et al, 2009; Brown et al, 2010).
Importantly, we were able to relate the total amount of piliation, sometimes referred to as a piliation index, to the actual number of pili on the bacterial surface seen in immunofluorescence. This study indicates that a relatively limited number of pili (5.5 ± 0.3) are present on the bacterial surface. We are confident that the fibrous structures observed by this approach are in fact individual pili rather than pili bundles. A previous study using electron microscopy has shown that overexpression of the pptB enzyme prevents the formation of bundles (Chamot-Rooke et al, 2011), and in these conditions, little effect on the number of visible fibers was observed using the fluorescence-based assay used here (data not shown). This approach also allowed measurement of the length of individual pili. These measurements revealed that pili expressed by the pilV and pilX mutants were slightly longer than those expressed by the wild-type strain. In principle, a steady state reduction in surface piliation as observed in pilV and pilX could be indicative of lower frequency of initiation, slower extension, or faster retraction. We observed fewer but longer pili in the mutants. This is in favor of a role for PilV and PilX in the initiation phase of pilus biogenesis. If they played a role in extension the prediction would be, they would express the same number of pili but shorter. If PilV and PilX proteins slowed down retraction, pili would retract faster in the mutants and thus be shorter as well on average. In this last scenario, motility due to retraction would be expected to be faster but it is not the case. Taken together, these observations strongly suggest a role of PilV and PilX in the initiation of pilus biogenesis.
Another argument in favor of a role for PilV and PilX in pilus biogenesis relates to the localization of these proteins. Based on previous publications, we attempted to visualize these proteins within fibers but failed to do so. Despite using a highly sensitive method, type IV pili-like fibers could not be detected, while PilV and PilX could be easily detected in the periplasm when the outer membrane was removed. An argument in favor of the association with type IV pili is the co-purification of these proteins with pili (Winther-Larsen et al, 2001; Helaine et al, 2005). Such approaches are based on highly sensitive Western blot detection of proteins, but the amount of detected protein was never related to the total amount of protein present in the bacteria. We provide evidence that the proportion of PilV and PilX that associates with pilus preparations is in fact very low and not significantly different from negative controls. Our results cannot exclude that small amounts of protein exit to the bacterial surface and localize in pili, which could explain observation of PilX inside pilus fibers by electron microscopy (Helaine et al, 2005). However, our results indicate that the vast majority of the protein stays in the periplasm. Periplasmic location is coherent with the fact that antibodies against PilV and PilX did not promote any bactericidal activity in the presence of complement (Cehovin et al, 2011). The exact location inside the periplasm remains to be determined. It is likely that PilV and PilX are mostly associated with the inner membrane although it is also possible that a certain proportion participate in the piliation machinery complex.
From the functional point of view, we show that the active fractions of PilV and PilX are in the periplasm. Blocking these proteins in the periplasm has strikingly little effect on type IV pili-dependent phenotypes. Current crystal structures indicate that the secretin pore tightly fits the pilus fiber (6 nm wide). Accordingly, fusion of the major pilin with the mCherry protein (3 × 5 nm) prevented its exit from the bacterial periplasm. In contrast, fusion of PilV or PilX with mCherry had surprisingly little impact on their function. Furthermore, mutation of the glutamate residue in position 5 that is known to be necessary for fiber assembly did not affect PilV or PilX function. This highlights the functional differences between the major pilin and the minor pilins PilV and PilX. This result implies that PilV and PilX participate in the pilus biogenesis machinery, likely in the periplasmic side of the inner membrane. This is confirmed by the fact that PilD-resistant mutations of PilV and PilX have a dominant-negative effect on piliation.
Our results point to similarities between the minor pilins PilV and PilX and the PilH, I, J and K proteins. The two groups of proteins have a hydrophobic alpha helix in common that is the hallmark of pilin-like proteins (Winther-Larsen et al, 2005). They were initially placed in different functional groups because PilV and PilX were considered to have no significant effect on pilus biogenesis (Brown et al, 2010). Although mutants of the pilH-K genes have a stronger phenotype as they do not display pili on their surface, the proteins could have similar modes of action. Our knowledge of the function of PilH-K proteins largely comes from homologous proteins in type II secretion systems. Structural work has shown that GspI, J and K form a complex, and it was suggested that these proteins participate in the initiation of pseudopilus formation and subsequently end up in the pseudopilus fiber (Korotkov & Hol, 2008; Cisneros et al, 2012). It is conceivable that PilV and PilX in some way assist the PilH-K complex in performing this function. The participation of PilV and PilX would not be absolutely required but somehow optimizes the process in order to reach a critical amount of pili to mediate efficient interaction with host cells. This hypothesis would explain why PilV and PilX are needed in the periplasm and occasionally end up in the extracellular pilus fiber (Korotkov et al, 2012). The presence of the pilX gene in the same operon as the pilH-K genes is also in favor of a functional link between these different proteins as previously proposed in N. gonorrheae (Winther-Larsen et al, 2005).
Perhaps, the most striking result of our study is that a relatively modest decrease in the amount of pili on the bacterial surface leads to a strong effect on the functions carried out by these structures (Fig7). The modest decrease in piliation observed in the pilV and pilX mutants is sufficient to explain all their phenotypes. Our study together with previous studies has shown that only low amounts of pili are sufficient to provide efficient natural transformation (Long et al, 2003). Competence rises with the amount of pili and quickly reaches a plateau around 20% of piliation. Interestingly, this corresponds to an average of one pilus per diplococcus (Fig7, ii). One pilus is thus likely sufficient to generate optimal competence. This is analogous to competence systems that frequently display only one pilus per bacterium (Laurenceau et al, 2013). Adhesion and aggregation require over 40% of piliation corresponding to about two pili per bacterium (Fig7, iii). It is easy to explain why at least two pili are necessary for aggregation. Once two bacteria are engaged via a first fiber, a second one is necessary to interact with a third bacterium and so on and so forth. Why more than two pili per bacterium are necessary for adhesion is unclear at this point although the adhesion assay performed here largely depends on aggregation as bacteria grow as three-dimensional microcolonies on the cellular surface. Perhaps, the result most difficult to understand is the requirement of over 70% of piliation to provide efficient plasma membrane reshaping. This corresponds to three to five pili per bacterium indicating that individual pili perhaps fail to trigger this cellular response but that the functional entity is a supramolecular organization of several pili. Considering that bacteria organize in arrays on the cellular surface with one bacterium surrounded by about 5 other bacteria, it is possible that pili need to be placed between each neighboring bacterium in order to trigger a response (Fig7, iv). It is noteworthy that because of the aggregation pattern, a given bacterium is also in contact with pili from the neighboring bacteria; hence, the actual number of pili around one bacterium is increased. Further work is needed to clarify this point.
Figure 7. Schematic representation of the impact of the amount of type IV pili on the different functions mediated by these organelles.
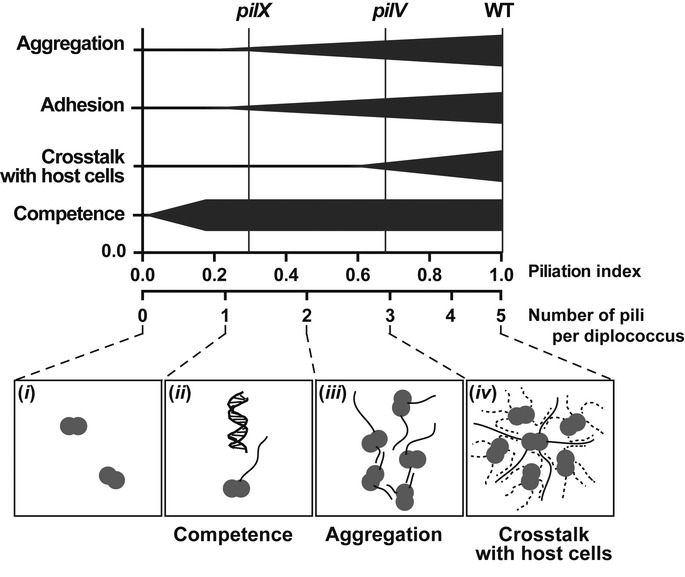
The top part of the schematic indicates the efficiency of the different pili-mediated properties as a function of bulk amount of pili and number of pili per bacterium. In the bottom part of the schematic, representative situations are depicted: (i) In the absence of pili, no related function is observed; (ii) A single pilus is required for competence; (iii) A minimum of 2 pili is necessary for aggregation; (iv) About 5 pili per bacterium are necessary for optimal bacteria-induced plasma membrane reshaping. Pili from neighboring bacteria are indicated as a dotted line.
Overall, the results described in this study indicate that optimal numbers of pili are necessary to promote efficient interaction with host cells. This necessity for optimal interaction with host cells has probably generated the evolutionary pressure required for N. meningitidis to select two proteins, PilV and PilX to optimize its piliation level. The exquisite sensitivity on the number of pili for their proper functions has practical implications. In terms of developing potential drugs that would target type IV pili, these results are encouraging, as even a partial effect on the amount or function of pili would result in a strong effect on pathogenesis.
Materials and Methods
Antibodies and chemicals
The following antibodies were used for Western blot, immunofluorescent labeling, or enzyme-linked immunosorbent assay (ELISA): anti-ezrin rabbit polyclonal antibody (generously provided by P. Mangeat, CNRS, UMR5539, Montpellier, France), anti-PilE mouse monoclonal antibody clone 20D9 (Pujol et al, 1997), anti-PilE polyclonal anti-serum (Morand et al, 2004), anti-PilX (Helaine et al, 2007), anti-PilV (Mikaty et al, 2009), and anti-Flag M2 monoclonal and polyclonal antibodies (Sigma). The polyclonal anti-PilU antiserum was generated by injecting rabbits with two peptides: YSQKKQSRSETPAEI (17–31) and PELLYDRVNGLNLIS (394–408). The following goat secondary antibodies were used for immunofluorescent labeling, Western blot or ELISA: anti-mouse or anti-rabbit IgG (H+L) coupled to horseradish peroxidase (Jackson ImmunoResearch Laboratories) and anti-rabbit or anti-mouse IgG (H+L) coupled to Alexa-488 (Life Technologies). 4′,6-diamidino-2-phenylindole (DAPI) was purchased from Life Technologies. Isopropyl β-D-1-thiogalactopyranoside (IPTG) was purchased from Promega.
Bacterial strains and growth conditions
Growth conditions
Neisseria meningitidis strains used in this study (Table2) were grown on GC agar base plates (Conda Laboratorios, Spain) containing Kellogg's supplements (Kellogg et al, 1968) and, when required, 100 μg/ml kanamycin, 2 μg/ml erythromycin, 50 μg/ml spectinomycin, or 5 μg/ml chloramphenicol at 37°C in moist atmosphere containing 5% CO2. Escherichia coli transformants were grown on liquid or solid Luria-Bertani medium (Difco) containing 20 μg/ml chloramphenicol, 50 μg/ml spectinomycin, 30 μg/ml kanamycin, or 200 μg/ml erythromycin when necessary.
Table 2.
Bacterial strains used in this study
| Strain | Genotype | References |
|---|---|---|
| pilE | ΔpilE::alphA-3 | Geoffroy et al (2003) |
| pilE pilEind | ΔpilE::alphA-3 pLac-pilE | This study |
| pilEpilEFlagind | ΔpilE::alphA-3 pLac-pilEFlag | This study |
| pilEFlagind | pLac-pilEFlag | This study |
| pilE pilEE5Aind | ΔpilE::alphA-3 pLac-pilEE5A | This study |
| pilE pilEmCherryind | ΔpilE::alphA-3 pLac-pilEmCherry | This study |
| pilE pilEG-1Nind | ΔpilE::alphA-3 pLac-pilEG-1N | This study |
| pilEG-1Nind | pLac-pilEG-1N | This study |
| pilX | ΔpilX::alphA-3 | Geoffroy et al (2003) |
| pilX pilXind | ΔpilX::alphA-3 pLac-pilX | Helaine et al (2005) |
| pilX pilXFlagind | ΔpilX::alphA-3 pLac-pilXFlag | This study |
| pilX pilXE5Aind | ΔpilX::alphA-3 pLac-pilXE5A | This study |
| pilX pilXmCherryind | ΔpilX::alphA-3 pLac-pilXmCherry | This study |
| pilX pilXG-1Nind | ΔpilX::alphA-3 pLac-pilXG-1N | This study |
| pilXG-1Nind | pLac-pilXG-1N | This study |
| pilV | ΔpilV::alphA-3 | Geoffroy et al (2003) |
| pilV pilVind | ΔpilV::alphA-3 pLac-pilV | Mikaty et al (2009) |
| pilV pilVFlagind | ΔpilV::alphA-3 pLac-pilVFlag | This study |
| pilV pilVE5Aind | ΔpilV::alphA-3 pLac-pilVE5A | This study |
| pilV pilVmCherryind | ΔpilV::alphA-3 pLac-pilVmCherry | This study |
| pilV pilVG-1Nind | ΔpilV::alphA-3 pLac-pilVG-1N | This study |
| pilVG-1Nind | pLac-pilVG-1N | This study |
| pilX pilV | ΔpilX::alphA-3 ΔpilV::cat | This study |
| pilU | ΔpilU::ermam | This study |
| pilF pilFind | ΔpilF::spec pLac-pilF | This study |
Gene inactivation and complementation
Mutations in the pilE, pilV, and pilX genes were described elsewhere (Nassif et al, 1993; Geoffroy et al, 2003; Helaine et al, 2005). To generate a mutation in the pilF gene, the entire ORF was amplified using PilF-F and PilF-R (see Table1 for primer sequences) and cloned into Topo-pCR2.1 (Invitrogen); a BamHI site was introduced inside the gene by site-directed mutagenesis using the megaprimer strategy (Ke & Madison, 1997) with primers PilF-BamHIrev and PilF-F; this construct was then subcloned into the pBluescript plasmid (Stratagene) using the SacI and SalI restriction sites; the spectinomycin resistance cassette (Klee et al, 2000) was introduced into the BamHI restriction site. The same strategy was used to generate the pilV mutant interrupted by a chloramphenicol cassette (Klee et al, 2000) using oligos PilV5′Fw and PilV3′rev for amplification of the pilV region and with PilV-BamHIrev and PilV5′Fw for mutagenesis. To delete the pilU gene, an upstream region was amplified with PilU-NF and PilU-NR primers, and a downstream region amplified with PilU-CF and PilU-CR primers were restricted by the corresponding enzymes and ligated in the pBluescript plasmid restricted with SalI and SacI (Stratagene), then the ERMAM erythromycin resistance cassette (Trieu-Cuot et al, 1988) was cloned in the BamHI site. Constructs allowing the complementation of the pilV and pilX mutations were described elsewhere (Helaine et al, 2005; Mikaty et al, 2009). For the complementation of the pilE and pilF mutations under the control of the lac promoter, ORFs were cloned into pCR2.1 (PilF-F/PilF-R for pilF and PilE-F/PilE-R for pilE) and the fragments were introduced into the pGCC4 vector (Mehr et al, 2000) using PacI and PmeI restriction.
Table 1.
Primers used in this study
| Primer | Sequencea | Enz |
|---|---|---|
| PilF-F | GTCGACTTAATTAAAGGAGTAATTTTATGAGCGTAGGTTTGCTGAG | SalI/PacI |
| PilF-R | GAGCTCGTTTAAACCTAATCGTTGGTATTTGCC | SacI/PmeI |
| PilV-F | TTAATTAAAGGAGTAATTTTATGAAAAACGTTCAAAAAG GC | PacI |
| PilV-R | GTTTAAACTTAGTCGAAGCCGGGGCAG | PmeI |
| PilX-F | TTAATTAAAGGAGTAATTTTATGATGAGTAATAAAATGG | PacI |
| PilX-R | GTTTAAACCTATTTTTTACGATTAGAG | PmeI |
| PilE-F | TTAATTAAAGGAGTAATTTTATGAACACCCTTCAAAAAGGTTTTACCC TTATCGAGCTGATG | PacI |
| PilE-R | GTTTAAACTTAGCTGGCAGATGAATCATCG | PmeI |
| PilE-Flagrev | GTTGATTTTGTCGTCGTCGTCCTTATAATCGGCGGCGACGTCGGT | |
| PilV-FlagCrev | GAAGCCGGGGCACTTATCGTCATCGTCCTTATAATCAATGGTACCGCC | |
| PilX-FlagCrev | GAAAGCTTCACACTTATCGTCATCGTCCTTATAATCCAAATGGGCTCG | |
| mch-F | CAGGTCGACGTGAGCAAGGGCGAGGAGGATAACATG | SalI |
| mch-R | CTAGTTTAAACCCTCTACAAATGTGGTATGGCTG | PmeI |
| PilE-mCh | CGCCCTTGTTTAAACGTCGACGCTGGCAGATGAATC | SalI |
| PilV-mCh | CGCCCTTGTTTAAACGTCGACGTCGAAGCCGGGGCA | SalI |
| PilX-mCh | CGCCCTTGTTTAAACGTCGACTTTTTTACGATTAGAGAA | SalI |
| PilEG-1N | AACACCCTTCAAAAAAACTTTACCCTTATCGAG | |
| PilVG-1N | AAAAACGTTCAAAAAAACTTTACGCTGCTCGAG | |
| PilXG-1N | AAAATGGAACAAAAAAACTTTACATTGATTGAGATGATG | |
| PilEE5A | TTTACCCTTATCGCACTGATGATTGTGATTGCC | |
| PilVE5A | CTTTACGCTGCTCGCACTGATGATTGCCGTC | |
| PilXE5A | GTGTTTACATTGATTGCAATGATGATAGTCGTC | |
| PilU-NF | GTCGACCCATCAAAGCAAAAAATCCCTG | SalI |
| PilU-NR | GGATCCAAGATGTCGTGCAGGTTATCGG | BamHI |
| PilU-CF | GGATCCGCGATTGCACATCCAACTGC | BamHI |
| PilU-CR | GAGCTCAAGTCGGCTTTGGCTTCGAG | SacI |
| PilV5′Fw | GTCGACCCAGGTTGGACGGCGAA | SalI |
| PilV3′Rev | GAGCTCGCCGCAATATTCGACTTCG | SacI |
| PilV-BamH1rev | TGATGAGCGTCAGGATCCGAGGATGGCGACG | BamHI |
| PilF-BamH1rev | CCTGCCGTCCTGCGGGATCCGTTTTTCGGAAATG | BamHI |
Inserted regions are underlined and restriction sites in bold.
Introduction of Flag tags
Generation of pilEFlag, pilXFlag, and pilVFlag fusions was done using the megaprimer technique (Ke & Madison, 1997). The first amplification for pilE (PilE-F and PilE-FlagRev), pilV (PilV-F and PilV-FlagCRev), and pilX (PilX-F and PilX-FlagCRev) substituted 5 to 8 residues found in the carboxy-terminal part of the D-region. This fragment (Mega-primer) was then used for the complete amplification of the vector. The mutated fragments were then subcloned in pGCC4.
Fusion with mCherry
The megaprimer strategy was used to remove the stop codons in pilV, pilX, and pilE and to insert a SalI site. The first amplification was done using PilV-F/PilV-mCh, PilX-F/PilX-mCh, and PilE-F/PilE-mCh primers using the corresponding ORFs cloned in Topo-pCR2.1 as a template. The mCherry ORF was amplified using primers mCh-F/mCh-R from the pmCherry-N1 vector (Clonetech) and cloned into Topo-pCR2.1. The mCherry fragment was then introduced carboxy-terminal of pilV, pilX, and pilE using SalI and PmeI restriction. The three mCherry fusions were then subcloned into the pGCC4 vector using PacI and PmeI restriction to allow chromosomal insertion.
Point mutations
All point mutations were done with the megaprimer strategy (Ke & Madison, 1997). The G-1N mutation was introduced in pilE, pilV and pilX using the following primer pairs PilEG-1N with PilE-R, PilVG-1N with PilV-R, and PilXG-1N with PilX-R using the corresponding ORFs cloned into Topo-pCR2.1 as a template. The E5A mutation was introduced in PilE, PilV, and PilX with the same strategy using the following primer pairs: PilEE5A with PilE-R, PilVE5A with PilV-R, and PilXE5A with PilX-R. The mutated fragments were subcloned into pGCC4.
Immunofluorescence on immobilized bacteria
A bacterial suspension at OD600 of 0.1 in PBS was placed on coverslips coated with Poly-L-Lysine (Sigma) and fixed in PBS containing 4% paraformaldehyde. Sample was blocked with PBS containing 0.2% gelatine (PBSG) for 10 min. Flag-tagged proteins were labeled with either the anti-Flag M2 monoclonal antibody at a concentration of 8 μg/ml or the anti-Flag polyclonal antiserum in PBSG. Native pili were detected with the 20D9 monoclonal antibody diluted at 2.5 μg/ml in PBSG. The secondary antibody was a goat anti-mouse antibody conjugated with Alexa Fluor 488 (Life technologies). To generate spheroplasts, bacteria were resuspended in 30 mM Tris (pH 7.5), 20% sucrose, 2 mM EDTA, 1 mg/ml lysozyme and incubated 10 min at room temperature under constant agitation. Cells were then pelleted by centrifugation at 9,000 g for 10 min and resuspended in an equal volume of cold 5 mM MgSO4. After a 10-min incubation on ice, bacteria were immobilized on coverslips coated with poly-L-lysine. Labeled bacteria were mounted in Mowiol (Valnes & Brandtzaeg, 1985).
Type IV pili quantification by ELISA
Bacteria were resuspended from fresh GCB plates in PBS at OD600 of 0.1. Serial twofold dilutions were added to the wells of a 96-well plate (Nunc, Germany). The plates were centrifuged for 10 min at 3,220 g, and the supernatant was carefully removed. Then, the plates were incubated without covers at 37°C for 10 min to allow drying. Bacteria were fixed with a solution of PBS containing 4% paraformaldehyde. Coated plates were washed 3 times with PBS and incubated in blocking solution (PBS containing 1% BSA and 0.1% Tween-20) for 10 min. The anti-PilE mouse monoclonal antibody 20D9, diluted at a concentration of 0.25 μg/ml in blocking solution, was added to the plates and incubated for 1 h. After several washes, a peroxidase-linked anti-mouse IgG, diluted at 1/10,000 in blocking solution, was added to the wells for 1 h. Finally, after three washes, the staining was revealed using TMB (3,3′,5,5′-tetramethylbenzidine) substrate and stop solution following the manufacturer's instructions (BD Bioscience). The absorbance was read at 450 nm using a microtiter plate reader (Flex station 3, Molecular Devices).
Cell culture
Human umbilical vein endothelial cells (HUVECs; PromoCell) were used between passages 1 and 8 and grown in endothelial serum-free medium (Endo-SFM; Gibco) supplemented with 10% heat-inactivated fetal bovine serum (PAA Laboratories GmbH) and 40 μg/ml of endothelial cell growth supplement (Sigma-Aldrich).
Immunofluorescence of infected cells
HUVECs were plated at a density of 105 cells/cm2 onto 12-mm diameter glass coverslips coated with 0.01% type I collagen (Sigma). Before the assay, bacteria grown on GCB agar plates were adjusted to OD600 of 0.05 and then incubated for 2 h at 37°C in prewarmed Endo-SFM supplemented with 10% FBS. Approximately 107 bacteria in culture medium were added to 105 cells/well in a 24-well (MOI of 100) and allowed to adhere for 30 min. Unbound bacteria were washed away, and infection was allowed to proceed for 2 h. Infected cells were fixed for 20 min in PBS containing 4% paraformaldehyde, permeabilized for 5 min in PBS containing 0.1% Triton X-100, and incubated with primary antibodies in PBS containing 0.2% gelatin (PBSG) for 1 h. After three washes with the same buffer, cells were incubated for 1 h in PBSG-containing DAPI and Alexa-conjugated secondary antibodies. After additional washes, coverslips were mounted in Mowiol. The recruitment efficiency was estimated by determining the proportion of colonies that efficiently recruit ezrin in a honeycomb shape under microcolonies. A minimum of 50 microcolonies was analyzed per coverslip. Each experiment was performed in duplicates and repeated three times.
Analysis of the length and numbers of pili on individual bacteria
Bacteria were cultured for 2 h in RPMI complemented with 10% FBS, coated on 96-well plates (μ-clear, Greiner BioOne) by centrifugation at 1,800 g for 10 min, fixed using 4% PFA for 20 min, and processed for immunostaining. Pili were stained with the 20D9 antibody diluted at 2.5 μg/ml in PBSG for 1 h. After several washes, samples were incubated in PBSG with anti-mouse secondary antibodies conjugated with Alexa 488 (5 μg/ml) and with DAPI (20 μg/ml). The number and the length of pili were determined on individual bacteria using the ImageJ software (Schneider et al, 2012). A minimum of 50 bacteria was analyzed per well. Each experiment was repeated three times.
Bacterial adhesion assay
Adhesion of meningococci to HUVECs was done as described previously (Eugene et al, 2002). In brief, monolayers of 105 cells in 24-well plates were infected with 107 bacteria (MOI of 100). The inoculum was characterized by CFU counts. After 30 min, unbound bacteria were removed by three washes, and the infection was continued for 4 h. Finally, after 3 washes, adherent bacteria were recovered by scraping the wells and counted by plating appropriate dilutions on GCB agar plates.
Bacterial aggregation assay
Bacteria grown on GCB agar plates were adjusted to OD600 of 0.05 and incubated for 2 h at 37°C in RPMI containing 10% FBS. The bacterial suspension was concentrated to OD600 of 0.3 by a centrifugation at 15,000 g for 1 min followed by resuspension in medium containing DAPI (0.1 μg/ml). Bacterial suspensions were briefly vortexed and transferred in a glass-bottom 96-well plate. After 30 min incubation, aggregates were observed microscopically with a 4× lens and size of aggregates and numbers of bacteria involved in aggregates relative to the total amount of bacteria were determined with the ImageJ software, as previously described (Chamot-Rooke et al, 2011).
SDS–PAGE and Western blot
Preparation of protein samples, SDS–PAGE separation, transfer to membranes, and immunoblotting were performed using standard biochemistry techniques (Harlow & Lane, 1988). Proteins were separated by SDS–PAGE in Tris-glycine gels containing 12–15% acrylamide, transferred onto PVDF membranes (Thermo Scientific) using semi-dry electrotransfer (Biorad). Membranes were blocked with 5% milk in PBS 0.1% Tween-20 and incubated with the specific antiserum (anti-PilE at 1/5,000, anti-PilX at 1/3,000, anti-PilV at 1/1,500, and anti-PilU at 1/1,000) diluted in blocking solution except for the anti-PilU which was diluted in PBS containing 0.1% Tween-20 and 1% BSA. Membranes were incubated horseradish peroxidase (HRP)-coupled anti-rabbit antibody (1/20,000) in the blocking solution. Peroxydase activity was detected by enhanced chemiluminescence (ECL-plus, Pierce) and recorded using the LAS-4000 ImageQuant (GE Healthcare). Protein quantifications were done using the ImageJ software.
Type IV pilus purification
Pili were prepared as previously described (Wolfgang et al, 1998; Helaine et al, 2005). The contents of ten Petri dishes were harvested in 10 ml of 150 mM ethanolamine at pH 10.5. Pili were sheared off by vortexing for 1 min. Bacteria were centrifuged at 3,000 g during 30 min. Supernatants were centrifuged an additional 30 min at 17,000 g. Pili were then precipitated by adding saturated ammonium sulfate to the supernatants to a final concentration of 10%, and samples were allowed to stand for 60 min. Precipitates were pelleted by centrifugation at 3,000 g for 30 min. Pellets were washed with PBS once and resuspended in distilled water.
Twitching motility assay
Twitching motility assays of N. meningitidis were performed inside a flow chamber (Ibidi GmbH, München, Germany). Bacteria (2.5 × 107) were introduced into the flow chamber and incubated for 30 min at 37°C. Unbound bacteria were removed by 3 washes. Bacterial motility was monitored by video microscopy over a 2-min period with the acquisition of two frames per second. Cell tracking was then manually analyzed with the ImageJ software. Velocities of single bacterial tracks were calculated in time intervals of 2 s.
Transformation assay
Neisseria meningitidis grown on agar plates were resuspended in transformation medium (15 mg/ml proteose peptone, 4 mg/ml K2HPO4, 1 mg/ml KH2PO4, 5 mg/ml NaCl, 2.5 mM MgCl2 and 2.5 mM MgSO4) at OD600 of 1. Two hundred microliters of the suspension were mixed with 100 ng of pBluescript SiaD:Cm plasmid in 24-well plates. After incubation at 37°C for 30 min with 5% CO2 on an orbital shaker, transformation medium (0.8 ml) was added to the wells and the plates were further incubated for 3 h at 37°C with shaking. Appropriate dilutions of bacteria were plated with and without chloramphenicol. Transformation frequency corresponds to the ratio of the number of chloramphenicol resistant CFUs to total CFUs.
Statistical analysis
For all quantitative measurements, mean ± SEM was determined and indicated on the figures. Statistical analysis was done by an ordinary one-way ANOVA and the Tukey's multiple comparisons test; *indicates P ≤ 0.05; **indicates P ≤ 0.01; ***indicates P ≤ 0.001; ****indicates P ≤ 0.0001.
Acknowledgments
Authors would like to thank Keira Melican, Arthur Charles-Orszag, Silke Machata, and Magali Soyer for critical reading of the manuscript; and Olivera Francetic for numerous and fruitful discussions. This work was supported by Fondation pour la Recherche Médicale and the French ministry for research and higher education (AFI); the Avenir INSERM Starting Grant; a CODDIM equipment grant (Région Ile de France); the Integrative Biology of Emerging Infectious Diseases (IBEID) laboratory of excellence and by a European Research Council starting grant.
Author contributions
GD and AFI designed the study and wrote the manuscript, and AFI conducted the experiments.
Conflict of interest
The authors declare that they have no conflict of interest.
Supplementary information for this article is available online: http://emboj.embopress.org
References
- Aas FE, Winther-Larsen HC, Wolfgang M, Frye S, Lovold C, Roos N, van Putten JP, Koomey M. Substitutions in the N-terminal alpha helical spine of Neisseria gonorrhoeae pilin affect Type IV pilus assembly, dynamics and associated functions. Mol Microbiol. 2007;63:69–85. doi: 10.1111/j.1365-2958.2006.05482.x. [DOI] [PubMed] [Google Scholar]
- Brissac T, Mikaty G, Dumenil G, Coureuil M, Nassif X. The meningococcal minor pilin PilX is responsible for type IV pilus conformational changes associated with signaling to endothelial cells. Infect Immun. 2012;80:3297–3306. doi: 10.1128/IAI.00369-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DR, Helaine S, Carbonnelle E, Pelicic V. Systematic functional analysis reveals that a set of seven genes is involved in fine-tuning of the multiple functions mediated by type IV pili in Neisseria meningitidis. Infect Immun. 2010;78:3053–3063. doi: 10.1128/IAI.00099-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos M, Nilges M, Cisneros DA, Francetic O. Detailed structural and assembly model of the type II secretion pilus from sparse data. Proc Natl Acad Sci USA. 2010;107:13081–13086. doi: 10.1073/pnas.1001703107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonnelle E, Helaine S, Nassif X, Pelicic V. A systematic genetic analysis in Neisseria meningitidis defines the Pil proteins required for assembly, functionality, stabilization and export of type IV pili. Mol Microbiol. 2006;61:1510–1522. doi: 10.1111/j.1365-2958.2006.05341.x. [DOI] [PubMed] [Google Scholar]
- Caugant DA, Hoiby EA, Magnus P, Scheel O, Hoel T, Bjune G, Wedege E, Eng J, Froholm LO. Asymptomatic carriage of Neisseria meningitidis in a randomly sampled population. J Clin Microbiol. 1994;32:323–330. doi: 10.1128/jcm.32.2.323-330.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cehovin A, Kroll JS, Pelicic V. Testing the vaccine potential of PilV, PilX and ComP, minor subunits of Neisseria meningitidis type IV pili. Vaccine. 2011;29:6858–6865. doi: 10.1016/j.vaccine.2011.07.060. [DOI] [PubMed] [Google Scholar]
- Cehovin A, Simpson PJ, McDowell MA, Brown DR, Noschese R, Pallett M, Brady J, Baldwin GS, Lea SM, Matthews SJ, Pelicic V. Specific DNA recognition mediated by a type IV pilin. Proc Natl Acad Sci USA. 2013;110:3065–3070. doi: 10.1073/pnas.1218832110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamot-Rooke J, Mikaty G, Malosse C, Soyer M, Dumont A, Gault J, Imhaus AF, Martin P, Trellet M, Clary G, Chafey P, Camoin L, Nilges M, Nassif X, Dumenil G. Posttranslational modification of pili upon cell contact triggers Neisseria meningitidis dissemination. Science. 2011;331:778–782. doi: 10.1126/science.1200729. [DOI] [PubMed] [Google Scholar]
- Cisneros DA, Bond PJ, Pugsley AP, Campos M, Francetic O. Minor pseudopilin self-assembly primes type II secretion pseudopilus elongation. EMBO J. 2012;31:1041–1053. doi: 10.1038/emboj.2011.454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coureuil M, Lecuyer H, Scott MG, Boularan C, Enslen H, Soyer M, Mikaty G, Bourdoulous S, Nassif X, Marullo S. Meningococcus Hijacks a beta2-adrenoceptor/beta-Arrestin pathway to cross brain microvasculature endothelium. Cell. 2010;143:1149–1160. doi: 10.1016/j.cell.2010.11.035. [DOI] [PubMed] [Google Scholar]
- Craig L, Volkmann N, Arvai AS, Pique ME, Yeager M, Egelman EH, Tainer JA. Type IV pilus structure by cryo-electron microscopy and crystallography: implications for pilus assembly and functions. Mol Cell. 2006;23:651–662. doi: 10.1016/j.molcel.2006.07.004. [DOI] [PubMed] [Google Scholar]
- Eugene E, Hoffmann I, Pujol C, Couraud PO, Bourdoulous S, Nassif X. Microvilli-like structures are associated with the internalization of virulent capsulated Neisseria meningitidis into vascular endothelial cells. J Cell Sci. 2002;115:1231–1241. doi: 10.1242/jcs.115.6.1231. [DOI] [PubMed] [Google Scholar]
- Geoffroy MC, Floquet S, Metais A, Nassif X, Pelicic V. Large-scale analysis of the meningococcus genome by gene disruption: resistance to complement-mediated lysis. Genome Res. 2003;13:391–398. doi: 10.1101/gr.664303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giltner CL, Nguyen Y, Burrows LL. Type IV pilin proteins: versatile molecular modules. Microbiol Mol Biol Rev. 2012;76:740–772. doi: 10.1128/MMBR.00035-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarner J, Greer PW, Whitney A, Shieh WJ, Fischer M, White EH, Carlone GM, Stephens DS, Popovic T, Zaki SR. Pathogenesis and diagnosis of human meningococcal disease using immunohistochemical and PCR assays. Am J Clin Pathol. 2004;122:754–764. doi: 10.1309/A7M2-FN2T-YE6A-8UFX. [DOI] [PubMed] [Google Scholar]
- Harlow E, Lane D. 1988. Antibodies: a laboratory manual.
- Helaine S, Carbonnelle E, Prouvensier L, Beretti JL, Nassif X, Pelicic V. PilX, a pilus-associated protein essential for bacterial aggregation, is a key to pilus-facilitated attachment of Neisseria meningitidis to human cells. Mol Microbiol. 2005;55:65–77. doi: 10.1111/j.1365-2958.2004.04372.x. [DOI] [PubMed] [Google Scholar]
- Helaine S, Dyer DH, Nassif X, Pelicic V, Forest KT. 3D structure/function analysis of PilX reveals how minor pilins can modulate the virulence properties of type IV pili. Proc Natl Acad Sci USA. 2007;104:15888–15893. doi: 10.1073/pnas.0707581104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbs M, Mattick JS. Common components in the assembly of type 4 fimbriae, DNA transfer systems, filamentous phage and protein-secretion apparatus: a general system for the formation of surface-associated protein complexes. Mol Microbiol. 1993;10:233–243. doi: 10.1111/j.1365-2958.1993.tb01949.x. [DOI] [PubMed] [Google Scholar]
- Ke SH, Madison EL. Rapid and efficient site-directed mutagenesis by single-tube “megaprimer” PCR method. Nucleic Acids Res. 1997;25:3371–3372. doi: 10.1093/nar/25.16.3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg DS, Jr, Cohen IR, Norins LC, Schroeter AL, Reising G. Neisseria gonorrhoeae. II. Colonial variation and pathogenicity during 35 months in vitro. J Bacteriol. 1968;96:596–605. doi: 10.1128/jb.96.3.596-605.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klee SR, Nassif X, Kusecek B, Merker P, Beretti JL, Achtman M, Tinsley CR. Molecular and biological analysis of eight genetic islands that distinguish Neisseria meningitidis from the closely related pathogen Neisseria gonorrhoeae. Infect Immun. 2000;68:2082–2095. doi: 10.1128/iai.68.4.2082-2095.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korotkov KV, Hol WG. Structure of the GspK-GspI-GspJ complex from the enterotoxigenic Escherichia coli type 2 secretion system. Nat Struct Mol Biol. 2008;15:462–468. doi: 10.1038/nsmb.1426. [DOI] [PubMed] [Google Scholar]
- Korotkov KV, Sandkvist M, Hol WG. The type II secretion system: biogenesis, molecular architecture and mechanism. Nat Rev Microbiol. 2012;10:336–351. doi: 10.1038/nrmicro2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurenceau R, Pehau-Arnaudet G, Baconnais S, Gault J, Malosse C, Dujeancourt A, Campo N, Chamot-Rooke J, Le Cam E, Claverys JP, Fronzes R. A type IV pilus mediates DNA binding during natural transformation in Streptococcus pneumoniae. PLoS Pathog. 2013;9:e1003473. doi: 10.1371/journal.ppat.1003473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long CD, Tobiason DM, Lazio MP, Kline KA, Seifert HS. Low-level pilin expression allows for substantial DNA transformation competence in Neisseria gonorrhoeae. Infect Immun. 2003;71:6279–6291. doi: 10.1128/IAI.71.11.6279-6291.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lory S, Strom MS. Structure-function relationship of type-IV prepilin peptidase of Pseudomonas aeruginosa–a review. Gene. 1997;192:117–121. doi: 10.1016/s0378-1119(96)00830-x. [DOI] [PubMed] [Google Scholar]
- Mairey E, Genovesio A, Donnadieu E, Bernard C, Jaubert F, Pinard E, Seylaz J, Olivo-Marin JC, Nassif X, Dumenil G. Cerebral microcirculation shear stress levels determine Neisseria meningitidis attachment sites along the blood-brain barrier. J Exp Med. 2006;203:1939–1950. doi: 10.1084/jem.20060482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehr IJ, Long CD, Serkin CD, Seifert HS. A homologue of the recombination-dependent growth gene, rdgC, is involved in gonococcal pilin antigenic variation. Genetics. 2000;154:523–532. doi: 10.1093/genetics/154.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melican K, Michea Veloso P, Martin T, Bruneval P, Dumenil G. Adhesion of Neisseria meningitidis to dermal vessels leads to local vascular damage and purpura in a humanized mouse model. PLoS Pathog. 2013;9:e1003139. doi: 10.1371/journal.ppat.1003139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikaty G, Soyer M, Mairey E, Henry N, Dyer D, Forest KT, Morand P, Guadagnini S, Prevost MC, Nassif X, Dumenil G. Extracellular bacterial pathogen induces host cell surface reorganization to resist shear stress. PLoS Pathog. 2009;5:e1000314. doi: 10.1371/journal.ppat.1000314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morand PC, Bille E, Morelle S, Eugene E, Beretti JL, Wolfgang M, Meyer TF, Koomey M, Nassif X. Type IV pilus retraction in pathogenic Neisseria is regulated by the PilC proteins. EMBO J. 2004;23:2009–2017. doi: 10.1038/sj.emboj.7600200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro S, Pelham HR. Use of peptide tagging to detect proteins expressed from cloned genes: deletion mapping functional domains of Drosophila hsp 70. EMBO J. 1984;3:3087–3093. doi: 10.1002/j.1460-2075.1984.tb02263.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassif X, Lowy J, Stenberg P, O'Gaora P, Ganji A, So M. Antigenic variation of pilin regulates adhesion of Neisseria meningitidis to human epithelial cells. Mol Microbiol. 1993;8:719–725. doi: 10.1111/j.1365-2958.1993.tb01615.x. [DOI] [PubMed] [Google Scholar]
- Parge HE, Forest KT, Hickey MJ, Christensen DA, Getzoff ED, Tainer JA. Structure of the fibre-forming protein pilin at 2.6 A resolution. Nature. 1995;378:32–38. doi: 10.1038/378032a0. [DOI] [PubMed] [Google Scholar]
- Pasloske BL, Paranchych W. The expression of mutant pilins in Pseudomonas aeruginosa: fifth position glutamate affects pilin methylation. Mol Microbiol. 1988;2:489–495. doi: 10.1111/j.1365-2958.1988.tb00055.x. [DOI] [PubMed] [Google Scholar]
- Peabody CR, Chung YJ, Yen MR, Vidal-Ingigliardi D, Pugsley AP, Saier MH., Jr Type II protein secretion and its relationship to bacterial type IV pili and archaeal flagella. Microbiology. 2003;149:3051–3072. doi: 10.1099/mic.0.26364-0. [DOI] [PubMed] [Google Scholar]
- Pelicic V. Type IV pili: e pluribus unum? Mol Microbiol. 2008;68:827–837. doi: 10.1111/j.1365-2958.2008.06197.x. [DOI] [PubMed] [Google Scholar]
- Pujol C, Eugene E, de Saint Martin L, Nassif X. Interaction of Neisseria meningitidis with a polarized monolayer of epithelial cells. Infect Immun. 1997;65:4836–4842. doi: 10.1128/iai.65.11.4836-4842.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Meth. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soyer M, Charles-Orszag A, Lagache T, Machata S, Imhaus AF, Dumont A, Millien C, Olivo-Marin JC, Dumenil G. Early sequence of events triggered by the interaction of Neisseria meningitidis with endothelial cells. Cell Microbiol. 2014;16:878–895. doi: 10.1111/cmi.12248. [DOI] [PubMed] [Google Scholar]
- Stephens DS, Hoffman LH, McGee ZA. Interaction of Neisseria meningitidis with human nasopharyngeal mucosa: attachment and entry into columnar epithelial cells. J Infect Dis. 1983;148:369–376. doi: 10.1093/infdis/148.3.369. [DOI] [PubMed] [Google Scholar]
- Trieu-Cuot P, Carlier C, Courvalin P. Conjugative plasmid transfer from Enterococcus faecalis to Escherichia coli. J Bacteriol. 1988;170:4388–4391. doi: 10.1128/jb.170.9.4388-4391.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valnes K, Brandtzaeg P. Retardation of immunofluorescence fading during microscopy. J Histochem Cytochem. 1985;33:755–761. doi: 10.1177/33.8.3926864. [DOI] [PubMed] [Google Scholar]
- Varga JJ, Nguyen V, O'Brien DK, Rodgers K, Walker RA, Melville SB. Type IV pili-dependent gliding motility in the gram-positive pathogen Clostridium perfringens and other Clostridia. Mol Microbiol. 2006;62:680–694. doi: 10.1111/j.1365-2958.2006.05414.x. [DOI] [PubMed] [Google Scholar]
- Weyand NJ, Wertheimer AM, Hobbs TR, Sisko JL, Taku NA, Gregston LD, Clary S, Higashi DL, Biais N, Brown LM, Planer SL, Legasse AW, Axthelm MK, Wong SW, So M. Neisseria infection of rhesus macaques as a model to study colonization, transmission, persistence, and horizontal gene transfer. Proc Natl Acad Sci USA. 2013;110:3059–3064. doi: 10.1073/pnas.1217420110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winther-Larsen HC, Hegge FT, Wolfgang M, Hayes SF, van Putten JP, Koomey M. Neisseria gonorrhoeae PilV, a type IV pilus-associated protein essential to human epithelial cell adherence. Proc Natl Acad Sci USA. 2001;98:15276–15281. doi: 10.1073/pnas.261574998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winther-Larsen HC, Wolfgang M, Dunham S, van Putten JP, Dorward D, Lovold C, Aas FE, Koomey M. A conserved set of pilin-like molecules controls type IV pilus dynamics and organelle-associated functions in Neisseria gonorrhoeae. Mol Microbiol. 2005;56:903–917. doi: 10.1111/j.1365-2958.2005.04591.x. [DOI] [PubMed] [Google Scholar]
- Wolfgang M, Lauer P, Park HS, Brossay L, Hebert J, Koomey M. PilT mutations lead to simultaneous defects in competence for natural transformation and twitching motility in piliated Neisseria gonorrhoeae. Mol Microbiol. 1998;29:321–330. doi: 10.1046/j.1365-2958.1998.00935.x. [DOI] [PubMed] [Google Scholar]
- Wolfgang M, van Putten JP, Hayes SF, Koomey M. The comP locus of Neisseria gonorrhoeae encodes a type IV prepilin that is dispensable for pilus biogenesis but essential for natural transformation. Mol Microbiol. 1999;31:1345–1357. doi: 10.1046/j.1365-2958.1999.01269.x. [DOI] [PubMed] [Google Scholar]
- Wolfgang M, van Putten JP, Hayes SF, Dorward D, Koomey M. Components and dynamics of fiber formation define a ubiquitous biogenesis pathway for bacterial pili. EMBO J. 2000;19:6408–6418. doi: 10.1093/emboj/19.23.6408. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.