Abstract
The positioning and the elongation of the mitotic spindle must be carefully regulated. In human cells, the evolutionary conserved proteins LGN/Gαi1-3 anchor the coiled-coil protein NuMA and dynein to the cell cortex during metaphase, thus ensuring proper spindle positioning. The mechanisms governing cortical localization of NuMA and dynein during anaphase remain more elusive. Here, we report that LGN/Gαi1-3 are dispensable for NuMA-dependent cortical dynein enrichment during anaphase. We further establish that NuMA is excluded from the equatorial region of the cell cortex in a manner that depends on the centralspindlin components CYK4 and MKLP1. Importantly, we reveal that NuMA can directly associate with PtdInsP (PIP) and PtdInsP2 (PIP2) phosphoinositides in vitro. Furthermore, chemical or enzymatic depletion of PIP/PIP2 prevents NuMA cortical localization during mitosis, and conversely, increasing PIP2 levels augments mitotic cortical NuMA. Overall, our study uncovers a novel function for plasma membrane phospholipids in governing cortical NuMA distribution and thus the proper execution of mitosis.
Keywords: dynein, NuMA, phosphoinositides, spindle elongation, spindle positioning
Introduction
Accurate positioning and elongation of the mitotic spindle are critical processes for error-free cell division. In animal cells, metaphase spindle positioning is dependent on an evolutionary conserved ternary complex consisting of a large coiled-coil protein, a GoLoCo domain containing protein, and a heterotrimeric G protein alpha subunit (reviewed in Kotak & Gönczy, 2013; Siller & Doe, 2009). In human cells, the members of this ternary complex are NuMA/LGN/Gαi1–3, and they serve to anchor the minus-end-directed motor protein complex dynein (hereafter referred to as dynein for simplicity) at the cell cortex during metaphase (Woodard et al, 2010; Kiyomitsu & Cheeseman, 2012; Kotak et al, 2012). Such cortically anchored dynein is thought to regulate spindle positioning by exerting a pull on the plus-end of astral microtubules and/or by its capability to remain associated with force-generating depolymerizing microtubules (reviewed in Kotak & Gönczy, 2013).
In addition to its role in spindle positioning, NuMA is also required for proper assembly and maintenance of the mitotic spindle (Yang & Snyder, 1992; Merdes et al, 1996). NuMA is a 2115 amino acid-long protein that comprises globular head and tail domains separated by a large coiled-coil moiety (Yang et al, 1992). An N-terminal fragment of NuMA can interact with dynein (Kotak et al, 2012), whereas the tail domain contains microtubule and LGN binding sites, as well as a nuclear localization signal (NLS) (Du & Macara, 2004). During metaphase, NuMA becomes weakly localized in the cortical regions situated above the two spindle poles as a result of its interaction with LGN/Gαi1-3 (Du & Macara, 2004; Woodard et al, 2010; Kiyomitsu & Cheeseman, 2012). Cortical levels of NuMA/dynein increase as cells progress from metaphase to anaphase, and such enrichment is crucial for robust spindle elongation (Collins et al, 2012; Kiyomitsu & Cheeseman, 2013; Kotak et al, 2013). NuMA can associate with several partners, including ERM 4.1 proteins (reviewed in Radulescu & Cleveland, 2010; Mattagajasingh et al, 1999), and it has been proposed that protein 4.1R and the related 4.1G together act as cortical anchors for NuMA during anaphase (Kiyomitsu & Cheeseman, 2013). During metaphase, the presence of Ran-GTP in the vicinity of chromosomes excludes NuMA from the equatorial cortical region and thus confines it to the polar regions (Kiyomitsu & Cheeseman, 2012). During anaphase, NuMA is also excluded from the equatorial cortical region (Kiyomitsu & Cheeseman, 2013; Kotak et al, 2013), but the mechanisms that operate at that stage to ensure continued equatorial exclusion remain to be discovered.
Centralspindlin is an evolutionary conserved complex that promotes cytokinesis in animal cells (reviewed in Glotzer, 2009). In human cells, this complex comprises two molecules of the Rho-family GTPase-activating protein CYK4 (also known as MgcRacGAP) and two molecules of the kinesin MKLP1 (Mishima et al, 2002; Pavicic-Kaltenbrunner et al, 2007; Loria et al, 2012). Centralspindlin promotes the activity of the small GTPases RhoA and Rac during furrow ingression, and is thus required for cytokinesis (Canman et al, 2008; Bastos et al, 2012; Loria et al, 2012). The centralspindlin component CYK4 directly associates with polyanionic phosphoinositides through a stretch of basic residues in its C1 domain, thus targeting CYK4 to the equatorial cortical region of the plasma membrane (Lekomtsev et al, 2012). Intriguingly, this localization is reciprocal to that of cortical NuMA, but whether there might be a relationship between these two components has not been investigated to date.
Here, we report that the ternary complex components LGN/Gαi1-3 are dispensable for NuMA/dynein localization to the cell cortex during anaphase. Furthermore, we uncover that the centralspindlin components CYK4 and MKLP1 are essential for excluding anaphase NuMA from the equatorial cortical region. We identify a small region within NuMA that targets the protein to the plasma membrane through interaction with the polyanionic phosphoinositides PIP and PIP2. Importantly, through chemical and enzymatic modulation, we demonstrate that the localization of NuMA is governed by such phosphoinositides during anaphase in vivo. Furthermore, we illustrate that premature increase in the levels of such membrane lipids during metaphase causes NuMA-dependent spindle positioning defects.
Results
LGN/Gαi1-3 are dispensable for cortical dynein localization during anaphase
In HeLa cells, cortical levels of dynein markedly increase in the polar regions of the cell during progression from metaphase to anaphase, and this in a NuMA-dependent manner (compare Fig1D with A; Kiyomitsu & Cheeseman, 2013; Kotak et al, 2013). Anaphase cortical NuMA/dynein contributes to spindle elongation during anaphase (Kotak et al, 2013) and is found in close proximity of astral microtubules at that stage (Supplementary Fig S1A). We discovered that cortical LGN, which is present in metaphase just like NuMA/dynein (Woodard et al, 2010; Kiyomitsu & Cheeseman, 2012), is barely detected in anaphase cells (compare Supplementary Fig S1D with B). This prompted us to investigate whether LGN and Gαi1-3 are required for anaphase NuMA/dynein cortical localization. We found that depleting LGN does not alter the anaphase cortical localization of NuMA, or of the dynein-associated dynactin component p150Glued (Fig1E, compare to 1B; see Supplementary Fig S1C and E for depletion efficiency). These results are consistent with recent findings by others (Kiyomitsu & Cheeseman, 2013; Zheng et al, 2014). Furthermore, we found that a GFP-NuMA fusion protein lacking a motif that is essential for interaction with LGN during metaphase [GFP-NuMA(ΔLGN)] (Du et al, 2001; Du & Macara, 2004) does not localize to the cortex in metaphase (Fig1G and I), but is cortical in anaphase (Fig1N).
Figure 1. Anaphase cortical NuMA/dynein localization is LGN/Gαi(1-3) independent.
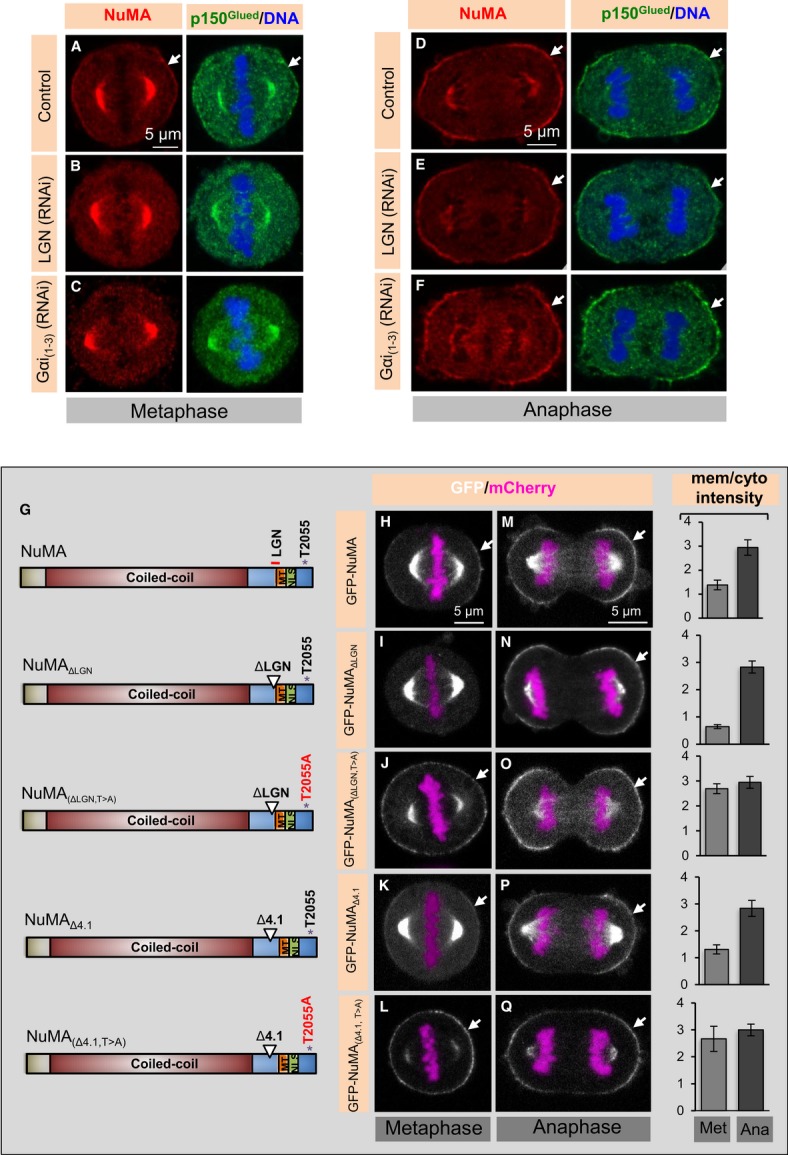
A–F HeLa cells in metaphase or anaphase, as indicated, transfected with control siRNAs (A, D), siRNAs against LGN (B, E) or Gαi(1-3) (C, F), fixed 72 h thereafter and stained for NuMA (red) as well as p150Glued (green). In this and other figures, DNA is visualized in blue and arrows point to cortical localization. In metaphase, 95% of control cells exhibited weak cortical p150Glued staining, as shown, but this was the case for only 2% of LGN (RNAi) and 5% of Gαi(1-3) (RNAi) cells, respectively. In anaphase, 100% of control cells, 98% of LGN (RNAi) cells, and 96% of Gαi(1-3) (RNAi) cells exhibited strong cortical NuMA/p150Glued signal as shown (n > 100 cells for all cases).
G Schematic representation of NuMA constructs used for the experiments shown on the right; the coiled-coil domain, the regions mediating interaction with LGN and microtubules (MT), the nuclear localization signal (NLS) and T2055 are represented.
H–Q Images from time-lapse microscopy of HeLa cells stably expressing mCherry-H2B and transfected with GFP-NuMA (H, M), GFP-NuMAΔLGN (I, N), GFP-NuMA(ΔLGN, T>A) (J, O), GFP-NuMAΔ4.1 (K, P) or GFP-NuMA(Δ4.1, T>A) (L, Q) in cells depleted of endogenous NuMA by RNAi for 72 h. The GFP signal is shown in white, the mCherry signal in pink. 4% of cells transfected with GFP-NuMA(ΔLGN) exhibited cortical GFP localization in metaphase, whereas 100% of such cells exhibited strong cortical GFP localization in anaphase. GFP-NuMA(ΔLGN, T>A) localizes to the cell cortex during metaphase in 100% of cells. Metaphase and anaphase cells expressing GFP-NuMA(Δ4.1) harbor GFP signal at the cortex similar to wild-type NuMA in 98% of the cells; note also that GFP-NuMA(Δ4.1, T>A) is strongly enriched at the plasma membrane already in metaphase. More than 50 cells were analysed by visual inspection in each case. Moreover, quantification of cortical enrichment is shown on the right for metaphase (Met) and anaphase (Ana) for 10 cells in each condition; see Materials and Methods (P < 0.0005 between metaphase and anaphase for GFP-NuMA, GFP-NuMAΔLGN, or GFP-NuMA(Δ4.1); P = 0.7 and P = 0.1 between metaphase and anaphase for GFP-NuMA(ΔLGN, T>A) and GFP-NuMA(Δ4.1, T>A), respectively; error bars, s.d.).
Because another GoLoco domain protein can substitute for LGN in regulating spindle positioning in a Gαi-dependent manner in some settings (Sanada & Tsai, 2005), we wondered whether an analogous situation may be at play during anaphase in HeLa cells. Therefore, we analysed the cortical distribution of NuMA/p150Glued upon compromised Gαi1-3 function. As shown in Fig1C and F and Supplementary Fig S1F–M, we found that the depletion of Gαi1-3 by siRNAs or their inactivation with Pertussis toxin does not affect NuMA/p150Glued cortical localization in anaphase, in contrast to their impact in metaphase.
NuMA is phosphorylated on T2055 by CDK1/cyclinB (hereafter referred as CDK1), and such phosphorylation negatively regulates cortical NuMA localization during metaphase (Kiyomitsu & Cheeseman, 2013; Kotak et al, 2013; Seldin et al, 2013). Accordingly, premature inhibition of CDK1 with RO-3306 or expression of a T2055A non-phosphorylatable version of NuMA [GFP-NuMA(T>A)] causes precocious strong enrichment of NuMA/dynein in metaphase, thus generating anaphase-like conditions (Kotak et al, 2013). Here, we found that depletion of LGN by siRNAs or inactivation of Gαi function by Pertussis toxin does not impair anaphase-like NuMA cortical enrichment in cells treated with RO-3306 (Supplementary Fig S1N–Q). The same holds upon expression of GFP-NuMA(ΔLGN, T>A), which in addition to bearing the T2055A mutation also lacks the LGN binding domain (Fig1G, J and O).
Overall, our results indicate that NuMA/dynein localization in the polar regions of the cell cortex during anaphase is LGN- and Gαi1–3-independent and that premature inactivation of CDK1 mimics anaphase-like cortical localization of NuMA/p150Glued.
Centralspindlin components negatively regulate cortical NuMA in the equatorial region during anaphase
Why is anaphase cortical NuMA distribution restricted to the polar regions? We set out to address this question by investigating how NuMA/p150Glued is prevented from occupying the equatorial cortical region (Fig1D; Collins et al, 2012; Kiyomitsu & Cheeseman, 2013; Kotak et al, 2013). During metaphase, Ran-GTP emanating from the centrally located chromatin negatively regulates NuMA distribution in the equatorial cortical region (Kiyomitsu & Cheeseman, 2012; Bird et al, 2013). We reasoned that this mechanism is unlikely to operate during anaphase, because chromosomes occupy a more polar position at that time (see Fig1D). Nonetheless, we investigated this possibility by analysing NuMA localization in anaphase cells expressing a dominant-negative form of Ran (GFP-RanT24N) or treated with Importazole, a small molecule inhibitor of Ran-GTP (Soderholm et al, 2011). Whereas both conditions influence the metaphase distribution of NuMA/dynein (Kiyomitsu & Cheeseman, 2012; Bird et al, 2013), we found by contrast that they do not alter cortical NuMA distribution in anaphase cells (Supplementary Fig S2A and B). We conclude that Ran-GTP does not modulate the spatial distribution of cortical NuMA during anaphase.
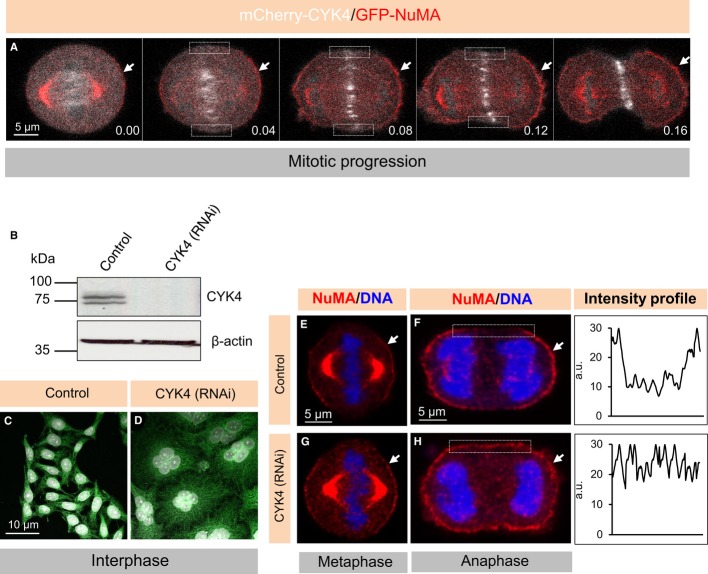
Since components of the cytokinetic machinery, including the centralspindlin components CYK4 and MKLP1, occupy the equatorial cortical region starting in anaphase (Fig2A and Supplementary Movie S1), we addressed whether centralspindlin is responsible for excluding NuMA from this region. Importantly, we found that cells depleted of CYK4 (Fig2B–D) or of MKLP1 fail to exclude NuMA from the equatorial cortical region during anaphase (Fig2E–H, Supplementary Fig S2C and D). To test whether this lack of exclusion is specific to the depletion of centralspindlin components or instead occurs irrespective of the cause of cytokinesis failure, we treated mitotic cells with the Rock-kinase inhibitor Y27632. Such treatment did not impair the exclusion of NuMA from the equatorial cortical region (Supplementary Fig S2E and F), indicating that impairing cleavage furrow contractility does not prevent NuMA equatorial exclusion. Although the biological significance of the lack of cortical NuMA/dynein in this area remains to be determined, we conclude that NuMA/dynein is excluded from the cortical equatorial region during anaphase in an CYK4- and MKLP1-dependent manner.
Figure 2. CYK4 prevents NuMA localization in the cortical equatorial region.
A Images from time-lapse recordings of a HeLa Kyoto cell transfected with GFP-NuMA and mCherry-CYK4 (see also corresponding Supplementary Movie S1). Arrows point to cortical NuMA localization and rectangles highlight weak CYK4 signal of CYK4 on the plasma membrane. Note that NuMA and CYK4 occupy mutually exclusive cortical regions. Time is indicated in [hours].[minutes], with t = 0 corresponding to the onset of the recording.
B Western blot with CYK4 antibodies of lysates from cells treated with control siRNAs or CYK4 siRNAs and synchronized in prometaphase with 100 nM nocodazole. β-actin was used as a loading control. Molecular weights are indicated in kiloDalton (kDa). Note that the specific CYK4 signal appears as a doublet.
C, D Interphase cells transfected with control siRNAs (C) or CYK4 siRNAs (D), fixed 72 h thereafter and stained for microtubules (green). DNA is visualized in white. Note accumulation of multi-nucleated cells in CYK4-depleted cells, indicating cytokinesis failure. 90% of the cells transfected with CYK4 are multi-nucleated in contrast to 2% in the control condition (n > 200).
E–H Metaphase and anaphase cells, as indicated, transfected with control siRNAs (E, F) or CYK4 siRNAs (G, H), fixed 48 h thereafter and stained for NuMA (red). Note the presence of NuMA in the cortical equatorial region in CYK4-depleted cells highlighted by the rectangle. The corresponding line scan of pixel intensities (in arbitrary units, a.u. on the y-axis) across the cortical region corresponding to the rectangle within the image is shown for one representative cell per condition. We found that 84% of the cells exhibit NuMA in the equatorial cortical region upon transfection with CYK4 siRNAs compared to 2% in control condition (n = 50 in each case).
Anaphase cortical localization of NuMA is mediated by a domain located in the C-terminal part, but not through an interaction with 4.1 proteins
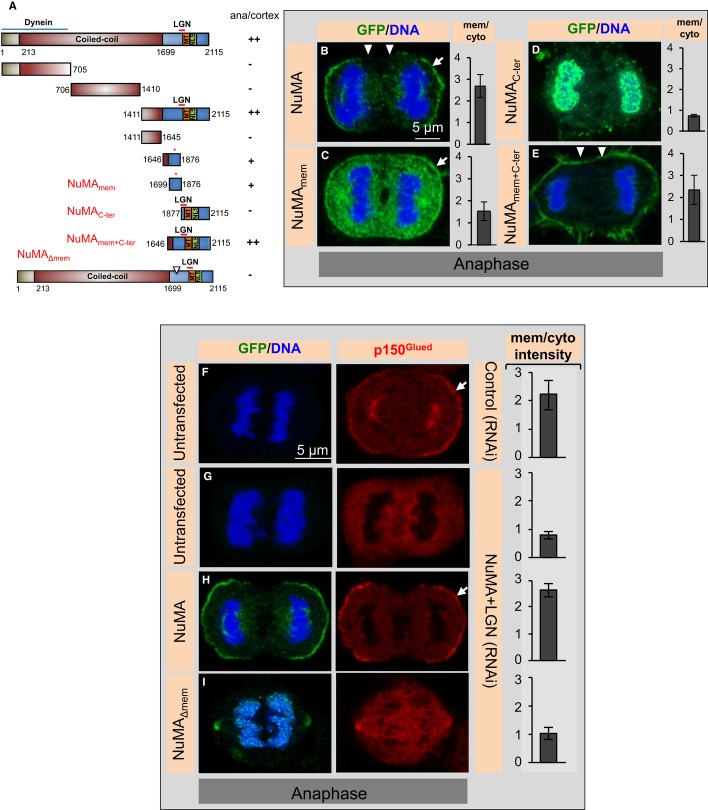
In order to decipher the mechanisms by which NuMA is targeted to the cell cortex in a LGN/Gαi1-3-independent manner during anaphase, we generated a series of truncation constructs fused with GFP and conducted transient transfection experiments to test which region is sufficient for cortical targeting. This revealed that a 178 amino acid-long region (aa 1699–1876) in the C-terminal part of NuMA (referred to as NuMAmem) can direct the fusion protein to the plasma membrane, albeit to a lesser extent than full-length NuMA or NuMAmem+C-ter (1646–2115) (Fig3A–C and E). Given that this 178 amino acid-long region does not contain the NLS, such plasma membrane localization is observed throughout the cell cycle (Figs 5B, 6A and Supplementary Fig S3A–D). Moreover, we found that fusing NuMAmem with NuMAC-ter, which itself cannot localize to the plasma membrane but that harbors the CDK1 phosphorylation site T2055, leads to robust plasma membrane localization during anaphase (Fig3D and E). This observation suggests that T2055 dephosphorylation during anaphase promotes robust NuMAmem enrichment at the plasma membrane. It was recently reported that a NuMA fragment spanning aa 1981–2060 can also localize to the plasma membrane during anaphase (Zheng et al, 2014), compatible with the notion that several stretches of NuMA may contribute to its presence at the membrane.
Figure 3. A small domain in the C-terminus of NuMA interacts with the plasma membrane.
A Schematic representation of NuMA constructs, as in Fig1G. ++ (strong), + (weak), or − (absent) indicates whether the respective GFP-tagged constructs localize to the cortex during anaphase (depicted as ana/cortex).
B–E Anaphase cells transfected with GFP-NuMA (aa 1–2115, B), GFP-NuMAmem (aa 1699–1876, C), GFP-NuMAC-ter (aa 1877–2115, D), or GFP-NuMAmem+C-ter (aa 1646–2115, E) for 36 h and stained for GFP (green). More than 20 cells were analysed by visual inspection in each case. Moreover, quantification of cortical enrichment is shown on the right for 10 cells in each condition (error bars, s.d.). Note localization of GFP-NuMAC-ter (aa 1877–2115) to chromosomes, which was observed in all cells for reasons that remain to be determined. Also note the equatorial localization of GFP-NuMAmem+C-ter (marked by arrowheads).
F–I Untransfected cells treated with control siRNAs (F) or NuMA siRNAs plus LGN siRNAs (G), as well as cells transfected with GFP-NuMA (H) or with GFP-NuMAΔmem (I) and treated with NuMA plus LGN siRNAs, all stained for GFP (green) and p150Glued (red). More than 23 cells were analysed by visual inspection in each case, and representative image is shown. Note that 60% of the GFP-NuMAΔmem-expressing cells exhibit no apparent GFP signal at the membrane, as shown, 30% weak GFP signal and 10% strong GFP signal, analogous to GFP-NuMA. Note also that GFP-NuMAΔmem-expressing cells show excess blebbing in mitosis. Quantification of anaphase cortical p150Glued enrichment is shown on the right for 10 cells in each condition (P < 0.0005 between GFP-NuMA transfected and untransfected cells treated with NuMA plus LGN siRNAs, P < 0.005 between GFP-NuMA-transfected and GFP-NuMAΔmem-transfected cells also treated with NuMA plus LGN siRNAs; error bars, s.d.).
What is the biological significance of abolishing such cortical NuMA localization? We have demonstrated that cortical dynein is needed for proper spindle elongation during anaphase (Kotak et al, 2013), and we thus addressed whether dynein distribution is affected in cells expressing GFP-NuMAΔmem and depleted of endogenous NuMA. We also depleted LGN in these experiments to eliminate the possibility that residual LGN that may be present in anaphase could mediate anchoring of GFP-NuMAΔmem when this fusion protein is overexpressed. Importantly, we found that whereas expression of GFP-NuMA in such a setting rescues anaphase cortical p150Glued localization, this is not the case in cells expressing GFP-NuMAΔmem, supporting the notion that NuMAmem is necessary for dynein localization during anaphase (Fig3F–I).
Intriguingly, a small domain within the 178 amino acid region is known to interact with the 4.1R protein (Mattagajasingh et al, 1999; Supplementary Fig S3E), which itself localizes to the cell cortex through an association with integral membrane proteins (Anderson & Lovrien, 1984). Therefore, we tested whether 4.1 proteins direct anaphase NuMA cortical localization. We found that simultaneous siRNA-mediated depletion of 4.1R and 4.1G (referred to as 4.1(R + G); Supplementary Fig S3F) not only decreases cortical localization of NuMA/p150Glued during anaphase, as reported (Kiyomitsu & Cheeseman, 2013), but also during metaphase (compare Supplementary Fig S3I and J with G and H). Since 4.1 proteins are essential for bridging the cortical actin cytoskeleton with membrane proteins (reviewed in Baines et al, 2014), we tested whether 4.1(R + G) depletion affects the cortical actin cytoskeleton during mitosis. As shown in Supplementary Fig S3K–N, we found that this is indeed the case. These results mirror the requirement of 4.1 proteins for cortical actin integrity in interphase migrating human cells (Ruiz-Saenz et al, 2011). We reasoned that if the consequence of 4.1(R + G) depletion on NuMA distribution during mitosis is through an impact on cortical actin, then cortical NuMA should be likewise affected by perturbing the actin cytoskeleton. To test this prediction, we treated mitotic cells with a range of concentrations of the actin depolymerizing drug Latrunculin A (50 nM to 1 μM; Supplementary Fig S4A–F and data not shown). We thus found that 200 nM of Latrunculin A impairs the cortical actin cytoskeleton and leads to a concomitant diminution of metaphase and anaphase NuMA/p150Glued cortical localization that resembles the effects observed upon 4.1(R + G) depletion (compare Supplementary Fig S4I and L with S3H and J). Therefore, an intact actin cytoskeleton is required for the presence of NuMA at the cell cortex during mitosis. Furthermore, to rigorously test whether interaction with 4.1 proteins is important, we generated a NuMA construct lacking the 4.1 binding region (GFP-NuMA(Δ4.1)). The resulting protein is unable to interact with either 4.1R or 4.1G in co-immunoprecipitation experiments, as anticipated (Supplementary Fig S4M). Importantly, we found that GFP-NuMA(Δ4.1) nevertheless localizes to the cell cortex, in a manner indistinguishable from wild-type GFP-NuMA, both in metaphase and anaphase (compare Fig1K and P with H and M). Moreover, we found that this is also the case for GFP-NuMA(Δ4.1) in LGN-depleted cells (Supplementary Fig S4N and O). Furthermore, GFP-NuMA(Δ4.1, T>A), which in addition to harboring a T2055A mutation also lacks the 4.1 binding region, localizes like GFP-NuMA(ΔLGN, T>A) (compare Fig1L and Q with J and O). In addition, as anticipated from the fact that GFP-NuMA(T>A) causes enrichment of cortical dynein in metaphase (Kotak et al, 2013), we found that GFP-NuMA(Δ4.1, T>A) causes excess cortical enrichment of p150Glued at the cell cortex (Supplementary Figs S4P–S).
Together, our findings indicate that 4.1R and 4.1G are not directly involved in mediating NuMA cortical localization during mitosis, a conclusion also reached by others (Seldin et al, 2013). Furthermore, our results demonstrate that a small fragment located in the C-terminal part of the protein is critical for anaphase cortical NuMA localization.
NuMA interacts with PtdInsP and PtdInsP2 in vitro
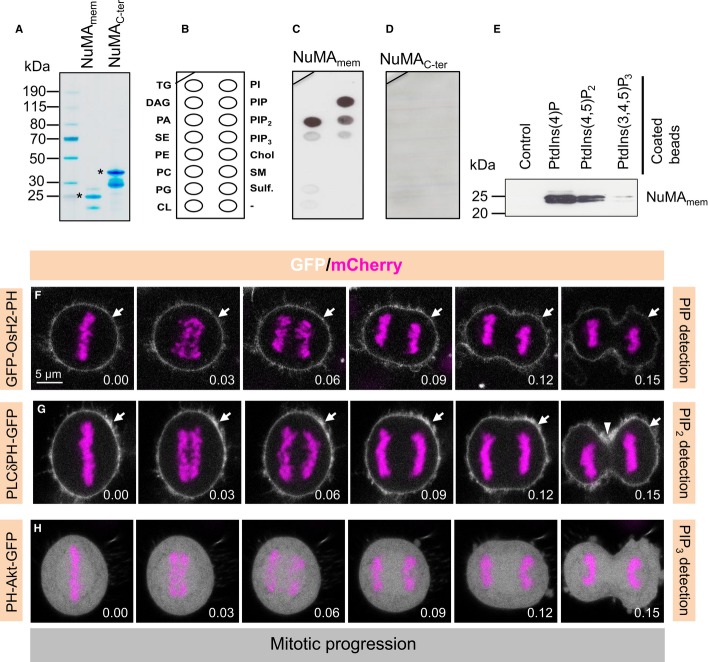
How can NuMA associate with the cell membrane during anaphase in a manner that is independent of LGN/Gαi1-3 and of 4.1 proteins? CYK4 associates directly with plasma membrane lipids in the cortical equatorial region (Lekomtsev et al, 2012), and since loss of CYK4 results in NuMA expanding into that region (see Fig2E–H), we considered whether NuMA could also directly associate with lipids. To investigate this possibility, we expressed and purified recombinant NuMAmem and tested its ability to bind various lipids species displayed on a nitrocellulose membrane (Fig4A and B). Interestingly, this analysis revealed that NuMAmem, but not another NuMA fragment that contains the LGN binding site (NuMAC-ter), can interact with polyanionic phosphoinositides, in particular PIP and PIP2, as well as phosphatidic acid (PA), and much more weakly PIP3 (Fig4C and D). Similar results were obtained in pull-down assays using beads coated with PIP, PIP2, or PIP3 (Fig4E). We investigated whether the position of the phosphate moiety on PIP and PIP2 is decisive for the interaction with NuMAmem, but found this not to be the case (Supplementary Fig S5A and B). Such lack of positional specificity has been observed for other lipid binding domains, including the PIP2-interacting PH domain (reviewed in Larijani & Poccia, 2012). Intriguingly, the two regions within NuMAmem that are outside of the 65 residues deleted in GFP-NuMA(Δ4.1) are well conserved amongst vertebrates (Supplementary Fig S3E); these regions contain basic amino acids that may mediate interaction with negatively charged polyanionic phosphoinositides.
Figure 4. NuMA interacts with polyanionic phosphoinositides in vitro and phosphoinositides distribution in mitosis.
A Recombinant purified hexa-histidine-tagged NuMAmem (aa 1699–1876) or NuMAC-ter (aa 1877–2115) run on SDS–PAGE and stained with Coomassie blue. Molecular mass is indicated in kiloDaltons (kDa). Note that bacterially expressed NuMAC-ter is unstable, thus explaining the presence of two major species. Asterisks mark expected molecular weight for each species.
B–D Lipid arrays (B) incubated with NuMAmem (C) or NuMAC-ter (D) and subsequently probed with anti-His antibodies. Key: triglyceride (TG), diacylglycerole (DAG), phosphatidic acid (PA), phosphatidylserine (PS), phosphatidylethanolamine (PE), phosphatidylcholine (PC), phosphatidylglycerole (PG), cardiolipin (CL), phosphatidylinositol (PI), phosphatidylinositol-4-phosphate PI(4)P, phosphatidylinositol-4,5-bisphosphate [PI(4,5)P2], phosphatidylinositol 3,4,5-triphosphate [PI(3,4,5)P3], cholesterol (Chol.), sphingomyelin (SM), sulfatide (Sulf.).
E Pull-down assay using recombinant NuMAmem, incubated with control beads or beads coated with indicated phosphoinositides. Recovered proteins were run on SDS–PAGE and subsequently probed with anti-His antibodies. Molecular mass is indicated in kiloDaltons (kDa).
F–H Images from time-lapse microscopy of HeLa cells stably expressing mCherry-H2B and transfected with OsH2-2xPH-GFP (F), PLCδPH-GFP (G), or PH-Akt-GFP (H) (see also corresponding Supplementary Movies S2, S3 and S4). The mCherry signal is shown in pink, the GFP signal in white. Time indicated in [hours].[minutes].
CYK4 interacts with PIP and PIP2 phosphoinositides in vivo (Lekomtsev et al, 2012), and since CYK4/MKLP1 depletion causes NuMA to occupy the equatorial cortical region, we focused on analysing the relevance of PIP and PIP2 in NuMA cortical localization. We explored whether these phosphoinositides are present at the right time and at the right place to potentially interact with NuMA during mitosis. We used time-lapse microscopy and visualized OsH2-2xPH-GFP to monitor PtdIns(4)P (Yu et al, 2004), PLCδ1PH-GFP to monitor PtdIns(4,5)P2 (Rescher et al, 2004), as well as PH-Akt-GFP to follow PtdIns(3,4,5)P3 (note that PH-Akt-GFP also detects PtdIns(3,4)P2; Watton & Downward, 1999). This analysis revealed that both PtdIns(4)P and PtdIns(4,5)P2 are present at the plasma membrane during metaphase and anaphase (Fig4F and G; Supplementary Movies S2 and S3). In addition, we found an increase in PtdIns(4,5)P2 at the cytokinesis furrow during late telophase, as reported previously (Field et al, 2005; Supplementary Movie S3). In contrast to the plasma membrane distributions of PtdIns(4)P and PtdIns(4,5)P2, PtdIns(3,4,5)P3 appears not to be enriched at the plasma membrane during mitosis (Fig4H and Supplementary Movie S4).
Taken together, the in vitro and in vivo experiments are both compatible with the notion that NuMA associates with PIP and/or PIP2 during mitosis.
Plasma membrane localization of NuMA depends on PIP and PIP2
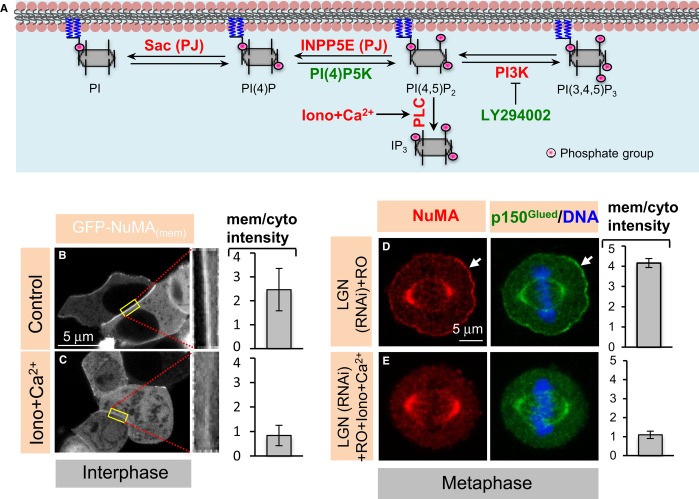
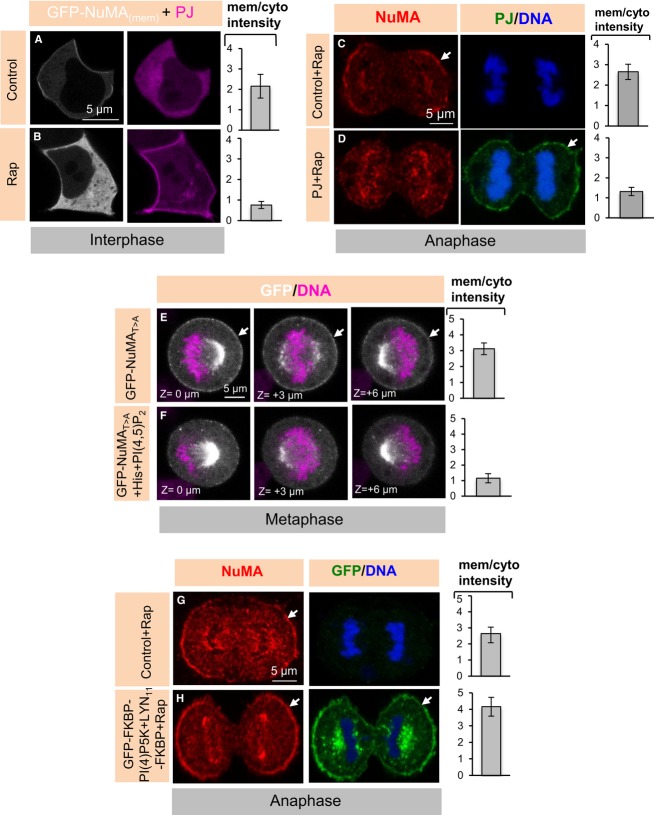
We set out to address whether PIP and PIP2 modulate NuMA distribution in vivo. To this end, we first depleted PIP and PIP2 from interphase cells by subjecting them to Ionomycin and Ca2+ for 200 s (Hammond et al, 2012; Fig5A). Importantly, we found that this treatment triggers the rapid release of NuMAmem from the plasma membrane to the cytosol in interphase cells (compare Fig5C with B). Because we found that such a short treatment with Ionomycin and Ca2+ does not alter the cortical signal of OsH2-2xPH-GFP and PLCδ1PH-GFP during mitosis (data not shown), possibly reflecting higher levels of PIP2, we sought to treat mitotic cells for a longer time. We found that exposure to Ionomycin and Ca2+ for 10 min is detrimental to the survival of anaphase cells (data not shown), precluding us from directly testing the impact of PIP and PIP2 depletion at this stage using this means. To circumvent this limitation, and given that cells earlier in mitosis survived such a 10-min treatment, we performed this experiment in metaphase cells that mimic anaphase-like conditions through LGN depletion and RO-3306 treatment. We found that membrane localization of PIP and PIP2, as monitored with OsH2-2xPH-GFP and PLCδ1PH-GFP, is substantially diminished in such cells upon Ionomycin and Ca2+ treatment (compare Supplementary Fig S5D and F with C and E). Strikingly in addition, we found that the cortical localization of NuMA/p150Glued is severely impaired under these conditions (compare Fig5E with D).
Figure 5. Membrane localization of NuMA depends on PtdIns(4)P and PtdIns(4,5)P2 in vivo.
A Schematic representation of the involvement of inhibitors and activators of PIP and PIP2 biosynthesis. Treatments that lead to increases in the levels of PIP and/or PIP2 are shown in green, those that result in corresponding decreases in red. Ionomycin and CaCl2 (Iono + Ca2+) treatment activates phospholipase C and thus decreases the levels of PIP and PIP2. INPP5E and Sac are part of the hybrid lipid phosphatase pseudojanin (PJ), which deplete PIP2 and PIP when targeted to the membrane using Rapamycin-based chemistry (see text for details). PI(4)P5K converts PI(4)P to PI(4,5)P2, thus resulting in increased PtdIns(4,5)P2 levels when targeted to the membrane. LY294002 is a PI3K inhibitor that enriches PIP2 upon PI3K inhibition.
B, C HEK293T interphase cells transfected with GFP-NuMAmem recorded before (control) and 200 s after treatment with 10 μM Ionomycin (Iono) and 1 mM CaCl2, with higher magnifications of the cortical regions highlighted by the yellow rectangle. Quantification of cortical enrichment is shown on the right in this and other panels of this figure and Fig6A; see Materials and Methods (P = 0.003; n = 10 cells each; error bars, s.d.).
D, E Metaphase HeLa cells transfected with siRNAs against LGN and treated with RO-3306 for 5 min (D) or treated with Ionomycin and CaCl2 (10 min) plus RO-3306 for 5 min (E) and stained for NuMA (red) as well as p150Glued (green) (P < 0.001; n = 10; error bars s.d.). We note also that 98% of LGN (RNAi) and RO-3306-treated cells exhibited strong cortical NuMA/p150Glued staining, as shown in (D), but this was the case for only 14% of LGN (RNAi); RO-3306; Ionomycin and CaCl2-treated cells (n > 50 cells for all cases).
Since phosphoinositides are important for maintaining the integrity of the actin cytoskeleton in some systems (reviewed by Yin & Janmey, 2003), it would be conceivable that the impact of Ionomycin and Ca2+ on NuMA/p150Glued localization occurs via their effect on the cortical actin cytoskeleton, a possibility that we set out to evaluate. We found that exposure to Ionomycin and Ca2+ leads to a dramatic loss of internal stress fibers in interphase cells (compare Supplementary Fig S5H with G). By contrast, we found that shortterm treatment with Ionomycin and Ca2+ has not apparent impact on the actin cytoskeleton in mitosis (compare Supplementary Fig S5J with I). We also performed a converse experiment by analysing PIP2 localization using the PLCδPH-GFP probe upon the addition of 1 μM Latrunculin A, and again found no apparent difference from the control (compare Supplementary Fig S5L with K). Overall, we conclude that phosphoinositides regulate NuMA/p150Glued localization independently of their influence on the actin cytoskeleton.
Next, we set out to selectively hydrolyse phosphoinositides at the plasma membrane instead of depleting them from the entire cell as upon Ionomycin and Ca2+ treatment. To this end, we utilized a construct in which Rapamycin induction targets the hybrid lipid phosphatase pseudojanin (PJ) to the plasma membrane, thus depleting PIP and PIP2 selectively in that location (Fig5A; Hammond et al, 2012). Importantly, we found that Rapamycin-mediated plasma membrane targeting of PJ causes the rapid release of NuMAmem from the plasma membrane (compare Fig6B with A). We also analysed the distribution of endogenous cortical NuMA by this means and found it to be drastically reduced following Rapamycin-mediated plasma membrane targeting of PJ (compare Fig6D with C). As anticipated from the experiments using Ionomycin and Ca2+, we found also that Rapamycin-mediated plasma membrane targeting of PJ does not alter the cortical actin cytoskeleton during anaphase or other stages of mitosis (Supplementary Fig S5M and N, data not shown). These experiments lead us to conclude that plasma membrane PIP and/or PIP2 phosphoinositides are essential for anaphase cortical NuMA localization.
Figure 6. Membrane localization of NuMA depends on PIP2 and/or PIP phosphoinositides in vivo.
A, B HEK293T cells co-transfected with GFP-NuMAmem, Lyn11–FRB, and monomeric red fluorescent protein (mRFP)-FKBP-tagged lipid phosphatases [pseudojanin (PJ); Hammond et al, 2012]. Images were taken before (control) and 200 s after treatment with 10 μM Rapamycin (Rap) (P < 0.001; n = 10; error bars: s.d.).
C, D Anaphase HeLa cells either untransfected (C) or co-transfected with Lyn11-CFP-FRB and mRFP-FKBP-tagged lipid phosphatases [pseudojanin (PJ); Hammond et al, 2012] for 36 h (D) and treated with Rapamycin for 10 min thereafter. Cells were fixed and stained for NuMA (red) and CFP to detect PJ (green) (P < 0.0005; n = 5 cells each; error bars, s.d.). Note that we could only obtain very few mitotic cells expressing PJ and also that we noticed a major incidence of apoptosis in PJ-expressing cells.
E, F Images from time-lapse microscopy of HeLa cells stably expressing mCherry-H2B and transfected with GFP-NuMAT>A, and imaged in three confocal sections either at the onset of the experiment (E) or 20 min after addition of PI(4,5)P2—histone complexes (F). The mCherry signal is shown in pink, the GFP signal in white (P = 0.0019; n = 5 cells in each condition; error bars, s.d.).
G, H Anaphase HeLa cells either untransfected (G) or co-transfected with YFP-FKBP-PI(4)P5K; LYN11-FRB for 24 h (H) and treated with 10 μM Rapamycin (Rap) for 10 min. Cells were fixed and stained for NuMA (red) and GFP (green) (P = 0.0006; n = 9 in each condition; error bars: s.d.). Note that only very few mitotic cells expressing GFP-FKBP-PI(4)P5K during mitosis could be obtained, suggesting that altering phosphoinositide levels in the cytoplasm, prior to Rap addition, impairs cell viability or cell cycle progression.
As a further test of the contribution of PIP2 for cortical NuMA localization, we set out to deliver synthetic PtdIns(4,5)P2 to the cytosol with a cationic histone carrier, a method previously used to alter lipid distribution (Ozaki et al, 2000; Weiner et al, 2002). We reasoned that if PIP2 can interact with NuMA at the cell membrane, then artificially elevating the levels of PIP2 in the cytosol should compete with and thus decrease the levels of cortical NuMA. As anticipated from this prediction, upon delivery of PtdIns(4,5)P2—histones complexes, we observed a diminution of the cortical GFP signal in cells expressing GFP-NuMA(T>A) (compare Fig6F with E), although cortical PLCδ1PH-GFP does not seem altered (data not shown). Perhaps this indicates tighter binding of PtdIns(4,5)P2—histones complexes to NuMA then to PLCδ1PH-GFP or, conversely, stronger affinity of PLCδ1PH-GFP than NuMA to the plasma membrane. By contrast, we found that delivery of carrier histone alone or complexed with PtdIns(3,4,5)P3 does not alter levels of cortical GFP-NuMA(T>A) (Supplementary Fig S5O–R). These results establish that NuMA can interact with PIP2 in vivo. Whether PIP may do so as well remains to be determined.
We then set out to address whether, conversely, increasing the levels of PIP2 at the cell membrane leads to an increase of cortical NuMA during anaphase. We utilized a distinct Rapamycin-based targeting method to enrich levels of PIP2 at the plasma membrane by delivering PI(4)P 5-kinase [PI(4)P5K], which converts PI(4)P to PI(4,5)P2, thus resulting in increased PtdIns(4,5)P2 levels (Ueno et al, 2011; Fig5A). Remarkably, we found that this markedly increases the levels of cortical NuMA during anaphase (compare Fig6H with G).
Overall, these results converge to support the notion that the plasma membrane localization of NuMA during anaphase is dictated by PIP2 and perhaps also by PIP.
Prematurely enriching PIP2 during metaphase perturbs spindle positioning
Experiments with drugs that impair the activity of phosphatidylinositide 3-kinases (PI3K) had implicated PtdIns(3,4,5)P3 in dynein-dependent spindle positioning during metaphase (Toyoshima et al, 2007). However, the nature of the protein whose distribution or function is affected by such drug treatment had not been determined. Because PI3K are involved in converting PIP2 to PIP3 (Fig5A) and since we found that NuMA can directly interact with PIP and/or PIP2, we reasoned that the reported metaphase spindle positioning phenotype upon PI3K inactivation may be due to excess cortical NuMA. We found that metaphase cells treated with the PI3K inhibitor LY294002 indeed exhibit increased PIP2 levels at the cell membrane as monitored with PLCδ1PH-GFP (Fig7A and B), in line with findings in interphase neuronal cells (Saengsawang et al, 2013). Interestingly, we found in addition that cortical NuMA levels in metaphase are increased under such conditions (compare Fig7D with C and Supplementary Movies S5 and S6). We then tested whether cells treated with LY294002 exhibit NuMA-dependent metaphase spindle positioning phenotypes. We monitored spindle positioning by analysing fixed cells on coverslips coated with an L-shaped fibronectin micropattern or on coverslips coated with uniform fibronectin (Supplementary Fig S6A–G; Théry et al, 2005; Toyoshima & Nishida, 2007). We found that spindle positioning is randomized in cells on both types of coverslips upon treatment with LY294002 (compare Supplementary Fig S6D and G with C and F; Toyoshima et al, 2007). Similarly, live-imaging experiments of cells treated with LY294002 uncovered dramatic chromosome oscillations, indicative of excess dynein-mediated pulling forces acting on the spindle poles, which are suppressed by NuMA depletion (Fig7E–G; Supplementary Movies S7, S8 and S9). Importantly in addition, we found that LY294002-induced chromosome oscillations are suppressed in cells also treated with Ionomycin and Ca2+ (compare Supplementary Fig S6I with H; Supplementary Movies S10 and S11), together suggesting that increases in PIP2/NuMA are responsible for excess movements upon LY294002 treatment.
Figure 7. Premature enrichment of PtdInsP2 (PIP2) causes NuMA-dependent spindle positioning defects.
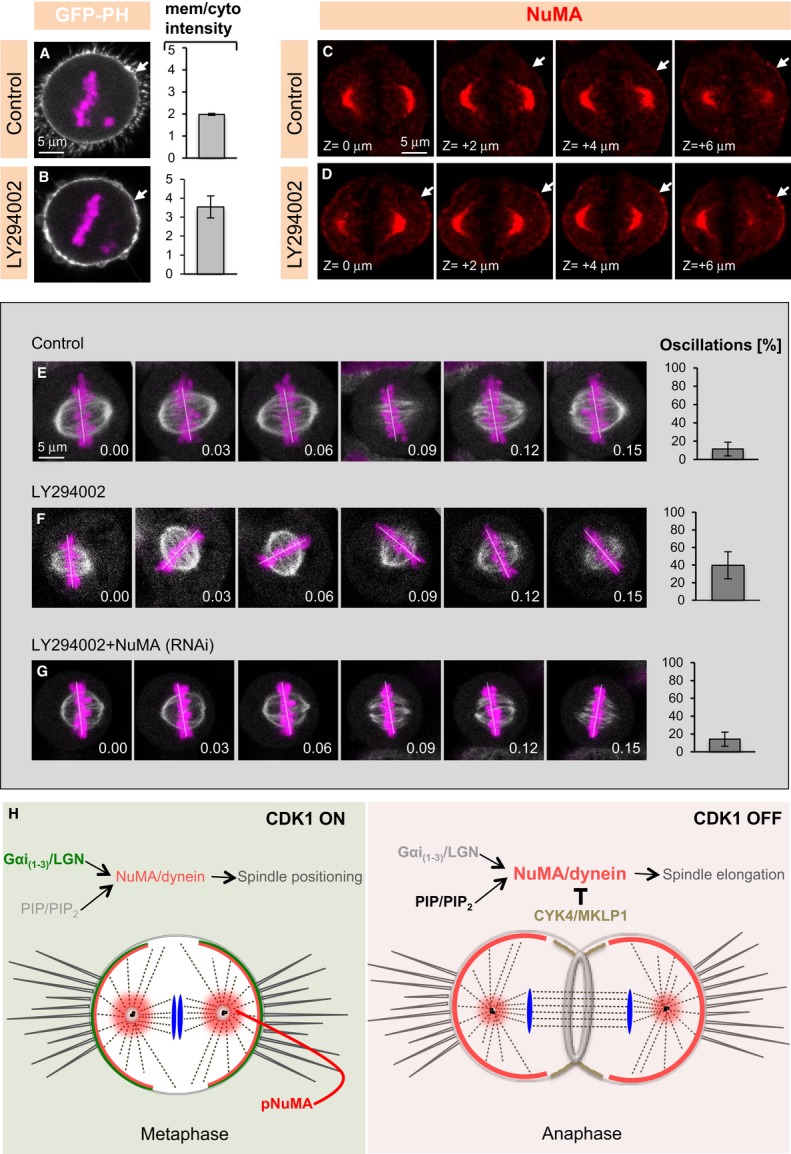
A, B Images from time-lapse microscopy of mitotic HeLa cells stably expressing mCherry-H2B and transfected with the PIP2-specific marker (PLCδPH-GFP) either treated with DMSO (Control) (A) or treated with 100 μM of the PI3K inhibitor LY294002 (B). Quantification of GFP cortical signal (right) was determined as mentioned in Materials and Methods. The mCherry signal is shown in pink and the GFP signal in white (P = 0.005; n = 10 cells in each condition; error bars, s.d.).
C, D Z-projection of images 2 μm apart of metaphase cell treated with DMSO (Control) or LY294002 and stained for NuMA (red). See corresponding Supplementary Movies S5 and S6 of the entire z-stack. 50 cells were analysed for each condition and representative results shown here.
E–G Images from time-lapse recordings of metaphase HeLa Kyoto cells stably expressing GFP-α-tubulin as well as mCherry-H2B (E) and treated with DMSO (Control) (E), LY294002 (F), and depleted of NuMA plus treated with LY294002 (G) (see also corresponding Supplementary Movies S7, S8, and S9). The white line indicates the position of chromosomes. Ten cells were imaged for each condition. The bar graphs on the right represent the frequency at which chromosome position changes > 10° between two frames, along with the standard deviation (s.d.). Time is indicated in [hours].[minutes], (P < 0.0005 between control (DMSO) and LY294002, as well as between LY294002 and LY294002 plus NuMA (RNAi); error bars, s.d.).
H Model for cortical localization of NuMA/dynein during metaphase, and anaphase. During metaphase, when CDK1 is active, the LGN/Gαi(1–3)-dependent pathway is responsible for cortical NuMA/dynein localization. By contrast, during anaphase, upon CDK1 inactivation, a PIP- and/or PIP2-based mechanism ensures cortical NuMA/dynein localization. Also note that CYK4/MKLP1 exclude NuMA/dynein from the equatorial cortical region in anaphase.
Discussion
The temporal and spatial regulation of dynein distribution is of critical importance for the proper execution of mitosis (reviewed in Kardon & Vale, 2009; Raaijmakers et al, 2013). Here, we propose a novel mechanism whereby NuMA directs dynein to the cell cortex during anaphase through direct association with phosphoinositides (Fig7H).
Switching from the ternary complex to phosphoinositides
An evolutionary conserved ternary complex is important during metaphase for ensuring that low levels of dynein are present in the polar regions of the cell cortex, from where the motor protein directs spindle positioning by exerting pulling forces on astral microtubules (Woodard et al, 2010; Kiyomitsu & Cheeseman, 2012; Kotak et al, 2012). The excess, as well as the lack, of cortical dynein during metaphase impairs spindle positioning, underscoring the importance of achieving proper levels of cortical dynein (Kotak et al, 2012). During metaphase, this is achieved by a tug-of-war between CDK1 kinase and PPP2CA phosphatase acting on T2055 of NuMA, whereby CDK1 negatively and PPP2CA positively regulate cortical NuMA/dynein levels (Kotak et al, 2013). At anaphase onset, CDK1 inactivation results in augmented levels of non-phosphorylated cortical NuMA and thus of dynein, which promotes spindle elongation during anaphase B (Collins et al, 2012; Kiyomitsu & Cheeseman, 2013; Kotak et al, 2013). As reported also by others (Kiyomitsu & Cheeseman, 2013; Seldin et al, 2013; Zheng et al, 2014), we found here that the ternary complex components LGN/Gαi1-3 are dispensable for NuMA-dependent enrichment of cortical dynein in the polar regions during anaphase.
We discovered instead that a region of NuMA encompassing aa 1699–1876 possesses the ability to bind phosphoinositides in vitro, including PIP and PIP2. A related observation has been reported recently for another NuMA fragment, encompassing aa 1981–2060, which can bind a partially overlapping set of phosphoinositides in vitro, including PIP and PIP2 (Zheng et al, 2014). Furthermore, that study reported that the plasma membrane localization of such a fragment fused to GFP depends on phosphoinositides in interphase (Zheng et al, 2014). However, whether this is also the case during mitosis, whether that fragment is not only sufficient but also necessary for such localization, and whether endogenous anaphase NuMA localization is dependent on phosphoinositides were not assessed in that study. Together with our findings, those observations raise the possibility that NuMA binds the plasma membrane through several basic residues present in the 1699–1876 and 1981–2060 fragments. Regardless, and importantly, by experimentally changing the cortical levels of phosphoinositides in vivo, we unequivocally demonstrate here that PIP and/or PIP2 regulate the cortical localization of both GFP-tagged and endogenous NuMA. Despite the fact that NuMA directly associates with PIP and PIP2 in vitro, we cannot formally exclude the possibility that another, as of yet unknown, protein would mediate interaction of NuMA with phosphoinositides in vivo.
Restricting NuMA/dynein to the polar regions of the cell
In addition to revealing a switch in the mechanism directing NuMA/dynein to the polar regions of the cell between metaphase and anaphase, our study uncovers a novel mechanism preventing their accumulation in the cortical equatorial region when comparing these two stages of the cell cycle. During metaphase, Ran-GTP emanating from the centrally located chromosomes prevents association of NuMA/dynein with the equatorial cortical regions (Kiyomitsu & Cheeseman, 2012; Bird et al, 2013). Such restriction of NuMA/dynein to the polar regions is important for proper spindle positioning. During anaphase, we show here that CYK4 and MKLP1, but not Ran-GTP, exert an analogous role. Preliminary results failed to reveal an impact on spindle elongation in cells depleted of centralspindlin components (data not shown), such that the potential importance of preventing NuMA/dynein from occupying the equatorial region remains to be determined.
Because CYK4 interacts with polyanionic phosphoinositides through its C1 domain (Lekomtsev et al, 2012), and because NuMAmem can also directly interact with polyanionic phosphoinositides, one tempting possibility is that both proteins compete for the same lipid moieties on the plasma membrane. However, fragments of NuMA fused to GFP that contain the anaphase membrane targeting sequences are not excluded from the equatorial region, suggesting that the postulated competition mechanism is more complex, with the N-terminal moiety of NuMA playing a role, perhaps owing to its ability to interact with dynein (Kotak et al, 2012). Furthermore, overexpression of full-length GFP-NuMA does not cause apparent cytokinesis abnormalities (data not shown). This suggests that excess NuMA is not sufficient to displace CYK4 from the equatorial region, perhaps because CYK4 has a higher local concentration and/or higher affinity toward phospholipids.
Anaphase-specific association with phosphoinositides
Why does the phosphoinositide-dependent mechanism that anchors NuMA to the polar regions of the cell normally not act during metaphase, since PIP and PIP2 are already available at the plasma membrane at that stage? One plausible explanation is that the little non-phosphorylated NuMA normally present during metaphase has a higher affinity toward LGN/Gαi1-3 than toward PIP and/or PIP2. In this scenario, only when a large excess of non-phosphorylated NuMA or an increase in PIP and/or PIP2 levels occurs could binding to phosphoinositides be revealed. This view is in line with the observation that either mimicking anaphase-like condition by acute inhibition of CDK1 or increasing the levels of cell membrane PIP2 through PI3K inhibition leads to premature cortical NuMA enrichment during metaphase, resulting in NuMA-dependent spindle positioning phenotypes. In C. elegans embryos, the polarity protein PAR-2, which can associate with phosphoinositides, is prevented from doing so if phosphorylated by PKC-3 (atypical protein kinase-3) (Motegi et al, 2011). By analogy, perhaps NuMA does not bind to phosphoinositides during metaphase because the bulk of NuMA is phosphorylated during that time. Irrespective of the actual mechanism, our work suggests that NuMA and thus dynein switch from a LGN/Gαi1-3-dependent to a phosphoinositide-dependent mode of targeting to the plasma membrane between metaphase and anaphase (Fig6H).
Intriguingly, it was proposed in C. elegans embryos that the cortical distribution of the ternary complex components GPR-1/2 and LIN-5, which are related to LGN and NuMA, respectively, is regulated by PI(4,5)P2 (Panbianco et al, 2008), indicating that the mechanism uncovered here may be evolutionary conserved.
Materials and Methods
Cell culture, siRNA treatment, and plasmid transfection
HeLa cells expressing GFP-Centrin 1 (Piel et al, 2000), HeLa Kyoto cells expressing EGFP-α-tubulin and mCherry-H2B (Schmitz et al, 2010), HeLa cells expressing mCherry-H2B as well as HEK293T cells were maintained as described previously (Kotak et al, 2013). Spindle positioning assays using fixed synchronized metaphase cells on coverslips uniformly coated with fibronectin (BD-Bioscience, 354088) were conducted as described earlier (Kotak et al, 2012, 2013). In addition to monitoring spindle positioning on uniform fibronectin as previously, here we also utilized fibronectin L-shape micropatterns (CYTOO SA), plating approximately 60,000 cells per chip in a 35-mm culture dish. After 1 h, cells that had not attached to these micropatterns were removed by gently washing with media. Cells were then fixed with PTEMF buffer (20 mM PIPES, pH 6.8, 10 mM EGTA, 1 mM MgCl2, 0.2% Triton X-100, 4% formaldehyde) for 10 min and stained with antibodies against γ-tubulin. siRNA experiments were performed as described (Kotak et al, 2012, 2013), In brief, 6 μl of 20 μM siRNAs in 100 μl OptiMEM medium (Invitrogen) and 4 μl of Lipofectamine RNAiMAX (Invitrogen) in 100 μl OptiMEM were incubated in parallel for 5 min, mixed for 20 min and then added to 2.5 ml medium per well containing approximately 100,000 cells. For plasmid transfections, 4 μg of plasmid DNA in 100 μl OptiMEM and 4 μl of Lipofectamine 2000 (Invitrogen) or 6 μl Turbofect (Thermo Scientific) in 100 μl OptiMEM were incubated in parallel for 5 min, mixed for 20 min, and added to each well.
Plasmids and RNAi
All NuMA clones were constructed using full-length NuMA as a template with appropriate PCR primer pairs (the sequences of all primers are available upon request). The amplified products were subcloned into pcDNA3-GFP (Merdes et al, 2000). The Rapamycin-regulated membrane-targeted system of lipid phosphatases is described in detail in Hammond et al, 2012. In brief, HEK293T cells were transfected with GFP-NuMAmem, Lyn11-FRB and a plasmid containing pseudojanin (PJ) (pmRFP- C1-FKBP-PJ); HeLa cells were transfected with plasmids containing PJ and LYN11-CFP-FRB. PJ contain polyphosphate-5-phosphatase E (INPP5E), which converts PtdIns(4,5)P2 to PtdIns(4)P, and Sac1 phosphatase (Sac), which dephosphorylates PtdIns(4)P. Similarly, the Rapamycin-based membrane targeted system of the lipid kinase PI(4)P5K is described in detail in Ueno et al (2011). In short, HeLa Kyoto cells were transfected with plasmids expressing GFP-FKBP-PI(4)P5K and LYN11-FRB. PI(4)P5K converts PI(4)P to PI(4,5)P2.
Double-stranded siRNA oligonucleotides were synthesized with the sequences:
UAGGAAAUCAUGAUCAAGCAA (LGN -siRNA, Qiagen)
GACUGCAGAACUUGCUGGAGUUAUA (Gαi1 -Stealth siRNA, Invitrogen)
GCCGUCACCGAUGUCAUCAUCAAGA (Gαi2 -Stealth siRNA, Invitrogen)
CCAAAGAAGUGAAGCUGCUGCUACU (Gαi3 -Stealth siRNA, Invitrogen)
GACGGUUAUUAAACCUGAAUCCUGU (CYK4 -Stealth siRNA, Invitrogen) and CAGGAUGUACAGAAGUUGAAGUGAA (MKLP1 -Stealth siRNA, Invitrogen).
In the cases where wild-type or mutant NuMA fusion constructs were expressed in NuMA-depleted cells, endogenous NuMA was depleted using the siRNAs sequence CCUCUGGAUCUAGAAGGGACCAUAA (NuMA -Stealth siRNA, Invitrogen) targeting the 3′UTR of the endogenous messenger, which is absent from the fusion constructs. Other sequences targeting NuMA and 4.1(R + G) are mentioned in Kotak et al (2013) as well as in Kiyomitsu and Cheeseman (2013).
Additional siRNAs were tested for LGN and CYK4, with a similar impact on anaphase NuMA/p150Glued localization. Cells were incubated with Pertussis toxin (List Biologicals, 181214A1) at 400 ng/ml for 4 h before analysis. For RanGFP/importin-β inhibition, cells were treated with 50 μM Importazole (Sigma-Aldrich, SML0341) for 10 min. CDK1 inhibition was performed by treating metaphase synchronized cells for 5 min with RO-3306 (Vassilev et al, 2006) (9 μM; Santa Cruz, sc-358700) For PIP and PIP2 inhibition, interphase and mitotic cells were treated with 10 μM Ionomycin (Calbiochem, 407950) and 1 mM Ca2+ for 200 s (interphase cells) and for 10 min (mitotic cells). Rapamycin (Sigma-Aldrich, R8781) was used at the concentration of 10 μM for 5 min for interphase cells and for 10 min for mitotic cells. For PI3K inhibition, cells were treated with 100 μM LY294002 (Cell Signaling, 9901). Actin, depolymerization was provoked with a range of concentrations (50 nM–1 μM) of Latranculin A (Sigma-Aldrich, L5613) for 10 min.
Indirect immunofluorescence and time-lapse imaging of HeLa cells
For immunofluorescence, cells were fixed in cold methanol and stained with the respective antibodies as described previously (Kotak et al, 2012, 2013). Primary antibodies were 1:200 rabbit anti-NuMA (Santa Cruz, sc-48773), 1:200 rabbit anti-LGN (Sigma-Aldrich, HPA007327), 1:200 mouse anti-p150Glued (Transduction Laboratories, 612709), 1:2,000 mouse anti-γ-tubulin (GTU88; Sigma-Aldrich), 1:300 mouse anti-GFP (MAB3580; Millipore), 1:500 rabbit anti-GFP (gift from V. Simanis), and 1:1,000 mouse anti-actin (Abnova, MAB8172).
Secondary antibodies were Alexa488 anti-mouse, Alexa488 anti-rabbit, Alexa568 anti-mouse, and Alexa568 anti-rabbit, all used at 1:500 (Invitrogen). Confocal images were acquired on a Zeiss LSM 710 confocal microscope equipped with a Axiocam MRm (B/W) CCD camera using a 63× NA 1.0 oil objective and processed in ImageJ and Adobe Photoshop, maintaining relative image intensities within a series.
Time-lapse microscopy was conducted on a Zeiss LSM 700 confocal microscope using a Axiocam MRm (B/W) CCD camera and a 40× NA 1.3 oil objective, in a Hi Q4 dish (Ibidi) at 5% CO2, 37°C, 90% humidity. Images were acquired every 3 min, capturing 4–5 sections, 7 μm apart at each time point. Time-lapse figures and movies were obtained using single confocal section of the Z-stack.
Spindle positioning assay
The angle of the metaphase spindle with respect to the fibronectin substratum was determined as described (Toyoshima & Nishida, 2007; Kotak et al, 2012, 2013). Briefly, cells were stained with γ-tubulin antibodies and counterstained with 1 μg/ml Hoechst 33342 (Sigma-Aldrich). Stacks of confocal images 0.4 μm apart were acquired and the distances between the two spindle poles in Z and in XY determined using Imaris (Bitplane Inc.); the spindle angle to the substratum was then calculated using inverse trigonometry.
Spindle positioning assay during live imaging
The bar graphs in live-spindle positioning assays are readouts of the extent of spindle oscillations, representing the frequency at which chromosome position changes > 10° between two frames (including when they move out of the imaging plane), along with the standard error of the mean (SEM). Two-tailed Student's t-tests were performed to determine the extent of spindle oscillations in various conditions.
Quantification of cortical intensity
Quantification of GFP cortical signal was determined by calculating the ratio of the mean intensity of the cortical signal divided by the mean intensity value in the cytoplasm (similar area) and correcting for background signal. Significance was determined using two-tailed Student's t-test for each condition.
Immunoprecipitation and immunoblotting
For co-immunoprecipitations, 3 mg of cell lysate were incubated with 30 μl GFP-Trap agarose beads (Chromotek, ACT-CM-GFA0050) in lysis buffer (10 mM Tris, pH 7.5; 150 mM NaCl; 0.5% NP-40; 0.5 mM EDTA; complete EDTA-free protease inhibitor tablet (11873580001, Roche)) for 4 h at 4°C. After extensive washing in wash buffer (10 mM Tris, pH 7.5; 150 mM NaCl; 0.5 mM EDTA; complete EDTA-free protease inhibitor tablet (Roche), the beads were denatured at 95°C in 2× SDS–PAGE buffer and analysed by SDS–PAGE and immunoblotting. For immunoblotting, 1:2,000 mouse anti-GFP (MAB3580, Millipore), 1:1,000 rabbit anti-CYK4 (A302-797A, Bethyl Laboratories), 1:500 rabbit anti-4.1G (A301-424A, Bethyl Laboratories),1:1,000 rabbit anti-4.1R (13014-1-AP, Protein Tech Group), and 1:10,000 mouse anti-actin (ABNOVA, MAB8172) primary antibodies were used; 1:5,000 anti-rabbit HRP (W4011, Promega) or 1:5,000 anti-mouse HRP (W4021, Promega) was used as secondary antibodies for Western blotting.
Lipid binding assay
His-NuMAmem (residues 1699–1876 with an hexa-histidine N-terminal fusion) and His-NuMAC-ter (residues 1877–2115 with an hexa-histidine N-terminal fusion) were expressed in the E. coli strain BL21. Membrane lipid arrays (P-6002 and P-6100, Echelon Biosciences) were incubated with 200 ng/ml of His-NuMAmem or His-NuMAC-ter in PBS containing 0.1% Tween-20 (PBST) and 3% BSA for 1 h at 25°C according to the manufacturer's protocol. After washing with PBST, proteins were detected using anti-His antibodies.
Cellular lipid delivery assay
The phosphoinositides—histone complexes were delivered intracellularly as described (Ozaki et al, 2000). Briefly, the histone—phosphoinositides complex was prepared using 300 μM of long-chain (Di-C16) phosphoinositides PI(4,5)P2 (P4516; Echelon Bioscience), PI(3,4,5)P3 (P-3916; Echelon Bioscience) with 100 μM of carrier histone (P-9C2; Echelon Bioscience). Twenty microlitre of each component were mixed, vortexed vigorously, and incubated at room temperature for 5–10 min before adding to the cells. Lipid delivery assays were performed in cells expressing GFP-NuMA(T>A), which mimics anaphase-like NuMA cortical localization (Kotak et al, 2013) and provides a time-window of up to 30 min to perform such experiments, in contrast to the much more rapid anaphase.
Acknowledgments
We thank Andreas Merdes, Daniel Gerlich, Arnaud Echard, Oliver Hantschel, Tamas Balla, and Takanari Inoue for precious reagents, as well as Marie Delattre, Fernando R. Balestra, Virginie Hachet, and Sveta Chakrabarti for critical remarks on the manuscript. We are grateful to the EPFL School of Life Sciences Microscopy Core Facility (PT-BiOP) for imaging advice. SK held a post-doctoral fellowship from the EMBO (ALTF-366-2009). This study was supported also by a grant to PG from the Swiss National Science Foundation (3100A0-122500/1).
Authors contributions
SK and PG conceived and designed the experiments; SK and CB executed experiments; SK and PG interpreted the results and wrote the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supplementary information for this article is available online: http://emboj.embopress.org
References
- Anderson RA, Lovrien RE. Glycophorin is linked by band 4.1 protein to the human erythrocyte membrane skeleton. Nature. 1984;307:655–658. doi: 10.1038/307655a0. [DOI] [PubMed] [Google Scholar]
- Baines AJ, Lu HC, Bennett PM. The Protein 4.1 family: hub proteins in animals for organizing membrane proteins. Biochim Biophys Acta. 2014;1838:605–619. doi: 10.1016/j.bbamem.2013.05.030. [DOI] [PubMed] [Google Scholar]
- Bastos RN, Penate X, Bates M, Hammond D, Barr FA. CYK4 inhibits Rac1-dependent PAK1 and ARHGEF7 effector pathways during cytokinesis. J Cell Biol. 2012;198:865–880. doi: 10.1083/jcb.201204107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird SL, Heald R, Weis K. RanGTP and CLASP1 cooperate to position the mitotic spindle. Mol Biol Cell. 2013;24:2506–2514. doi: 10.1091/mbc.E13-03-0150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canman JC, Lewellyn L, Laband K, Smerdon SJ, Desai A, Bowerman B, Oegema K. Inhibition of Rac by the GAP activity of centralspindlin is essential for cytokinesis. Science. 2008;322:1543–1546. doi: 10.1126/science.1163086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins ES, Balchand SK, Faraci JL, Wadsworth P, Lee WL. Cell cycle-regulated cortical dynein/dynactin promotes symmetric cell division by differential pole motion in anaphase. Mol Biol Cell. 2012;23:3380–3390. doi: 10.1091/mbc.E12-02-0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Q, Stukenberg PT, Macara IG. A mammalian Partner of inscuteable binds NuMA and regulates mitotic spindle organization. Nat Cell Biol. 2001;3:1069–1075. doi: 10.1038/ncb1201-1069. [DOI] [PubMed] [Google Scholar]
- Du Q, Macara IG. Mammalian Pins is a conformational switch that links NuMA to heterotrimeric G proteins. Cell. 2004;119:503–516. doi: 10.1016/j.cell.2004.10.028. [DOI] [PubMed] [Google Scholar]
- Field SJ, Madson N, Kerr ML, Galbraith KA, Kennedy CE, Tahiliani M, Wilkins A, Cantley LC. PtdIns(4,5)P2 functions at the cleavage furrow during cytokinesis. Curr Biol. 2005;15:1407–1412. doi: 10.1016/j.cub.2005.06.059. [DOI] [PubMed] [Google Scholar]
- Glotzer M. The 3Ms of central spindle assembly: microtubules, motors and MAPs. Nat Rev Mol Cell Biol. 2009;10:9–20. doi: 10.1038/nrm2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond GR, Fischer MJ, Anderson KE, Holdich J, Koteci A, Balla T, Irvine RF. PI4P and PI(4,5)P2 are essential but independent lipid determinants of membrane identity. Science. 2012;337:727–730. doi: 10.1126/science.1222483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardon JR, Vale RD. Regulators of the cytoplasmic dynein motor. Nat Rev Mol Cell Biol. 2009;10:854–865. doi: 10.1038/nrm2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyomitsu T, Cheeseman IM. Chromosome- and spindle-pole-derived signals generate an intrinsic code for spindle position and orientation. Nat Cell Biol. 2012;14:311–317. doi: 10.1038/ncb2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyomitsu T, Cheeseman IM. Cortical dynein and asymmetric membrane elongation coordinately position the spindle in anaphase. Cell. 2013;154:391–402. doi: 10.1016/j.cell.2013.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotak S, Busso C, Gönczy P. Cortical dynein is critical for proper spindle positioning in human cells. J Cell Biol. 2012;199:97–110. doi: 10.1083/jcb.201203166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotak S, Busso C, Gönczy P. NuMA phosphorylation by CDK1 couples mitotic progression with cortical dynein function. EMBO J. 2013;32:2517–2529. doi: 10.1038/emboj.2013.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotak S, Gönczy P. Mechanisms of spindle positioning: cortical force generators in the limelight. Curr Opin Cell Biol. 2013;25:741–748. doi: 10.1016/j.ceb.2013.07.008. [DOI] [PubMed] [Google Scholar]
- Larijani B, Poccia DL. Effects of phosphoinositides and their derivatives on membrane morphology and function. Curr Top Microbiol Immunol. 2012;362:99–110. doi: 10.1007/978-94-007-5025-8_5. [DOI] [PubMed] [Google Scholar]
- Lekomtsev S, Su KC, Pye VE, Blight K, Sundaramoorthy S, Takaki T, Collinson LM, Cherepanov P, Divecha N, Petronczki M. Centralspindlin links the mitotic spindle to the plasma membrane during cytokinesis. Nature. 2012;492:276–279. doi: 10.1038/nature11773. [DOI] [PubMed] [Google Scholar]
- Loria A, Longhini KM, Glotzer M. The RhoGAP domain of CYK-4 has an essential role in RhoA activation. Curr Biol. 2012;22:213–219. doi: 10.1016/j.cub.2011.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattagajasingh SN, Huang SC, Hartenstein JS, Snyder M, Marchesi VT, Benz EJ. A nonerythroid isoform of protein 4.1R interacts with the nuclear mitotic apparatus (NuMA) protein. J Cell Biol. 1999;145:29–43. doi: 10.1083/jcb.145.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merdes A, Ramyar K, Vechio JD, Cleveland DW. A complex of NuMA and cytoplasmic dynein is essential for mitotic spindle assembly. Cell. 1996;87:447–458. doi: 10.1016/s0092-8674(00)81365-3. [DOI] [PubMed] [Google Scholar]
- Merdes A, Heald R, Samejima K, Earnshaw WC, Cleveland DW. Formation of spindle poles by dynein/dynactin-dependent transport of NuMA. J Cell Biol. 2000;149:851–862. doi: 10.1083/jcb.149.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishima M, Kaitna S, Glotzer M. Central spindle assembly and cytokinesis require a kinesin-like protein/RhoGAP complex with microtubule bundling activity. Dev Cell. 2002;2:41–54. doi: 10.1016/s1534-5807(01)00110-1. [DOI] [PubMed] [Google Scholar]
- Motegi F, Zonies S, Hao Y, Cuenca AA, Griffin E, Seydoux G. Microtubules induce self-organization of polarized PAR domains in Caenorhabditis elegans zygotes. Nat Cell Biol. 2011;13:1361–1367. doi: 10.1038/ncb2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozaki S, DeWald DB, Shope JC, Chen J, Prestwich GD. Intracellular delivery of phosphoinositides and inositol phosphates using polyamine carriers. Proc Natl Acad Sci U S A. 2000;97:11286–11291. doi: 10.1073/pnas.210197897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panbianco C, Weinkove D, Zanin E, Jones D, Divecha N, Gotta M, Ahringer J. A casein kinase 1 and PAR proteins regulate asymmetry of a PIP(2) synthesis enzyme for asymmetric spindle positioning. Dev Cell. 2008;15:198–208. doi: 10.1016/j.devcel.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavicic-Kaltenbrunner V, Mishima M, Glotzer M. Cooperative assembly of CYK-4/MgcRacGAP and ZEN-4/MKLP1 to form the centralspindlin complex. Mol Biol Cell. 2007;18:4992–5003. doi: 10.1091/mbc.E07-05-0468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piel M, Meyer P, Khodjakov A, Rieder CL, Bornens M. The respective contributions of the mother and daughter centrioles to centrosome activity and behavior in vertebrate cells. J Cell Biol. 2000;149:317–330. doi: 10.1083/jcb.149.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaijmakers JA, Tanenbaum ME, Medema RH. Systematic dissection of dynein regulators in mitosis. J Cell Biol. 2013;201:201–215. doi: 10.1083/jcb.201208098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radulescu AE, Cleveland DW. NuMA after 30 years: the matrix revisited. Trends Cell Biol. 2010;20:214–222. doi: 10.1016/j.tcb.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rescher U, Ruhe D, Ludwig C, Zobiack N, Gerke V. Annexin 2 is a phosphatidylinositol (4,5)-bisphosphate binding protein recruited to actin assembly sites at cellular membranes. J Cell Sci. 2004;117(Pt 16):3473–3480. doi: 10.1242/jcs.01208. [DOI] [PubMed] [Google Scholar]
- Ruiz-Saenz A, Kremer L, Alonso MA, Millan J, Correas I. Protein 4.1R regulates cell migration and IQGAP1 recruitment to the leading edge. J Cell Sci. 2011;124(Pt 15):2529–2538. doi: 10.1242/jcs.083634. [DOI] [PubMed] [Google Scholar]
- Saengsawang W, Taylor KL, Lumbard DC, Mitok K, Price A, Pietila L, Gomez TM, Dent EW. CIP4 coordinates with phospholipids and actin-associated proteins to localize to the protruding edge and produce actin ribs and veils. J Cell Sci. 2013;126(Pt 11):2411–2423. doi: 10.1242/jcs.117473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanada K, Tsai LH. G protein betagamma subunits and AGS3 control spindle orientation and asymmetric cell fate of cerebral cortical progenitors. Cell. 2005;122:119–131. doi: 10.1016/j.cell.2005.05.009. [DOI] [PubMed] [Google Scholar]
- Schmitz MH, Held M, Janssens V, Hutchins JR, Hudecz O, Ivanova E, Goris J, Trinkle-Mulcahy L, Lamond AI, Poser I, Hyman AA, Mechtler K, Peters JM, Gerlich DW. Live-cell imaging RNAi screen identifies PP2A-B55alpha and importin-beta1 as key mitotic exit regulators in human cells. Nat Cell Biol. 2010;12:886–893. doi: 10.1038/ncb2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seldin L, Poulson ND, Foote HP, Lechler T. NuMA localization, stability, and function in spindle orientation involve 4.1 and Cdk1 interactions. Mol Biol Cell. 2013;24:3651–3662. doi: 10.1091/mbc.E13-05-0277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siller KH, Doe CQ. Spindle orientation during asymmetric cell division. Nat Cell Biol. 2009;11:365–374. doi: 10.1038/ncb0409-365. [DOI] [PubMed] [Google Scholar]
- Soderholm JF, Bird SL, Kalab P, Sampathkumar Y, Hasegawa K, Uehara-Bingen M, Weis K, Heald R. Importazole, a small molecule inhibitor of the transport receptor importin-beta. ACS Chem Biol. 2011;6:700–708. doi: 10.1021/cb2000296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Théry M, Racine V, Pepin A, Piel M, Chen Y, Sibarita JB, Bornens M. The extracellular matrix guides the orientation of the cell division axis. Nat Cell Biol. 2005;7:947–953. doi: 10.1038/ncb1307. [DOI] [PubMed] [Google Scholar]
- Toyoshima F, Matsumura S, Morimoto H, Mitsushima M, Nishida E. PtdIns(3,4,5)P3 regulates spindle orientation in adherent cells. Dev Cell. 2007;13:796–811. doi: 10.1016/j.devcel.2007.10.014. [DOI] [PubMed] [Google Scholar]
- Toyoshima F, Nishida E. Integrin-mediated adhesion orients the spindle parallel to the substratum in an EB1- and myosin X-dependent manner. EMBO J. 2007;26:1487–1498. doi: 10.1038/sj.emboj.7601599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno T, Falkenburger BH, Pohlmeyer C, Inoue T. Triggering actin comets versus membrane ruffles: distinctive effects of phosphoinositides on actin reorganization. Sci Signal. 2011;4:ra87. doi: 10.1126/scisignal.2002033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilev LT, Tovar C, Chen S, Knezevic D, Zhao X, Sun H, Heimbrook DC, Chen L. Selective small-molecule inhibitor reveals critical mitotic functions of human CDK1. Proc Natl Acad Sci U S A. 2006;103:10660–10665. doi: 10.1073/pnas.0600447103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watton SJ, Downward J. Akt/PKB localisation and 3′ phosphoinositide generation at sites of epithelial cell-matrix and cell-cell interaction. Curr Biol. 1999;9:433–436. doi: 10.1016/s0960-9822(99)80192-4. [DOI] [PubMed] [Google Scholar]
- Weiner OD, Neilsen PO, Prestwich GD, Kirschner MW, Cantley LC, Bourne HR. A PtdInsP(3)- and Rho GTPase-mediated positive feedback loop regulates neutrophil polarity. Nat Cell Biol. 2002;4:509–513. doi: 10.1038/ncb811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodard GE, Huang NN, Cho H, Miki T, Tall GG, Kehrl JH. Ric-8A and Gi alpha recruit LGN, NuMA, and dynein to the cell cortex to help orient the mitotic spindle. Mol Cell Biol. 2010;30:3519–3530. doi: 10.1128/MCB.00394-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CH, Lambie EJ, Snyder M. NuMA: an unusually long coiled-coil-related protein in the mammalian nucleus. J Cell Biol. 1992;116:1303–1317. doi: 10.1083/jcb.116.6.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CH, Snyder M. The nuclear-mitotic apparatus protein is important in the establishment and maintenance of the bipolar mitotic spindle apparatus. Mol Biol Cell. 1992;3:1259–1267. doi: 10.1091/mbc.3.11.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin HL, Janmey PA. Phosphoinositide regulation of the actin cytoskeleton. Annu Rev Physiol. 2003;65:761–789. doi: 10.1146/annurev.physiol.65.092101.142517. [DOI] [PubMed] [Google Scholar]
- Yu JW, Mendrola JM, Audhya A, Singh S, Keleti D, DeWald DB, Murray D, Emr SD, Lemmon MA. Genome-wide analysis of membrane targeting by S. cerevisiae pleckstrin homology domains. Mol Cell. 2004;13:677–688. doi: 10.1016/s1097-2765(04)00083-8. [DOI] [PubMed] [Google Scholar]
- Zheng Z, Wan Q, Meixiong G, Du Q. Cell cycle-regulated membrane binding of NuMA contributes to efficient anaphase chromosome separation. Mol Biol Cell. 2014;25:606–619. doi: 10.1091/mbc.E13-08-0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.