Abstract
Living cells compartmentalize materials and enzymatic reactions to increase metabolic efficiency. While eukaryotes use membrane-bound organelles, bacteria and archaea rely primarily on protein-bound nanocompartments. Encapsulins constitute a class of nanocompartments widespread in bacteria and archaea whose functions have hitherto been unclear. Here, we characterize the encapsulin nanocompartment from Myxococcus xanthus, which consists of a shell protein (EncA, 32.5 kDa) and three internal proteins (EncB, 17 kDa; EncC, 13 kDa; EncD, 11 kDa). Using cryo-electron microscopy, we determined that EncA self-assembles into an icosahedral shell 32 nm in diameter (26 nm internal diameter), built from 180 subunits with the fold first observed in bacteriophage HK97 capsid. The internal proteins, of which EncB and EncC have ferritin-like domains, attach to its inner surface. Native nanocompartments have dense iron-rich cores. Functionally, they resemble ferritins, cage-like iron storage proteins, but with a massively greater capacity (˜30,000 iron atoms versus ˜3,000 in ferritin). Physiological data reveal that few nanocompartments are assembled during vegetative growth, but they increase fivefold upon starvation, protecting cells from oxidative stress through iron sequestration.
Keywords: cryo-electron microscopy, encapsulin, ferritin, HK97 fold, oxidative stress
Introduction
Compartmentation allows cells to confine enzymes to increase metabolic efficiency, to protect them from proteolysis or other challenges, and to sequester toxic products (Diekmann & Pereira-Leal, 2013). In bacteria and archaea, cytosolic compartments are mainly protein-bound (Kerfeld et al, 2010; Yeates et al, 2008, 2011). One such class is encapsulin nanocompartments whose common feature is that their shell protein encapsulin (formerly referred to as linocin-like protein) has an HK97-like fold (Sutter et al, 2008). This fold was first observed in the capsid protein of HK97, a lambdoid bacteriophage with a triangulation number (T-number) of 7 (Wikoff et al, 2000) (see below, Discussion). Since its discovery, the HK97 fold has been observed in the capsid proteins of several other tailed bacteriophages (Agirrezabala et al, 2007; Fokine et al, 2005; Jiang et al, 2006; Parent et al, 2010; Spilman et al, 2011; White et al, 2012), and cryo-EM data strongly suggest that it is also present in herpesvirus capsids (Baker et al, 2005; Cardone et al, 2012; Homa et al, 2013). Recently, it was found by X-ray crystallography that an archaeal encapsulin and a bacterial encapsulin have similar folds arrayed, respectively, in T = 3 (180-subunit) and T = 1 (60-subunit) icosahedral shells (Akita et al, 2007; Sutter et al, 2008). On this basis, it has been proposed that encapsulins and the capsid proteins of tailed phages share a common evolutionary origin (Abrescia et al, 2012; Akita et al, 2007; Forterre & Prangishvili, 2009; Heinemann et al, 2011; Sutter et al, 2008), a hypothesis supported by the presence of genes encoding phage-like proteins (e.g., a replicative helicase) nearby an archaeal encapsulin gene (Heinemann et al, 2011). However, viral capsids and encapsulin nanocompartments have different roles: While the former transport viral genomes from one cell to another, the latter are involved in metabolism. Importantly, despite the identification of encapsulin proteins in a variety of bacteria, including medically important species such as Mycobacterium tuberculosis (Rosenkrands et al, 1998), the in vivo functions of encapsulin nanocompartments have not yet been demonstrated.
Myxococcus xanthus is a social bacterium characterized by a complex developmental life cycle. Actively growing cells are motile and, upon amino acid starvation, aggregate to form elaborate spore-filled fruiting bodies (Zusman et al, 2007). These developmental processes make M. xanthus an ideal model organism to study cellular adaptation to environmental signals. While studying vegetatively growing M. xanthus cells, we noted the production of round, 32-nm-diameter, seemingly proteinaceous particles. Although these particles were sparse, amino acid starvation resulted in a fivefold increase, enabling purification of sufficient quantities for biochemical and structural characterization. In this study, we have characterized these particles: In particular, they contain a dense core composed mainly of iron and phosphorus. Using a variety of electron microscopic techniques, we compiled a detailed account of the molecular architecture of their encapsulin shell and of the composition and organization of its dense core. Exploring the functional correlates of these particles, we found that in disruption and deletion mutants of the shell-forming protein EncA, whereby the iron-sequestering ability is lost, more cells die when exposed to oxidative stress in the form of H2O2 compared to wild-type cells. Taken together, these observations lead to a model for the iron-sequestering encapsulin nanocompartments of M. xanthus and support the hypothesis that their function involves the regulated uptake of iron to protect the cells from oxidative stress and its subsequent release when needed by iron-requiring cytosolic enzymes.
Results
Protein composition of native M. xanthus encapsulin nanocompartments
Electron microscopy of negatively stained specimens confirmed that purified material comprised a homogeneous population of spherical particles, ˜32 nm in diameter (Fig1A). SDS–PAGE (Fig1B) showed that the particles are composed of four proteins, which were identified by mass spectrometry and named EncA (MXAN_3556), EncB (MXAN_3557), EncC (MXAN_4464), and EncD (MXAN_2410). Gel quantitation yielded the estimate that EncA accounts for ˜72% of the total protein content and the minor proteins EncB, EncC, and EncD for ˜7, ˜15, and ˜6%, respectively. Taken together, the minor proteins amount to ˜40% of the EncA mass. Since each nanocompartment has 180 copies of EncA (see below), this is accompanied by, on average, ˜36 copies of EncB, ˜92 of EncC, and ˜47 of EncD. Edman degradation analysis indicated that three of the proteins were incorrectly annotated in the NCBI database: EncA is seven aa shorter, EncB is six aa shorter, and EncC is fourteen aa longer.
Figure 1. Native encapsulin nanocompartments are composed of four proteins and contain a dense iron-rich core.
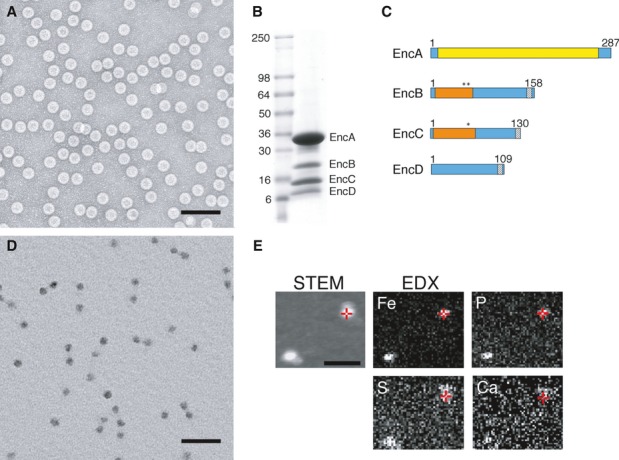
- EM of negatively stained nanocompartments. Scale bar, 100 nm.
- SDS–PAGE of purifed native encapsulin nanocompartments. Mass markers are indicated (in kDa).
- Domain maps of the four encapsulin proteins. Yellow, HK97-like domain; orange, rubrerythrin/ferritin-like domain; asterisks, ExxH metal coordination motifs; gray, encapsulation signal.
- EM of unstained/air-dried nanocompartments. Scale bar, 100 nm.
- STEM and energy-dispersive X-ray spectroscopy (EDX) images of two representative particles. In the STEM image (unstained specimen), positive signals from mass scattering identify two nanocompartments, while the corresponding EDX images map elemental concentrations of Fe and P (strongly above background), and S and Ca (at or close to background). Red crosshairs indicate identical positions on one nanocompartment. Scale bar, 100 nm.
Source data are available online for this figure.
Sequence analysis showed that EncA shares 24% sequence identity and 43% sequence similarity with the encapsulin shell protein of Thermotoga maritima (Fig1C and Supplementary Fig S1). Furthermore, EncB, EncC, and EncD were found each to have a C-terminal motif (LTVGSLRR; Fig1C and Supplementary Figs S2, S3 and S4) similar to those proposed to serve as encapsulation signals in other systems (Fan et al, 2010; Sutter et al, 2008). No other protein encoded in the M. xanthus genome (Goldman et al, 2006) contains this motif. Using the Pfam server (Finn et al, 2014), we found that EncB and EncC—but not EncD—possess rubrerythrin/ferritin-like domains. Bioinformatic analysis of EncB and EncC shows that in both proteins, this domain is highly conserved, including their copies of the ExxH motif (Fig1C and Supplementary Figs S2 and S3), which is known to bind iron and other metals (Liu & Theil, 2004; Sieker et al, 2000).
Native nanocompartments have dense cores rich in iron and phosphorus
During purification, we observed that encapsulin particles sediment at an unusually high density in cesium chloride gradients. Moreover, they have an electron-dense core that is conspicuous in electron micrographs of unstained preparations (Fig1D). This core is significantly smaller— ˜24 nm versus ˜32 nm—than the protein shell. To investigate its elemental composition, we analyzed these particles by energy-dispersive X-ray spectroscopy and found that their core is highly enriched for iron and phosphorus (Fig1E and Supplementary Fig S5). No other element was detected in comparable amounts. To estimate the relative abundances of iron and phosphorus, similar preparations were also analyzed by inductively coupled plasma mass spectrometry (ICP-MS) (Table1) which revealed a molar ratio of approximately 4:1 for Fe:Pi.
Table 1.
Quantitation of iron and phosphorus in M. xanthus encapsulin nanocompartments and horse spleen ferritin by inductively coupled plasma mass spectrometry.
| Sample | Iron (μg/l) | Phosphorus (μg/l) | Fe:Pi ratio |
|---|---|---|---|
| Control | 73.40 ± 75.20 (102) | < LOD | |
| Encapsulin | 276,600 ± 7,764 (2.81) | 72,390 ± 1,495 (2.07) | 4:1 |
| Ferritin | 269,200 ± 21,370 (7.94) | 30,160 ± 726 (2.41) | 9:1 |
Values for each element are the average concentration plus or minus the standard deviation in μg/l, calculated based on at least 5 experimental replicates per sample. All protein samples were dissolved in deionized water, which was also used as control solution. Note that the measured Fe:Pi ratio of the used horse spleen ferritin is in agreement with published data (Michaelis et al, 1943). Parentheses, coefficient of variation, in % (standard deviation divided by the average concentration for each sample and multiplied by 100).
Native encapsulin nanocompartments and recombinant EncA shells are T = 3 icosahedra
Visualized by cryo-EM, native nanocompartments from M. xanthus appear as round 32-nm-diameter particles (Fig2A). These images confirmed that a large majority— ˜94% in this preparation—have electron-dense cores (a minority particle lacking such a core is marked with a black arrow in Fig2A). When EncA was expressed recombinantly in E. coli, most (˜80%) of the particles produced were of the same size as native nanocompartments, that is, ˜32 nm (Fig2B), while ˜20% were smaller, mostly ˜18 nm in diameter (e.g., white arrow in Fig2B). These observations implied that EncA is capable of self-assembly, but with somewhat lower fidelity in size determination than native M. xanthus particles.
Figure 2. Cryo-EM of encapsulin nanocompartments and EncA shells.
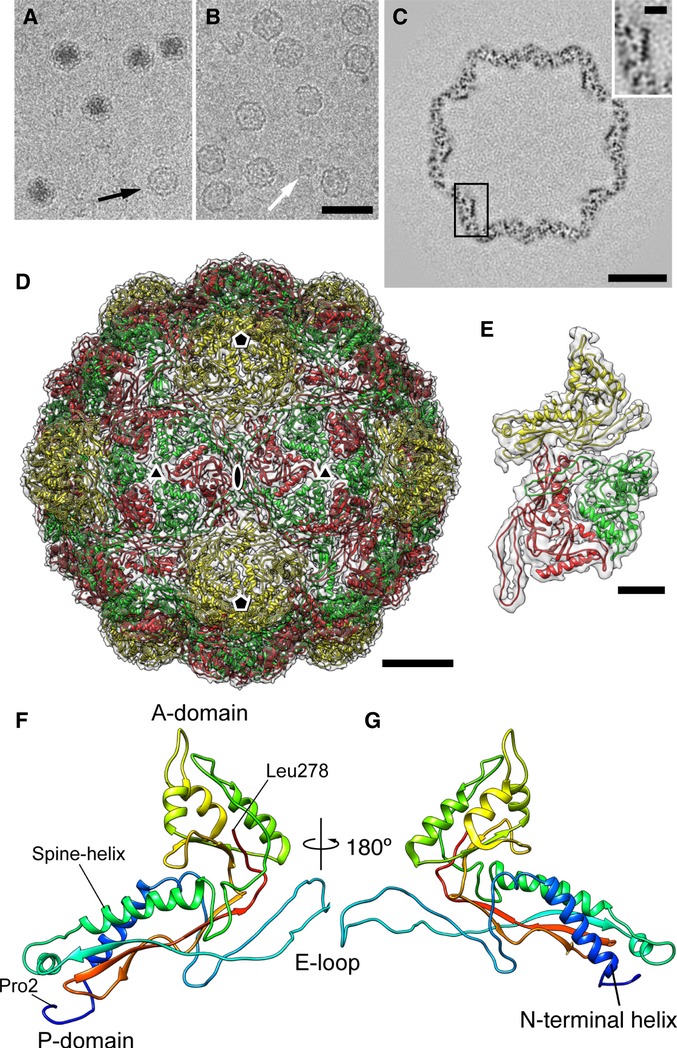
A, B Cryo-micrographs of (A) native nanocompartments and (B) recombinantly expressed EncA capsids.
C Section through a reconstruction of the EncA capsid. An enlargement of the boxed area showing a longitudinal section through an α-helix is inset (upper right).
D Atomic model of the T = 3 EncA capsid. The three quasi-equivalent subunits are colored yellow, green, and red. Symmetry axes are marked.
E The 3-subunit asymmetric unit fitted into electron density (transparent).
F, G Atomic model of EncA, rainbow-colored with the N-terminus in blue, and the C-terminus in red.
Data information: Scale bars: (A, B) 50 nm; (C) 10 nm; (D) 5 nm; (E, C insert) 2 nm.
To obtain a high-resolution reconstruction of the shell, we focused on the 32-nm recombinant EncA shells. A data set of ˜14,000 images yielded a density map at 4.6 Å resolution that depicts a shell with icosahedral symmetry and a triangulation number of T = 3 (180-subunits) (Fig2C–E and Supplementary Fig S6). In it, α-helices are patent in slices through the map (Fig2C), and landmark features of the HK97 fold (Wikoff et al, 2000), including the “spine” α-helix, the A- and P-domains, and the β-hairpin E-loop, are evident. Accordingly, its information content was considered sufficient to justify the construction of an atomic model (Fig2D–G, Supplementary Figs S7 and S8 and Supplementary Table S2). To do so, we first generated a homology model using the encapsulin protein from T. maritima (pdb 3DKT) as template and docked it as a rigid body into the EM map. This structure was then adapted by “flexible fitting” (Trabuco et al, 2008), whereby the starting model was systematically modified to optimize the fit. Finally, the structure was refined using X-ray crystallographic procedures, applying constraints that compensate in part for the limited resolution (Adams et al, 2010). Starting with other references, for example, the encapsulin protein from Pyrococcus furiosus (pdb 2E0Z), led to essentially the same end-product.
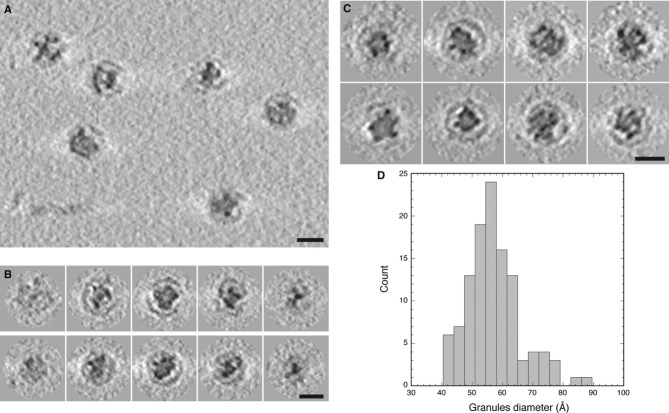
The dense cores of native nanocompartments consist of multiple 5-nm granules
From the similar diameters and scalloped peripheries of both the 32-nm EncA shells from E. coli and the dense-cored encapsulin particles from M. xanthus, we expected the latter to have an icosahedral shell similar to that of the EncA shell. However, the native particles posed a challenge for reconstruction in that the images are dominated by the core material, which is not icosahedrally ordered, as we initially inferred from visual inspection and subsequently confirmed by cryo-ET (see below). This property made it difficult to determine the particles' orientations, a necessary step for calculating a three-dimensional reconstruction. This problem was solved by limiting the correlated areas used for orienting the particles to a peripheral annulus to which the core material does not contribute (Fig3A). The fact that only a small fraction (˜10%) of the image area was used restricted the information content, leaving greater uncertainties in the Euler angles used to define orientations; in turn, this limited the resolution. Nevertheless, this approach yielded a reconstruction at a resolution of 17 Å (Fig3B and C): It depicts an icosahedral T = 3 surface shell whose outer surface is essentially indistinguishable from that of the EncA capsid when the latter is limited to the same resolution. The reconstruction shows the internal contents as three layers of density of which the densest is the middle one, a 3-nm-thick shell peaking at a radius of 6 nm.
Figure 3. Cryo-EM of encapsulin nanocompartments with electron-dense cores.
- Cryo-EM image of a native encapsulin nanocompartment without (top) and with (bottom) masking out of the electron-dense core.
- Sections through the single-particle reconstruction of the native Myxococcus xanthus encapsulin nanocompartment.
- Isosurface representation of the reconstruction color-coded according to radial distance from the center (given in Å). Left, viewed from the outside; right, cutaway view of the internal structure.
Data information: Scale bars, 10 nm in (A, C); 25 nm in (B).
However, this reconstruction conveys the core structure after it has been subjected to extensive inter-particle averaging and icosahedral symmetrization. In order to obtain 3-dimensional structures of individual particles that had not been symmetrized, we imaged them by cryo-ET (Fig4A–C), which yielded density maps with a resolution of 40–50 Å. The tomograms revealed that the core is not a continuous structure but consists of discrete granules. The number of granules per particle ranges from 11 to 19, averaging 14. They are typically round and 5–6 nm in diameter (Fig4D). The granules are not regularly distributed—in particular, neither in number nor in their spatial distribution do they comply with icosahedral symmetry. Most of the granules are offset from the capsid, with the commonest offset (˜5 nm) matching the radial position of the densest layer in the icosahedral reconstruction (Fig 4B and C). The combined volume of granules within a single nanocompartment is variable but averages ˜1,600 nm3.
Figure 4. Cryo-ET of encapsulin nanocompartments with electron-dense cores.
- Tomographic slice showing several native Myxococcus xanthus encapsulin nanocompartments.
- Tomographic slices through two nanocompartments (top and bottom rows).
- Gallery of tomographic central sections of eight nanocompartments.
- Histogram of the diameters of electron-dense granules in the nanocompartment cores.
Data information: Scale bars, 25 nm.
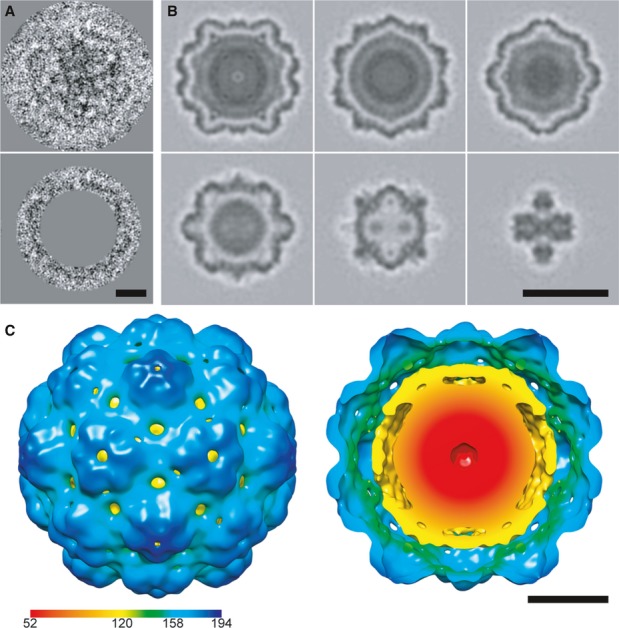
The minor proteins line the interior surface of the EncA shell
A minor fraction (˜6%) of the particles isolated from M. xanthus lack dense cores (black arrow in Fig2A). To investigate whether they nevertheless contain the putative internal proteins (EncB, EncC and EncD), we calculated a cryo-EM reconstruction (Fig5A). Although the resolution was limited to 24 Å by their paucity (N = 370), it does indeed show internal densities underlying the EncA shell (Fig5A). These internal densities are most evident at the three-fold axes, and they are not present in a similarly calculated reconstruction of the EncA shell (Fig5B). The locations and shapes of these densities are better seen in a difference map (Fig5C and D, and Supplementary Fig S9). However, they are less dense than the shell, indicative of partial occupancy. We interpret these data to mean that these proteins bind sub-stoichiometrically to defined sites on the inner surface of the capsid.
Figure 5. Cryo-EM visualization of internal proteins.
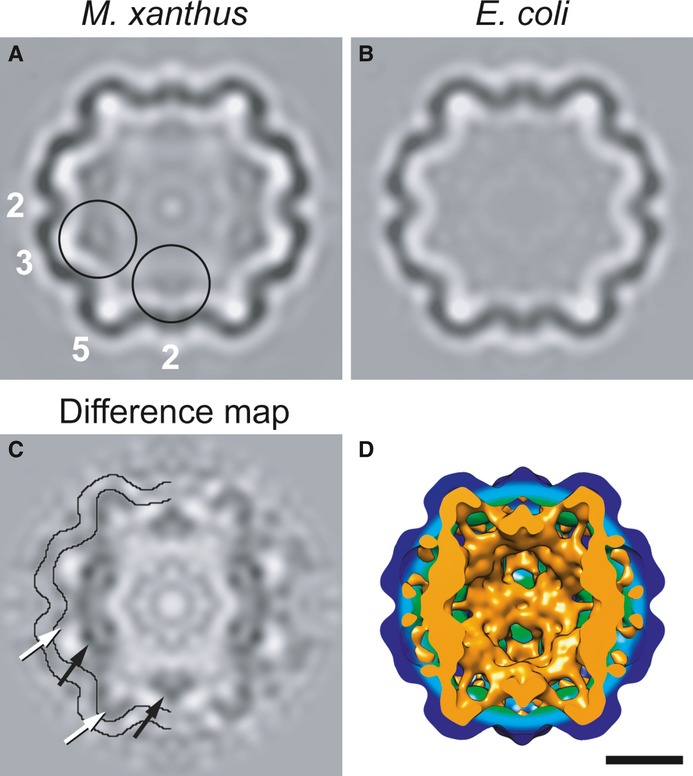
A–C Central sections through reconstructions of (A) native nanocompartments lacking a dense core; (B) EncA capsids; (C) the difference map between (A) and (B). In (C), contours delineating the outer and inner surfaces of the EncA capsid are superimposed in half of the section to aid in locating densities of internal proteins. White arrows label densities in direct contact with the inner surface, and black arrows label offset densities.
D Isosurface rendering of a cutaway view of the EncA capsid (green-blue) and the positive difference density (orange) attributable to the internal proteins. Scale bar, 10 nm.
The dense cores contain on average ˜30,000 iron atoms
To measure the amount of cargo in the native encapsulin particles, we imaged them by STEM, a technique in which the signal recorded at each pixel is proportional to the mass locally sampled by the electron probe (Goldsbury et al, 2011; Thomas et al, 1994) (Fig6). These experiments were carried out with a preparation that was essentially devoid of empty particles, as ascertained by cryo-EM (data not shown). All of the particles had electron-dense cores, albeit of variable sizes (Fig6A and B). Their masses give a symmetric unimodal distribution with a mean of 10.0 MDa and a standard deviation (SD; σ) of 0.74 MDa (Fig6C). The SD reflects both genuine particle-to-particle variability (σ1) and the spread (σ2) to be expected in a set of STEM mass measurements from homogeneous protein particles of the same size. As these sources of variability are independent, they are related by σ2 = σ12 + σ22. We estimated σ2 from the SD of a set of mass measurements made on 56-nm segments of tobacco mosaic virus (TMV), which contain approximately the same amount of protein as the encapsulin nanocompartments. The TMV virions have a known and uniform mass density (131.4 kDa/nm) and serve as mass calibrators. Present on the same grid as the encapsulin particles, they were prepared and imaged under identical conditions. These data gave 7.4 ± 0.43 MDa (Fig6D); accordingly, σ1 = 0.60 MDa.
Figure 6. STEM of native encapsulin nanocompartments.
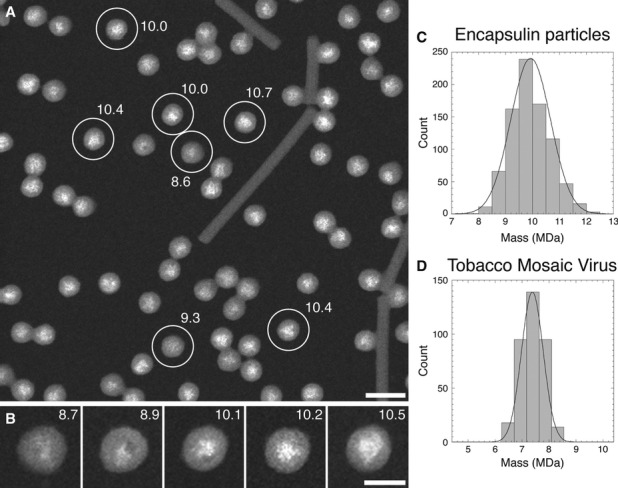
A, B Dark-field STEM micrograph of unstained encapsulin nanocompartments. The masses (in MDa) of some particles are indicated. The rod-like structures are TMV virions, used as a mass standard. Scale bars, 50 nm in (A); 25 nm in (B).
C, D Histograms of the masses measured for (C) the encapsulin nanocompartments and (D) 56-nm-long segments of TMV (mass/length = 131.4 kDa/nm), which contain approximately the same amount of protein as the nanocompartments.
We used these data to calculate the mean and range of the cargo mass and its payload of iron atoms. Since an EncA monomer has a mass of 32.5 kDa, that of the shell (180 subunits) is 5.85 MDa. We estimated the amount of internal proteins (EncB, EncC, and EncD) (see above and Fig1B) to be 40% of EncA, that is, 2.34 MDa. This estimate is consistent with one obtained from the 3D reconstruction of iron-free native nanocompartments (Fig5A) in which the number of voxels in the shell is compared with that occupied by the internal proteins. It follows that the mass of dense material (total particle mass minus combined mass of the EncA shell and internal proteins) is, on average, 1.81 MDa. The SD calculated above gives a range (mean ± σ1) of 1.21 to 2.41 MDa.
If we infer from the ICP-MS data (see above) that iron and phosphorus are the only significant elemental components and they are present in a molar ratio of 4 (Fe; 56 Da) to 1 (P; 31 Da), then the average nanocompartment contains ˜28,500 iron atoms. If the phosphorus is in the form of phosphate (PO4, 95 Da), this number falls to ˜22,500. However, for particles at the upper end of the mass range (mass = mean + σ1), these numbers increase to ˜38,000 and ˜30,000 iron atoms, respectively. Given the residual ambiguity in this calculation, we take 30,000 as a round number for the average payload of iron atoms. In any case, this payload is an order of magnitude larger that that of ferritins, which has been estimated to average ˜3,000 iron atoms (Andrews, 2010).
Encapsulin nanocompartment assembly is induced under starvation conditions
A genetic study of M. xanthus found the EncA gene to be required for fruiting body formation, though it was not recognized as an encapsulin homolog at the time (Kim et al, 2009). Fruiting body formation is a developmental program triggered by starvation and resulting in sporulation (Zusman et al, 2007). Since starvation leads to increased assembly of loaded nanocompartments (see above), we measured the production of EncA protein and of encapsulin nanocompartments during starvation and sporulation. Actively growing and starved cells contained comparable amounts of EncA (Fig7A). However, isolation of nanocompartments from starved cells yielded a fivefold increase in yield, compared to actively growing cells (Fig7B). The enhanced production of nanocompartments by starved cells was confirmed by direct visualization of the iron cores of assembled particles in EM of unstained thin sections (Fig7C). This suggests that encapsulin production is regulated not only at the level of gene expression, but also post-translationally at the level of assembly. Neither nanocompartments nor EncA were found at significant levels in mature spores after 48 or 96 h of development (Fig7A and B, and Supplementary Fig S10), raising the possibility that pre-existing nanocompartments were eliminated by proteolytic degradation to recycle their iron and phosphorus contents.
Figure 7. Encapsulin nanocompartments are upregulated during starvation and protect the cells from oxidative stress.
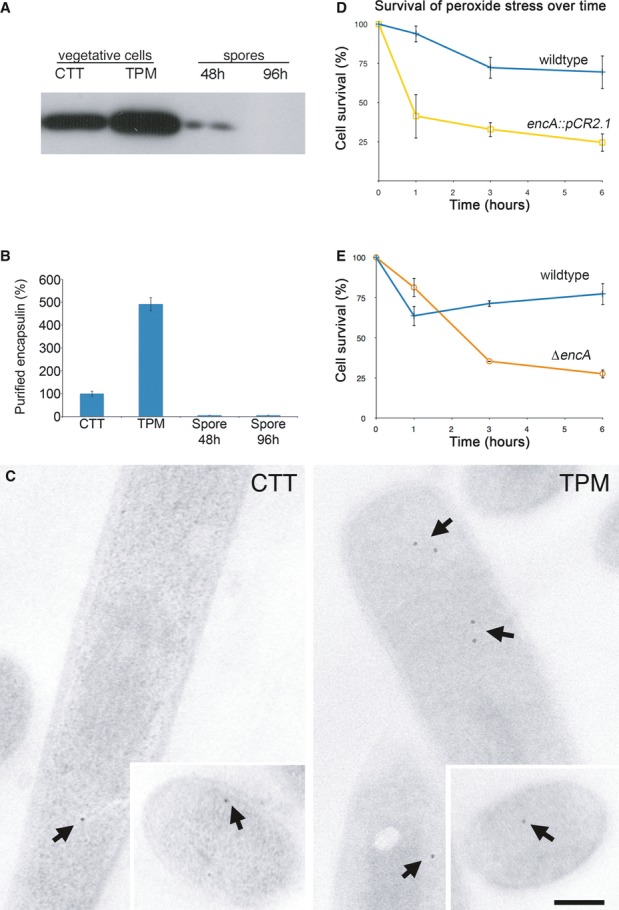
A Western blot of EncA from lysate of 1 × 108 vegetative cells in rich CTT medium or starvation TPM medium, and spores after 48 or 96 h of development.
B Amounts of purified nanocompartments recovered from different cell types, compared to yield recovered from cells grown in CTT (100%).
C EM of unstained longitudinal thin sections of vegetative cells grown in CTT (left panel) or TPM medium (right panel). Arrows mark dense-cored encapsulin nanocompartments. Insets, transverse thin cross-sections. Scale bar, 0.5 μm.
D, E Survival of wild-type (blue curve) and encA::pCR2.1 disruption (yellow curve) strains (D) and of wild-type (blue curve) and ΔencA in-frame deletion (orange curve) (E) after exposure to 0.5 mM hydrogen peroxide.
Source data are available online for this figure.
Encapsulin nanocompartments protect cells from oxidative stress caused by peroxide
Fruiting body formation is accompanied by a massive die-off of cells whereby ˜80% of vegetative cells lyse and a small proportion proceed to sporulation (Lee et al, 2012a; Wireman & Dworkin, 1977). Since iron can catalyze the production of toxic reactive oxygen species through the Fenton reaction (Cornelis et al, 2011), the assembly of encapsulin nanocompartments during starvation may serve a protective role against cellular death due to oxidative stress by reducing the amount of iron free in the cytosol. To test this hypothesis, we investigated the effects of mutations in EncA (Supplementary Fig S11) on the survival of M. xanthus under oxidative stress caused by peroxide (H2O2) exposure. Both the independently generated disruption mutant encA::pCR2.1 and the in-frame deletion mutant ΔencA showed increased sensitivity to 0.5 mM H2O2 confirming that these structures are indeed important for oxidative stress survival (Fig7D and E).
Discussion
Encapsulin nanocompartments encapsulate iron
Iron is essential to life because it serves as a cofactor of many enzymes. However, it can also produce highly reactive hydroxyl radicals that cause cellular damage (Harrison & Arosio, 1996). To regulate the supply of free intracellular iron, cells have evolved protein assemblies that act as iron storage containers. The best known are the three subfamilies of ferritins: classical ferritins, bacterioferritins, and “DNA-binding proteins from starved cells” (Dps proteins), of which classical ferritins and bacterioferritins function in iron storage and Dps proteins in iron detoxification (Andrews, 2010; Theil et al, 2013). Ferritins are multimeric complexes whose subunits, each a four-helix bundle, assemble into hollow shells with cubic symmetry (Zeth, 2012). These “molecular cages” can accommodate substantial quantities of iron (Andrews, 2010). The shells are penetrated by channels through which iron ions can enter to be stored inside the complex and subsequently released under iron-requiring conditions.
Encapsulin nanocompartments appear to be operationally similar to ferritins, despite marked differences in size (32 nm versus 11 nm), storage capacity for iron—which is greater by an order of magnitude, shell symmetry (icosahedral vs. octahedral), the fold of the building block (HK97-like versus a 4-helix bundle), and in that the iron deposits nucleate on cargo proteins rather than directly on to the shell protein.
Although there is, to our knowledge, no prior history of studies into the occurrence of ferritins in M. xanthus, genome analysis reveals that it encodes several putative ferritin-like proteins (MXAN_2878, MXAN_2342, MXAN_1008, and Dps proteins MXAN_0866 and MXAN_1562) besides those packaged into encapsulin particles. Pending further information, it appears plausible that M. xanthus may use its ferritins for iron homeostasis during vegetative growth and, upon starvation, induce a second iron storage system that functions independently alongside the ferritin system. The second (encapsulin-based) system would allow the cells to temporarily sequester iron and phosphorus as an antioxidant response or to be used later for nutritional purposes (Theil, 2011).
Comparison of M. xanthus encapsulin with other encapsulins and with HK97 gp5*
A growing body of evidence indicates that encapsulin proteins generically possess HK97-like folds, as do the capsid proteins of other tailed bacteriophages as well as herpesviruses, often in the complete absence of sequence similarity. This mutual resemblance is apparent on comparing the encapsulin structures of M. xanthus, T. maritima, and P. furiosus (Fig8A–D). Differences between EncA and the other encapsulins are observed primarily in the E-loop (expected, as these are flexible regions) and in the insertion loop anterior to the spine helix (which is absent in P. furiosus). Of note, these three molecules have an N-terminal α-helix (blue in Fig8A–C) that is absent from HK97 but present in the capsid proteins of some other bacteriophages (e.g., T4 (Fokine et al, 2005), P22 (Parent et al, 2010), and 80α (Spilman et al, 2011)). Since this helix faces the interior of the encapsulin shell, it might be involved in interactions with the internal proteins. On comparing EncA with HK97 gp5* (Fig8A, D and E), the most pronounced differences, other than the absence of the N-terminal α-helix in gp5*, are in the A-domains where EncA has an extra α-helix (yellow in Fig8A), and in the E-loops and the tips of the P-domains which differ slightly.
Figure 8. Comparison of encapsulin proteins and HK97 gp5* in different maturation states.
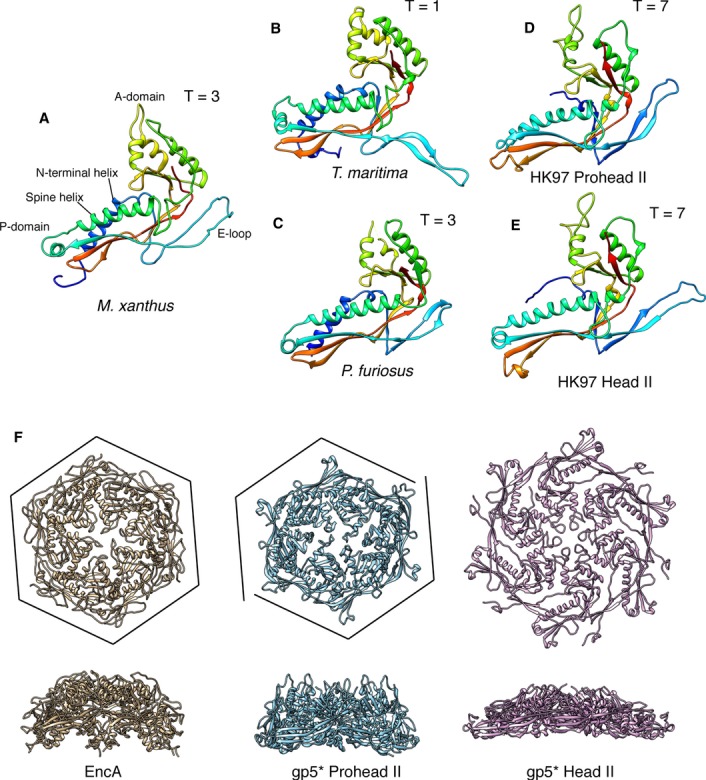
A EncA from this study (pdb 4PT2; EMDataBank EMD-5917).
B Encapsulin from Thermotoga maritima (pdb 3DKT).
C Encapsulin from Pyrococcus furiosus (pdb 2E0Z).
D, E gp5* from HK97 in the Prohead II (D; pdb 3E8K) and Head II(E; pdb 2FT1) conformations. Models are rainbow-colored, with the N-termini of the solved structures in blue and the C-termini in red.
F Top views (upper row) and side views (lower row) of EncA and gp5* hexamers. The EncA hexamer is more similar to Prohead II gp5* than to Head II gp5* despite lacking the skew disclination of the Prohead II gp5* hexamer (cf. the respective frames).
Two distinctive features of HK97 gp5 are: (i) that it undergoes maturational proteolysis in which the N-terminal Δ-domain is excised as gp5 (41 kDa) is converted to gp5* (31 kDa) (Duda et al, 1995); and (ii) the maturing capsid undergoes a sequence of large-scale conformational changes en route from the first assembled precursor Prohead I to the fully mature Head II (reviewed by (Hendrix & Johnson, 2012; Steven et al, 2005)). To what extent do encapsulins share these properties? Here, we restrict the discussion to M. xanthus encapsulin.
The Δ-domain appears to serve as a scaffolding protein that guides assembly (Huang et al, 2011a). Essentially, all phage capsids with T-numbers of 7 or greater employ a Δ-domain in this role, either appended as an N-terminal fusion (as in HK97) or as a separate gene product. The morphologically correct particles generated upon expressing EncA in E. coli indicate that this protein does not need a scaffolding protein. The reason may reflect the lower T-number (3), there being numerous examples of T = 3 capsids that self-assemble efficiently without a scaffolding protein, although few if any examples have been documented for viral capsids with the HK97 fold. In this context, the 20% of smaller particles among the recombinant expression products in E. coli (see above) are ˜18 nm in diameter and conform to T = 1 (60-subunit) icosahedral symmetry (unpublished results).
The conformational state of EncA shells resembles that of HK97 in its immature form
The successive states of the HK97 maturation pathway are distinguished primarily by rigid-body rotations of the subunits as the intermolecular interactions are shifted to different surfaces (Conway et al, 2001), although there are also some local adjustments (Gertsman et al, 2009). Which state of the maturing HK97 capsid does the encapsulin shell most closely resemble? We addressed this question in two ways: by comparing the fold of EncA monomer with the various HK97 gp5* conformers and by comparing the respective capsomers. These calculations placed the EncA monomer marginally closer to the Head II gp5* monomer (the RMSDs obtained were 2.17 Å for Head II and 2.76 Å for Prohead II; alignment was performed using the superpose program from CCP4 (Collaborative Computational Project, 1994)) (compare Fig8A with D and E). On the other hand, EncA capsomers are clearly most similar to those of Prohead II (Fig8F), although the EncA hexamers are not skewed (those of Prohead II are skewed, resembling two dislocated trimers (Conway et al, 1995)). Of note, we did not find any evidence of a similar expansion process in EncA, and such a transition has yet to be reported for any other encapsulin. We speculate that this may reflect the absence of an evolutionary mandate to increase the encapsulin shell's capacity for cargo and to confer additional stability, as applies to HK97 and other phage capsids.
A functional model of the encapsulin nanocompartment
Taken together, current observations led us to the nanocompartment model shown in Fig9. It envisages substoichiometric binding of EncB, EncC, and EncD to specific sites (of which there are 180) on the inner surface of an EncA shell via their common C-terminal binding motif. We anticipate that iron atoms may enter the shell via narrow channels through the capsid wall, of which those located on the fivefold axes or between the A-domain and the spine helix of adjacent subunits from the same capsomer (Fig9B) are the most likely candidates. Entering iron atoms are then incorporated into nascent granules nucleated on and around the metal-binding motifs on the rubrerythrin domains of EncB and EncC. Known structures for rubrerythrin domains depict a bundle of four lengthy α-helices. As the corresponding sequences in EncB and EncC are only long enough to accommodate two such helices (Supplementary Figs S2 and S3), it is plausible that they dimerize. Between the rubrerythrin domains and the C-terminal binding motifs, there are tracts that may afford flexible linkers long enough to account for the observed, non-icosahedrally symmetric distribution of iron-containing granules. As the internal proteins are reasonably well ordered and shell associated in iron-free shells (Fig5), it appears likely that the buildup of iron deposits on the EncB and EncC proteins is accompanied by detachment from the EncA shell.
Figure 9. Schematic model of an Myxococcus xanthus encapsulin nanocompartment.
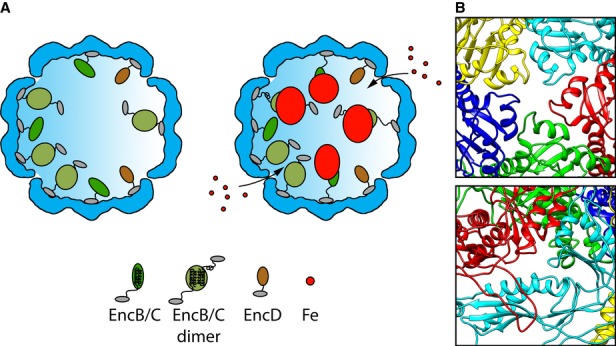
- Shown are cutaway views of the EncA shell (blue) for the particles with (left) and without (right) iron-rich granules. The C-terminal anchor motif common to EncB, EncC, and EncD is depicted as a small gray oval. In EncB and EncC, this motif is connected by an extended flexible linker to the rubrerythrin domain (green ovals). EncD's domain is shown as a brown oval. Since EncB and EncC are predicted to have only two α-helices each, we hypothesize that they dimerize forming a 4-helix bundle in homo- or hetero-dimers with the iron-binding motifs midway along the bundle. However, monomers may also exist, and they may be capable of nucleating iron deposition. The model envisages iron atoms entering the shell through narrow channels, to be incorporated into a nascent granule nucleated on the rubrerythrin domains. Interaction of the iron with the internal proteins probably induces a conformational change that causes the rubrerythrin domains to detach from the EncA shell.
- The most likely candidates for the entry channels are shown.
Granulation of the iron-containing core of native M. xanthus nanocompartments as opposed to the formation of a single mineralized nugget may be advantageous in terms of rapidity of uptake and subsequent release. If mineralization initiates at 15 sites (the average number of granules) instead of one, iron uptake can proceed proportionately more rapidly. Conversely, if there are 15 granules of 5.5 nm in diameter (taken to be spherical for the sake of this discussion) instead of the same amount of iron packed in a single super-granule of 13.5 nm in diameter, the amount of surface area exposed, from which iron atoms may presumably be shed, is almost three times greater (˜1,425 nm2 versus 572 nm2).
A diversity of encapsulin functions, natural and synthetic
Only recently has it been recognized that the cytoplasms of many bacteria are populated with protein-bound nanocompartments (Kerfeld et al, 2010; Yeates et al, 2008, 2011). One such class is the encapsulins whose shell proteins possess a fold previously found only in viral capsids. However, unlike virus capsids, which serve as vehicles for genome transport, encapsulins are involved in metabolism. To date, only a few have been identified on the basis of structural similarity but we anticipate that this set will grow considerably. One subset of encapsulin operons also code for a dye-decolorizing peroxidase (DyP) or for a ferritin-like protein (Flp), which was taken to suggest a role in the oxidative-stress response (Sutter et al, 2008), now amply confirmed in the M. xanthus system. However, other encapsulin nanocompartments have been suggested to perform different functions, including antibacterial activity in Brevibacterium linens (Valdes-Stauber & Scherer, 1994), proteolytic activity in T. maritima (Hicks et al, 1998), and lignin degradation activity in Rhodococcus jostii RHA1 (Rahmanpour & Bugg, 2013).
The encapsulation signal at the C-termini of EncB, EncC, and EncD suggests the possibility of appending this short amino acid sequence to other exogenous proteins for the purpose of engineering novel encapsulin particles housing a variety of activities with a broad range of applications ranging from imaging to therapeutics (Huang et al, 2011b; Lee et al, 2012b; Theil et al, 2013). Moreover, close scrutiny of the differences between HK97 gp5, which assembles into 48 nm T = 7 shells, may make it possible to adapt EncA in order to assemble EncA nanocompartments larger than the native 32 nm T = 3 form.
Materials and Methods
Strains and media
The wild-type strain was M. xanthus DK1622 (Kaiser, 1979). Vegetative cells were grown at 32°C in rich CTT medium (1% casitone, 1 mM K2PO4, 8 mM MgSO4, 10 mM Tris/HCl, final pH 7.6) or in starvation TPM medium (1 mM K2PO4, 8 mM MgSO4, 10 mM Tris/HCl, final pH 7.6) (Hodgkin & Kaiser, 1979). For preparation of agar plates and maintenance of cells on solid medium, 1.5% agar was added to CTT before autoclaving.
The encaA::pCR2.1 (EH410) disruption strain was constructed by recombination of the suicide plasmid pCR2.1 containing a 396-bp homology region into the MXAN_3556 (encA) gene. The in-frame deletion strain ΔencA (EH411) was constructed using an allelic exchange by homologous recombination strategy, with plasmid pBJ113 as described previously (Ueki et al, 1996). Both procedures are described in Supplementary Materials and Methods.
Large-scale purification
DK1622 cells grown in CTT medium to an Abs550 of 0.2 were transferred into TPM medium and grown for 18–24 h to induce nanocompartment assembly. Cells were harvested, re-suspended in water or Tris buffer, chilled, and broken by ultrasonication. Cellular debris was removed by pelleting at 16,000 × g and again after adding 15% (w/v) ammonium sulfate. Polyethylene glycol was added to the supernatant and incubated on ice for 45 min. Precipitated proteins were harvested at 47,800 × g, re-suspended in water or Tris buffer, and CsCl added to a concentration of 0.3 g/ml. Ten milliliter of this solution was layered over a 2-ml cushion of 5.7 M CsCl and spun for 16 h at 35,000 rpm in a Beckman SW55 rotor. The nanocompartments formed a whitish band near the top of the CsCl cushion and were harvested and stored at 4°C. For more concentrated preparations, the sample was dialyzed against water or buffer to remove the CsCl, then layered over a 10–40% sucrose gradient, and centrifuged for 2.5 h at 20,000 rpm. Nanocompartment-containing fractions were combined and re-centrifuged for 1.5 h at 55,000 rpm in a Beckman W55 rotor. The nanocompartments formed a small brownish pellet that could easily be re-dissolved in water or buffer and was stored at 4°C. Negative staining EM and SDS–PAGE were used to confirm the purity of the preparations.
Identification of protein components of native encapsulin nanocompartments
The proteins in several independent isolates were identified by two approaches: MALDI/TOF mass spectrometry and Edman degradation analysis (Supplementary Table S1). Purified nanocompartments at ˜0.1 mg/ml protein were run on a SDS–PAGE gel from which bands were excised and analyzed by MALDI/TOF mass spectrometry by Protea Biosciences, Inc. (Morgantown, West Virginia) or the Johns Hopkins University High Throughput Center Mass Spectrometry Facility. To identify the N-termini, bands separated on SDS–PAGE were blotted onto PVDF membranes, stained with colloidal Coomassie and subjected to Edman degradation at the Biochemistry Core Facility of the Max Planck Institute of Biochemistry (Martinsried, Germany).
Inductively coupled plasma mass spectrometry
Purified nanocompartments were dissolved in reverse osmosis-purified water, which was also used as control. Analysis was performed on samples extracted with 1% HNO3 and 0.5% HCl, using an Agilent 7500ce ICP-MS. Elemental standards were used to determine the limit of detection for each element (0.066 μg/l for iron and 6.77 μg/l for phosphorus). Final measurements consisted of 5 independent isolations done in triplicate for the nanocompartments, water control, and 150 μl of a horse spleen ferritin solution of 0.8 mg/ml as internal standard (Fitzgerald Industries International).
Energy dispersive X-ray analysis
Samples consisting of either purified nanocompartments or adenovirus (strain Ad5.CMV-GFP 10MR2000, Quantum) were applied to carbon-coated copper grids. STEM and EDX data were acquired at the High-Resolution Analytical Electron Microbeam Facility of the Integrated Imaging Center in the Departments of Earth and Planetary Sciences and Biology of Johns Hopkins University using a Philips CM 300 FEG microscope operating at 300 kV equipped with an Oxford light element energy dispersive X-ray spectroscopy detector. Gatan DigitalMicrograph and ES Vision software packages were used for subsequent analyses. Iron was consistently found at high levels in the core of the encapsulin particles and was not present in the proteinaceous rim of the particles (the EncA shell) nor in background areas.
Cloning of the encA gene in E. coli and preparation of EncA capsids
The expression vector was constructed by amplifying the coding region of the encA (MXAN_3556) gene and cloning this fragment into plasmid pET151-d/TOPO (Invitrogen), thus generating plasmid pET151-encA, as described in Supplementary Materials and Methods. For expression, E. coli BL21 cells harboring the plasmid were grown to log phase in LB medium and then vigorously shaken for one min in 500 mM sodium chloride with 0.05% chloroform. Cells and large debris were removed by centrifugation, and polyethylene glycol was added to the supernatant on ice and mixed for 45 min, after which precipitated proteins were pelleted. The pellets were re-suspended in Tris-Triton X buffer (10 mM Tris pH 7.6, 10 mM MgCl2, 0.1% Triton X-100), and cesium chloride added to give a density of 1.3 g/ml. Tubes were centrifuged for 16 h at 35,000 rpm. Capsids formed a whitish band near the top of the gradient, which was recovered and analyzed by SDS–PAGE and Western blot and checked by negative stain EM. To remove detergent for cryo-EM, samples were diluted in 2 ml of 10 mM Tris pH 7.6 and 10 mM MgCl2 and centrifuged for 3 h at 360,000 × g. The pellet was re-suspended in 30 μl of Tris buffer and stored at 4°C.
Cryo-electron microscopy and image reconstruction
Purified nanocompartments or EncA shells were applied to glow-discharged Quantifoil grids bearing a thin carbon film, blotted, vitrified, and imaged with a CM200 FEG microscope, as described (Cheng et al, 1999). For high-resolution analysis, the same preparation was imaged on an FEI Polara at 59,000× magnification. Micrographs recorded on Kodak SO163 film at 1- to 2-μm (CM200) or 0.5- to 1.6-μm (Polara) defocus were digitized at 1.23 Å/pixel (CM200) or 1.10 Å/pixel (Polara). Image processing was performed with the Bsoft package (Heymann & Belnap, 2003), as described (Nemecek et al, 2012). Images were CTF corrected by phase flipping in the initial stages and by phase and baseline compensation in later stages. Particles were picked manually, and origins and orientations were determined by projection matching. To obtain an unbiased initial reference, four sets, each of ˜200 EncA capsids, were reconstructed, initially assigning random orientations (Yan et al, 2007). The resulting density maps were quite consistent and were further validated by the variance-based procedure VIVA (Cantele et al, 2003). One of these maps was used as initial reference, leading to a reconstruction at ˜8 Å resolution from ˜8,000 CM200 images. The hand of the reconstruction was established by the tilt-pair method (Belnap et al, 1997; Cheng et al, 2002). For higher resolution, ˜14,000 particles from Polara micrographs were reconstructed in similar fashion, using the CM200 EncA map filtered to 15 Å as initial reference. It had a resolution of 4.6 Å according to the 0.143 FSC criterion (Scheres & Chen, 2012). Fourier amplitudes were “sharpened” by using the P. furiosus crystal structure as reference.
To reconstruct dense-cored nanocompartments, only the rims of the shell were used for particle alignment and orientation determination. The initial reference was generated as described above. The final reconstruction, combining information from ˜900 particles, had ˜17 Å resolution, at 0.3 FSC cutoff.
To visualize the internal proteins, a reconstruction of the EncA shell was subtracted from a reconstruction of native nanocompartments lacking electron-dense cores. The latter reconstruction, calculated from ˜400 images, had a resolution of ˜25 Å at 0.3 FSC cutoff. The former reconstruction was calculated using ˜400 randomly selected EncA images for a resolution of ˜23 Å. A difference map calculated after scaling the two maps consistently, using a purpose-built algorithm (Heymann et al, 2007). To estimate the cumulative mass of internal proteins, the reconstruction of native nanocompartments lacking electron-dense cores was segmented into two regions, the shell and the internal content, using Bsoft, and the number of voxels corresponding to each region were integrated using MAPMAN.
Atomic modeling of EncA
An initial homology model was generated using I-TASSER (Zhang, 2008) with the T. maritima encapsulin as reference (pdb 3DKT). This model was flexibly fitted into the EM map, using MDFF (Trabuco et al, 2008). The resulting atomic coordinates were then used to generate the three quasi-equivalent subunits of an asymmetric unit by rigid-body fitting into the corresponding densities and propagated over the icosahedral shell, using Chimera (Pettersen et al, 2004). Additionally, the EM density map was placed in a cubic cell using Bsoft, MAPMAN (Kleywegt & Jones, 1996), and MAPMASK tools (Collaborative Computational Project, 1994). Structure factors and Hendrickson–Lattman coefficients, describing phases, were calculated using the phenix.map_to_structure_factors program from PHENIX (Adams et al, 2010), and the atomic model was refined with phased maximum-likelihood target as implemented in phenix.refine (Afonine et al, 2005). To compensate for the limited resolution, icosahedral restraints, Ramachandran restraints, and weighting of the stereochemical restraints were employed. Manual model building in Coot (Emsley et al, 2010) alternated with automatic refinement. Refinement statistics are shown in Supplementary Table S2. R-free is not reported. If calculated, the R-free would be similar to the R-value because of the high non-crystallographic symmetry (Kleywegt & Brunger, 1996).
Cryo-electron tomography
Grids bearing thin films of native nanocompartment suspension were mixed with 10-nm colloidal gold particles coated with bovine serum albumin (BSA) (Aurion, Wageningen, the Netherlands) to serve as fiducial markers and vitrified as described above. Single-axis tilt series were recorded on a Tecnai-12 TEM (FEI) equipped with a GIF 2002 energy filter (Gatan, Warrendale, PA) operated in zero-loss mode with an energy slit width of 20 eV. Images were acquired on a 2048 × 2048 pixel CCD camera using SerialEM (Mastronarde, 2005) at a magnification of 38,500× (0.78 nm/pixel) and 4-μm nominal defocus. The tilt range was from −66° to +66° with images recorded every 2°, for a total electron dose of ˜70 e-/Å2. According to the NLOO-2D method (Cardone et al, 2005), the in-plane resolution of the tomograms was 40–50 Å.
Scanning transmission electron microscopy
Samples of nanocompartments mixed with TMV were adsorbed for 1 min to a titanium grid bearing a thick holey carbon film, washed 10 times with water, and then blotted and plunged into liquid nitrogen slush. Freeze-drying, grid transfer into the microscope, and imaging were performed as previously described (Sen et al, 2007). Dark-field micrographs were recorded at raster steps of 1.0 or 2.0 nm per pixel. Masses for individual nanocompartments and TMV segments were calculated using PCMass (available from the Brookhaven STEM resource). The nanocompartment preparation used for this analysis was different than the one used for cryo-EM/cryo-ET. Both preparations were found to be consistent by negative staining EM and SDS–PAGE except for the fact that the cryo-EM/cryo-ET preparation contained ˜6% of the core-lacking particles while the STEM one contained < 1%.
Quantitative analysis of encapsulin expression and assembly
For Western blotting, 1 × 108 cells grown in CTT and TPM medium and 1 × 108 purified spores were collected by centrifugation. Cells were lysed by adding 30 μl of SDS–PAGE loading buffer to this pellet and boiling for 10 min. Spores were re-suspended in 30 μl loading buffer and broken by ultrasonication, before boiling. Breakage was confirmed by light microscopy, using a Zeiss AxioImager. The lysates were loaded onto a 4–20% Tris-Glycine gel (Invitrogen) and transferred to a membrane. The primary antibody, G3576, was generated using a standard immunization protocol for rabbit against the KLH-conjugated target peptide RLRGESGVVVSTGR, and the secondary antibody (goat anti-rabbit) was from Jackson ImmunoResearch Laboratories Inc. Reactive bands were visualized with Pierce ECL Western Blotting Chemiluminescent Substrate (Thermo Scientific).
To quantify yields of dense-cored nanocompartments, equal samples of the respective isolates were applied for 15 s to carbon-coated 400 mesh copper grids and the numbers of particles per area counted. At least three independent fields were analyzed in each experiment. Means and standard deviations were determined, and all values normalized against the count of nanocompartments from CTT-grown cells. The total number of particles counted was 735 for wild-type cells grown in CTT, and 3,899 for wild-type cells grown in TPM.
Isolation of spores
Spots containing ˜4 × 109 wild-type cells/ml were placed on TPM agar and incubated for 48 or 96 h to induce fruiting body formation and spore development. Fruiting bodies were harvested using a cell scraper and re-suspended in Tris buffer (10 mM Tris/HCl pH 7.5). Vegetative cells were disrupted by ultrasonication and heating (60 min 50°C), and spores were concentrated by centrifugation (10 min, 14,000 × g). To remove more cellular debris, the spores were re-centrifuged using a 15% (w/v) Percoll gradient (GE Healthcare Sciences). Upon centrifugation (30 min 47,800 × g), membranes and cellular debris floated on top, while the spores formed a pellet at the bottom of the tube. After removal of the supernatant, the spores were washed twice with buffer, before being centrifuged and stored at 4°C.
Embedding of cells for thin sectioning
Vegetative cells and spores were briefly centrifuged and then high pressure-frozen, using a Leica EM HPM100 system. Frozen cells were transferred into pure acetone containing 2% glutaraldehyde and cryo-substituted at −80°C. After 72 h, the samples were gradually warmed and embedded in low-viscosity Spurr medium. Polymerized resin blocks were trimmed and thin sectioned using a diamond knife (Diatome). Silver-to-gold sections were mounted on copper grids and viewed without staining on a CM120 Philips electron microscope. Images were taken with a 4 × 4 ORCA ER CCD camera (Hamamatsu).
Oxidative stress survival assays
Cells were grown in rich medium and then treated with hydrogen peroxide. A protocol similar to that used to assay hydrogen peroxide stress survival in M. tuberculosis was performed, with modifications as described (Wu et al, 1997; Lu et al, 2010). DK1622 and encA cells were grown to mid-log phase in CTT medium and then diluted to an Abs550 of 0.2 units with fresh CTT. Next, 10-ml samples of each culture were transferred to sterile 25-ml glass flasks (Fisher Scientific) and were either left untreated or treated with a final concentration of 0.5 mM hydrogen peroxide, with each treatment done in triplicate. Cultures were incubated at 32°C with shaking at 250 rpm. Cell survival was monitored by measuring the Abs550 of each sample at 0, 1, 3, and 6 h after the addition of peroxide, and the values were normalized and expressed as a percentage of the starting absorbance in each tube. As described in Supplementary Materials and Methods, the disruption and deletion strains used in these experiments were generated completely independently.
Data deposition
The EncA refined coordinates were deposited in the protein data bank (PDB ID 4PT2). The cryo-EM maps of the T = 3 EncA shells and of the core-lacking M. xanthus encapsulin particles were deposited in the Electron Microscopy Data Bank (accession codes EMD-5917 and EMD-5953).
Acknowledgments
We thank G. Ketner, I. Coppens, K. McLean, D. Sullivan, A. Rule, D. Zusman, H. Engelhardt, and J. Kellermann for contributions of reagents and advice and P. Afonine for advice on the use of phenix.refine and phenix.map_to_structure_factors programs. Funding for this work was provided by the Intramural Research Program of the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health (ACS), a Ruth L. Kirschstein National Research Service Award T32 ES07141 (CAM), and National Institutes of Health, National Institute of General Medical Sciences grant R01 GM085024 (EH).
Author contributions
CAM, JF, DN, ACS, and EH designed research; CAM, JF, DN, NC, AAA, DW, ASL, JSW, and EH performed research; JBH contributed to analytic tools; CAM, JF, DN, ACS, and EH analyzed data; and JF, CAM, EH, and ACS wrote the paper with contributions from all other authors.
Conflict of interest
The authors declare that they have no conflict of interest.
Supplementary information for this article is available online: http://emboj.embopress.org
References
- Abrescia NG, Bamford DH, Grimes JM, Stuart DI. Structure unifies the viral universe. Annu Rev Biochem. 2012;81:795–822. doi: 10.1146/annurev-biochem-060910-095130. [DOI] [PubMed] [Google Scholar]
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonine PV, Grosse-Kunstleve RW, Adams PD. 2005. The Phenix refinement framework. CCP4 Newsl 42, contribution 8.
- Agirrezabala X, Velazquez-Muriel JA, Gomez-Puertas P, Scheres SH, Carazo JM, Carrascosa JL. Quasi-atomic model of bacteriophage T7 procapsid shell: insights into the structure and evolution of a basic fold. Structure. 2007;15:461–472. doi: 10.1016/j.str.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Akita F, Chong KT, Tanaka H, Yamashita E, Miyazaki N, Nakaishi Y, Suzuki M, Namba K, Ono Y, Tsukihara T, Nakagawa A. The crystal structure of a virus-like particle from the hyperthermophilic archaeon Pyrococcus furiosus provides insight into the evolution of viruses. J Mol Biol. 2007;368:1469–1483. doi: 10.1016/j.jmb.2007.02.075. [DOI] [PubMed] [Google Scholar]
- Andrews SC. The Ferritin-like superfamily: evolution of the biological iron storeman from a rubrerythrin-like ancestor. Biochim Biophys Acta. 2010;1800:691–705. doi: 10.1016/j.bbagen.2010.05.010. [DOI] [PubMed] [Google Scholar]
- Baker ML, Jiang W, Rixon FJ, Chiu W. Common ancestry of herpesviruses and tailed DNA bacteriophages. J Virol. 2005;79:14967–14970. doi: 10.1128/JVI.79.23.14967-14970.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belnap DM, Olson NH, Baker TS. A method for establishing the handedness of biological macromolecules. J Struct Biol. 1997;120:44–51. doi: 10.1006/jsbi.1997.3896. [DOI] [PubMed] [Google Scholar]
- Cantele F, Lanzavecchia S, Bellon PL. The variance of icosahedral virus models is a key indicator in the structure determination: a model-free reconstruction of viruses, suitable for refractory particles. J Struct Biol. 2003;141:84–92. doi: 10.1016/s1047-8477(02)00577-4. [DOI] [PubMed] [Google Scholar]
- Cardone G, Grünewald K, Steven AC. A resolution criterion for electron tomography based on cross-validation. J Struct Biol. 2005;151:117–129. doi: 10.1016/j.jsb.2005.04.006. [DOI] [PubMed] [Google Scholar]
- Cardone G, Heymann JB, Cheng N, Trus BL, Steven AC. Procapsid assembly, maturation, nuclear exit: dynamic steps in the production of infectious herpesvirions. Adv Exp Med Biol. 2012;726:423–439. doi: 10.1007/978-1-4614-0980-9_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng N, Conway JF, Watts NR, Hainfeld JF, Joshi V, Powell RD, Stahl SJ, Wingfield PE, Steven AC. Tetrairidium, a four-atom cluster, is readily visible as a density label in three-dimensional cryo-EM maps of proteins at 10–25 Å resolution. J Struct Biol. 1999;127:169–176. doi: 10.1006/jsbi.1999.4120. [DOI] [PubMed] [Google Scholar]
- Cheng N, Trus BL, Belnap DM, Newcomb WW, Brown JC, Steven AC. Handedness of the herpes simplex virus capsid and procapsid. J Virol. 2002;76:7855–7859. doi: 10.1128/JVI.76.15.7855-7859.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Computational Project. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Conway JF, Duda RL, Cheng N, Hendrix RW, Steven AC. Proteolytic and conformational control of virus capsid maturation: the bacteriophage HK97 system. J Mol Biol. 1995;253:86–99. doi: 10.1006/jmbi.1995.0538. [DOI] [PubMed] [Google Scholar]
- Conway JF, Wikoff WR, Cheng N, Duda RL, Hendrix RW, Johnson JE, Steven AC. Virus maturation involving large subunit rotations and local refolding. Science. 2001;292:744–748. doi: 10.1126/science.1058069. [DOI] [PubMed] [Google Scholar]
- Cornelis P, Wei Q, Andrews SC, Vinckx T. Iron homeostasis and management of oxidative stress response in bacteria. Metallomics. 2011;3:540–549. doi: 10.1039/c1mt00022e. [DOI] [PubMed] [Google Scholar]
- Diekmann Y, Pereira-Leal JB. Evolution of intracellular compartmentalization. Biochem J. 2013;449:319–331. doi: 10.1042/BJ20120957. [DOI] [PubMed] [Google Scholar]
- Duda RL, Hempel J, Michel H, Shabanowitz J, Hunt D, Hendrix RW. Structural transitions during bacteriophage HK97 head assembly. J Mol Biol. 1995;247:618–635. doi: 10.1006/jmbi.1995.0168. [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan C, Cheng S, Liu Y, Escobar CM, Crowley CS, Jefferson RE, Yeates TO, Bobik TA. Short N-terminal sequences package proteins into bacterial microcompartments. Proc Natl Acad Sci USA. 2010;107:7509–7514. doi: 10.1073/pnas.0913199107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, Sonnhammer EL, Tate J, Punta M. Pfam: the protein families database. Nucleic Acids Res. 2014;42:D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fokine A, Leiman PG, Shneider MM, Ahvazi B, Boeshans KM, Steven AC, Black LW, Mesyanzhinov VV, Rossmann MG. Structural and functional similarities between the capsid proteins of bacteriophages T4 and HK97 point to a common ancestry. Proc Natl Acad Sci USA. 2005;102:7163–7168. doi: 10.1073/pnas.0502164102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forterre P, Prangishvili D. The origin of viruses. Res Microbiol. 2009;160:466–472. doi: 10.1016/j.resmic.2009.07.008. [DOI] [PubMed] [Google Scholar]
- Gertsman I, Gan L, Guttman M, Lee K, Speir JA, Duda RL, Hendrix RW, Komives EA, Johnson JE. An unexpected twist in viral capsid maturation. Nature. 2009;458:646–650. doi: 10.1038/nature07686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman BS, Nierman WC, Kaiser D, Slater SC, Durkin AS, Eisen JA, Ronning CM, Barbazuk WB, Blanchard M, Field C, Halling C, Hinkle G, Iartchuk O, Kim HS, Mackenzie C, Madupu R, Miller N, Shvartsbeyn A, Sullivan SA, Vaudin M, et al. Evolution of sensory complexity recorded in a myxobacterial genome. Proc Natl Acad Sci USA. 2006;103:15200–15205. doi: 10.1073/pnas.0607335103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldsbury C, Baxa U, Simon MN, Steven AC, Engel A, Wall JS, Aebi U, Muller SA. Amyloid structure and assembly: insights from scanning transmission electron microscopy. J Struct Biol. 2011;173:1–13. doi: 10.1016/j.jsb.2010.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison PM, Arosio P. The ferritins: molecular properties, iron storage function and cellular regulation. Biochim Biophys Acta. 1996;1275:161–203. doi: 10.1016/0005-2728(96)00022-9. [DOI] [PubMed] [Google Scholar]
- Heinemann J, Maaty WS, Gauss GH, Akkaladevi N, Brumfield SK, Rayaprolu V, Young MJ, Lawrence CM, Bothner B. Fossil record of an archaeal HK97-like provirus. Virology. 2011;417:362–368. doi: 10.1016/j.virol.2011.06.019. [DOI] [PubMed] [Google Scholar]
- Hendrix RW, Johnson JE. Bacteriophage HK97 capsid assembly and maturation. Adv Exp Med Biol. 2012;726:351–363. doi: 10.1007/978-1-4614-0980-9_15. [DOI] [PubMed] [Google Scholar]
- Heymann JB, Cheng N, Newcomb WW, Trus BL, Brown JC, Steven AC. Dynamics of herpes simplex virus capsid maturation visualized by time-lapse cryo-electron microscopy. Nat Struct Biol. 2003;10:334–341. doi: 10.1038/nsb922. [DOI] [PubMed] [Google Scholar]
- Heymann JB, Belnap DM. Bsoft: image processing and molecular modeling for electron microscopy. J Struct Biol. 2007;157:3–18. doi: 10.1016/j.jsb.2006.06.006. [DOI] [PubMed] [Google Scholar]
- Hicks PM, Rinker KD, Baker JR, Kelly RM. Homomultimeric protease in the hyperthermophilic bacterium Thermotoga maritima has structural and amino acid sequence homology to bacteriocins in mesophilic bacteria. FEBS Lett. 1998;440:393–398. doi: 10.1016/s0014-5793(98)01451-3. [DOI] [PubMed] [Google Scholar]
- Hodgkin J, Kaiser D. Genetics of gliding motility in Myxococcus xanthus (Myxobacterales): two gene systems control movement. Mol Gen Genet. 1979;171:177–191. [Google Scholar]
- Homa FL, Huffman JB, Toropova K, Lopez HR, Makhov AM, Conway JF. Structure of the pseudorabies virus capsid: comparison with herpes simplex virus type 1 and differential binding of essential minor proteins. J Mol Biol. 2013;425:3415–3428. doi: 10.1016/j.jmb.2013.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang RK, Khayat R, Lee KK, Gertsman I, Duda RL, Hendrix RW, Johnson JE. The Prohead-I structure of bacteriophage HK97: implications for scaffold-mediated control of particle assembly and maturation. J Mol Biol. 2011a;408:541–554. doi: 10.1016/j.jmb.2011.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang RK, Steinmetz NF, Fu CY, Manchester M, Johnson JE. Transferrin-mediated targeting of bacteriophage HK97 nanoparticles into tumor cells. Nanomedicine (Lond) 2011b;6:55–68. doi: 10.2217/nnm.10.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Chang J, Jakana J, Weigele P, King J, Chiu W. Structure of epsilon15 bacteriophage reveals genome organization and DNA packaging/injection apparatus. Nature. 2006;439:612–616. doi: 10.1038/nature04487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser D. Social gliding is correlated with the presence of pili in Myxococcus xanthus. Proc Natl Acad Sci USA. 1979;76:5952–5956. doi: 10.1073/pnas.76.11.5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerfeld CA, Heinhorst S, Cannon GC. Bacterial microcompartments. Annu Rev Microbiol. 2010;64:391–408. doi: 10.1146/annurev.micro.112408.134211. [DOI] [PubMed] [Google Scholar]
- Kim D, Chung J, Hyun H, Lee C, Lee K, Cho K. Operon required for fruiting body development in Myxococcus xanthus. J Microbiol Biotechnol. 2009;19:1288–1294. doi: 10.4014/jmb.0903.0112. [DOI] [PubMed] [Google Scholar]
- Kleywegt GJ, Brunger AT. Checking your imagination: applications of the free R value. Structure. 1996;4:897–904. doi: 10.1016/s0969-2126(96)00097-4. [DOI] [PubMed] [Google Scholar]
- Kleywegt GJ, Jones TA. xdlMAPMAN and xdlDATAMAN – programs for reformatting, analysis and manipulation of biomacromolecular electron-density maps and reflection data sets. Acta Crystallogr D Biol Crystallogr. 1996;52:826–828. doi: 10.1107/S0907444995014983. [DOI] [PubMed] [Google Scholar]
- Lee B, Holkenbrink C, Treuner-Lange A, Higgs PI. Myxococcus xanthus developmental cell fate production: heterogeneous accumulation of developmental regulatory proteins and reexamination of the role of MazF in developmental lysis. J Bacteriol. 2012a;194:3058–3068. doi: 10.1128/JB.06756-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, DeLoache WC, Dueber JE. Spatial organization of enzymes for metabolic engineering. Metab Eng. 2012b;14:242–251. doi: 10.1016/j.ymben.2011.09.003. [DOI] [PubMed] [Google Scholar]
- Liu X, Theil EC. Ferritin reactions: direct identification of the site for the diferric peroxide reaction intermediate. Proc Natl Acad Sci USA. 2004;101:8557–8562. doi: 10.1073/pnas.0401146101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu LD, Sun Q, Fan XY, Zhong Y, Yao YF, Zhao GP. Mycobacterial MazG is a novel NTP pyrophosphohydrolase involved in oxidative stress response. J Biol Chem. 2010;285:28076–28085. doi: 10.1074/jbc.M109.088872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Michaelis L, Coryell CD, Granick S. Ferritin: III. The magnetic properties of ferritin and some other colloidal ferric compounds. J Biol Chem. 1943;148:463–480. [Google Scholar]
- Nemecek D, Qiao J, Mindich L, Steven AC, Heymann JB. Packaging accessory protein P7 and polymerase P2 have mutually occluding binding sites inside the bacteriophage φ6 procapsid. J Virol. 2012;86:11616–11624. doi: 10.1128/JVI.01347-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent KN, Khayat R, Tu LH, Suhanovsky MM, Cortines JR, Teschke CM, Johnson JE, Baker TS. P22 coat protein structures reveal a novel mechanism for capsid maturation: stability without auxiliary proteins or chemical crosslinks. Structure. 2010;18:390–401. doi: 10.1016/j.str.2009.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Rahmanpour R, Bugg TD. Assembly in vitro of Rhodococcus jostii RHA1 encapsulin and peroxidase DypB to form a nanocompartment. FEBS J. 2013;280:2097–2104. doi: 10.1111/febs.12234. [DOI] [PubMed] [Google Scholar]
- Rosenkrands I, Rasmussen PB, Carnio M, Jacobsen S, Theisen M, Andersen P. Identification and characterization of a 29-kilodalton protein from Mycobacterium tuberculosis culture filtrate recognized by mouse memory effector cells. Infect Immun. 1998;66:2728–2735. doi: 10.1128/iai.66.6.2728-2735.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SH, Chen S. Prevention of overfitting in cryo-EM structure determination. Nature Meth. 2012;9:853–854. doi: 10.1038/nmeth.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen A, Baxa U, Simon MN, Wall JS, Sabate R, Saupe SJ, Steven AC. Mass analysis by scanning transmission electron microscopy and electron diffraction validate predictions of stacked beta-solenoid model of HET-s prion fibrils. J Biol Chem. 2007;282:5545–5550. doi: 10.1074/jbc.M611464200. [DOI] [PubMed] [Google Scholar]
- Sieker LC, Holmes M, Le Trong I, Turley S, Liu MY, LeGall J, Stenkamp RE. The 1.9 A crystal structure of the “as isolated” rubrerythrin from Desulfovibrio vulgaris: some surprising results. J Biol Inorg Chem. 2000;5:505–513. doi: 10.1007/pl00021450. [DOI] [PubMed] [Google Scholar]
- Spilman MS, Dearborn AD, Chang JR, Damle PK, Christie GE, Dokland T. A conformational switch involved in maturation of Staphylococcus aureus bacteriophage 80alpha capsids. J Mol Biol. 2011;405:863–876. doi: 10.1016/j.jmb.2010.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steven AC, Heymann JB, Cheng N, Trus BL, Conway JF. Virus maturation: dynamics and mechanism of a stabilizing structural transition that leads to infectivity. Curr Opin Struct Biol. 2005;15:227–236. doi: 10.1016/j.sbi.2005.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutter M, Boehringer D, Gutmann S, Gunther S, Prangishvili D, Loessner MJ, Stetter KO, Weber-Ban E, Ban N. Structural basis of enzyme encapsulation into a bacterial nanocompartment. Nat Struct Mol Biol. 2008;15:939–947. doi: 10.1038/nsmb.1473. [DOI] [PubMed] [Google Scholar]
- Theil EC. Ferritin protein nanocages use ion channels, catalytic sites, and nucleation channels to manage iron/oxygen chemistry. Curr Opin Chem Biol. 2011;15:304–311. doi: 10.1016/j.cbpa.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theil EC, Behera RK, Tosha T. Ferritins for Chemistry and for Life. Coord Chem Rev. 2013;257:579–586. doi: 10.1016/j.ccr.2012.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas D, Schultz P, Steven AC, Wall JS. Mass analysis of biological macromolecular complexes by STEM. Biol Cell. 1994;80:181–192. doi: 10.1111/j.1768-322x.1994.tb00929.x. [DOI] [PubMed] [Google Scholar]
- Trabuco LG, Villa E, Mitra K, Frank J, Schulten K. Flexible fitting of atomic structures into electron microscopy maps using molecular dynamics. Structure. 2008;16:673–683. doi: 10.1016/j.str.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueki T, Inouye S, Inouye M. Positive-negative KG cassettes for construction of multi-gene deletions using a single drug marker. Gene. 1996;183:153–157. doi: 10.1016/s0378-1119(96)00546-x. [DOI] [PubMed] [Google Scholar]
- Valdes-Stauber N, Scherer S. Isolation and characterization of Linocin M18, a bacteriocin produced by Brevibacterium linens. Appl Environ Microbiol. 1994;60:3809–3814. doi: 10.1128/aem.60.10.3809-3814.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White HE, Sherman MB, Brasiles S, Jacquet E, Seavers P, Tavares P, Orlova EV. Capsid structure and its stability at the late stages of bacteriophage SPP1 assembly. J Virol. 2012;86:6768–6777. doi: 10.1128/JVI.00412-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikoff WR, Liljas L, Duda RL, Tsuruta H, Hendrix RW, Johnson JE. Topologically linked protein rings in the bacteriophage HK97 capsid. Science. 2000;289:2129–2133. doi: 10.1126/science.289.5487.2129. [DOI] [PubMed] [Google Scholar]
- Wireman JW, Dworkin M. Developmentally induced autolysis during fruiting body formation by Myxococcus xanthus. J Bacteriol. 1977;129:798–802. doi: 10.1128/jb.129.2.798-802.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu QL, Kong D, Lam K, Husson RN. A mycobacterial extracytoplasmic function sigma factor involved in survival following stress. J Bacteriol. 1997;179:2922–2929. doi: 10.1128/jb.179.9.2922-2929.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X, Dryden KA, Tang J, Baker TS. Ab initio random model method facilitates 3D reconstruction of icosahedral particles. J Struct Biol. 2007;157:211–225. doi: 10.1016/j.jsb.2006.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeates TO, Kerfeld CA, Heinhorst S, Cannon GC, Shively JM. Protein-based organelles in bacteria: carboxysomes and related microcompartments. Nat Rev Microbiol. 2008;6:681–691. doi: 10.1038/nrmicro1913. [DOI] [PubMed] [Google Scholar]
- Yeates TO, Thompson MC, Bobik TA. The protein shells of bacterial microcompartment organelles. Curr Opin Struct Biol. 2011;21:223–231. doi: 10.1016/j.sbi.2011.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeth K. Dps biomineralizing proteins: multifunctional architects of nature. Biochem J. 2012;445:297–311. doi: 10.1042/BJ20120514. [DOI] [PubMed] [Google Scholar]
- Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zusman DR, Scott AE, Yang Z, Kirby JR. Chemosensory pathways, motility and development in Myxococcus xanthus. Nat Rev Microbiol. 2007;5:862–872. doi: 10.1038/nrmicro1770. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.