Abstract
In this issue of The EMBO Journal, mechanistic analyses of substrate cleavage by rhomboid intramembrane proteases suggest that catalytic efficiency towards natural, transmembrane substrates is allosterically stimulated by initial substrate interaction with an intramembrane exosite, whose formation depends on rhomboid dimerisation. In the realm of intramembrane proteolysis, dimerisation and allosteric cooperativity represent new concepts that, once confirmed more broadly, should radically alter our view of how these proteases work.
See also: E Arutyunova et al (September 2014)
Intramembrane proteolysis has emerged as an important and widespread biological regulatory mechanism with an increasing number of medical implications, but our mechanistic understanding of the unusual enzymes catalysing proteolysis in the membrane is still rudimentary. Although crystal structures of four classes of intramembrane proteases (serine-, metallo-, aspartyl- and glutamyl) have hitherto been solved (reviewed in Strisovsky, 2013), insights into their mechanism and regulation remains limited since we lack structures of their complexes with substrates. Over the past 8 years, rhomboid proteases have become the most tractable mechanistic and structural model intramembrane proteases, as reflected by a flurry of published reports. Interestingly, the rhomboid-like superfamily includes proteins that lack proteolytic activity, some of which (e.g. iRhoms, Derlins and RHBDDs) have already been found to have important biological functions, raising the attractive hypothesis that the basic function of the rhomboid-like domain is to bind transmembrane domains (reviewed in Adrain & Freeman, 2012). Structural and functional information about how rhomboids interact with their substrates should therefore also illuminate aspects of the functions of non-protease members of the superfamily.
In the new work, Arutyunova et al (2014) study the mechanism of substrate recognition and catalysis by rhomboid proteases, a topic intensely debated over the last few years. The authors conducted detailed initial rate analyses of three bacterial rhomboid proteases in detergent micelles, GlpG from Escherichia coli (ecGlpG), GlpG from H. influenzae (hiGlpG), and AarA from P. stuartii (psAarA), for cleavage of the substrate TatA from P. stuartii (psTatA). Unexpectedly, a gel-based assay revealed that these model reactions, including the physiological psAarA/psTatA pair, deviate from ideal Michaelis–Menten kinetics, instead showing positive cooperativity. In layman's terms, this means that the enzymes switch into a more active form upon binding to substrate—a kind of positive feedback (see Fig1). In contrast, cleavage of fluorescently labelled casein, a soluble, non-membranous (and non-physiological) substrate, lacked cooperativity, suggesting that allosteric cooperativity depends on the presence of a transmembrane domain (TMD), assumed to be a critical feature of all natural rhomboid substrates.
Figure 1. Schematic representation of substrate binding and allosteric regulation of rhomboid protease dimers and monomers.
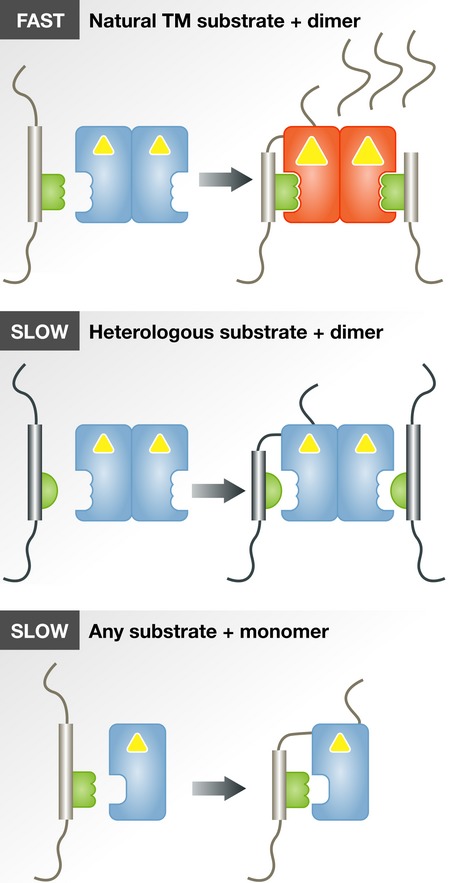
Yellow triangles represent protease active sites. See text for details.
Beyond the discovery of cooperativity, the authors make a strong case for reversible dimerisation of ecGlpG. While it is monomeric in decylmaltoside (DM) detergent, it is predominantly dimeric in dodecylmaltoside (DDM), and it is only in DDM that rhomboids exhibit the cooperativity described above. Monomeric forms of all three rhomboids were significantly less active in vitro than the dimeric ones against the transmembrane substrate psTatA, although both monomers and dimers retain baseline activity against soluble casein (Arutyunova et al, 2014). Since ecGlpG is also mainly dimeric in lipid membranes of E. coli cells (Sampathkumar et al, 2012; Lazareno-Saez et al, 2013), this suggests that dimers are the biologically relevant active form of rhomboids.
Continuing the theme that classical enzymological approaches still have value, Arutyunova et al provided further mechanistic insights by analysing the nature of competition between substrates. The transmembrane substrate psTatA inhibited cleavage by rhomboids of the soluble substrate casein. This inhibition was competitive in the case of ecGlpG and hiGlpG, but non-competitive for psTatA's physiological enzyme psAarA. This implied that psTatA and casein compete for the same catalytic binding site on heterologous enzymes, while on its cognate enzyme, psTatA may bind to an additional binding site (exosite) before binding the catalytic site. Again, non-competitive inhibition of casein cleavage by psTatA was only observed for dimeric psAarA in DDM detergent but not for its monomeric form in DM. From this, the authors conclude that the natural, but not a heterologous, substrate elicits a homotropic allosteric activation effect by binding to an intramembrane exosite on a rhomboid dimer. In other words, binding of the natural substrate primes the dimeric enzyme for higher affinity or catalytic efficiency, which makes it more selective for its natural substrate (see Fig1). These results support an earlier proposal of the existence of an intramembrane exosite on rhomboids (Strisovsky et al, 2009), to which substrate TMD binds prior to engaging the catalytic site, and are consistent with other recent observations (Fleig et al, 2012). Conceptually, the presence of two recognition elements in substrates, each by itself with relatively non-stringent recognition preferences, is an elegant mechanism to sharpen rhomboid specificity (Strisovsky, 2013), especially if they allosterically influence each other as shown in the new work.
It is worth pointing out some considerable differences between the current work and another recent study by Dickey et al (2013), which also used enzymological approaches to infer mechanism of rhomboid function. Both papers used the psTatA substrate, but Dickey et al focused largely on the heterologous E. coli enzyme ecGlpG, assayed not in detergent micelles but in liposomes. The most striking discrepancy is that Dickey et al did not observe any cooperativity in the kinetics of the ecGlpG/psTatA reaction, but this might simply be explained by Arutyunova et al's finding that this particular enzyme/substrate pair exhibited only very weak cooperativity. Another difference is that Arutyunova et al measured a approximately 30-fold higher affinity of ecGlpG for psTatA; the reasons for this large difference are less clear, but they may be due to the different assay methods or the exact substrate construct employed. Finally, whereas Dickey et al find that the major distinction between different rhomboids is in their catalytic rate (kcat), Arutyuonova et al observe significant differences also in substrate affinity (KD and KM); this distinction may reflect the consequence of measuring the reaction in the lipid bilayer or detergents, respectively. Unsurprisingly, these differences led the two teams to interpret their data in qualitatively different ways, but direct comparison shows that the data are, in fact, compatible in several important respects: Both studies confirm earlier results showing that rhomboids display substrate specificity (Urban et al, 2002; Urban & Freeman, 2003; Strisovsky et al, 2009). Furthermore, the results of Dickey et al do not exclude the dimerisation of rhomboids revealed by Arutyunova et al, and the observation by Dickey et al that mutations in the P1 or P4 and P2′ positions of the “recognition motif” of psTatA severely inhibit its cleavage by ecGlpG is also compatible with the results of Arutyunova et al (and both are consistent with an earlier report of this substrate determining motif, Strisovsky et al, 2009). In conclusion, although it is too early to describe an emerging consensus, the differences between these recent papers may not be as great as they at first appear.
Overall, both recent papers make a strong case for the power of enzymological analysis, combined with prior structural information, for revealing mechanistic details about intramembrane proteases. Unsurprisingly, even after these publications, we are left with as many questions as answers. For example, what constitutes the proposed intramembrane exosite? Is dimerisation a common property of rhomboids and, if so, which parts of the rhomboid molecule form the dimerisation interface? And how is the exosite influenced by rhomboid dimerisation? Stepping back from the details, however, these papers represent substantial progress and we now have several clear hypotheses that can be tested further by biochemical and structural methods. Given the biological and medical importance of intramembrane proteolysis, as well as the recently discovered importance of the related rhomboid-like pseudoproteases, which may share such substrate/client recognition principles, rhomboid mechanisms are likely to remain centre stage for quite a while.
Acknowledgments
KS acknowledges support by Czech Ministry of Education (projects no. LK11206 and LO1302), EMBO (Installation Grant No. 2329) and the National Subvention for Development of Research Organizations (RVO: 61388963) to the Institute of Organic Chemistry and Biochemistry (IOCB). MF is supported by a Wellcome Trust Investigator Award.
References
- Adrain C, Freeman M. New lives for old: evolution of pseudoenzyme function illustrated by iRhoms. Nat Rev Mol Cell Biol. 2012;13:489–498. doi: 10.1038/nrm3392. [DOI] [PubMed] [Google Scholar]
- Arutyunova E, Panwar P, Skiba PM, Gale N, Mak MW, Lemieux MJ. Allosteric regulation of rhomboid intramembrane proteolysis. EMBO J. 2014;33:1869–1881. doi: 10.15252/embj.201488149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickey SW, Baker RP, Cho S, Urban S. Proteolysis inside the membrane is a rate-governed reaction not driven by substrate affinity. Cell. 2013;155:1270–1281. doi: 10.1016/j.cell.2013.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleig L, Bergbold N, Sahasrabudhe P, Geiger B, Kaltak L, Lemberg MK. Ubiquitin-dependent intramembrane rhomboid protease promotes ERAD of membrane proteins. Mol Cell. 2012;47:558–569. doi: 10.1016/j.molcel.2012.06.008. [DOI] [PubMed] [Google Scholar]
- Lazareno-Saez C, Arutyunova E, Coquelle N, Lemieux MJ. Domain swapping in the cytoplasmic domain of the Escherichia coli rhomboid protease. J Mol Biol. 2013;425:1127–1142. doi: 10.1016/j.jmb.2013.01.019. [DOI] [PubMed] [Google Scholar]
- Sampathkumar P, Mak MW, Fischer-Witholt SJ, Guigard E, Kay CM, Lemieux MJ. Oligomeric state study of prokaryotic rhomboid proteases. Biochim Biophys Acta. 2012;1818:3090–3097. doi: 10.1016/j.bbamem.2012.08.004. [DOI] [PubMed] [Google Scholar]
- Strisovsky K, Sharpe HJ, Freeman M. Sequence-specific intramembrane proteolysis: identification of a recognition motif in rhomboid substrates. Mol Cell. 2009;36:1048–1059. doi: 10.1016/j.molcel.2009.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strisovsky K. Structural and mechanistic principles of intramembrane proteolysis – lessons from rhomboids. FEBS J. 2013;280:1579–1603. doi: 10.1111/febs.12199. [DOI] [PubMed] [Google Scholar]
- Urban S, Schlieper D, Freeman M. Conservation of intramembrane proteolytic activity and substrate specificity in prokaryotic and eukaryotic rhomboids. Curr Biol. 2002;12:1507–1512. doi: 10.1016/s0960-9822(02)01092-8. [DOI] [PubMed] [Google Scholar]
- Urban S, Freeman M. Substrate specificity of rhomboid intramembrane proteases is governed by helix-breaking residues in the substrate transmembrane domain. Mol Cell. 2003;11:1425–1434. doi: 10.1016/s1097-2765(03)00181-3. [DOI] [PubMed] [Google Scholar]


