Abstract
Neuronal histone acetylation has been linked to memory consolidation, and targeting histone acetylation has emerged as a promising therapeutic strategy for neuropsychiatric diseases. However, the role of histone-modifying enzymes in the adult brain is still far from being understood. Here we use RNA sequencing to screen the levels of all known histone acetyltransferases (HATs) in the hippocampal CA1 region and find that K-acetyltransferase 2a (Kat2a)—a HAT that has not been studied for its role in memory function so far—shows highest expression. Mice that lack Kat2a show impaired hippocampal synaptic plasticity and long-term memory consolidation. We furthermore show that Kat2a regulates a highly interconnected hippocampal gene expression network linked to neuroactive receptor signaling via a mechanism that involves nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB). In conclusion, our data establish Kat2a as a novel and essential regulator of hippocampal memory consolidation.
Keywords: epigenetics, histone acetylation, histone acetyltransferases, learning, memory
Introduction
Numerous findings across species have established that long-term memory consolidation critically depends on tightly controlled gene expression (Kandel, 2001). Recent studies suggest that epigenetic mechanisms of gene expression such as histone acetylation also play a role in memory formation and may provide a novel therapeutic avenue to treat cognitive diseases (Fischer et al, 2007; Ricobaraza et al, 2009; Kilgore et al, 2010; Peleg et al, 2010; Fischer, 2014). Histone acetylation is mediated by the counteracting activity of histone acetyltransferases (HATs) and histone deacetylases (HDACs), but the role of these enzymes during memory formation is only beginning to emerge. This is especially true for the 18 HATs encoded in the human and mouse genomes (Allis et al, 2007). While there is substantial evidence that manipulating the levels of CREB-binding protein (Crebbp; CBP; KAT3A, ENSMUSG00000022521) affects memory formation in mice (Alarcon et al, 2004; Korzus et al, 2004; Wood et al, 2005; Wood et al, 2006; Vecsey et al, 2007; Chen et al, 2010; Barrett et al, 2011), the current knowledge on the role of other HATs in the adult brain (Maurice et al, 2008; Oliveira et al, 2007) and especially the knowledge on the corresponding gene expression programs controlled by HAT activity is quite limited.
We therefore performed RNA sequencing to generate a quantitative transcriptome of the hippocampal CA1 region, which is a key brain structure for memory consolidation. These data allowed us to compare expression of all HATs within the CA1 region. Highest expression was observed for Kat2a, formerly known as General control of amino acid synthesis protein 5-like 2 (Gcn5l2). Since also the Kat2a protein was highly expressed in the adult mouse hippocampus and the function of this protein in memory formation has not been studied before, we decided to further investigate the role of Kat2a in the adult brain.
To this end, we generated conditional Kat2a KO mice. Deletion of Kat2a from excitatory neurons of the adult forebrain resulted in impaired hippocampus-dependent memory consolidation as well as impaired synaptic and nuclear plasticity. In addition, our data suggest that Kat2a controls hippocampal memory function by regulating a gene expression network linked to neuroactive ligand–receptor interaction via mechanisms that involve the activity of nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB).
In conclusion, we establish Kat2a as an important regulator of transcriptome plasticity essential for hippocampus-dependent memory formation.
Results
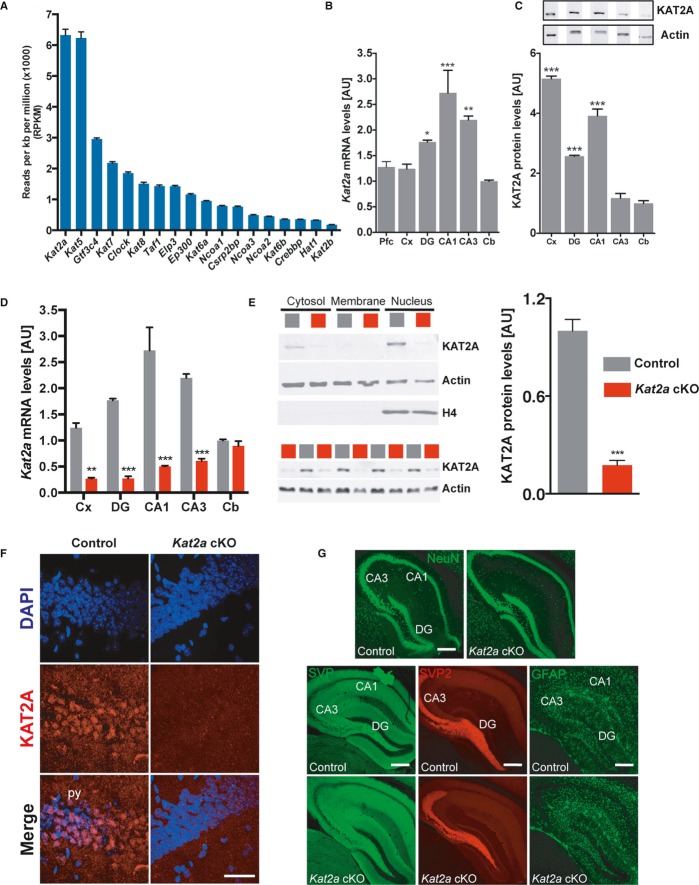
Kat2a is highly expressed in the hippocampal CA1 region
The hippocampal CA1 region is critical for memory function in mice and in humans and has been linked to learning-induced epigenetic gene expression (Levenson et al, 2004). As a starting point to further elucidate the role of HATs in memory function, we therefore performed RNA sequencing analysis of the hippocampal CA1 region from adult mice. In contrast to other methods for gene expression analysis, RNA sequencing allows for proportional quantification of the transcriptome, thereby enabling us to directly compare transcript abundance, including levels of different HATs within a given sample. We were able to detect 16,218 genes that show more than 10 reads per kilobase per million reads (RPMK). As expected, we found genes that are strongly associated with learning and memory processes to be enriched among the most highly expressed genes (Supplementary Table S1). When we specifically analyzed the expression of the 18 mammalian HATs, the highest read count was observed for the Kat2a gene (Fig1A). Kat2a belongs to the GNAT family of HATs and has been implicated in acetylation of histones as well as non-histone proteins (Rodríguez-Navarro, 2009). A role for Kat2a in memory function has not been investigated so far. However, Kat2a has been implicated in stimulus-dependent gene expression in other cell types and organisms (Hargreaves et al, 2009; Johnsson et al, 2009), making it a promising candidate for regulation of neuronal activity-driven gene expression. Furthermore, Kat2a was identified as one of the genes that are up-regulated in response to fear-conditioning training using a microarray approach (Peleg et al, 2010). Thus, we decided to further study this enzyme in the adult brain. We started our analysis by measuring the expression of Kat2a in different brain regions. Quantitative real-time PCR (qPCR) revealed expression of Kat2a in the hippocampal subregions CA1, CA3, and the dentate gyrus as well as in the cortex and prefrontal cortex along with relatively low expression in the cerebellum (Fig1B). Within the hippocampus, the highest expression of Kat2a was seen in the CA1 region. Next, we measured Kat2a protein levels. Quantitative immunoblot analysis revealed highest Kat2a production in the hippocampal CA1 region and the cortex, while low amounts of Kat2a were observed in the cerebellum and the CA3 region (Fig1C).
Figure 1. Kat2a is highly expressed in the adult forebrain and dispensable for maintenance of gross brain morphology.
- RNA was isolated from the hippocampal CA1 region of wild-type mice (n = 6) and subjected to RNA sequencing. Quantitative expression analysis revealed that of all HATs, Kat2a showed the highest expression.
- qPCR analysis for Kat2a in the adult brain of wild-type mice (n = 5). Data were normalized and compared to the levels in the cerebellum (one-way ANOVA with Holm–Sidak-corrected multiple comparisons, ***P < 0.001; **P < 0.01, *P < 0.05).
- Quantitative immunoblot analysis of Kat2a in the adult brain of wild-type mice (n = 3). Representative immunoblot images (upper panel) and quantification (lower panel) are shown. Data were normalized and compared to Kat2a levels in the cerebellum. Actin served as a loading control (one-way ANOVA with Holm–Sidak-corrected multiple comparisons, ***P < 0.001 versus cerebellum).
- qPCR analysis for Kat2a in the adult brain of Kat2a cKO and control mice (two-way ANOVA with Sidak-corrected multiple comparisons, **P < 0.01, ***P < 0.001 versus control).
- Representative images showing immunoblot analysis for Kat2a in subcellular protein fractions (upper panel) and nuclear KAT2A levels (lower panel) in the hippocampus of control and Kat2a cKO mice. Actin and nuclear histone H4 were used for quality control. Quantification (n = 4, two-sided t-test, ***P < 0.001) is shown on the right.
- Representative images showing immunostaining for Kat2a in the hippocampal CA1 region of control and Kat2a cKO mice. DAPI was used to stain nuclei. Note the nuclear signal for Kat2a in control mice, which is absent in Kat2a cKO mice. Scale bar, 150 μm.
- Representative images showing NeuN, SVP2, SVP, and GFAP immunoreactivity in control and Kat2a cKO mice (n = 4/group). Scale bar, 250 μm. CA1, hippocampal subfield CA1; CA3, hippocampal subfield CA3; DG, dentate gyrus; Cb, cerebellum, Cx, cortex, py, pyramidal cell layer of CA1 region, NeuN, neuronal nuclei (Rbfox3), SVP2, synaptoporin; SVP, synaptophysin; GFAP, glia fibrillary acidic protein.
Data information: Error bars indicate s.e.m.
To study the role of Kat2a in greater detail, we generated mice that lack Kat2a in excitatory neurons of the adult forebrain. To this end, animals, in which exons 3–17 of the Kat2a gene are flanked by loxP sites (Lin et al, 2008), were crossed to mice expressing CRE recombinase under the control of the CamKIIα promoter (Kat2a cKO). Mice that carry the floxed Kat2a allele but do not express CRE recombinase were used as control (control) (Kuczera et al, 2010; Kerimoglu et al, 2013). As expected, qPCR analysis revealed a significant reduction of Kat2a expression in all hippocampal regions of Kat2a cKO mice while Kat2a mRNA levels in the cerebellum were unaffected (Fig1D). Analyzing the subcellular localization of Kat2a in the hippocampus by immunoblot analysis, we found Kat2a predominantly localized to the nucleus, which is in line with its role in histone acetylation (Fig1E). Quantitative analysis confirmed a highly significant reduction of Kat2a protein levels in the hippocampal CA1 region of Kat2a cKO mice (Fig1E), which was also observed using immunohistochemical analysis (Fig1F). While the constitutive deletion of Kat2a in mice is lethal (Bu et al, 2007), mice lacking Kat2a in the adult forebrain were viable and displayed home-cage behavior that was indistinguishable from control mice. In line with this observation, the brain weight of Kat2a cKO mice (Supplementary Fig S1) as well as immunoreactivity for NeuN, a marker for neuronal integrity (Fischer et al, 2005), were similar to control mice (Fig1G). Gross brain morphology was unaffected in cKO mice, as shown by immunohistochemical staining for the synaptic marker proteins synaptophysin (SVP), synaptoporin (SVP2), and microtubule-associated protein 2 (MAP2) as well as the oligodendrocyte marker PLP1 and the astrocyte marker GFAP (Fig1G and Supplementary Fig S1).
In conclusion, these data show that Kat2a is highly expressed in the adult hippocampus but deletion of Kat2a from adult forebrain neurons does not lead to obvious detrimental phenotypes.
Kat2a is necessary for long-term memory consolidation and synaptic plasticity
To test for a role of Kat2a in cognitive function more specifically, we subjected Kat2a cKO and control mice to behavior testing. Explorative behavior and basal anxiety, as assessed in the open field test, were found to be similar among groups (Fig2A and B). The observation that basal anxiety was unaffected in Kat2a cKO mice could be confirmed in the elevated plus maze test (Fig2C). Moreover, the rotarod test demonstrated no difference in physical strength, endurance, and motor coordination (Fig2D). Next, we measured working memory function by employing the cross maze test, where Kat2a cKO and control mice performed equally well (Fig2E). Intact short-term memory function in Kat2a cKO mice was confirmed in the novel object recognition test, as no difference in the preference for a novel object was detectable among groups when mice were exposed to the novel object 5 min after the training session (Fig2F). While short-term memory is independent of de novo gene expression, the formation of stable long-term memories depends on differential gene expression, a process that critically involves chromatin plasticity (Day & Sweatt, 2011; Gräff & Tsai, 2013). Thus, we also analyzed long-term memory consolidation in Kat2a cKO mice. First, we subjected mice to a long-term memory version of the novel object recognition task. Interestingly, although both groups performed better than chance level in this test, Kat2a cKO mice showed a slight, but significant, reduction in preference for the novel object (Fig2G). To further investigate the role of Kat2a in long-term memory function, we subjected mice to the hippocampus-dependent contextual fear-conditioning paradigm, which measures associative memory consolidation. Activity during training and response to the electric foot shock were similar among groups (Fig2H). Freezing behavior, a quantitative measure of memory consolidation, was significantly reduced in Kat2a cKO mice when compared to control littermates 24 h after training, indicating impaired associative memory consolidation (Fig2H).
Figure 2. Kat2a is required for long-term memory consolidation.
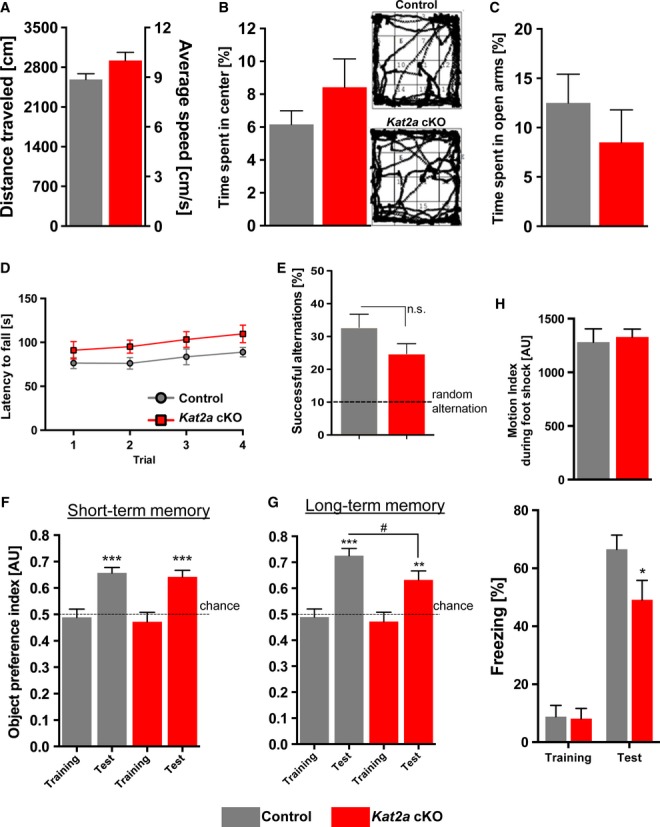
- No difference was observed when the distance traveled, and the average speed during a 5-min exposure to an open field was compared between Kat2a cKO (n = 11) and control mice (n = 16).
- Basal anxiety in the open field test. The time spent in the center of the open field arena was similar among Kat2a cKO (n = 16) and control mice (n = 24). Representative tracking plots during the 5-min open field exposure are shown on the right.
- When exposed to the elevated plus maze test, the time spent in the open arms was similar in Kat2a cKO (n = 11) and control mice (n = 16).
- During the rotarod test, Kat2a cKO (n = 11) and control mice (n = 10) spent a similar amount of time on the rotating rod in all trials, with both groups increasing performance during trials.
- The number of successful alternations in the cross maze test was similar when we compared Kat2a cKO (n = 10) and control mice (n = 14) and higher than chance level in both groups (one-sample t-test, ***P < 0.001).
- In a novel object recognition test protocol designed to test for short-term memory, Kat2a cKO (n = 16) and control mice (n = 24) showed a similar preference for the novel object (one-sample t-test, ***P < 0.001) when comparing the preference index to chance level for each genotype. There was no significant difference among genotypes for the performance during the test sessions (two-sided t-test, P = 0.6446).
- When Kat2a cKO (n = 16) and control mice (n = 24) were exposed to a novel object recognition paradigm designed to test long-term memory consolidation, the object preference index was significantly decreased in Kat2a cKO compared to control mice (two-sided t-test, #P < 0.05), while preference for the novel object was significant in both groups (one-sample t-test, ***P < 0.001, **P < 0.01).
- Upper panel: Response to the electric foot shock during contextual fear-conditioning training was similar among groups (control mice, n = 15; Kat2a cKO mice, n = 11). Lower panel: Baseline-freezing behavior during the training was similar among groups. Kat2a cKO mice showed reduced freezing behavior during the memory test, when compared to control group (*P < 0.05).
Data information: Error bars indicate s.e.m.
To further test the role of Kat2a in hippocampus-dependent long-term memory function, we subjected mice to the Morris water maze test. When compared to the control group, Kat2a cKO mice showed a significantly higher escape latency to find the hidden platform throughout the 9 days of training (Fig3A). No such difference was observed when mice had to locate a visible platform (Fig3A), indicating that this effect is not due to impaired vision or motor coordination. The latter was further confirmed by the finding that swim speed was similar among groups (Fig3B). On day 10, memory consolidation was tested in a probe test. While control mice spent significantly more time in the target quadrant when compared to the other quadrants, Kat2a cKO mice only performed at chance level (Fig3C). In line with this observation, the number of platform crossings was also impaired in Kat2a cKO mice (Fig3D). While the CamKIIα-Cre line employed to delete functional Kat2a has been used previously to postnatally delete target genes and to analyze hippocampal memory formation (Minichiello et al, 1999; Kuczera et al, 2010; Görlich et al, 2012; Kerimoglu et al, 2013), CRE expression in these mice is driven in the forebrain and not exclusively in the hippocampal formation. To confirm that the observed memory impairment in Kat2a cKO mice is due to the role of Kat2a in the hippocampal CA1 region, we used a viral-mediated approach to delete Kat2a in 3-month-old mice. Adeno-associated virus (AAV) particles mediating synapsin-promoter-driven expression of GFP-fused CRE recombinase were injected into the dorsal CA1 region of mice homozygous for the floxed Kat2a allele (Kat2afl/fl AAV-CRE mice). Wild-type littermates injected with the same AAV were used as control (Kat2a+/+ AAV-CRE). qPCR and immunohistochemical analysis confirmed the deletion of Kat2a in CA1 neurons of Kat2afl/fl AAV-CRE mice (Fig3E and Supplementary Fig S2). Viral-mediated deletion of Kat2a in the CA1 region did not affect explorative behavior nor basal anxiety, and we could rule out an effect of the floxed allele on water maze performance between wild-type controls (Kat2a+/+) and Kat2afl/fl animals (Supplementary Fig S2). Next, we subjected mice to the water maze test to assess hippocampus-specific long-term memory. Mice lacking Kat2a in the CA1 region showed severely impaired escape latency throughout the training (Fig3F) and performed significantly worse than the control group in the probe test (Fig3G and H), demonstrating that Kat2a in the hippocampal CA1 region is essential for spatial memory consolidation.
Figure 3. Kat2a expressed in the CA1 region is necessary for plasticity and memory formation.
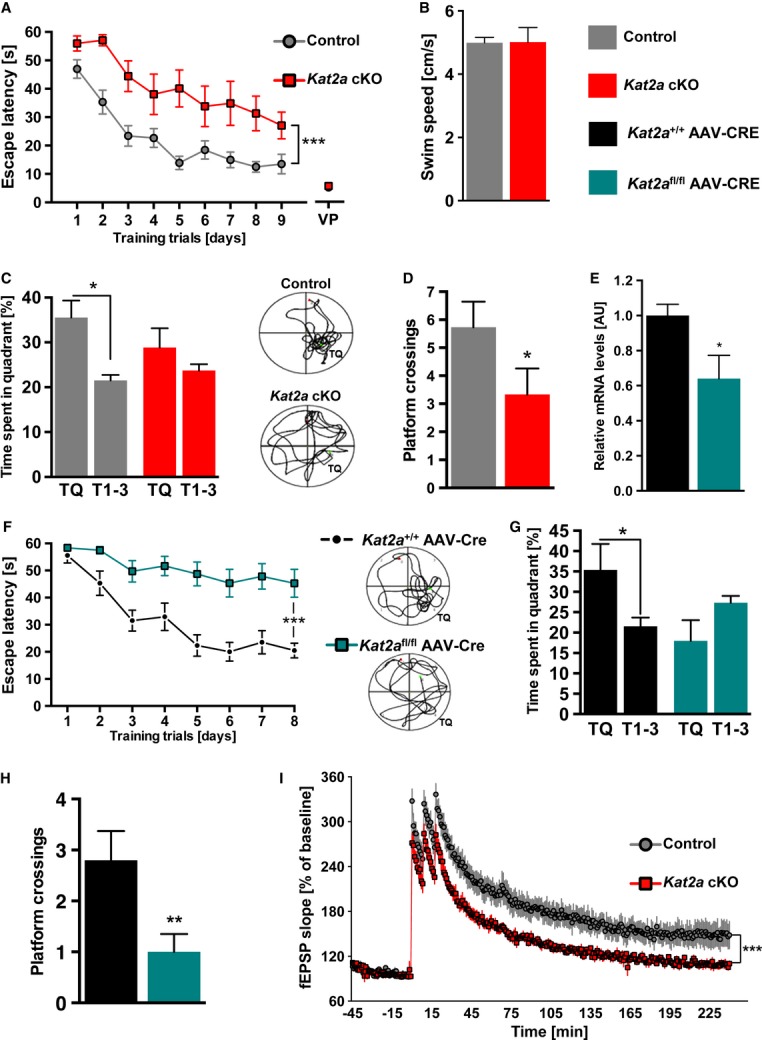
- Escape latency to find the hidden platform was significantly impaired throughout the training days in Kat2a cKO mice (n = 9) when compared to the control group (n = 15; repeated-measures (rm) ANOVA, ***P < 0.001, F(1,22) = 18.04). Escape latency to a visual platform (VP) was similar among groups (n = 10/group).
- Average swim speed during the water maze test was similar among groups (control mice, n = 15; Kat2a cKO mice, n = 9).
- During probe test, only control mice spent significantly more time on the target quadrant (TQ), when compared to the other three quadrants (T1-3) (two-sided t-test, *P < 0.05). Representative swim paths during probe test are shown on the right.
- During probe test, the number of platform crossings was significantly lower in Kat2a cKO mice when compared to the control group (two-sided t-test, *P < 0.05).
- qPCR analysis for Kat2a shows the reduction of Kat2a expression in Kat2afl/fl AAV-CRE mice (n = 3) when compared to wild-type littermates transfected with Kat2a+/+ AAV-CRE (n = 6) (*P < 0.05).
- In the water maze test, escape latency was significantly impaired in Kat2afl/fl AAV-CRE mice (n = 14) when compared to the Kat2a+/+ AAV-CRE group (n = 15; rmANOVA, ***P < 0.001, F(1,27) = 19.22). Representative swim paths are shown on the right.
- The Kat2a+/+ AAV-CRE group showed a significant preference for the target quadrant (two-sided t-test, *P < 0.05 for TQ versus T1-3).
- The number of platform crossings during probe test was significantly decreased in Kat2afl/fl AAV-CRE mice when compared to the control group (*P < 0.05).
- Hippocampal LTP was significantly impaired in slices from Kat2a cKO mice when compared to control slices (n = 5/group) (rmANOVA, main effect of time: F(285, 2280) = 116.9, P < 0.001, main effect of genotype: F(1, 8) = 8.3, P = 0.021, interaction: F(285, 2280) = 3.3, P < 0.001).
Data information: Error bars indicate s.e.m.
To further explore the cellular mechanisms by which Kat2a regulates long-term memory formation, we analyzed the electrophysiological properties at the CA3-CA1 synapse. To this end, CA3-originating Schaffer collateral fibers were externally stimulated, and excitatory post-synaptic potentials (fEPSPs) were recorded in the apical dendritic layer of the CA1 region of Kat2a cKO and control littermates. In both groups, stimulation strength was well correlated with the elicited fEPSP slope, yielding similar linear input/output curves (Supplementary Fig S3). This indicated normal basal synaptic transmission at the CA3-CA1 synapse. Also paired-pulse facilitation, a form of short-term plasticity that reflects the probability of presynaptic transmitter release, was indistinguishable between Kat2a cKO and control mice (Supplementary Fig S3). To measure long-term plasticity, we analyzed long-term potentiation (LTP) in Kat2a cKO and control littermates. Baseline fEPSP slopes were similar in both groups during the first 45 min of the measurement (Fig3I). While robust LTP was induced in control mice, the corresponding fEPSP slope in Kat2a cKO was significantly reduced and showed a faster decay back to baseline (Fig3I). These data indicate that basal synaptic transmission in Kat2a cKO mice is intact but long-term plasticity at the CA3-CA1 synapse is impaired, which is most likely due to the deregulation of postsynaptic processes in the CA1 cells. These findings are in line with our data showing high levels of KAT2A in the CA1 region when compared to CA3, and the fact that impaired spatial memory formation is observed in Kat2a cKO mice.
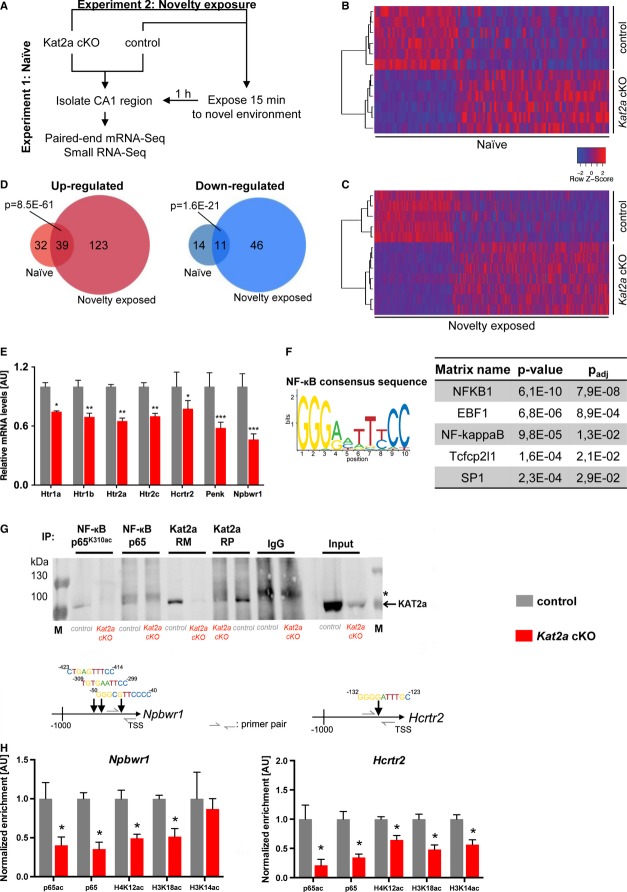
Kat2a regulates a hippocampal gene expression network linked to neuroactive ligand–receptor signaling
To further elucidate the molecular mechanisms by which Kat2a contributes to memory formation, we employed paired-end RNA sequencing. This allowed us to analyze the Kat2a-dependent gene expression program in the hippocampal CA1 region of adult mice in an unbiased and quantitative manner. We designed a 4-arm approach comparing naïve Kat2a cKO and control mice as well as Kat2a cKO and control mice 1 h after a 15-min exposure to a novel context. Using this experimental setup, we were able to capture both, basal and stimulus-induced Kat2a-dependent gene expression (Fig4A). When comparing the expression of genes in naïve control versus naïve Kat2a cKO mice, we detected 96 differentially expressed genes, including the gene coding for Kat2a that was—as expected—down-regulated in Kat2a cKO mice (Fig4B, Supplementary Table S2). When we analyzed the gene expression pattern in response to novelty exposure, we observed a total of 209 genes that were differentially expressed between Kat2a cKO and control mice (Fig4C, Supplementary Table S2). Although 50 of these genes were attributed to the deletion of Kat2a independent of novelty exposure, these data suggest that deregulation of the hippocampal CA1 transcriptome in mice lacking Kat2a is increased in response to novelty exposure (Fig4D). Pathway analysis revealed that the genes affected by the loss of Kat2a form a tightly interconnected network (Supplementary Fig S4) in which we identified the neuroactive ligand–receptor signaling pathway as a major component (Supplementary Fig S4, Supplementary Table S3). Since this was true both for naïve or novelty-exposed mice, we decided to validate down-regulation of selected genes in the hippocampal CA1 region of naïve Kat2a cKO and control littermates via qPCR. We could confirm differential expression of key genes linked to neuroactive ligand–receptor signaling, including down-regulation of different serotonin receptor genes (5-hydroxytryptamine receptor; Htr genes) such as Htr1a, Htr1b, Htr2a, and Htr2c (Fig4E). We furthermore confirmed down-regulation of the Hcrtr2 gene coding for the Hypocretin/Orexin receptor 2, the Penk gene coding for the Proenkephalin and the Npbwr1 gene encoding the neuropeptide B/W receptor 1 (Fig4E). All of these genes have been linked to memory function (see Discussion), and thus, deregulation of these genes provides a reasonable explanation for the observed phenotypes in Kat2a cKO mice.
Figure 4. Kat2a regulates hippocampal gene expression.
A Experimental design.
B, C Heat map showing the clustering of differentially expressed genes comparing the CA1 region of naïve Kat2a cKO to naïve control mice (B) or after novelty exposure (C).
D Venn diagram showing the up- and down-regulated genes in Kat2a cKO mice, grouped by treatment. The probability (P) to observe the respective overlaps by chance is given.
E qPCR analysis shows validation of down-regulation of selected genes in the hippocampal CA1 region of Kat2a cKO mice compared to the control group (n = 5/group; two-sided t-test versus control, ***P < 0.001, **P < 0.01, *P < 0.05).
F Consensus sequence for NF-κB DNA-binding (JASPAR ID: MA0061.1) used to screen genes down-regulated in Kat2a cKO mice (left panel). The Top 5 results from promoter analysis of differentially expressed genes using pScan (right panel).
G CA1 tissue from control and Kat2a cKO mice was subjected to co-immunoprecipitation using different antibodies. RM: rabbit monoclonal antibody; RP: rabbit polyclonal antibody. * indicates unspecific band just above the Kat2a signal that is visible in the IgG samples and in the IP for non-acetylated-p65.
H ChIP analysis from CA1 tissue was performed for p65 binding and histone modifications at the promoter regions for Nbpwr1 and Hcrtr2 genes. The upper panels represent the NF-κB biding sites with the corresponding promoter regions for which primers were designed (n = 4/group) (two-sided t-test, *P < 0.05 versus control).
Data information: Error bars indicate s.e.m.
In addition to protein-coding transcriptome analysis, we also investigated the microRNAome (miRNome) in the hippocampal CA1 region of Kat2a cKO and control mice using small RNA sequencing. While the miRNome of the whole hippocampus has been reported before (Pena et al, 2009; Zovoilis et al, 2011), to our knowledge this is the first miRNome analysis of a hippocampal subfield. We detected the expression of 495 miRNAs (Supplementary Table S4). When compared to the miRNome of the entire hippocampus, we found substantial differences, indicating region-specific functions for hippocampal miRNAs (Supplementary Fig S5A). Among the Top 50 miRNAs, there were many candidates that have been linked to memory function in previous studies. However, when we compared miRNA expression in Kat2a cKO and control mice, we did not detect any significant differences (Supplementary Fig S5B; Supplementary Table S4) suggesting that the observed phenotype in Kat2a cKO mice is independent of miRNA regulation and that Kat2a is not involved in miRNA expression.
Kat2a associates with NF-κB
The question remained how histone-modifying enzymes such as Kat2a acquire specificity toward certain functional genomic loci. We thus explored the possibility that Kat2a regulates selected genes in concert with the action of DNA-binding enzymes such as transcription factors (TF) and analyzed the promoter regions of genes differentially regulated in Kat2a cKO mice (Supplementary Table S2) for common motifs and TF binding sites. This revealed a significant enrichment for the binding motif of nuclear factor kappa-light-chain-enhancer of activated B-cell complex (NF-κB), a TF complex linked to hippocampus-dependent memory function (Fig4F) (Meffert et al, 2003; Ahn et al, 2008). These data suggested that NF-κB could be a key player involved in the recruitment of Kat2a to its target genes. Transcriptional activation by the prototypical NF-κB dimeric complex critically involves the RelA subunit (p65) (Schmitz et al, 2004; Chen & Greene, 2004), and concomitant promoter binding of Kat2a and p65 has been linked to active gene expression before (Lusic et al, 2003; Meffert & Baltimore, 2005). Moreover, p65 function is regulated by acetylation of lysine residue 310 (Chen et al, 2001; Schmitz et al, 2004) and has been implicated in hippocampus-dependent memory formation (Meffert et al, 2003). On the basis of this evidence, we further investigated the role of p65 in Kat2a-mediated gene expression during memory formation. Co-immunoprecipitation experiments showed that Kat2a interact with acetylated as well as non-acetylated p65 in the hippocampal CA1 region of wild-type mice (Fig4G). Tissue obtained from Kat2a cKO mice as well as IgG precipitations served as negative controls. Because these data further supported a role of p65 in Kat2a-mediated gene expression in the hippocampus, we analyzed the levels of p65K310ac, p65 as well as histone 3 (H3) acetylated at lysine 14 (H3K14ac) and lysine 18 (H3K18ac) and histone 4 (H4) acetylated at lysine 12 (H4K12ac) at predicted NF-κB binding motifs within the promoters of genes that are likely to be direct Kat2a targets (Fig4H, Supplementary Fig S4). Chromatin immunoprecipitation (ChIP) revealed that promoter binding of p65, p65K310ac, and corresponding histone acetylation was significantly reduced in Kat2a cKO mice when compared to the control group. This was not the case for selected known NF-κB target genes, which were not differentially expressed between control and Kat2a cKO mice and are therefore unlikely to be Kat2a-regulated target genes (Supplementary Fig S4). In conclusion, these data provide further evidence that NF-κB plays a role in Kat2a-mediated gene expression.
Discussion
The role of histone acetylation in the adult brain is gaining increasing attention because targeting HATs and HDACs has emerged as a promising therapeutic strategy to treat neurodegenerative diseases (Fischer et al, 2010; Fischer, 2014). As such, pharmacological inhibition of HDAC proteins (Fischer et al, 2007; Kilgore et al, 2010; Ricobaraza et al, 2009; Govindarajan et al, 2011; Govindarajan et al, 2013) or activation of HATs (Selvi et al, 2010; Chatterjee et al, 2013; Schneider et al, 2013) was found to facilitate cognitive function in preclinical studies. The development of more specific pharmacological compounds is, however, hindered by the fact that knowledge about the 11 HDACs and 18 HATs encoded in the human and mouse genome is still limited. This is particularly true for the HATs. A role in memory function has been investigated for only three of the existing 18 HATs, namely KAT3A (CBP) (Alarcon et al, 2004; Korzus et al, 2004; Wood et al, 2005; Chen et al, 2010; Valor et al, 2011), the related KAT3B (p300) (Oliveira et al, 2007), and KAT2B (PCAF) (Maurice et al, 2008). By employing RNA sequencing on tissue obtained from the mouse hippocampal CA1 region, we were able to detect expression of all HATs. However, highest expression was observed for Kat2a, more widely known as Gcn5. We chose the CA1 region because of its well-established role in memory consolidation and neurodegenerative diseases (Gallagher & Koh, 2011; Howard & Eichenbaum, 2013). Notably, also Kat5 showed very high expression in the CA1, which was recently implicated in neuronal function in a Drosophila model of Alzheimer's disease (Pirooznia et al, 2012). More research to investigate the function of this and the other HATs in memory formation is therefore warranted.
When Kat2a levels were compared in different brain regions, highest expression was seen in the hippocampal formation, which is in congruence with data available via the Allen Brain Atlas (http://mouse.brain-map.org/). While the level of gene expression does not necessarily translate directly into protein levels, we could detect high levels of Kat2a protein in the hippocampus, especially in the CA1 region, when compared to other brain regions. Immunohistochemical as well as immunoblot analysis showed that Kat2a is localized to the nucleus, which is in line with its documented role in histone acetylation and gene expression. These data suggested a possible role of Kat2a in memory function. Indeed, deletion of Kat2a from excitatory forebrain neurons caused memory impairment in three different tests all requiring intact hippocampal function. Consistent with previous studies in which CamKIIα-driven expression of CRE was used to generate forebrain-specific knockout of target genes (Minichiello et al, 1999; Kuczera et al, 2010), loss of Kat2a in the forebrain was not complete. About 18% of the corresponding mRNA and protein levels were still present, most likely due to Kat2a expression in glia cells and in inhibitory neurons. In turn, these data suggest that the vast majority of Kat2a is expressed in excitatory neurons. Notably, memory impairment was specific for the consolidation of long-term memories since short-term memory was unaffected in Kat2a cKO mice as measured in two different tests. Impairment in memory consolidation was paralleled by impaired hippocampal plasticity, namely reduced LTP at the CA3-CA1 synapse, which is considered to be a molecular correlate of memory function. These data further suggested a specific role of hippocampal Kat2a in memory formation. We could confirm this hypothesis by showing that the specific deletion of Kat2a from the hippocampal CA1 region by viral-mediated expression of CRE causes a similarly severe impairment in spatial long-term memory consolidation as in Kat2a cKO mice. Together with our finding that Kat2a expression was highest in the hippocampus, specifically in the CA1 region, this experiment suggests that lack of Kat2a from the CA1 significantly contributes to the phenotype observed in Kat2a cKO mice. However, our results do not rule out the possibility that Kat2a has important functions in other brain regions.
Interestingly, Kat3a (Cbp, Crebbp), which has been previously studied for its role in memory formation, showed rather low expression levels in the hippocampal CA1 region. It is therefore interesting to note that the studies investigating KAT3A function in memory performance have often focused on the whole hippocampus and consistently showed that loss of KAT3A leads to impaired object recognition learning or fear conditioning while water maze learning—that highly depends on the hippocampal CA1 region—was impaired in some but not all studies (Korzus et al, 2004; Alarcon et al, 2004; Chen et al, 2010; Josselyn, 2005). These data might reflect specific expression patterns of HATs. In our study, we find that loss of Kat2a leads to very robust impairment in the water maze task while novel object recognition learning and fear conditioning are rather mildly impaired. We speculate that this could be explained in part by a region-specific role of HATs in memory formation, with Kat2a being particularly important for cognitive tasks linked to the CA1 region.
Since long-term memory consolidation critically depends on tightly regulated gene expression programs, our data suggest that the observed cognitive phenotypes are—at least in part—mechanistically linked to Kat2a-mediated gene expression. On this basis, we decided to study the role of Kat2a in hippocampal gene expression by performing paired-end RNA sequencing of the CA1 region in naïve Kat2a cKO and control mice as well as in mice that were exposed to a novelty stimulus. We took this approach because there is evidence that counteracting HATs and HDACs do not only regulate basal gene expression, but their activities become increasingly important during stimulus-induced gene expression changes (Peleg et al, 2010; Lopez-Atalaya et al, 2011). We could detect a number of differentially expressed genes in naïve and in novelty-exposed Kat2a cKO mice. The Kat2a gene was the most significantly down-regulated gene, which validated our analysis pipeline. Importantly, we did not detect compensatory expression of other HATs or HDACs in any of the experimental groups, suggesting a non-redundant function of Kat2a. Pathway enrichment analysis revealed that under all conditions, the loss of Kat2a affects neuroactive ligand–receptor signaling. We could confirm the down-regulation of key genes within this pathway in Kat2a cKO mice. Specifically, down-regulation of serotonin receptors Htr1a, Htr1b, Htr2a, and Htr2c was detected. The serotonergic system in the adult brain is complex, and thus, the existing pharmacological and genetic data are not always conclusive. Yet there is sufficient evidence for a critical role of serotonin receptor signaling in memory formation, and its deregulation has been linked to cognitive dysfunction (Bert et al, 2008). The Htr2a gene has been linked to hippocampal novelty processing and memory formation in humans (De Quervain et al, 2003; Reynolds et al, 2006; Sigmund et al, 2008; Schott et al, 2011). Moreover, Htr2a expression was decreased in rats that were treated with amyloid beta peptides that cause memory impairments (Christensen et al, 2008), and HTR2A binding capacity is decreased in the brains of AD patients (Meltzer et al, 1998; Marner et al, 2012). Furthermore, activation of HTR2A facilitates hippocampus-dependent memory function in rodents while pharmacological inhibition had the opposite effect (Alhaider et al, 1993; Zhang et al, 2013). Thus, our data showing down-regulation of Htr2a in Kat2a cKO mice are in full agreement with published reports. Altered editing and increased expression of the Htr2c mRNA was observed in response to water maze training in mice (Cavallaro et al, 2002; Du et al, 2007) which is in line with the previously reported transiently increased expression of Kat2a after a learning stimulus (Peleg et al, 2010). In fact, we found that Htr2c editing was altered in Kat2a cKO mice (Supplementary Fig S6, Supplementary Table S6). Also Htr1a has been implicated with cognitive function (Bert et al, 2008), and in line with our data, Htr1a knockout mice display impaired hippocampus-dependent spatial memory consolidation (Sarnyai et al, 2000). Moreover, we observed down-regulation of Hcrtr2, encoding the receptor 2 for Hypocretin/Orexin. The Hypocretin/Orexin system has been implicated with different cognitive functions and various brain diseases (Mochizuki et al, 2011; Scott et al, 2011), and in agreement with our findings, pharmacological inhibition of orexin receptors in the hippocampal CA1 region was shown to impair memory consolidation in the Morris water maze test in rodents (Akbari et al, 2006; Akbari et al, 2007). Other genes that were down-regulated in Kat2a cKO mice were Penk, which encodes the endogenous opioid Proenkephalin, and Npbwr1, which encodes the neuropeptides B/W receptor 1. The endogenous opioid system has been linked to multiple cognitive processes (Bodnar, 2012) including hippocampus-dependent memory consolidation (Jamot et al, 2003; Jang et al, 2003; Meilandt et al, 2004). Moreover, the expression of Penk in the rodent hippocampus is induced in response to NMDA-receptor-mediated LTP (Thompson et al, 2003). Finally, mice that lack Npbwr1 display impaired hippocampus-dependent memory formation (Nagata-Kuroiwa et al, 2011). While the collective down-regulation of the above-described genes linked to neuroactive ligand–receptor signaling already provides a reasonable explanation for the memory impairment observed in Kat2a cKO mice, it is likely that the observed phenotypes are due to the deregulation of multiple genes, reflecting a systems deregulation. Indeed, our network analysis showed that genes deregulated in Kat2a cKO mice are tightly connected and most likely represent direct as well as indirect Kat2a targets. It is also important to note that we did not observe any difference in microRNA expression in Kat2a cKO mice. Since microRNAs have been implicated with memory function (Zovoilis et al, 2011; Rajasethupathy et al, 2009; Gao et al, 2010; Konopka et al, 2010), these data further confirm a rather specific role of Kat2a in the regulation of gene expression in the hippocampus.
A recent study investigated the role of Kat2a in neuronal stem cells (NSC) in the developing mouse brain. Loss of Kat2a led to reduced NCS proliferation and a severe microcephaly phenotype (Martínez-Cerdeño et al, 2012). Gene expression analysis revealed that loss of Kat2a in these cells resulted in the down-regulation of 2,729 genes which is about 25 times more genes than we identified in the CA1 region of adult mice that lack Kat2a in excitatory neurons. There was no overlap among the Kat2a-regulated gene in NSC and excitatory neurons of the adult CA1 region. While these data are likely reflecting the different roles of Kat2a during development and in the adult brain, it is interesting to note that the list of genes regulated by Kat2a in the adult CA1 region shows no overlap to genes identified by microarray approaches searching for hippocampal genes regulated by Kat3a/Cbp (Chen et al, 2010; Lopez-Atalaya et al, 2013). In fact, it appears that in contrast to our data on Kat2a, calcium signaling is a major pathway regulated by Kat3a/Cbp (Chen et al, 2010). These data suggest that in the adult hippocampus, Kat2a regulates a transcriptional program linked to memory formation that is non-overlapping with genes regulated by Kat3a/Cbp, which further supports a functional distinction of Kat2a and Kat3a/Cbp as observed on the behavioral level. Since compared to microarray data, RNA sequencing does not depend on probe design and is readily available for comparisons of different experiments, our data can be viewed as the starting point for a database that shall help to decipher the regulatory gene expression networks of HATs in the adult brain.
To learn more about the putative mechanisms by which Kat2a regulates hippocampal gene expression, we performed a promoter analysis for the differentially regulated genes in the CA1 region of Kat2a cKO mice. Interestingly, there was a statistically significant overrepresentation of NF-κB binding sites in this gene list. This is of particular importance since NF-κB function has been linked to hippocampus-dependent memory formation and plasticity (Meffert et al, 2003; Meffert & Baltimore, 2005; Boccia et al, 2007; Ahn et al, 2008; Boersma et al, 2011). Notably, when the same analysis was performed using a random list of genes, no enrichment for NF-κB was observed. Consistent with a role of NF-κB in Kat2a-mediated gene expression, we found that Kat2a physically interacts with the acetylated and non-acetylated NF-κB subunit p65 which is in line with the fact that a complex including p65 and Kat2a has been linked to gene activation (Lusic et al, 2003), and the fact that Kat2a can bind acetylated proteins via its bromodomain (Owen et al, 2000). For selected genes that were down-regulated in Kat2a cKO mice, we performed a detailed promoter analysis via ChIP and observed reduced levels of the acetylated p65 protein and histone acetylation marks linked to active gene expression at the corresponding promoter. Thus, our findings suggest a scenario in which Kat2a interacts with NF-κB at the promoter region of target genes, which activates gene expression via acetylation of the p65 subunit and nearby histone tails. An important role for this proposed mechanisms is further supported by a recent study demonstrating that NF-κB-dependent histone acetylation is required for hippocampal plasticity and memory formation (Lopez-Atalaya et al, 2013; Federman et al, 2013). It is, however, very likely that Kat2a also regulates genes independent of NF-κB activity and that moreover other mechanisms controlled by Kat2a function also contribute to the observed phenotypes. In fact, we also provide evidence that Kat2a affects editing of the Htr2c mRNA (Supplementary Fig S6). In conclusion, our data show that Kat2a controls a gene expression network that is essential for hippocampal synaptic plasticity and long-term memory formation and established Kat2a as a novel regulator of memory function.
Material and Methods
Animals
Specific-pathogen-free (SPF) C57Bl6/J wild-type mice were obtained from Janvier SAS. Mice were kept in individually ventilated cages (32 × 16 × 14 cm, Techniplast) on a 12-h light/dark cycle with food and water ad libitum. To obtain Kat2a conditional knockout (cKO) mice, mice carrying a heterozygous ‘floxed’ allele of Kat2a (recognized as Kat2atm3.2Roth in the MGI database) on a 129/Sv-C57BL/6J-mixed background (Lin et al, 2008) were crossed to mice carrying a transgenic construct for expressing the Cre recombinase under control of the CamKIIα promoter on a C57BL/6J background (recognized as Tg(CamKIIα-cre)159Kln). This driver line is described as expressing the Cre recombinase in adulthood with expression is limited to the forebrain (Minichiello et al, 1999; Kuczera et al, 2010; Kerimoglu et al, 2013). Unless otherwise stated, young adult mice at the age of 3–5 months were used for behavioral and molecular analyses. Male and female mice were used in the different groups where appropriate to avoid gender bias. For none of the groups, sex-specific differences were observed. All procedures were performed by experienced experimenters and according to protocols approved by the Lower Saxony State Office for Consumer Protection and Food Safety.
Behavioral analysis
Tracking and analysis was done using the Videomot2 tracking system (Versions 7.02, TSE Systems) for all tests except Rotarod and fear conditioning. Before and during testing, animals were housed individually (males) or in groups of 2–4 (females) for at least 1 week.
Open field test
Animals were placed individually in a uniform-gray plastic arena (50 × 50 × 40 cm) and allowed to explore the arena for 5 min.
Rotarod performance test
Motor function and performance was tested on the TSE RotaRod system. Animals were placed in individual chambers on the rotating rod and habituated for four trials at 10 rpm, 3 min each. Falling animals were placed back on the rod to continue training. During four testing trials, rotation speed was linearly increasing from 5 to 40 rpm for 3 min and kept at 40 rpm for another 1 min. Time between all trials was 6–12 h. Time until fall was recorded by the supplier's software for each animal during all testing trials.
Elevated plus maze
Animals were placed individually in a uniform-gray plastic arena consisting of two non-walled (open) and two walled (closed) arms (10 × 40 cm each, walls were 40 cm high). Time spent in open versus closed arms was measured.
Four-armed cross maze-exploration test
Spatial working memory was tested in a uniform-gray plastic arena consisting of four numbered, walled arms (10 × 40 cm, angled 90°, walls were 40 cm high). Animals were allowed to explore the arena for 10 min. The sequence of arm entries was extracted, and successful trials were counted. A successful trial was defined as entering an arm that was not entered in one the last three trials.
Novel object recognition
Animals were habituated individually to a uniform-gray plastic arena (50 × 50 cm, walls were 40 cm high) for 5 min on two subsequent days. Animals were then further habituated to two equal objects (objects A and A) placed in opposing corners of the arena for 5 min on the next 2 days. On day 5, objects A and A were exchanged by two new but equal objects (B), and animals were allowed to explore the objects for 5 min. Then, mice were sent back to their home cages for 5 min (for short-term memory assessment, STM) and reintroduced to the arena after one object was exchanged (objects B and C). After 24 h, object C was exchanged for object D for long-term memory assessment (LTM). Duration of object contacts was measured. Mice that only showed summed contact time of < 1 s were excluded from the analysis of this test. Object preference was defined as (novel object)/sum (both objects).
Fear conditioning
Fear conditioning was performed, recorded, and analyzed on the NIR Video Fear Conditioning system (Med Associates Inc.) using Video Freeze software. Mice were placed in a sound-protected box supplied with white noise and allowed to explore the new context, while baseline freezing was monitored. After 3 min, an electrical foot shock (0.7 mA, 2 s) was delivered through the grid floor. Twenty-four hours later, animals were reintroduced to the cage and contextual freezing behavior was recorded for 3 min. Freezing was counted in linear analysis mode if the Motion Index was below a threshold of 50 for at least 1 s.
Morris water maze
Animals were placed in a circular pool (1.2 m diameter) containing opaque water at ambient temperature (20–22°C) and a platform submerged 1 cm below the water surface. The pool was equipped with four visual cues for orientation. Escape latency was measured on four trials per day. If they did not find it within 1 min, mice were guided to the platform by the experimenter. Mice not finding the platform within 1 min in any trial were removed from the analysis. On the day of probe testing, the platform was removed and mice were allowed to swim for 1 min. The percentage of time spent in four virtual quadrants and the number of crossings were calculated. For a visual form of the test, the submerged platform was flagged with a visual cue. Please note that each experiment can only be quantitatively compared to its own control group.
Novelty exposure
A plastic arena (50 × 50 × 40 cm) was enriched with four objects, different in shape, size, color, and material. Mice were allowed to explore the objects for 15 min while movements were monitored. One hour after they were sent back to their home cages, mice were sacrificed and tissue was isolated. Naïve mice, taken directly from their home cage, served as controls.
Immunohistochemistry
Fluorescent staining of target proteins was performed as previously described (Peleg et al, 2010). In brief, mice were transcardially perfused with 4% PFA, brains isolated, and post-fixed for another 16 h in 4% PFA. Free-floating cryosections (30 μm) were incubated with 5% goat serum for blocking and followed by incubation with target-specific primary antibodies (anti-Kat2a [C26A10, #3305, Cell Signaling, 1:100], anti-NeuN [A60, MAB377, Merck Millipore, 1:1,000], anti-MAP2 [188002, Synaptic Systems, 1:1,000], anti-Synaptoporin [1023002, Synaptic Systems, 1:1,000], anti-Synaptophysin [S5768, Sigma, 1:1,000], anti-GFAP [G5601, Promega, 1:1,000], anti-PLP1 [gift, self-raised, 1:1,000]. For Kat2a stainings, heat-mediated antigen retrieval was achieved by incubating the sections in boiling-hot citrate buffer (10 mM sodium citrate, 0.1% Triton X-100, pH 6.0) for 10 min before the blocking step. Corresponding secondary antibodies were from Life Technologies (anti-mouse Alexa-488 labeled, A11029; anti-rabbit Cy3 labeled, A10520). Images were taken on a Leica SP2 confocal microscope.
Immunoblotting
Protein extracts from subcellular compartments were obtained using the ProteoExtract Subcellular Proteome Extraction Kit (Merck Chemicals/Calbiochem). 10–40 μg of protein sample was loaded for SDS-PAGE and blotted on nitrocellulose membrane, blocked, and incubated with primary target-specific antibodies (anti-Kat2a [C26A10, #3305, Cell Signaling, 1:1,000], anti-β-actin [AC-15, sc-69879, Santa Cruz, 1:1,000], anti-Histone H4 [ab10158-100, Abcam, 1:1,000]) as described before (Peleg et al, 2010). Membranes were then incubated with corresponding secondary antibody (IRDye 800 anti-mouse or anti-rabbit), scanned, and results quantified using the Odyssey Imaging system (LI-COR). Of note, we have tested 10 Kat2a antibodies that all detected a band in the wild-type and Kat2a cKO mice and were therefore considered to be unspecific (NBP1-00845, Novus Biologicals; ab1831, ab61174, ab137515, ab136235, ab71227 all Abcam; 07-1545, Millipore; 607201 Biolegend; 607302, Biolegend; sc-365321 (A-11), Santa Cruz).
RNA extraction
Total RNA was extracted using TRI Reagent (Sigma-Aldrich) as described previously (Peleg et al, 2010). Following DNase1 (Life Technologies) treatment for 20 min at 37°C, RNA was again purified using phenol–chloroform extraction.
qPCR
Quantitative real-time PCR (qPCR) was performed as described before (Peleg et al, 2010). In summary, 1 μg of total RNA was used for cDNA synthesis, and resulting cDNA was diluted 1:10. qPCR was carried out using probes from the Universal Probe Library (Roche) on a LightCycler 480 II (Roche) and analyzed using suppliers software. Primers used can be found in Supplementary Table S5.
RNA sequencing and analysis
RNA was quality controlled using the 2100 Bioanalyzer (Agilent Technologies). Library preparation and cluster generation for mRNA sequencing was performed according to Illumina standard protocols (TruSeq, Illumina). Libraries were quality controlled and quantified using a Nanodrop 2000 (Thermo Scientific), Agilent 2100 Bioanalyzer (Agilent Technologies), and Qubit (life Technologies). Small RNA library preparation and sequencing was performed analogously.
A streamlined, customized bioinformatics approach was used. Base calling from raw images and file conversion to fastq-files was achieved by Illumina pipeline scripts. Subsequent steps included quality control (FastQC, http://www.bioinformatics.babraham.ac.uk/projects/fastqc/), mapping to reference genome (STAR aligner v2.3.0, (Djebali et al, 2012) non-default parameters). For comparison of expression levels of the different HATs, SeqMonk (v0.24.1 http://www.bioinformatics.babraham.ac.uk/projects/seqmonk/) was used. For each sample, reads falling into the gene locus of each HAT were counted, and this number was divided by the length of the gene and the total number of reads in the sample to calculate RPKM. For calling of differentially expressed genes, mapped reads were counted with HTSeq v0.5.4p2 (http://www-huber.embl.de/users/anders/HTSeq) (non-default parameters: -m intersection non-empty), and count tables were analyzed in pairwise comparisons using the DESeq2 v1.2.5 R-package (Anders & Huber, 2010). Genes with a log2(fold-change) = 0.5 and adjusted P-value ≤ 0.1 were considered differentially regulated. Functional annotation and category analysis was carried out using the Database for Annotation, Visualization and Integrated Discovery (DAVID, v6.7) (Huang et al, 2009). Network analysis was performed using commercially available Ingenuity® software (Ingenuity Systems/Qiagen).
Compressed fastq-files and primary analysis tables from mRNA and small RNA sequencing pipelines are made publicly available at:
mRNA-seq: GEO (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=cvsrouyyjhabbwp&acc=GSE53380)
smallRNAseq: GEO (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=sfmdseyktzgdbyl&acc=GSE53427)
Promoter analysis to search for overrepresented transcription factor binding sites was done using the Pscan web interface (Zambelli et al, 2009) (http://159.149.160.51/pscan/), scanning the promoter region from 1000-bp upstream to the TSS comparing to JASPAR descriptors. All Gene ID conversion was done using BioMart database queries (http://www.ensembl.org/biomart/).
Chromatin immunoprecipitation
ChIP was carried out using a LowCell# ChIP kit (Diagenode) according to the manufactures with the following changes as described before (Kerimoglu et al, 2013). Homogenized tissue was fixed for 15 min in 1% formaldehyde. 3.5 μg of target-specific antibody (anti-p65, ab7970, Abcam; anti-p65K310ac, ab52175, Abcam; anti-H3K14ac, 07-353, Merck Millipore; anti-H3K18ac, ab1191, Abcam; anti-H4K12ac, ab61238, Abcam) or control (IgG) was used per reaction. 10 μl of cross-linked homogenate was used as input. Incubation of antibody-coupled Protein G-covered beads with target chromatin was done over night. Reverse-crosslinking was done for 45 min. qPCR assays were performed on eluted DNA from precipitated samples and corresponding input samples using SYBR Green I Master (Roche) with primers specific for the predicted NF-κB binding region. Quantification was done relative to input samples and then normalized to average levels of the control group (relative enrichment). We have tested the following antibodies for KAT2 ChIP (NBP1-00845, Novus Biologicals; ab1831, Abcam; 07-1545, Millipore; 3305, Cell Signaling; sc-20698 (H-75), Santa Cruz; 607201 Biolegend), but none of the antibodies showed any difference between tissue obtained from wild-type of Kat2a cKO mice and were therefore considered to be not suitable for ChIP experiments.
Co-immunoprecipitation
Target-specific antibodies (anti-p65K310ac, ab52175, Abcam; anti-Kat2a, C26A10, #3305, Cell Signaling) and non-specific control (IgG) were pre-coupled to surface-activated magnetic Dynabeads using an antibody coupling kit (life Technologies) at 20 μg of antibody per 1 mg of beads. After coupling beads were equilibrated with IP buffer (50 mM HEPES, 100mM NaCl, 0.5% NP-40, pH 7.8, complete protease inhibitors (Roche)).
Nuclear protein extract was obtained from dissected cortical brain tissue using ProteoExtract Subcellular Proteome Extraction Kit (Merck Chemicals/Calbiochem). 1 mg of nuclear protein was incubated with the pre-coupled beads for 20 h. Elution was achieved by incubation of the antibody-antigen-bead complex with non-reducing SDS-PAGE loading buffer (1 M Tris–HCl, 50% glycerol, 10% SDS, 5 mM EDTA, 0.001% bromophenol blue for 10 min at 70°C). qPCR was performed using SYBR green dye. Primers used can be found in Supplementary Table S5. Primer binding specificity was monitored by melting curve analysis.
Adeno-associated virus (AAV) injection
Viral particles for Cre expression under control of the hSyn promoter were obtained from Penn Vector Core (AAV2/1.hsynapsin.hGHintron.GFP-Cre.WPRE.SV40, University of Pennsylvania). For Cre-AAV, stock was diluted 1:100 to result in 1.76 × 108 genome copies per μl; 1 μl was injected per hemisphere. Coordinates for injection were as follows (relative to bregma): anteroposterior −1.75 mm, lateral −1 mm, dorsoventral −1.3 mm, targeting the dorsal CA1 region. Behavioral analysis was performed not before 2 weeks after injection.
Electrophysiology
Mice were euthanized by cervical dislocation. After decapitation, the brain was quickly removed and placed into ice-cold artificial cerebrospinal fluid (ACSF) with the following composition (in mM): NaCl 124, KCl 4.9, MgSO4 1.3, CaCl2 2.5, KH2PO4 1.2, NaHCO3 25.6, D-glucose 10, saturated with 95% O2/5% CO2, pH 7.4). Transverse slices (400 μm) were prepared using a tissue chopper with a cooled stage and immediately transferred into a pre-chamber containing 8 ml of permanently carbogen-gasified ACSF for 2 h to allow recovery from preparation stress. Slices were then transferred into a submerged-type recording chamber and were allowed to adapt for at least 30 min before the experiment started. The chamber was constantly perfused with artificial cerebrospinal fluid (ACSF) at a rate of 2.5 ml/min at 33 ± 1°C. Synaptic responses were elicited by stimulation of the Schaffer collateral–commissural fibers in the stratum radiatum of the CA1 region using lacquer-coated stainless steel stimulating electrodes. Glass electrodes (filled with ACSF, 1–4 MΩ) were placed in the apical dendritic layer to record field excitatory postsynaptic potentials (fEPSPs). The initial slope of the fEPSP was used as a measure of this potential. The stimulus strength of the test pulses was adjusted to 30% of the fEPSP maximum. During baseline recording, single stimuli were applied every minute (0.0166 Hz). Once a stable baseline had been established, LTP was induced by applying 100 pulses at an interval of 10 ms and a width of the single pulses of 0.2 ms (strong tetanus) three times at 10-min intervals. The input–output relationship was obtained from a curve of fEPSP amplitude versus stimulation intensity. For paired-pulse stimulation, the stimulus strength of the test pulses was adjusted to 50% of the fEPSP maximum. Paired-pulse stimulation was delivered with inter-stimulus interval of 20 ms, and the paired-pulse ratio (PPR) was calculated as the ratio of the second fEPSP amplitude to the first.
Statistics
Statistical analysis for behavioral and molecular analyses was performed using GraphPad Prism 6d. Details of statistical analysis, methods, and parameters are given in the figure legends. Statistical tests performed were based on data structure and included as follows: one-way, two-way, or repeated-measures analysis of variance (1-way/2-way/rmANOVA), (Holm-)Šidák's multiple testing correction, the Benjamini–Hochberg procedure (FDR) (Benjamini & Hochberg, 1995), Student's t-test. Probability of overlap in Venn diagrams was computed using an online tool (http://nemates.org/MA/progs/overlap_stats.html).
Sample sizes used were based on the literature and previous experience with the experiments. In addition to specific outlier definitions for some behavioral tests, outliers were defined as values deviating more than two standard deviations from the mean. In all experiments, groups were processed in an alternating order, and in behavioral experiments, the experimenter was blinded toward the genotype to avoid bias. Replicates were defined as biological replicates. In some cases, technical replicates from the same biological sample were averaged before performing statistical tests.
Acknowledgments
This project was partly supported by the following funds to AF: the Euryi award (DFG grant FI 981), the ERA NET Neuron Project EPITHERAPY, the Schram Foundation, the Hans and Ilse Breuer Foundation and the German Center for Neurodegenerative Diseases (DZNE) Göttingen. FS is funded by the DFG grant (SA 1050). RMS was a student of the Göttingen Graduate School for Neurosciences, Biophysics, and Molecular Biosciences (GGNB). The antibodies for GFAP and PLP1 were a kind gift from Anja Schneider, University Medical Center, Göttingen.
Author contributions
RMS, KGS and AF designed experiments. SB, ALS, AMH, CK and SYRD performed experiments. RMS, RR, EB, HU, SBJ and FS performed experiments and analysed data. VC, JB, JD, SB, KGR and AF analysed data. RMS and AF contributed in writing the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supplementary information for this article is available online: http://emboj.embopress.org
References
- Ahn HJ, Hernandez CM, Levenson JM, Lubin FD, Liou HC, Sweatt JD. c-Rel, an NF-kappaB family transcription factor, is required for hippocampal long-term synaptic plasticity and memory formation. Learn Mem. 2008;15:539–549. doi: 10.1101/lm.866408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbari E, Naghdi N, Motamedi F. Functional inactivation of orexin 1 receptors in CA1 region impairs acquisition, consolidation and retrieval in Morris water maze task. Behav Brain Res. 2006;173:47–52. doi: 10.1016/j.bbr.2006.05.028. [DOI] [PubMed] [Google Scholar]
- Akbari E, Naghdi N, Motamedi F. The selective orexin 1 receptor antagonist SB-334867-A impairs acquisition and consolidation but not retrieval of spatial memory in Morris water maze. Peptides. 2007;28:650–656. doi: 10.1016/j.peptides.2006.11.002. [DOI] [PubMed] [Google Scholar]
- Alarcon JM, Malleret G, Touzani K, Vronskaya S, Ishii S, Kandel ER, Barco A. Chromatin acetylation, memory, and LTP are impaired in CBP+/- mice: a model for the cognitive deficit in Rubinstein-Taybi syndrome and its amelioration. Neuron. 2004;42:947–959. doi: 10.1016/j.neuron.2004.05.021. [DOI] [PubMed] [Google Scholar]
- Alhaider AA, Ageel AM, Ginawi OT. The quipazine- and TFMPP-increased conditioned avoidance response in rats: role of 5HT1C/5-HT2 receptors. Neuropharmacology. 1993;32:1427–1432. doi: 10.1016/0028-3908(93)90040-a. [DOI] [PubMed] [Google Scholar]
- Allis CD, Berger SL, Cote J, Dent S, Jenuwein T, Kouzarides T, Pillus L, Reinberg D, Shi Y, Shiekhattar R, Shilatifard A, Workman J, Zhang Y. New nomenclature for chromatin-modifying enzymes. Cell. 2007;131:633–636. doi: 10.1016/j.cell.2007.10.039. [DOI] [PubMed] [Google Scholar]
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett RM, Malvaez M, Kramar E, Matheos DP, Arrizon A, Cabrera SM, Lynch G, Greene RW, Wood MA. Hippocampal focal knockout of CBP affects specific histone modifications, long-term potentiation, and long-term memory. Neuropsychopharmacology. 2011;36:1545–1556. doi: 10.1038/npp.2011.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300. [Google Scholar]
- Bert B, Fink H, Rothe J, Walstab J, Bönisch H. Learning and memory in 5-HT(1A)-receptor mutant mice. Behav Brain Res. 2008;195:78–85. doi: 10.1016/j.bbr.2008.02.028. [DOI] [PubMed] [Google Scholar]
- Boccia M, Freudenthal R, Blake M, de la Fuente V, Acosta G, Baratti C, Romano A. Activation of hippocampal nuclear factor-kappa B by retrieval is required for memory reconsolidation. J Neurosci. 2007;27:13436–13445. doi: 10.1523/JNEUROSCI.4430-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodnar RJ. Endogenous opiates and behavior: 2011. Peptides. 2012;38:463–522. doi: 10.1016/j.peptides.2012.09.027. [DOI] [PubMed] [Google Scholar]
- Boersma MC, Dresselhaus EC, De Biase LM, Mihalas AB, Bergles DE, Meffert MK. A requirement for nuclear factor-kappaB in developmental and plasticity-associated synaptogenesis. J Neurosci. 2011;31:5414–5425. doi: 10.1523/JNEUROSCI.2456-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu P, Evrard YA, Lozano G, Dent SY. Loss of Gcn5 acetyltransferase activity leads to neural tube closure defects and exencephaly in mouse embryos. Mol Cell Biol. 2007;27:3405–3416. doi: 10.1128/MCB.00066-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavallaro S, D'Agata V, Manickam P, Dufour F, Alkon DL. Memory-specific temporal profiles of gene expression in the hippocampus. Proc Natl Acad Sci USA. 2002;99:16279–16284. doi: 10.1073/pnas.242597199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee S, Mizar P, Cassel R, Neidl R, Selvi BR, Mohankrishna DV, Vedamurthy BM, Schneider A, Bousiges O, Mathis C, Cassel JC, Eswaramoorthy M, Kundu TK, Boutillier AL. A novel activator of CBP/p300 acetyltransferases promotes neurogenesis and extends memory duration in adult mice. J Neurosci. 2013;33:10698–10712. doi: 10.1523/JNEUROSCI.5772-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LF, Fischle W, Verdin E, Greene WC. Duration of nuclear NF-kappaB action regulated by reversible acetylation. Science. 2001;293:1653–1657. doi: 10.1126/science.1062374. [DOI] [PubMed] [Google Scholar]
- Chen LF, Greene WC. Shaping the nuclear action of NF-kappaB. Nat Rev Mol Cell Biol. 2004;5:392–401. doi: 10.1038/nrm1368. [DOI] [PubMed] [Google Scholar]
- Chen G, Zou X, Watanabe H, van Deursen JM, Shen J. CREB binding protein is required for both short-term and long-term memory formation. J Neurosci. 2010;30:13066–13070. doi: 10.1523/JNEUROSCI.2378-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen R, Marcussen AB, Wörtwein G, Knudsen GM, Aznar S. Abeta(1-42) injection causes memory impairment, lowered cortical and serum BDNF levels, and decreased hippocampal 5-HT(2A) levels. Exp Neurol. 2008;210:164–171. doi: 10.1016/j.expneurol.2007.10.009. [DOI] [PubMed] [Google Scholar]
- Day JJ, Sweatt JD. Epigenetic mechanisms in cognition. Neuron. 2011;70:813–829. doi: 10.1016/j.neuron.2011.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Quervain DJ-F, Henke K, Aerni A, Coluccia D, Wollmer MA, Hock C, Nitsch RM, Papassotiropoulos A. A functional genetic variation of the 5-HT2a receptor affects human memory. Nat Neurosci. 2003;6:1141–1142. doi: 10.1038/nn1146. [DOI] [PubMed] [Google Scholar]
- Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin WSF, Xue C, Marinov GK, Khatun J, Williams BA, Zaleski C, Rozowsky J, Röder M, Kokocinski F, Abdelhamid RF, Alioto T, Antoshechkin I, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y, Stasko M, Costa AC, Davisson MT, Gardiner KJ. Editing of the serotonin 2C receptor pre-mRNA: effects of the Morris Water Maze. Gene Regul Syst Bio. 2007;391:186–197. doi: 10.1016/j.gene.2006.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Federman N, de la Fuente V, Zalcman G, Corbi N, Onori A, Passananti C, Romano A. Nuclear factor κB-dependent histone acetylation is specifically involved in persistent forms of memory. J Neurosci. 2013;33:7603–7014. doi: 10.1523/JNEUROSCI.4181-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer A, Sananbenesi F, Pang PT, Lu B, Tsai LH. Opposing roles of transient and prolonged expression of p25 in synaptic plasticity and hippocampus-dependent memory. Neuron. 2005;48:825–838. doi: 10.1016/j.neuron.2005.10.033. [DOI] [PubMed] [Google Scholar]
- Fischer A, Sananbenesi F, Wang X, Dobbin M, Tsai LH. Recovery of learning & memory after neuronal loss is associated with chromatin remodeling. Nature. 2007;447:178–182. doi: 10.1038/nature05772. [DOI] [PubMed] [Google Scholar]
- Fischer A, Sananbenesi F, Mungenast A, Tsai LH. Targeting the right HDAC(s) to treat cognitive diseases. Trends Pharmacol Sci. 2010;31:605–617. doi: 10.1016/j.tips.2010.09.003. [DOI] [PubMed] [Google Scholar]
- Fischer A. Epigenetic Memory: the Lamarckian brain. EMBO J. 2014;33:945–967. doi: 10.1002/embj.201387637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher M, Koh MT. Episodic memory on the path to Alzheimer's disease. Curr Opin Neurobiol. 2011;21:929–934. doi: 10.1016/j.conb.2011.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Wang WY, Mao YW, Gräff J, Guan JS, Pan L, Mak G, Kim D, Su SC, Tsai LH. A novel pathway regulates memory and plasticity via SIRT1 and miR-134. Nature. 2010;466:1105–1109. doi: 10.1038/nature09271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlich A, Zimmermann AM, Schober D, Böttcher RT, Sassoè-Pognetto M, Friauf E, Witke W, Rust MB. Preserved morphology and physiology of excitatory synapses in profilin1-deficient mice. PLoS ONE. 2012;7:e300068. doi: 10.1371/journal.pone.0030068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govindarajan N, Agis-Balboa C, Walter J, Sananbenesi F, Fischer A. Sodium butyrate improves memory function in an alzheimer's disease mouse model when administered at an advanced stage of disease progression. J Alzheimer's Dis. 2011;24:1–11. doi: 10.3233/JAD-2011-110080. [DOI] [PubMed] [Google Scholar]
- Govindarajan N, Rao P, Burkhardt S, Sananbenesi F, Schlüter OM, Bradke F, Lu J, Fischer A. Reducing HDAC6 ameliorates cognitive deficits in a mouse model for Alzheimer's disease. EMBO Mol Med. 2013;5:52–63. doi: 10.1002/emmm.201201923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gräff J, Tsai LH. Histone acetylation: molecular mnemonics on the chromatin. Nat Rev Neurosci. 2013;14:97–111. doi: 10.1038/nrn3427. [DOI] [PubMed] [Google Scholar]
- Hargreaves DC, Horng T, Medzhitov R. Control of inducible gene expression by signal-dependent transcriptional elongation. Cell. 2009;138:129–145. doi: 10.1016/j.cell.2009.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard MW, Eichenbaum H. The Hippocampus, Time, and Memory Across Scales. J Exp Psychol Gen. 2013;142:1211–1230. doi: 10.1037/a0033621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Jamot L, Matthes HW, Simonin F, Kieffer BL, Roder JC. Differential involvement of the mu and kappa opioid receptors in spatial learning. Genes Brain Behav. 2003;2:80–92. doi: 10.1034/j.1601-183x.2003.00013.x. [DOI] [PubMed] [Google Scholar]
- Jang CG, Lee SY, Yoo JH, Yan JJ, Song DK, Loh HH, Ho IK. Impaired water maze learning performance in mu-opioid receptor knockout mice. Brain Res Mol Brain Res. 2003;117:68–72. doi: 10.1016/s0169-328x(03)00291-2. [DOI] [PubMed] [Google Scholar]
- Johnsson A, Durand-Dubief M, Xue-Franzén Y, Rönnerblad M, Ekwall K, Wright A. HAT-HDAC interplay modulates global histone H3K14 acetylation in gene-coding regions during stress. EMBO Rep. 2009;10:1009–1014. doi: 10.1038/embor.2009.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josselyn SA. What's right with my mouse model? New insights into the molecular and cellular basis of cognition from mouse models of Rubinstein-Taybi Syndrome. Learn Mem. 2005;12:80–83. doi: 10.1101/lm.93505. [DOI] [PubMed] [Google Scholar]
- Kandel ER. The molecular biology of memory storage: a dialogue between genes and synapses. Science. 2001;294:1030–1038. doi: 10.1126/science.1067020. [DOI] [PubMed] [Google Scholar]
- Kerimoglu C, Agis-Balboa RC, Kranz A, Stilling R, Bahari-Javan S, Benito-Garagorri E, Halder R, Burkhardt S, Stewart AF, Fischer A. Histone-methyltransferase mll2 (kmt2b) is required for memory formation in mice. J Neurosci. 2013;33:3452–3464. doi: 10.1523/JNEUROSCI.3356-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilgore M, Miller CA, Fass DM, Hennig KM, Haggarty SJ, Sweatt JD, Rumbaugh G. Inhibitors of class 1 histone deacetylases reverse contextual memory deficits in a mouse model of Alzheimer's disease. Neuropsychopharmacology. 2010;35:870–880. doi: 10.1038/npp.2009.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka W, Kiryk A, Novak M, Herwerth M, Parkitna JR, Wawrzyniak M, Kowarsch A, Michaluk P, Dzwonek J, Arnsperger T, Wilczynski G, Merkenschlager M, Theis FJ, Köhr G, Kaczmarek L, Schütz G. MicroRNA loss enhances learning and memory in mice. J Neurosci. 2010;30:14835–14842. doi: 10.1523/JNEUROSCI.3030-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korzus E, Rosenfeld MG, Mayford M. CBP histone acetyltransferase activity is a critical component of memory consolidation. Neuron. 2004;42:961–972. doi: 10.1016/j.neuron.2004.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuczera T, Stilling RM, Hsia HE, Bahari-Javan S, Irniger S, Nasmyth K, Sananbenesi F, Fischer A. The anaphase promoting complex is required for memory function in mice. Learn Mem. 2010;18:49–57. doi: 10.1101/lm.1998411. [DOI] [PubMed] [Google Scholar]
- Levenson JM, O'Riordan KJ, Brown KD, Trinh MA, Molfese DL, Sweatt JD. Regulation of histone acetylation during memory formation in the hippocampus. J Biol Chem. 2004;279:40545–40559. doi: 10.1074/jbc.M402229200. [DOI] [PubMed] [Google Scholar]
- Lin W, Zhang Z, Srajer G, Chen YC, Huang M, Phan HM, Dent SYR. Proper expression of the Gcn5 histone acetyltransferase is required for neural tube closure in mouse embryos. Dev Dyn. 2008;237:928–940. doi: 10.1002/dvdy.21479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Atalaya JP, Ciccarelli A, Viosca J, Valor LM, Jimenez-Minchan M, Canals S, Giustetto M, Barco A. CBP is required for environmental enrichment-induced neurogenesis and cognitive enhancement. EMBO J. 2011;30:4287–4298. doi: 10.1038/emboj.2011.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Atalaya JP, Ito S, Valor LM, Benito E, Barco A. Genomic targets, and histone acetylation and gene expression profiling of neural HDAC inhibition. Nucleic Acids Res. 2013;41:8072–8084. doi: 10.1093/nar/gkt590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusic M, Marcello A, Cereseto A, Giacca M. Regulation of HIV-1 gene expression by histone acetylation and factor recruitment at the LTR promoter. EMBO J. 2003;22:6550–6561. doi: 10.1093/emboj/cdg631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marner L, Frokjaer VG, Kalbitzer J, Lehel S, Madsen K, Baare WF, Knudsen GM, Hasselbalch SG. Loss of serotonin 2A receptors exceeds loss of serotonergic projections in early Alzheimer's disease: a combined [11C]DASB and [18F]altanserin-PET study. Neurobiol Aging. 2012;33:479–487. doi: 10.1016/j.neurobiolaging.2010.03.023. [DOI] [PubMed] [Google Scholar]
- Martínez-Cerdeño V, Lemen JM, Chan V, Wey A, Lin W, Dent SR, Knoepfler PS. N-Myc and GCN5 Regulate Significantly Overlapping Transcriptional Programs in Neural Stem Cells. PLoS ONE. 2012;7:e39456. doi: 10.1371/journal.pone.0039456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurice T, Duclot F, Meunier J, Naert G, Givalois L, Meffre J, Célérier A, Jacquet C, Copois V, Mechti N, Ozato K, Gongora C. Altered memory capacities and response to stress in p300/CBP-associated factor (PCAF) histone acetylase knockout mice. Neuropsychopharmacology. 2008;33:1584–1602. doi: 10.1038/sj.npp.1301551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meffert MK, Chang JM, Wiltgen BJ, Fanselow MS, Baltimore D. NF-kappa B functions in synaptic signaling and behavior. Nat Neurosci. 2003;6:1072–1078. doi: 10.1038/nn1110. [DOI] [PubMed] [Google Scholar]
- Meffert MK, Baltimore D. Physiological functions for brain NF-kappaB. Trends Neurosci. 2005;28:37–43. doi: 10.1016/j.tins.2004.11.002. [DOI] [PubMed] [Google Scholar]
- Meilandt WJ, Barea-Rodriguez E, Harvey SA, Martinez JLJ. Role of hippocampal CA3 mu-opioid receptors in spatial learning and memory. J Neurosci. 2004;24:2953–2962. doi: 10.1523/JNEUROSCI.5569-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meltzer CC, Smith G, DeKosky ST, Pollock BG, Mathis CA, Moore RY, Kupfer DJ, Reynolds CF., 3rd Serotonin in aging, late-life depression, and Alzheimer's disease: the emerging role of functional imaging. Neuropsychopharmacology. 1998;18:407–430. doi: 10.1016/S0893-133X(97)00194-2. [DOI] [PubMed] [Google Scholar]
- Minichiello L, Korte M, Wolfer D, Kühn R, Unsicker K, Cestari V, Rossi-Arnaud C, Lipp HP, Bonhoeffer T, Klein R. Essential role for TrkB receptors in hippocampus-mediated learning. Neuron. 1999;24:401–414. doi: 10.1016/s0896-6273(00)80853-3. [DOI] [PubMed] [Google Scholar]
- Mochizuki T, Arrigoni E, Marcus JN, Clark EL, Yamamoto M, Honer M, Borroni E, Lowell BB, Elmquist JK, Scammell TE. Orexin receptor 2 expression in the posterior hypothalamus rescues sleepiness in narcoleptic mice. Proc Natl Acad Sci USA. 2011;108:4471–4476. doi: 10.1073/pnas.1012456108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata-Kuroiwa R, Furutani N, Hara J, Hondo M, Ishii M, Abe T, Mieda M, Tsujino N, Motoike T, Yanagawa Y, Kuwaki T, Yamamoto M, Yanagisawa M, Sakurai T. Critical role of neuropeptides B/W receptor 1 signaling in social behavior and fear memory. PLoS ONE. 2011;6:e16972. doi: 10.1371/journal.pone.0016972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira AM, Wood MA, McDonough CB, Abel T. Transgenic mice expressing an inhibitory truncated form of p300 exhibit long-term memory deficits. Learn Mem. 2007;14:564–572. doi: 10.1101/lm.656907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen DJ, Ornaghi P, Yang JC, Lowe N, Evans PR, Ballario P, Neuhaus D, Filetici P, Travers AA. The structural basis for the recognition of acetylated histone H4 by the bromodomain of histone acetyltransferase gcn5p. EMBO J. 2000;19:6141–6149. doi: 10.1093/emboj/19.22.6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peleg S, Sananbenesi F, Zovoilis A, Burkhardt S, Bahari-Java S, Agis-Balboa RC, Cota P, Wittnam J, Gogul-Doering A, Opitz L, Salinas-Riester G, Dettenhofer M, KAng H, Farinelli L, Chen W, Fischer A. Altered histone acetylation is associated with age-dependent memory impairment in mice. Science. 2010;328:753–756. doi: 10.1126/science.1186088. [DOI] [PubMed] [Google Scholar]
- Pena JT, Sohn-Lee C, Rouhanifard SH, Ludwig J, Hafner M, Mihailovic A, Lim C, Holoch D, Berninger P, Zavolan M, Tuschl T. miRNA in situ hybridization in formaldehyde and EDC-fixed tissues. Nat Methods. 2009;6:139–141. doi: 10.1038/nmeth.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirooznia SK, Sarthi J, Johnson AA, Toth MS, Chiu K, Koduri S, Elefant F. Tip60 HAT activity mediates APP induced lethality and apoptotic cell death in the CNS of a Drosophila Alzheimer's disease model. PLoS ONE. 2012;7:e41776. doi: 10.1371/journal.pone.0041776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajasethupathy P, Fiumara F, Sheridan R, Betel D, Puthanveettil SV, Russo JJ, Sander C, Tuschl T, Kandel E. Characterization of small RNAs in Aplysia reveals a role for miR-124 in constraining synaptic plasticity through CREB. Neuron. 2009;63:803–817. doi: 10.1016/j.neuron.2009.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds CA, Jansson M, Gatz M, Pedersen NL. Longitudinal change in memory performance associated with HTR2A polymorphism. Neurobiol Aging. 2006;27:150–154. doi: 10.1016/j.neurobiolaging.2004.12.009. [DOI] [PubMed] [Google Scholar]
- Ricobaraza A, Cuadrado-Tejedor M, Pérez-Mediavilla A, Frechilla D, Del Río J, García-Osta A. Phenylbutyrate ameliorates cognitive deficit and reduces tau pathology in an Alzheimer's disease mouse model. Neuropsychopharmacology. 2009;34:1721–1732. doi: 10.1038/npp.2008.229. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Navarro S. Insights into SAGA function during gene expression. EMBO Rep. 2009;10:843–850. doi: 10.1038/embor.2009.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarnyai Z, Sibille EL, Pavlides C, Fenster RJ, McEwen BS, Toth M. Impaired hippocampal-dependent learning and functional abnormalities in the hippocampus in mice lacking serotonin(1A) receptors. Proc Natl Acad Sci USA. 2000;97:14731–14736. doi: 10.1073/pnas.97.26.14731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz ML, Mattioli I, Buss H, Kracht M. NF-kappaB: a multifaceted transcription factor regulated at several levels. ChemBioChem. 2004;4:1348–1358. doi: 10.1002/cbic.200400144. [DOI] [PubMed] [Google Scholar]
- Schneider A, Chatterjee S, Bousiges O, Selvi BR, Swaminathan A, Cassel R, Blanc F, Kundu TK, Boutillier A. Acetyltransferases (HATs) as Targets for Neurological Therapeutics. Neurotherapeutics. 2013;10:569–588. doi: 10.1007/s13311-013-0204-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schott BH, Seidenbecher CI, Richter S, Wüstenberg T, Debska-Vielhaber G, Schubert H, Heinze HJ, Richardson-Klavehn A, Düzel E. Genetic variation of the serotonin 2a receptor affects hippocampal novelty processing in humans. PLoS ONE. 2011;6:e15984. doi: 10.1371/journal.pone.0015984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott MM, Marcus JN, Pettersen A, Birnbaum SG, Mochizuki T, Scammell TE, Nestler EJ, Elmquist JK, Lutter M. Hcrtr1 and 2 signaling differentially regulates depression-like behaviors. Behav Brain Res. 2011;222:289–294. doi: 10.1016/j.bbr.2011.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvi BR, Cassel JC, Kundu TK, Boutillier AL. Tuning acetylation levels with HAT activators: therapeutic strategy in neurodegenerative diseases. Biochim Biophys Acta. 2010;1799:840–853. doi: 10.1016/j.bbagrm.2010.08.012. [DOI] [PubMed] [Google Scholar]
- Sigmund JC, Vogler C, Huynh KD, de Quervain DJ, Papassotiropoulos A. Fine-mapping at the HTR2A locus reveals multiple episodic memory-related variants. Biol Psychiatry. 2008;79:239–242. doi: 10.1016/j.biopsycho.2008.06.002. [DOI] [PubMed] [Google Scholar]
- Thompson KJ, Orfila JE, Achanta P, Martinez JLJ. Gene expression associated with in vivo induction of early phase-long-term potentiation (LTP) in the hippocampal mossy fiber-Cornus Ammonis (CA)3 pathway. Cell Mol Biol (Noisy-le-grand) 2003;49:1281–1287. [PubMed] [Google Scholar]
- Valor LM, Pulopulos MM, Jimenez-Minchan M, Olivares R, Lutz B, Barco A. Ablation of CBP in Forebrain Principal Neurons Causes Modest Memory and Transcriptional Defects and a Dramatic Reduction of Histone Acetylation But Does Not Affect Cell Viability. J Neurosci. 2011;11:1676–1678. doi: 10.1523/JNEUROSCI.4737-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vecsey CG, Hawk JD, Lattal KM, Stein JM, Fabian SA, Attner MA, Cabrera SM, McDonough CB, Brindle PK, Abel T, Wood MA. Histone deacetylase inhibitors enhance memory and synaptic plasticity via CREB:CBP-dependent transcriptional activation. J Neurosci. 2007;27:6128–6140. doi: 10.1523/JNEUROSCI.0296-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood MA, Kaplan MP, Park A, Blanchard EJ, Oliveira AM, Lombardi TL, Abel T. Transgenic mice expressing a truncated form of CREB-binding protein (CBP) exhibit deficits in hippocampal synaptic plasticity and memory storage. Learn Mem. 2005;12:111–119. doi: 10.1101/lm.86605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood MA, Attner MA, Oliveira AM, Brindle PK, Abel T. A transcription factor-binding domain of the coactivator CBP is essential for long-term memory and the expression of specific target genes. Learn Mem. 2006;13:609–617. doi: 10.1101/lm.213906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambelli F, Pesole G, Pavesi G. Pscan: finding over-represented transcription factor binding site motifs in sequences from co-regulated or co-expressed genes. Nucleic Acids Res. 2009;37:W247–W252. doi: 10.1093/nar/gkp464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang GÁH, Cohen SJ, Munchow AH, Barrera MP, Stackman RW., Jr Stimulation of serotonin 2A receptors facilitates consolidation and extinction of fear memory in C57BL/6J mice. Neuropharmacology. 2013;64:403–413. doi: 10.1016/j.neuropharm.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zovoilis A, Agbemenyah HY, Agis-Balboa RC, Stilling RM, Edbauer D, Rao P, Farinelli L, Delalle I, Schmitt A, Falkai P, Bahari-Javan S, Burkhardt S, Sananbenesi S, Fischer A. microRNA-34c is a novel target to treat dementias. EMBO J. 2011;30:4299–4308. doi: 10.1038/emboj.2011.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.