Abstract
Primary cilia are solitary, microtubule-based organelles that serve as signaling hubs for the Hedgehog (Hh) pathway, which regulates embryonic development and adult tissue homeostasis. While protein localization studies have suggested that the dynamic trafficking of Hh components at cilia plays an important role, the molecular basis of Hh signal transduction at cilia is not well understood. In a recent study published in Nature Cell Biology (He et al, 2014), He and colleagues demonstrate that the kinesin KIF7, a conserved regulator of Hh signaling, limits ciliary length by acting at the plus-ends of microtubules to both reduce growth rate and increase catastrophe frequency. They propose that this biochemical activity establishes a specialized compartment at the tip of the cilia where the activity the Gli family of Hh transcription factors is regulated.
See also: M He et al (July 2014)
Cilia are microtubule-based structures that are present on almost all cells in our bodies. They are classified into two types—motile and immotile. Motile cilia function in processes such as cleaning dirt from airway tracts and transporting oocytes in the oviduct. Non-motile or primary cilia, poorly studied for decades, have emerged as key signaling organelles in development. Defects in the structure and function of cilia are associated with a wide spectrum of developmental disorders collectively referred to as the ciliopathies. The central structural feature of the primary cilium is the membrane-ensheathed axoneme, which is composed of 9 pairs of doublet microtubules templated on the 9 triplets of the basal body (Fig1A). These microtubules are all oriented with their plus-ends abutting the membrane at the tips of cilia, with electron microscopy studies showing the presence of stereotyped structures that seem to cap the microtubules and tether them to the membrane (Dentler, 1980). Several distinct compartments in cilia have been defined (Fig1A), either based on morphology or on the localization of specific proteins. Other than the tip and the basal body, these include the ciliary pocket, a membrane cleft surrounding the ciliary base that serves as a station for vesicle trafficking, and the transition zone (TZ), a region of tight membrane–microtubule contacts near the base that functions as a barrier between the cilium and the cell.
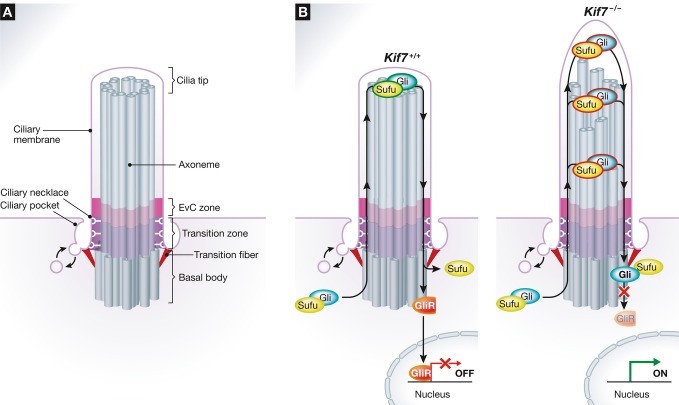
Figure 1. Ciliary compartments in Hh signaling.
(A) Depiction of ciliary compartments where many Hh pathway components are localized. Ptch1 is found at the base of cilia, around the basal body and the pocket. Both Ptch1 and Smo are localized in the ciliary membrane. Smo interacts with the Ellis-van Creveld syndrome (EvC) membrane protein complex at the EvC Zone, located just distal to the Transition Zone and Transition Fibers. Gli and Sufu are localized at cilia tips. (B) A model for the regulation of Gli–Sufu complexes with (left) or without (right) KIF7 in the absence of Hh signal activation. Trafficking of Gli–Sufu complexes through the cilium licenses them for repressor (GliR) production, leading to inactivation of target genes in the nucleus. In the absence of KIF7, GliR production is impaired, and thus, its target genes are derepressed.
The Hh pathway regulates tissue patterning and cell-fate determination during embryonic development and its deregulation is often associated with pathological outcomes such as birth defects and cancer. A subset of the phenotypes seen in ciliopathy patients can be attributed to defective Hh signaling. Some Hh signaling components are concentrated in specific ciliary compartments. In the absence of signaling, Ptch1 resides at the base of cilia, perhaps in the pocket or in nearby vesicles, in addition to in punctate structures along the ciliary membrane. Ptch1 inhibits Smo, preventing its high-level accumulation in the ciliary membrane. In this OFF state, the production of transcriptionally active Gli proteins (Gli2/3A) is restrained by their association with Suppressor of Fused (Sufu) and by Protein Kinase A (PKA) activity. Instead, Sufu association and PKA promote the conversion of full-length Gli3 (Gli3FL) into a transcriptional repressor (Gli3R, Fig1B). While the former process, inhibition of Gli2/3A formation, is independent of primary cilia and likely occurs in the cytoplasm, the latter, Gli3R formation, depends on an intact cilium. Indeed, Gli2/3 and Sufu can be found localized at cilia tips even in the absence of signaling. Hh ligands induce a stereotyped reorganization in the localization of these proteins—Ptch1 leaves cilia, Smo accumulates in the ciliary membrane to high levels with a distinct concentration just distal to the TZ, and Gli2/3 and Sufu levels at the ciliary tip increase. This results in the dissociation of Sufu from Gli2 and Gli3—thereby preventing Gli3R formation and allowing Gli2A to enter the nucleus to induce target genes. A parsimonious way to conceptualize the above is that ciliary transport of Gli–Sufu complexes has distinct outcomes in the absence and presence of Hh ligands—conversion to Gli repressors (GliR) or Gli activators (GliA), respectively. The switch between the two is likely operated by the high-level accumulation of active Smo in the ciliary membrane. Thus, the loss of cilia leads to defects in both GliR and GliA formation.
In Drosophila, where Hh signal transduction does not require primary cilia, the kinesin-like protein Costal 2 (Cos2) scaffolds a signaling complex that also controls whether cubitus interruptus (Ci), the single homolog of the Gli proteins, is converted to repressor or activator forms. Studies in both zebrafish (Tay et al, 2005) and mice (Cheung et al, 2009; Endoh-Yamagami et al, 2009; Liem et al, 2009) established a role for the Cos2 homolog KIF7 in vertebrate Hh signal transduction. Loss of KIF7 or inactivating mutations in its motor domain (KIF7L130P) in mice results in exencephaly and polydactyly due to ectopic Hh signaling. KIF7 mutations in humans have been implicated in ciliopathies, such as Jouberts, hydrolethalus, and acrocallosal syndromes, characterized by some Hh-related phenotypes. KIF7 physically associates with the Gli proteins and is itself enriched at the tips of cilia in response to Hh pathway activation. In the absence of KIF7, Gli3FL conversion to Gli3R is impaired, and the Hh-induced increase in Gli2/3 and Sufu staining at cilia tips is abrogated. At least in some contexts, KIF7 has also been implicated in a positive role in signaling, promoting the dissociation of Gli2/3 from Sufu (Hsu et al, 2011; Maurya et al, 2013). The dual roles of KIF7 (like those of Cos2) in both GliA and GliR formation led to the hypothesis that KIF7, as a potential plus-end directed motor, may transport Gli–Sufu complexes to the tips of cilia.
He et al (2014) analyzed the biochemical properties of KIF7 toward microtubules using a purified N-terminal fragment of the protein containing the motor domain and the first coiled-coil segment. This fragment could bind to microtubules but failed to move along them, a unique feature compared to canonical N-kinesin family members that usually exhibit plus-end directed motilities. Using assays based on Total Internal Reflection Fluorescence (TIRF) microscopy, they find that this KIF7 fragment associates with the plus-ends of microtubules, perhaps because it has some preference for the GTP-Tubulin cap at growing plus-ends, and both decreases the growth rate and increases catastrophe frequency (the rapid disassembly seen at plus-ends when the GTP-Tubulin cap is lost). These activities are dependent on ATP hydrolysis, suggesting that they are regulated by an active nucleotide hydrolysis cycle of the KIF7 motor domain.
The microtubule destabilizing property of KIF7 seen in the reconstituted system nicely explains some of the structural abnormalities in cilia from both mouse embryonic fibroblasts (MEFs) and mouse embryos lacking KIF7 activity. Cilia in the absence of KIF7 are longer. This is not an indirect consequence of abnormalities in the rates of anterograde and retrograde intraflagellar transport (IFT), motor-driven mechanisms that ferry cargo from the base to the tip or the tip to the base, respectively. In the absence of KIF7, cilia show a twisted morphology suggestive of abnormalities in the structure of the axoneme. The axoneme is also more unstable, based on sensitivity to nocodazole or cold exposure or based on stability markers such as acetylation and glutamylation. Inexplicably, a previous study (Dafinger et al, 2011) using RNAi-mediated knockdown of KIF7 reported the opposite phenotype—a stabilization of acetylated cytoplasmic microtubules. These results suggest that KIF7 likely cannot be the motor that transports Gli proteins from the base to the tip of cilia. Taken together, the cellular observations and biochemical assays provide a mechanistic basis for ciliary length control by KIF7.
What is the functional consequence of the depolymerizing activity of KIF7 on microtubules at the tips of cilia? To address this question, the authors performed trafficking experiments to analyze IFT movement and the ciliary accumulation of Gli2 and Sufu. The ciliary tip compartment is the site where IFT trains reverse directions. One would expect a single spot where this event takes place at the positive ends of axonemal microtubules. However, in Kif7−/− cells, this reversal occurred at multiple spots along the length of the axoneme, implying the existence of ectopic “cilia tip-like” compartments. The authors speculate that these ectopic tips might be generated due to a loss in the synchrony of the growth of the nine doublets in the axoneme (Fig1B). Remarkably, the authors also observed the Hh-regulated accumulation of Gli2 and Sufu at these ectopic cilia tips.
These results raise several interesting questions. Since proteolysis of Gli3FL to Gli3R is impaired in the absence of KIF7 (Endoh-Yamagami et al, 2009; Liem et al, 2009), these ectopic tip compartments must be defective in executing the yet unknown biochemical step required to license repressor production (Fig1B). Gli and Sufu also accumulated at the distal tip of the cilium in the absence of KIF7; thus, the trapping of Gli–Sufu complexes away from the anatomical tip cannot solely explain the phenotype. An alternate possibility is that the derangement of axonemal structure in the absence of KIF7 prevented the assembly of complexes that directly or indirectly regulate Gli3R formation. The authors' results also seem to exclude a role for KIF7 in the ciliary accumulation of Gli2/3 and Sufu in response to Hh signals. So how might Gli–Sufu complexes accumulate in cilia if not through KIF7? One possibility is that they are ferried along cilia by the IFT machinery, explaining why they accumulate at sites of IFT reversal. Alternatively, they might enter by simple diffusion and get selectively trapped at tip-like sites via as yet known protein interactions. Clearly, understanding what happens to Gli–Sufu complexes at these tips, ectopic or otherwise, is necessary to make progress in understanding mechanism. The cilia tip, where plus-ends of microtubules contact the ciliary membrane, has some analogies to the interface between the plus-end of spindle microtubules and the kinetochore in mitosis (Miller et al, 1990). The latter interface is used to organize signaling to the spindle assembly checkpoint and other events in mitosis; it is tempting to speculate the cilia tip serves a similar role in Hh and other ciliary signaling pathways. The ectopic cilia tips seen in the absence of KIF7 may lack mature contacts with the membrane, hence failing to appropriately regulate Gli–Sufu complexes.
In summary, the authors' work represents an important advance in understanding the biochemical activity of KIF7 and its relationship to cilia length control and Hh signal transduction. A major goal moving forward is to understand the composition and specific biochemical function of these ciliary compartments (Fig1A) where Hh proteins localize and to understand how signals are communicated between these compartments to properly regulate the ratio of GliR to GliA, which ultimately regulates the outcome of signaling.
References
- Cheung HO, Zhang X, Ribeiro A, Mo R, Makino S, Puviindran V, Law KK, Briscoe J, Hui CC. The kinesin protein Kif7 is a critical regulator of Gli transcription factors in mammalian hedgehog signaling. Sci Signal. 2009;2:ra29. doi: 10.1126/scisignal.2000405. [DOI] [PubMed] [Google Scholar]
- Dafinger C, Liebau MC, Elsayed SM, Hellenbroich Y, Boltshauser E, Korenke GC, Fabretti F, Janecke AR, Ebermann I, Nurnberg G, Nurnberg P, Zentgraf H, Koerber F, Addicks K, Elsobky E, Benzing T, Schermer B, Bolz HJ. Mutations in KIF7 link Joubert syndrome with Sonic Hedgehog signaling and microtubule dynamics. J Clin Invest. 2011;121:2662–2667. doi: 10.1172/JCI43639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dentler WL. Structures linking the tips of ciliary and flagellar microtubules to the membrane. J Cell Sci. 1980;42:207–220. doi: 10.1242/jcs.42.1.207. [DOI] [PubMed] [Google Scholar]
- Endoh-Yamagami S, Evangelista M, Wilson D, Wen X, Theunissen JW, Phamluong K, Davis M, Scales SJ, Solloway MJ, de Sauvage FJ, Peterson AS. The mammalian Cos2 homolog Kif7 plays an essential role in modulating Hh signal transduction during development. Curr Biol. 2009;19:1320–1326. doi: 10.1016/j.cub.2009.06.046. [DOI] [PubMed] [Google Scholar]
- He M, Subramanian R, Bangs F, Omelchenko T, Liem KF, Jr, Kapoor TM, Anderson KV. The kinesin-4 protein KIF7 regulates mammalian Hedgehog signaling by organizing the cilia tip compartment. Nat Cell Biol. 2014;16:663–672. doi: 10.1038/ncb2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu SH, Zhang X, Yu C, Li ZJ, Wunder JS, Hui CC, Alman BA. Kif7 promotes hedgehog signaling in growth plate chondrocytes by restricting the inhibitory function of Sufu. Development. 2011;138:3791–3801. doi: 10.1242/dev.069492. [DOI] [PubMed] [Google Scholar]
- Liem KF, Jr, He M, Ocbina PJ, Anderson KV. Mouse Kif7/Costal2 is a cilia-associated protein that regulates Sonic hedgehog signaling. Proc Natl Acad Sci USA. 2009;106:13377–13382. doi: 10.1073/pnas.0906944106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurya AK, Ben J, Zhao Z, Lee RT, Niah W, Ng AS, Iyu A, Yu W, Elworthy S, van Eeden FJ, Ingham PW. Positive and negative regulation of Gli activity by Kif7 in the zebrafish embryo. PLoS Genet. 2013;9:e1003955. doi: 10.1371/journal.pgen.1003955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JM, Wang W, Balczon R, Dentler WL. Ciliary microtubule capping structures contain a mammalian kinetochore antigen. J Cell Biol. 1990;110:703–714. doi: 10.1083/jcb.110.3.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay SY, Ingham PW, Roy S. A homologue of the Drosophila kinesin-like protein Costal2 regulates Hedgehog signal transduction in the vertebrate embryo. Development. 2005;132:625–634. doi: 10.1242/dev.01606. [DOI] [PubMed] [Google Scholar]