Abstract
Plant resistance proteins of the class of nucleotide-binding and leucine-rich repeat domain proteins (NB-LRRs) are immune sensors which recognize pathogen-derived molecules termed avirulence (AVR) proteins. We show that RGA4 and RGA5, two NB-LRRs from rice, interact functionally and physically to mediate resistance to the fungal pathogen Magnaporthe oryzae and accomplish different functions in AVR recognition. RGA4 triggers an AVR-independent cell death that is repressed in the presence of RGA5 in both rice protoplasts and Nicotiana benthamiana. Upon recognition of the pathogen effector AVR-Pia by direct binding to RGA5, repression is relieved and cell death occurs. RGA4 and RGA5 form homo- and hetero-complexes and interact through their coiled-coil domains. Localization studies in rice protoplast suggest that RGA4 and RGA5 localize to the cytosol. Upon recognition of AVR-Pia, neither RGA4 nor RGA5 is re-localized to the nucleus. These results establish a model for the interaction of hetero-pairs of NB-LRRs in plants: RGA4 mediates cell death activation, while RGA5 acts as a repressor of RGA4 and as an AVR receptor.
Keywords: Magnaporthe oryzae, pathogen recognition, plant immunity, resistance protein, rice
Introduction
Plants possess a highly efficient, two-layered innate immune system that renders them resistant against most microbial pathogens (Jones & Dangl, 2006; Dodds & Rathjen, 2010; Dangl et al, 2013). The first layer of defense relies on the recognition of conserved microbe-associated molecular patterns (MAMPs) by the so-called pattern recognition receptors (PRRs). PRRs are generally plasma membrane receptors which are often coupled to intracellular kinase domains (Monaghan & Zipfel, 2012). Adapted pathogens are able to suppress or to bypass this first layer of resistance through the secretion of effector proteins that interfere with plant immunity. The second layer of plant immunity relies on the specific recognition of pathogen effectors by disease resistance proteins (R). This recognition leads to a strong immune response which is often associated with a localized programmed cell death termed the hypersensitive response (HR). Since effectors are generally species or isolate specific, this second layer of immunity is only efficient against isolates that carry the recognized effector which is then called an avirulence protein (AVR).
Most R-proteins belong to the nucleotide-binding and leucine-rich repeat (NB-LRR) family and comprise a multidomain architecture with a C-terminal leucine-rich repeat domain (LRR) and a central nucleotide-binding domain (NB-ARC). In most cases, they also carry at their N-terminus a coiled-coil (CC) domain or a domain with homologies to the TOLL/interleukin-1 receptor (TIR) (Takken & Goverse, 2012).
The molecular mechanisms and the structural bases governing NB-LRR function are still insufficiently understood, but the detailed investigation of a limited number of NB-LRR proteins generated a conceptual framework for the role of the different domains (Bernoux et al, 2011a). The LRR domain often confers recognition specificity to NB-LRR proteins and is supposed to act in certain cases as a receptor domain (Ellis et al, 1999, 2007; Jia et al, 2000; Wang et al, 2007; Krasileva et al, 2010). In addition, it is involved in complex intramolecular interactions that contribute to proper regulation of NB-LRR proteins (Leister, 2005; Ade et al, 2007; Van Ooijen et al, 2008a; Slootweg et al, 2013). N-termini of NB-LRR proteins, including the CC, NB-ARC, or TIR domains, are believed to be important for the activation of resistance signaling and HR induction (Frost et al, 2004; Leister, 2005; Ade et al, 2007; Van Ooijen et al, 2008a; Swiderski et al, 2009; Krasileva et al, 2010). Recently, the CC domain of MLA10 from barley and the TIR domain of L6 from flax were demonstrated to be necessary and sufficient for cell death induction and to mediate immune receptor homodimerization (Bernoux et al, 2011b; Maekawa et al, 2011). The central NB-ARC domain shows homologies to the metazoan apoptosis regulators Apaf-1 and CED-4 and consists of the subdomains NB, ARC1 (for Apaf-1, R-protein and CED-4), and ARC2. These proteins form the NB-ARC family within the class of signal transduction ATPases with numerous domains (STAND) proteins. The NB-ARC domain was shown to bind and hydrolyze nucleotides (Tameling, 2002) and, in analogy to other STAND proteins, nucleotide binding and exchange is supposed to control the activity of NB-LRR proteins. ADP binding is believed to stabilize the inactive state, while ATP binding stabilizes the signaling competent state (Bernoux et al, 2011a; Williams et al, 2011).
AVR recognition by NB-LRR proteins can occur in a direct manner by physical binding of the AVR effector to the NB-LRR protein or in an indirect manner (Dangl & Jones, 2001; Van der Hoorn & Kamoun, 2008; Collier & Moffett, 2009). In the latter case, a plant protein modified by the AVR effector or the complex between the AVR effector, and the plant protein binds to the NB-LRR protein and triggers resistance. This plant protein may be either the operational target of the effector and in this case it is called a guardee since it is guarded by the NB-LRR, or a mimic of the operational target in which case it is called a decoy (Dangl & Jones, 2001; Van der Hoorn & Kamoun, 2008).
In most cases, individual NB-LRR proteins seem sufficient to mediate AVR effector recognition and to confer resistance. However, in recent years, an increasing number of cases have been reported where two NB-LRR proteins are required for resistance in both mono- and dicotyledonous plants (Sinapidou et al, 2004; Ashikawa et al, 2008; Lee et al, 2009; Loutre et al, 2009; Narusaka et al, 2009; Eitas & Dangl, 2010; Okuyama et al, 2011; Yuan et al, 2011; Brotman et al, 2012; Cesari et al, 2013; Wang et al, 2013). Why pairs of NB-LRR proteins are required for resistance in these cases, and how these paired proteins interact functionally and mechanistically, is largely unknown.
Blast disease caused by the fungus Magnaporthe oryzae is the most important disease in rice (Pennisi, 2010; Dean et al, 2012). It causes frequent and devastating epidemics in all rice-producing areas of the world and is therefore a severe threat for the cultivation of the most important crop for human nutrition. More than 20 rice blast resistance genes have been cloned, and with one exception, they all code for CC-NB-LRR proteins (Liu et al, 2010). A particular feature of rice blast NB-LRR proteins is that in a relatively high proportion (6 cases: Pik, Pikp, Pikm, Pi-a, Pi-CO39, and Pi-5), resistance is conferred by pairs of CC-NB-LRRs (Ashikawa et al, 2008; Lee et al, 2009; Okuyama et al, 2011; Yuan et al, 2011; Cesari et al, 2013; Wang et al, 2013). Examples of this are Pik-1 and Pik-2 which confer resistance to AVR-Pik-expressing M. oryzae strains (Zhai et al, 2011; Kanzaki et al, 2012) and RGA4 and RGA5 which recognize two unrelated effectors AVR-Pia and AVR1-CO39 (Okuyama et al, 2011; Cesari et al, 2013). The genes coding these matching NB-LRR pairs show a conserved type of organization in the rice genome consisting of extremely tight physical linkage and arrangement in inverted orientation.
To date, seven M. oryzae AVR proteins have been cloned and six of them code for small secreted proteins translocated into host cells suggesting that the majority of AVRs of the rice blast fungus are cytoplasmic effectors. For 4 of these AVRs, AVR-Pia, AVR1-CO39, AVR-Pita, and AVR-Pik, recognition by a matching CC-NB-LRR protein or a CC-NB-LRR protein pair has been described and was found in all cases to be direct (Jia et al, 2000; Kanzaki et al, 2012; Cesari et al, 2013). AVR-Pita interacts with the LRR domain of its R-protein Pi-ta (Jia et al, 2000), while AVR-Pik, AVR-Pia, and AVR1-CO39 bind physically to only one member of their respective CC-NB-LRR pair, Pik-1 and RGA5, respectively (Kanzaki et al, 2012; Cesari et al, 2013). Interestingly, the AVR interaction domain is, in both cases, a conserved domain with homologies to ATX1, a copper chaperone present in most eukaryotic organisms and containing a heavy metal-associated (HMA) domain (Cesari et al, 2013). This RATX1 domain (Related to ATX1), which is uncommon in NB-LRR proteins, is located between the CC and NB-ARC domains in Pik-1 and after the LRR domain in RGA5. In the cases of Pik-1/Pik-2 and RGA4/RGA5, it therefore seems that one of the members of the CC-NB-LRR pair acts as an AVR receptor, while the function of the other partner as well as the functional and molecular interactions between the two partners remain unknown.
To better understand the molecular mechanism by which RGA4 and RGA5 interact and trigger pathogen recognition, we undertook a functional analysis of this pair of R-proteins using both the homologous rice system and the heterologous Nicotiana benthamiana system. This study reveals a novel mechanism involving physical and functional interactions between the two CC-NB-LRR proteins. We show that RGA4 and RGA5 interact through their CC domains and form homo- and hetero-complexes. Furthermore, constitutive RGA4 expression triggers an effector-independent cell death response that is repressed by the presence of RGA5. Upon recognition of AVR-Pia, repression is relieved and HR-like cell death occurs.
Therefore, it seems that evolution has led each CC-NB-LRR partner in this and perhaps other hetero-pairs to undertake distinct functions (receptor or signaling inducer) in an immune receptor complex that mediates pathogen recognition and defense signaling.
Results
RGA4 acts as a constitutively active cell death inducer and is regulated by RGA5
To investigate functional and molecular interactions between RGA4 and RGA5, these proteins were transiently expressed in N. benthamiana leaves by Agrobacterium tumefaciens-mediated transformation. Several constructs driven by the 35S promoter and allowing expression of RGA4 or RGA5 with or without fusion to the HA epitope tag or fluorescent proteins (CFP or YFP) were tested. Western blot analysis showed that all fusion proteins were properly expressed (Fig1A and Supplementary Fig S1A). Expression of RGA4:HA triggered strong and rapid HR-like cell death which became visible 2 days after inoculation (Fig1B and Supplementary Fig S2A). RGA4 and RGA4:CFP induced the same response (Supplementary Fig S2A). These results suggest that RGA4 acts as a constitutively active cell death inducer.
Figure 1. RGA4 auto-activates HR-like cell death that is repressed by RGA5.
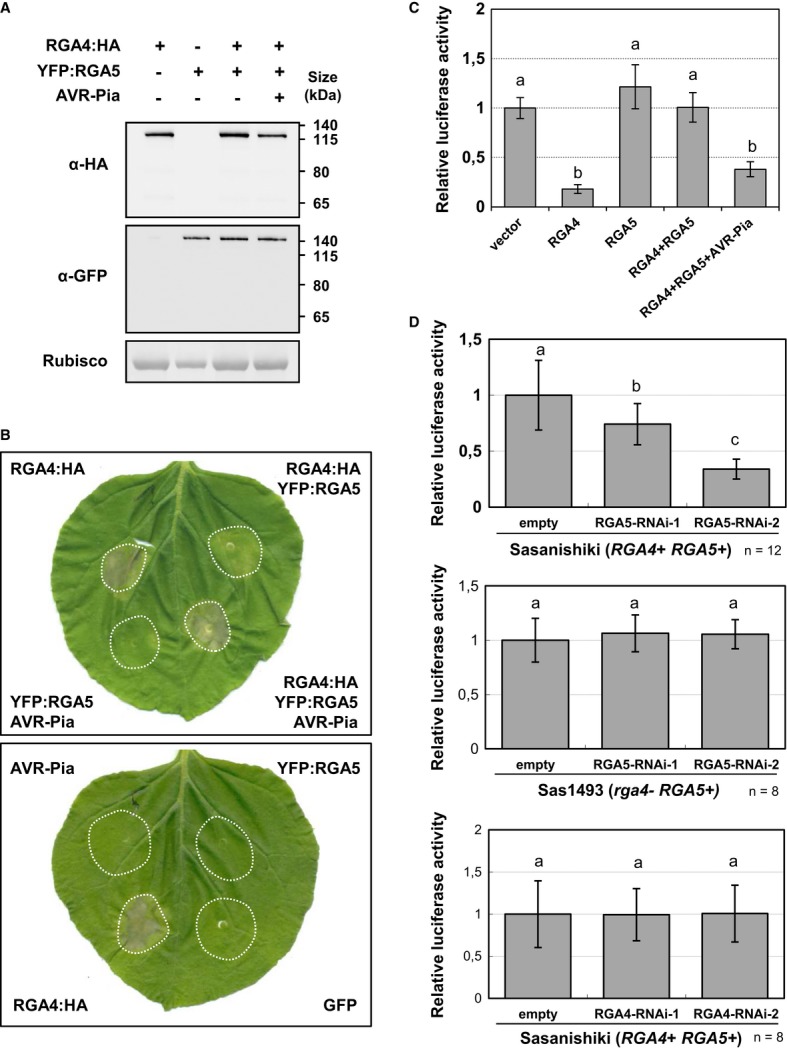
- RGA4:HA, YFP:RGA5, and AVR-Pia constructs were transiently expressed in Nicotiana benthamiana leaves by A. tumefaciens infiltration. Proteins were extracted 24 h after infiltration, and protein extracts were analyzed by immunoblotting with anti-GFP (α-GFP) and anti-HA (α-HA) antibodies. Ponceau staining of Rubisco small subunit was used to verify equal protein loading.
- The indicated combinations of fusion proteins were transiently expressed in N. benthamiana. The RGA4:HA strain was infiltrated at OD600 of 0.2, while strains carrying all other constructs were infiltrated at OD600 of 0.4. A bacterial strain carrying the GFP construct was used to equilibrate the total OD600 to 1 before infiltration. Cell death induced by the different combinations was visualized 3 days after infiltration. Equivalent results were obtained in at least three independent experiments.
- Leaf protoplasts of the susceptible rice cultivar Himenomochi (pia) were transfected with a plasmid for constitutive LUC expression in combination with constructs allowing expression of RGA4, RGA5, RGA4+RGA5, or RGA4+RGA5+AVR-Pia. LUC activity was determined in protein extracts of protoplast samples harvested 40 h after transfection. Average values and standard deviations were calculated from three replicate samples, and values were normalized with respect to the average values of the empty vector samples. The experiment was repeated three times with equivalent results. Different letters indicate groups that are significantly different (P < 0.05 in pairwise t-test).
- Rice protoplast of the resistant rice variety Sasanishiki or of the rga4 mutant line Sas1493 was co-transfected with RNAi constructs directed against RGA4 or RGA5 and the luciferase reporter gene. Luc activity was determined and analyzed as in (C). Average values and standard deviations were calculated from the indicated number of replicate samples (n). The experiment was repeated three times with equivalent results.
Source data are available online for this figure.
Unlike RGA4, expression of RGA5 alone did not induce any response in N. benthamiana leaves, suggesting that it has no cell death-inducing activity (Fig1B). However, co-expression of RGA4 with RGA5, HA:RGA5, or YFP:RGA5 attenuated or completely abolished RGA4-triggered cell death (Fig1B and Supplementary Fig S2B). The extent of cell death suppression was dependent on the ratio between 35S::RGA4- and 35S::RGA5-carrying agrobacteria in the infiltration inoculum. Using a ratio of 1:2, no symptom or only slight chlorosis was observed. These findings suggest that RGA5 negatively regulates RGA4 and acts as an inhibitor of RGA4-triggered cell death.
To assess the specificity of RGA5 repression activity, RGA5 was transiently co-expressed with the auto-active rice CC-NB-LRR Orin1 (Kawano et al, 2010) and an auto-active mutant of the flax TIR-NB-LRR L6 (L6MHV, Howles et al, 2005) in N. benthamiana and N. tabacum. Neither Orin1- nor L6MHV-mediated cell death was suppressed by RGA5 showing that its inhibition activity is specific to RGA4 (Supplementary Fig S3).
To test whether the RGA4-RGA5 pair is responsive to AVR-Pia and AVR1-CO39 in the heterologous N. benthamiana system, both NB-LRRs were co-expressed with AVR1-CO39, AVR-Pia, or the inactive H3 allele of AVR-Pia which does not interact physically with RGA5 and does not confer avirulence to M. oryzae (Cesari et al, 2013). Co-expression of AVR-Pia, RGA4, and RGA5 induced rapid and strong cell death indistinguishable in timing and appearance from the response induced by RGA4 alone (Fig1B). On the contrary, expression of AVR-Pia and RGA5 alone induced no symptoms, supporting the idea that RGA5 has no cell death-inducing activity (Fig1B). Expression of AVR-Pia_H3, RGA4, and RGA5 induced no response or only slight chlorosis, resembling co-expression of RGA4 and RGA5 (Supplementary Fig S2C). Taken together, these results suggest that AVR-Pia removes RGA5-mediated repression of RGA4-triggered cell death. The lack of activity of AVR-Pia_H3 indicates that binding of AVR-Pia to RGA5 is crucial to relieve RGA5 repressor activity. N- or C-terminally tagged versions of AVR-Pia did not activate cell death in the presence of RGA4 and RGA5, although they accumulated to high levels according to Western blot experiments (Supplementary Fig S1B) and interacted with RGA5 co-immunoprecipitation experiments (Cesari et al, 2013). The same was true for AVR1-CO39 or tagged variants of AVR1-CO39, which were detected in Western blot experiments but did not trigger cell death when co-expressed with RGA4 and RGA5 (Supplementary Figs S1B and S2C).
To validate the functional interactions observed in N. benthamiana in rice, combinations of RGA4, RGA5, and AVR-Pia were co-expressed with the luciferase reporter protein in leaf protoplasts of the susceptible rice variety Himenomochi or protoplasts from Oc rice suspension culture (Baba et al, 1986). Compared to the empty vector control, expression of RGA4 alone or co-expression of RGA4, RGA5, and AVR-Pia resulted in significant reduction of luciferase reporter activity, indicating induction of HR-like cell death (Fig1C and Supplementary Fig S4A, B and D). Expression of RGA5 alone had no effect as luciferase activity was not significantly reduced compared to the empty vector control (Fig1C and Supplementary Fig S4A and C). However, when RGA5 was co-expressed with RGA4, RGA4-triggered reduction of luciferase activity was abolished, indicating that RGA5 also suppresses RGA4-induced cell death in rice (Fig1C and Supplementary Fig S4A and D). RGA4 and RGA5 seem therefore to interact in the same manner in rice and in the heterologous N. benthamiana system.
To exclude that cell death induction by RGA4 is a consequence of overexpression and does not reflect its activity at normal physiological levels, RGA5 was silenced in rice protoplasts of the resistant Sasanishiki variety by transfection with RNAi constructs (Fig1D and Supplementary Fig S5). Depletion of RGA5 by two different RNAi constructs induced cell death, while depletion of RGA4 had no significant effect. In the rga4 loss-of-function-mutant line Sas1493 (Okuyama et al, 2011), RGA5 depletion had no effect indicating that the cell death induced by RGA5 depletion relies entirely on RGA4.
Taken together, these results suggest that RGA4 acts as an HR executer which is regulated in its activity by RGA5. RGA5 can be regarded as an AVR receptor since it physically binds AVR1-CO39 and AVR-Pia (Cesari et al, 2013) and transduces AVR signal perception into a relief of RGA4 repression, resulting in resistance induction and HR.
A functional nucleotide-binding pocket is required for RGA4 but not for RGA5 activity
To further test this model, mutant alleles of RGA4 and RGA5 in motifs required for the nucleotide-binding and exchange functions of NB-LRR proteins were generated and tested, respectively, for cell death-inducing or repressing activity. The central lysine (K) in the P-loop, a highly conserved motif in all nucleotide-binding proteins, was replaced by an arginine (R), giving rise to rga4K209R (rga4K/R) and rga5K210R (rga5K/R). Such P-loop mutations strongly impair nucleotide binding and abolish resistance and cell death induction in many NB-LRR proteins (Dinesh-Kumar et al, 2000; Tao et al, 2000; Tameling, 2002; Howles et al, 2005; Ade et al, 2007; Williams et al, 2011; Bai et al, 2012).
Tagged versions of rga4K/R and rga5K/R, with a C-terminal HA tag (rga4K/R:HA) or an N-terminal YFP tag (YFP:rga5K/R), were transiently expressed in N. benthamiana, while untagged rga4K/R and rga5K/R proteins were tested in rice protoplasts. Western blotting confirmed proper expression of the fusion proteins in N. benthamiana (Fig2A and B). Expression of rga4K/R alone or in combination with RGA5 and AVR-Pia did not trigger cell death in N. benthamiana nor in rice (Fig2C, D and E). On the contrary, rga5K/R still repressed RGA4-triggered cell death (Fig2D, E and F) and co-expression of RGA4, rga5K/R, and AVR-Pia triggered cell death to the same extent as co-expression of RGA4, RGA5, and AVR-Pia (Fig2E and F). Thus, an intact RGA4 nucleotide-binding pocket is required for cell death induction by RGA4, either on its own or in the presence of RGA5 and AVR-Pia. However, an intact RGA5 P-loop motif is not necessary for repression of RGA4-induced cell death or for removal of this repression after AVR-Pia recognition. This supports the notion that cell death is exclusively activated by RGA4 while RGA5 acts as an AVR receptor and negative regulator of RGA4. Binding of AVR-Pia to RGA5 may either not lead to nucleotide exchange and RGA5 activation or such nucleotide exchange may not be required for AVR-induced de-repression of RGA4.
Figure 2. An intact nucleotide-binding pocket is required for RGA4 but not for RGA5 activity.
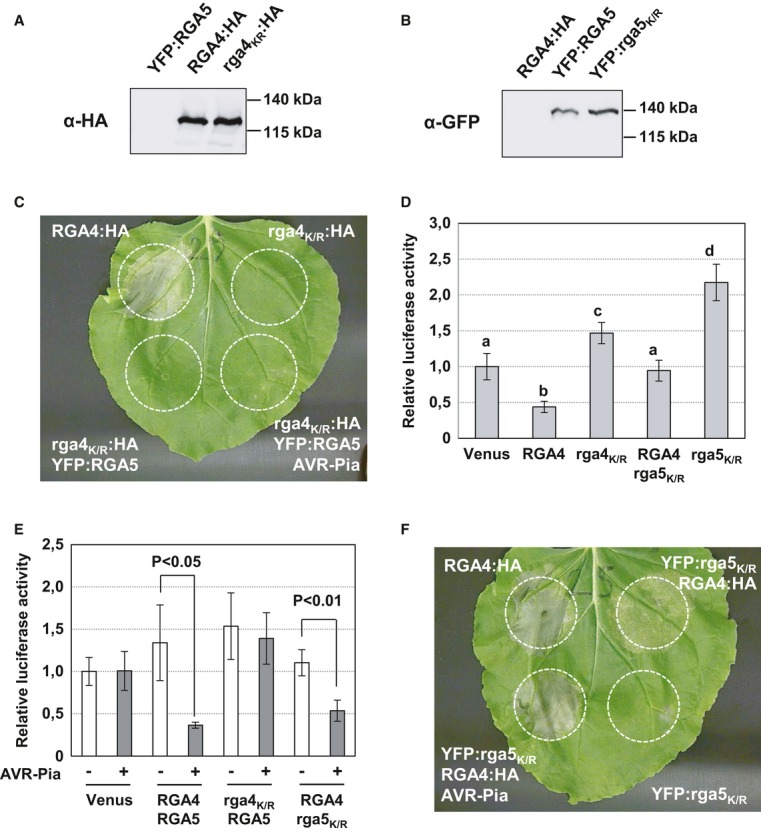
A, B Constructs were transiently expressed in N. benthamiana, and protein extracts were analyzed by immunoblotting with anti-HA (α-HA) and anti-GFP (α-GFP) antibodies.
C The rga4K/R:HA strain carrying a construct for the rga4K209R P-loop mutant was infiltrated alone and in combination with YFP:RGA5 or YFP:RGA5 and AVR-Pia strains in N. benthamiana leaves. The RGA4:HA strain was infiltrated alone as a positive control. Cell death was visualized 3 days after infiltration. Equivalent results were obtained in at least three independent experiments.
D, E Rice protoplasts from Oc suspension culture were transfected with plasmids for constitutive LUC expression and different combinations of constructs for expression of: Venus, RGA4, rga4K209R, RGA5, rga5K210R, and AVR-Pia. LUC activity was determined in protoplast protein extract samples 40 h after transfection. Average values and standard deviations were calculated from three replicate samples, and values were normalized with respect to the average values of the corresponding Venus samples. The experiments were repeated three times with equivalent results. Average values significantly different from the average value of the corresponding control (Venus) samples are indicated (P < 0.05 in Student's t-test).
F The YFP:rga5K/R strain carrying a construct for the rga5K210R P-loop mutant was infiltrated alone and in combination with RGA4:HA or RGA4:HA and AVR-Pia strains. The RGA4:HA strain was infiltrated alone as a positive control. Cell death was visualized 3 days after infiltration. Equivalent results were obtained in at least 3 independent experiments.
Source data are available online for this figure.
A non-canonical MHD motif is crucial for RGA4 but not for RGA5 activity
Within the ARC2 domain, the conserved MHD motif is particularly important for the regulation of NB-LRR proteins. Replacement of the conserved histidine or aspartate in the MHD core sequence by other amino acids leads in most cases to auto-active NB-LRR proteins, which constitutively activate immune responses and cell death (Bendahmane et al, 2002; De La Fuente van Bentem et al, 2005; Howles et al, 2005; Van Ooijen et al, 2008b; Kawano et al, 2010; Gao et al, 2011; Maekawa et al, 2011; Bai et al, 2012; Zhang et al, 2012; Roberts et al, 2013). Sequence analysis of RGA4 and RGA5 ARC2 domains revealed that both NB-LRR proteins contain non-canonical MHD motifs (Fig3A and Supplementary Fig S6). In RGA4, the motif is highly degenerated, and all three amino acids differ from the consensus MHD sequence (T500Y501G502). The hydrophobic methionine is substituted by a hydrophilic threonine, while the hydrophilic positively charged histidine and the negatively charged aspartate are replaced by a hydrophobic tyrosine and an uncharged glycine, respectively. In RGA5, the motif is less disturbed (L504H505H506) since the methionine is replaced by a leucine, which has similar biophysical properties, and the central histidine is conserved. However, the acidic, negatively charged aspartate is substituted by a basic, positively charged histidine.
Figure 3. A disturbed MHD motif in RGA4 is crucial for its function.
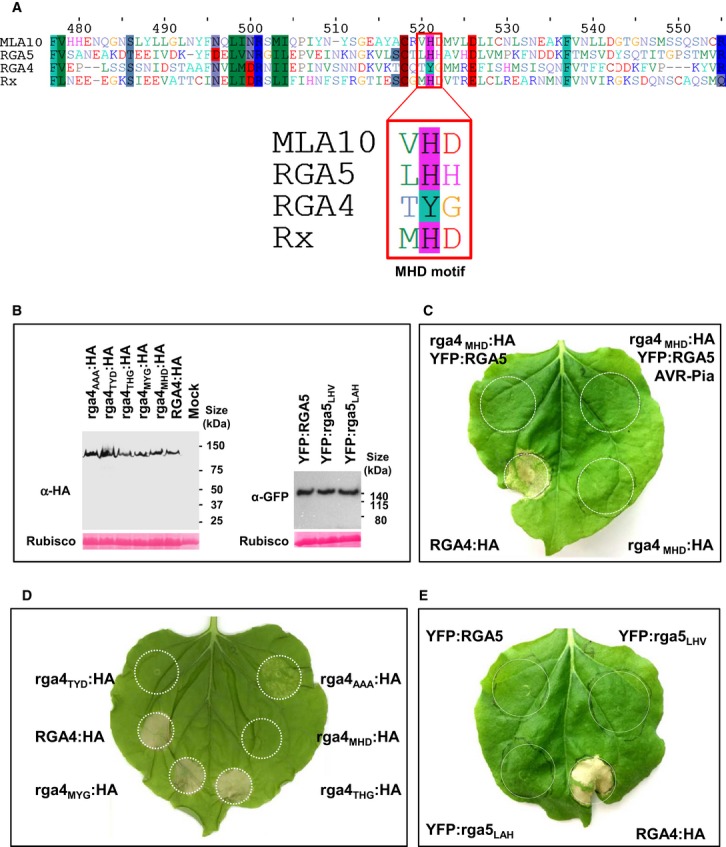
- Alignment of a portion of the ARC2 domain sequences of RGA4, RGA5, Rx, and MLA10 containing the MHD motif (red box).
- Proteins were extracted 24 h after infiltration of the indicated RGA4:HA or YFP:RGA5 constructs in N. benthamiana and analyzed by immunoblotting with anti-HA or anti-GFP antibodies (α-HA and α-GFP). Ponceau staining of Rubisco small subunit was used to verify equal protein loading.
- The rga4MHD:HA strain harboring a construct for expression of an RGA4 variant with the MHD consensus motif was infiltrated alone and in combination with YFP:RGA5 or YFP:RGA5 and AVR-Pia strains in N. benthamiana. Cell death was visualized 3 days after infiltration. The same results were obtained in at least three independent experiments.
- The RGA4 MHD mutant constructs rga4MHD:HA, rga4MYG:HA, rga4THG:HA, rga4TYD:HA and rga4AAA:HA were expressed in N. benthamiana and analyzed for cell death induction.
- YFP:RGA5, YFP:rga5LHV, YFP:rga5LAH, and RGA4:HA were expressed in N. benthamiana, and cell death was visualized 3 days after infiltration. Similar results were obtained in at least three independent experiments.
Source data are available online for this figure.
To determine whether RGA4 auto-activity could be due to its non-canonical TYG motif, point mutations were introduced in the RGA4 sequence to generate a MHD consensus sequence. This construct was fused to a C-terminal HA tag (rga4MHD:HA) and transiently expressed in N. benthamiana, alone or in combination with YFP:RGA5 or YFP:RGA5 and AVR-Pia (Fig3B, C and D). None of these combinations were effective in triggering cell death, whereas RGA4:HA, used as a positive control, induced a strong cell death response (Fig3C). To test whether single amino acids in the non-canonical core sequence TYG are required for auto-activity, they were individually replaced by M, H, or D, respectively (Fig3B). In addition, an AAA mutant was generated. The mutant forms rga4MYG and rga4THG retained auto-activity, while rga4TYD lost auto-activity indicating that the threonine and the tyrosine are neither required nor sufficient for auto-activity (Fig3D) and that the glycine is crucial. The rga4AAA mutant still induced cell death, however, to a much lower rate than wild-type RGA4. Similar strong reduction of cell death after replacement of multiple amino acids in the MHD sequence was also observed for the tomato I-2 NB-LRR protein and was attributed to a potential disturbance of the ATP-binding pocket (Van Ooijen et al, 2008b).
These results suggest that the disturbed MHD motif of RGA4 is crucial for its activity as an auto-active cell death inducer. In addition, they indicate that recognition of AVR-Pia by RGA5 does not lead to activation of rga4MHD supporting the hypothesis that auto-activity of RGA4 is necessary for AVR-Pia-induced cell death.
To determine whether RGA5 can function as an activator of cell death signaling, point mutations leading generally to NB-LRR auto-activity were introduced in its MHD motif. The RGA5 LHH core motif was replaced by an LHV or an LAH sequence, and the resulting constructs, N-terminally fused to YFP (YFP:rga5LHV and YFP:rga5LAH), were transiently expressed in N. benthamiana. Both rga5LHV and rga5LAH were expressed at similar levels as the RGA5 wild-type protein (Fig3B), but they did not trigger any cell death when expressed alone and are therefore not auto-active (Fig3E). Nevertheless, they were fully functional in repressing RGA4-mediated cell death and in recognizing AVR-Pia (Supplementary Fig S7A and B). The RGA5 MHD motif contains like several other NB-LRR proteins (Van Ooijen et al, 2008b) a second MHD-like sequence, VHD, separated by one amino acid from the MHD core sequence (Fig3A). To test whether mutation of this sequence leads to auto-activity, the aspartate in the VHD sequence was replaced by valine. The corresponding rga5D510V mutant behaved like the wild-type RGA5. It did not activate cell death but repressed RGA4 and recognized AVR-Pia (Supplementary Fig S8).
These results support the conclusion that cell death signaling is activated exclusively by RGA4 and that AVR-Pia binding does not lead to RGA5 activation but solely to RGA4 de-repression.
RGA4 and RGA5 form hetero- and homo-complexes
To investigate whether the functional interaction between RGA4 and RGA5 may rely on the formation of RGA4-RGA5 hetero-complexes, co-immunoprecipitation experiments with in planta-expressed tagged variants of RGA4 and RGA5 were performed. In addition, the formation of RGA4 and RGA5 homo-complexes was investigated since NB-LRR proteins have been reported to form homo-dimers (Mestre & Baulcombe, 2006; Ade et al, 2007; Gutierrez et al, 2010; Bernoux et al, 2011b; Maekawa et al, 2011).
In rice protoplasts, full-length RGA4 fused to a C-terminal T7 tag (RGA4:T7) was co-expressed with full-length RGA5 fused to an N-terminal 10×Myc tag (10×Myc:RGA5). These fusion proteins were fully functional as shown by cell death assays in rice protoplasts (Supplementary Fig S9). As a negative control, RGA4:T7 was expressed with 10×Myc-tagged GFP (10×Myc:GFP) and un-tagged RGA5 which served to prevent cell death. All fusion proteins were detected in protoplast extracts by immunoblotting with anti-Myc or anti-T7 antibodies (Fig4A). 10×Myc:RGA5 and 10×Myc:GFP were expressed at similar levels and were precipitated by anti-Myc antibodies. RGA4 co-precipitated specifically with RGA5 but not with GFP (Fig4A).
Figure 4. RGA4 and RGA5 form hetero- and homo-complexes in planta.
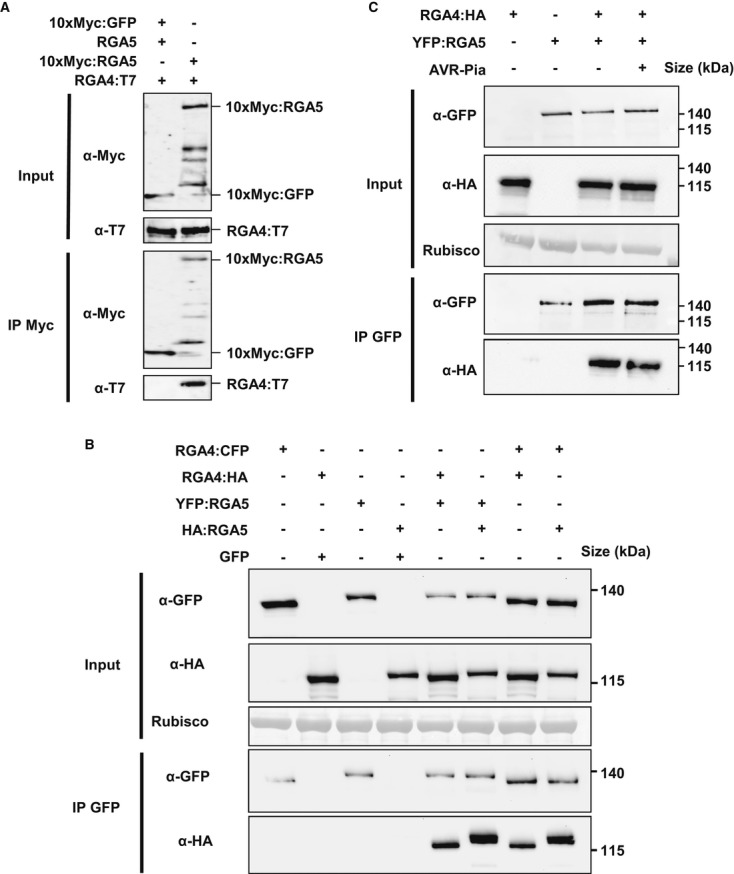
A RGA4:T7, 10×Myc:RGA5, RGA5, and 10×Myc:GFP were transiently expressed in rice protoplasts from Oc cells. Proteins were extracted 16 h after transfection and analyzed by immunoblotting with anti-c-Myc (α-c-Myc) and anti-T7 antibodies (α-T7) (Input). In addition, immunoprecipitations were performed with anti-c-Myc beads (IP c-Myc) and analyzed by immunoblotting with anti-c-Myc antibodies for the detection of immunoprecipitated 10×Myc:RGA5 and 10×Myc:GFP and with anti-T7 antibodies for the detection of co-precipitated RGA4:T7.
B, C Different combinations of RGA4:CFP, RGA4:HA, YFP:RGA5, HA:RGA5, GFP, and AVR-Pia constructs were transiently expressed in N. benthamiana. Protein extracts were analyzed by immunoblotting with anti-HA (α-HA) and anti-GFP antibodies (α-GFP) (Input). In addition, immunoprecipitations were performed with anti-GFP beads (IP GFP) and analyzed by immunoblotting with anti-GFP antibodies for the detection of immunoprecipitated RGA4:CFP, YFP:RGA5, and GFP proteins and with anti-HA antibodies for the detection of co-precipitated RGA4:HA and HA:RGA5 proteins.
Source data are available online for this figure.
Similar results were obtained using the N. benthamiana system where different combinations of RGA4:CFP, RGA4:HA, YFP:RGA5, HA:RGA5, and GFP (used as a negative control) were expressed. Immunoblotting using anti-GFP and anti-HA antibodies showed proper expression of all proteins (Fig4B). Immunoprecipitation with anti-GFP antibodies resulted in enrichment of RGA4:CFP, YFP:RGA5 (Fig4B), and GFP (Supplementary Fig S10). RGA4:HA specifically co-precipitated with YFP:RGA5 but not with GFP (Fig4B), and HA:RGA5 was efficiently co-precipitated by RGA4:CFP but not by GFP. Interestingly, HA:RGA5 was also co-precipitated by YFP:RGA5, and RGA4:HA co-precipitated with RGA4-CFP. Taken together, these results indicate that in planta RGA4-RGA5 hetero-complexes exist and suggest that RGA4 binds RGA5 in a specific manner. In addition, RGA4 and RGA5 may interact with themselves and form homo-complexes.
To investigate whether AVR-Pia disrupts RGA4-RGA5 hetero-complexes, additional co-immunoprecipitation experiments were performed where RGA4:HA and YFP:RGA5 were co-expressed in N. benthamiana in the presence or in the absence of AVR-Pia. As observed previously, AVR-Pia expression had no influence on the abundance of RGA4 or RGA5 (Fig4C). In addition, AVR-Pia had no influence on the co-precipitation of RGA4:HA by YFP:RGA5 (Fig4C). This indicates that RGA4–RGA5 hetero-complexes still exist in the presence of AVR-Pia.
Taken together, these results suggest that RGA5 regulates RGA4 through direct interaction. However, complete disruption of RGA4–RGA5 hetero-complexes is not induced by AVR-Pia recognition and seems not necessary for HR.
The CC domains of RGA4 and RGA5 form homo- and hetero-complexes and the RATX1 domain of RGA5 homo-complexes
To investigate whether formation of RGA4–RGA5 hetero-complexes and RGA4 and RGA5 homo-complexes involves direct physical binding and to identify the protein domains involved, pairwise interactions of isolated CC, NB-ARC, and LRR domains of RGA4 and RGA5 and of the C-terminal RATX1 domain of RGA5 (Supplementary Fig S11A and B) were analyzed in the yeast two-hybrid system. NB-ARC domains and LRR domains showed no interactions with the exception of the RGA5 NB-ARC domain which interacted weakly with the RGA5 RATX1 domain (Fig5A and Supplementary Fig S12A and B). Expression of the different domains was confirmed by Western blot analysis (Supplementary Fig S12C). RGA4 and RGA5 CC domains (RGA41–176 and CC-RGA51–177) interacted with themselves but did not interact with one another (Fig5A and Supplementary Fig S12A and B). The RATX1 domain showed also homo-dimerization.
Figure 5. The CC domains of RGA4 and RGA5 form homo- and hetero-complexes and the RATX1 domain of RGA5 self-interacts in yeast.
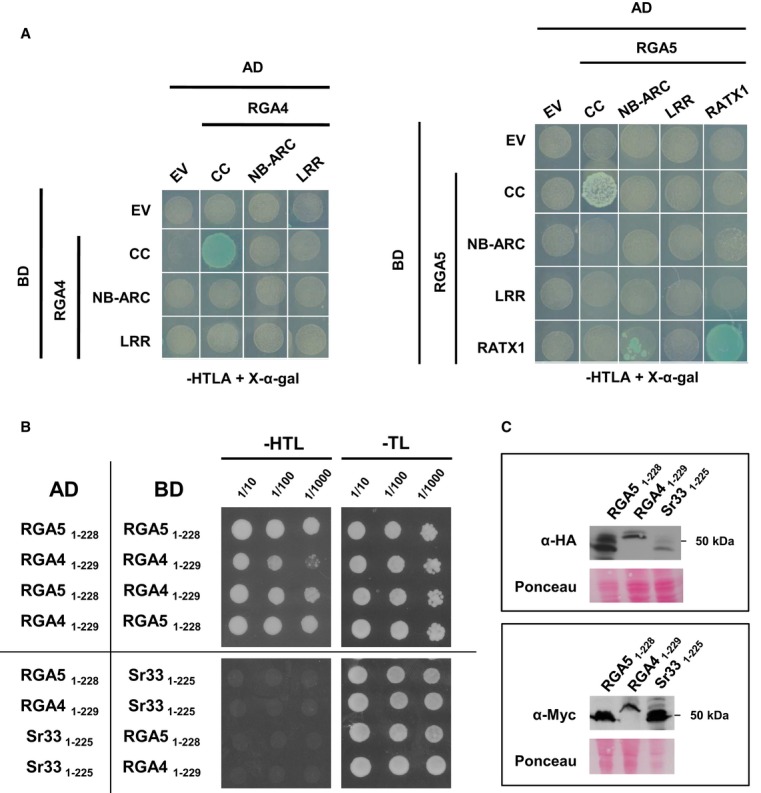
- Pairwise interaction of CC, NB-ARC, and LRR domains from RGA4 and RGA5, as well as the RGA5 RATX1 domain (see Supplementary Fig S9A for domain borders) was evaluated by blue coloration and growth of yeast cells on basal media lacking Trp, Leu, Ade, and His and supplemented with 5-bromo-4-chloro-3-indolyl α-D-galactopyranoside (X-a-gal). Pictures were taken after 3 days of growth.
- RGA4 and RGA5 CC domain homo- and hetero-complexes. Dilution series of yeast cells expressing GAL4-AD and GAL4-BD fusions of RGA5 1–228 and/or RGA4 1–229 CC domains on non-selective synthetic media lacking Trp and Leu (-TL) and selective media additionally lacking His (-HTL). The wheat Sr33 (1–225) CC domain was used as a negative control. Pictures were taken after 3 days of growth. Growth on -HTL media is indicative of interaction.
- GAL4-AD and -BD fusions of CC domains of RGA4, RGA5, and Sr33 were detected by immunoblotting of yeast protein extracts with, respectively, anti-HA and anti-Myc antibodies. Equal loading was verified by red Ponceau staining.
Source data are available online for this figure.
To further investigate possible hetero-interactions of CC-RGA4 and CC-RGA5, three additional fragments were analyzed for each protein (Supplementary Fig S13C). CC-RGA41–130 and CC-RGA51–133 correspond to a fragment of the MLA10 CC domain that homo-dimerizes in vitro and has been structurally characterized by crystallography (Maekawa et al, 2011). CC-RGA41–130 and CC-RGA51–133 showed homo- and hetero-interactions (Supplementary Fig S13). CC-RGA41–164 and CC-RGA51–167 correspond to a minimal MLA10 fragment sufficient for cell death induction (Maekawa et al, 2011; Bai et al, 2012). CC-RGA41–164 interacted with itself but CC-RGA51–167 interacted neither with itself nor with CC-RGA41–164 (Supplementary Fig S13). CC-RGA41–229 and CC-RGA51–228 contain, in addition to the CC domain, a part of the NB domain including the P-loop motif, and correspond to an MLA10 fragment that homo-dimerizes in the yeast two-hybrid system (Maekawa et al, 2011). CC-RGA51–228 interacted with itself and with CC-RGA41–229, and CC-RGA41–229 interacted with itself although interactions seemed much weaker than CC-RGA41–130 and CC-RGA41–164 homo-interactions (Fig5B and Supplementary Fig S13). CC-homo- and hetero-interactions were specific since no interaction with the CC domain of the sequence-unrelated wheat rust resistance NB-LRR protein Sr33 (Periyannan et al, 2013) was detected (Fig5B). Proper expression of RGA4, RGA5, and Sr33 CC domains was verified by immunoblotting (Fig5C).
Taken together, the yeast two-hybrid analyses suggest that CC domain interactions are involved in RGA4-RGA5 hetero-complex and RGA4 and RGA5 homo-complex formation. However, it becomes evident that CC domain interactions depend strongly on the length of the CC fragments. In addition, RATX1 self-interaction may contribute to RGA5 homo-complex formation.
To validate the interactions of the RGA4 and RGA5 CC domains, co-immunoprecipitation experiments were performed. CC-RGA41–229:HA, CC-RGA41–229:CFP, HA:CC-RGA51–228, CC-RGA51–228:CFP, and GFP (used as negative control) were co-expressed in N. benthamiana. Immunoblotting using anti-GFP and anti-HA antibodies showed proper expression of all constructs in the input (Fig6). Immunoprecipitation with anti-GFP antibodies resulted in enrichment of CC-RGA41–229:CFP, CC-RGA51–228:CFP, and GFP (Fig6). CC-RGA41–229:HA and HA:CC-RGA51–228 specifically co-precipitated with CC-RGA41–229:CFP and CC-RGA51–228:CFP but not with GFP (Fig6). Taken together, these results indicate that in planta RGA4 and RGA5 CC domains can form hetero- and homo-complexes.
Figure 6. RGA4 and RGA5 CC domains form homo- and hetero-complexes in planta.
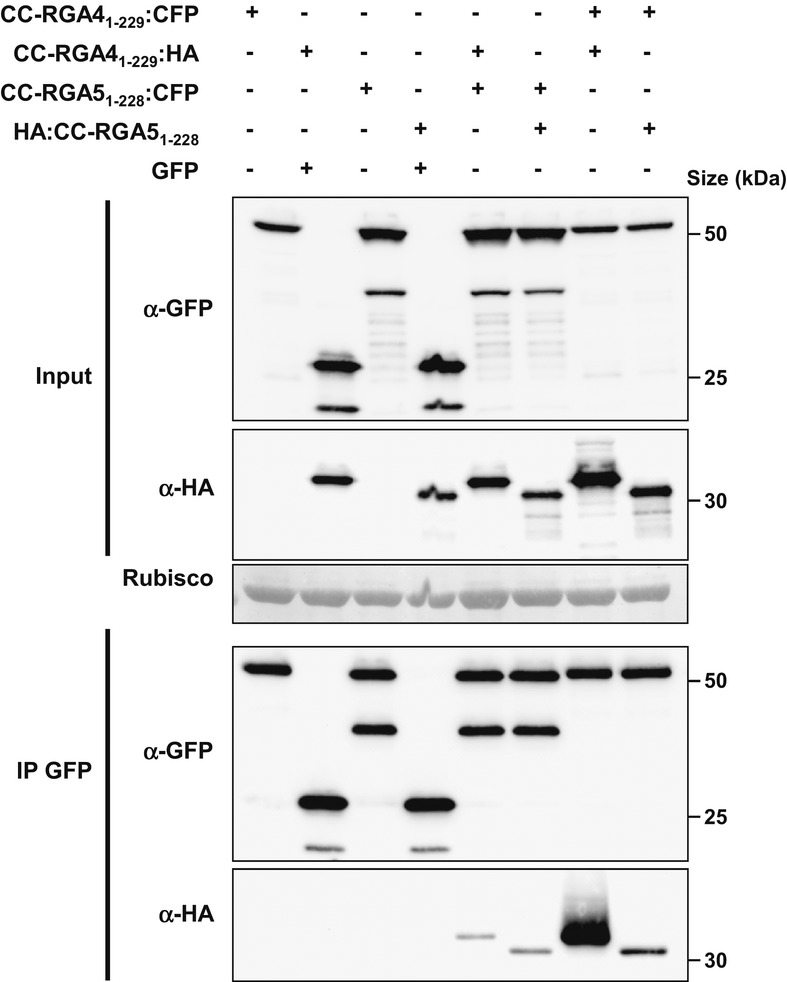
RGA41–229:CFP, RGA41–229:HA, YFP:RGA51–228, HA:RGA51–228, and GFP constructs were transiently expressed in N. benthamiana. Protein extracts were analyzed by immunoblotting with anti-HA (α-HA) and anti-GFP antibodies (α-GFP) (Input). Immunoprecipitations were performed with anti-GFP beads (IP GFP) and analyzed by immunoblotting with anti-GFP antibodies for the detection of immunoprecipitated RGA41–229:CFP, YFP:RGA51–228, and GFP proteins and with anti-HA antibodies for the detection of co-precipitated RGA41–229:HA and HA:RGA51–228 proteins.
Source data are available online for this figure.
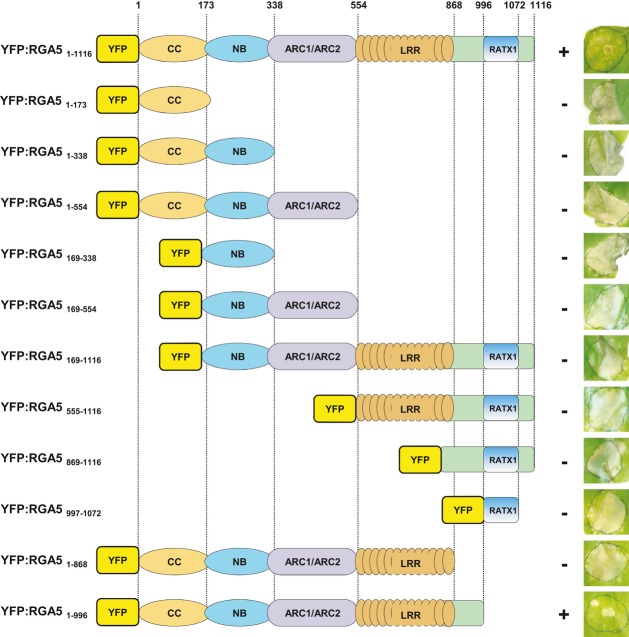
The RGA5 CC domain is necessary but not sufficient for repression of RGA4-mediated cell death, while its RATX1 domain is dispensable
To elucidate the role of individual domains of RGA5- in RGA4-mediated cell death repression and to identify eventually a minimal domain for this activity, a deletion series of RGA5 was analyzed. Individual CC, NB, ARC, LRR, and RATX1 domains (the latter with and without flanking sequences) as well as combinations of two, three, or four of these five domains were expressed together with RGA4 in N. benthamiana (Fig7 and Supplementary Fig S14). None of the individual domains or combinations of two or three domains was sufficient to repress RGA4-mediated cell death. In addition, deletion of the CC domain alone abolished repressor activity indicating that the CC domain is important for RGA4 repression but not sufficient (Fig7). Only the RGA5 variant where the RATX1 domain but not the sequence between the LRR domain and RATX1 domain is deleted retained RGA4 repressor activity indicating that the RATX1 domain is not required for RGA4 repression. These results further highlight a role of the RATX1 domain in AVR recognition but not in RGA4 repression and indicate that the CC domain is crucial for functional interactions between RGA5 and RGA4.
Figure 7. The RGA5 CC domain is required but not sufficient for repression of RGA4-mediated cell death, while the RATX1 domain is dispensable.
The indicated RGA5 deletion constructs were co-expressed with RGA4 in N. benthamiana, and development of cell death was visualized 4 days after infiltration.
RGA4 and RGA5 are mainly localized in the cytosol of rice protoplasts
To determine whether RGA4 and RGA5 act in the same cellular compartments, translational fusions of RGA4 or RGA5 with fluorescent proteins (RGA4:GFP and Venus:RGA5) were expressed in rice protoplasts (Supplementary Fig S15) and visualized by confocal laser scanning microscopy (Fig8). Protoplast cell death assays demonstrated that the fusion proteins are functional (Supplementary Fig S9). Green fluorescence of RGA4:GFP was observed mainly in the cytosol labeled by mCherry, whereas no or only weak signal could be detected in nuclei labeled by mOrange fused to a nuclear localization signal (NLS:2xmOrange; Fig8A). Similar results were obtained for Venus:RGA5 which co-localized with Cerulean in the cytosol but not with NLS:Cerulean in the nuclei (Fig8B). These results suggest that RGA4 and RGA5 localize to the cytosol and are excluded from the nucleus. Co-expression of RGA4 and RGA5 or RGA4, RGA5, and AVR-Pia did not change the localization patterns of the NB-LRR proteins and did not induce their re-localization to the nucleus (Fig9).
Figure 8. RGA4 and RGA5 are mainly localized in the cytosol of rice protoplasts.
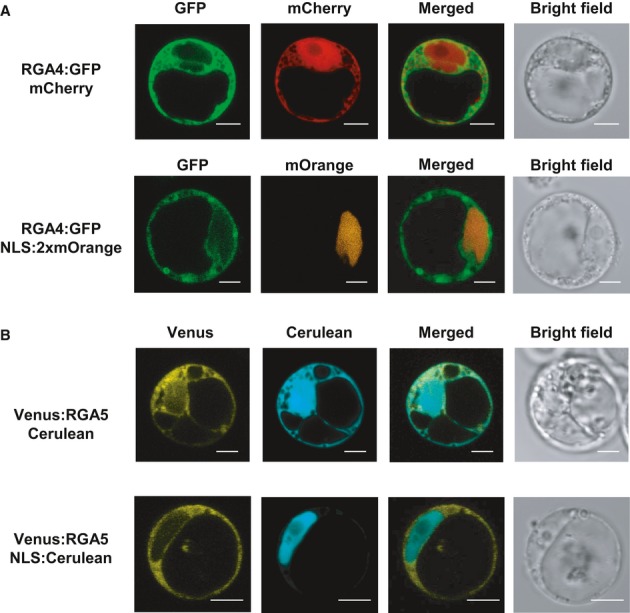
A, B Rice protoplasts derived from Oc cells (Baba et al, 1986) were transiently transformed with combinations of constructs allowing the expression of RGA4:GFP, Venus:RGA5, NLS:2×mOrange, NLS:Cerulean, mCherry, and Cerulean. Confocal images of protoplasts were taken 16–22 h after transfection. Scale bars, 5 μm.
Figure 9. Localization patterns of RGA4 and RGA5 are not affected by the presence of AVR-Pia or RGA4-RGA5 co-expression.
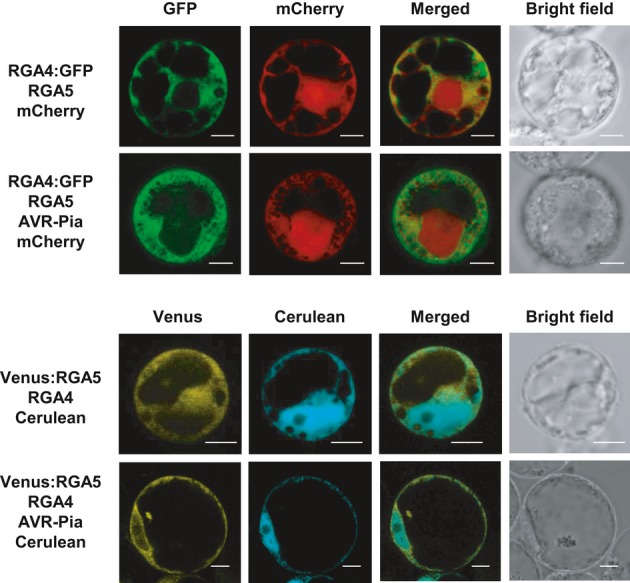
Rice protoplasts derived from Oc cells (Baba et al, 1986) were transfected with combinations of constructs for the expression of RGA4:GFP, Venus:RGA5, AVR-Pia, mCherry and Cerulean, and un-labeled RGA4 and RGA5. Confocal images of protoplasts were taken 16–22 h after transfection. Scale bars, 5 μm.
Discussion
Plant resistance is often conferred by single NB-LRR proteins that are polymorphic between different accessions/genotypes of a plant species. It is therefore generally supposed that these NB-LRRs do not need to interact with other NB-LRR proteins. However, in recent years, an increasing number of plant resistances conferred by pairs of two distinct NB-LRR proteins, and not individual NB-LRRs, have been described. The first example was the TIR-NB-LRRs RPP2A and RPP2B which are both required for A. thaliana resistance against the Hyaloperonospora parasitica isolate Cala2 (Sinapidou et al, 2004). More recently, A. thaliana TIR-NB-LRRs RPS4 and RRS1 were found to be both required for resistance to multiple pathogens, by recognizing the unrelated effectors AvrRps4 from Pseudomonas syringae, PopP2 from Ralstonia solanacearum, and an uncharacterized factor produced by Colletotrichum higginsianum (Gassmann et al, 1999; Deslandes et al, 2002; Narusaka et al, 2009). Fom1 and Prv, from melon, were recently described to provide resistance against Fusarium oxysporum and papaya ring-spot virus (Brotman et al, 2012). Interestingly, several R gene pairs, encoding CC-NB-LRRs, have also been characterized in monocotyledonous plants. For instance, Lr10 and RGA2 are both required for rust resistance in wheat (Loutre et al, 2009), and Rpg5 and HvRGA1 confer, together with HvAdf3, rust resistance to barley (Wang et al, 2013). In the rice blast pathosystem, resistance protein pairs seem to be particularly frequent (Sinapidou et al, 2004; Ashikawa et al, 2008; Lee et al, 2009; Okuyama et al, 2011; Yuan et al, 2011; Cesari et al, 2013; Wang et al, 2013).
Why, in certain cases, two distinct plant NB-LRR proteins are required for the functions often executed by individual R-proteins and how those paired NB-LRR proteins interact functionally are important questions. Our previous investigation of RGA4/RGA5 and Pik-1/Pik-2 indicated that RGA5 and Pik-1 mediate AVR protein recognition through physical binding to their RATX1 domains and thus act as AVR receptors, while RGA4 and Pik-2 have no AVR-binding property (Kanzaki et al, 2012; Cesari et al, 2013). This suggested that each pair partner accomplishes on its own a subset of the functions frequently executed by one individual NB-LRR protein rather than cooperating with its partner to accomplish together R-protein tasks such as AVR recognition or resistance activation.
RGA4 is an activator of cell death and HR
This hypothesis is strengthened in the present study, which shows that cell death activation is mediated exclusively by RGA4. RGA4 but not RGA5 acts as a constitutively active inducer of HR-like cell death as demonstrated by transient overexpression experiments in rice and N. benthamiana. Constitutive activity is an intrinsic property of RGA4 and not a consequence of overexpression since silencing of RGA5 leads to RGA4 de-repression and cell death when driven by its own promotor. Interestingly, previously generated transgenic rice lines expressing RGA4 in the Kanto51 variety lacking Pi-a and Pi-CO39 resistance do not show spontaneous cell death (Okuyama et al, 2011; Cesari et al, 2013). The lack of cell death induction may be due to the fact that rice varieties lacking Pi-a and Pi-CO39 resistance carry at the Pi-a/Pi-CO39 locus, a highly homologous allele of RGA5 and a pseudogene homologous to RGA4 (respectively, Os11g11810 and Os11g11790 in the Nipponbare reference variety). This RGA5 allele Os11g11810 lacks the RATX1 sequence and is non-functional in recognizing AVR-Pia and AVR1-CO39 but may be active in repressing the product of the RGA4 transgene since it shows high overall similarity to RGA5 (> 70% identity).
Mutation of the conserved P-loop motif abolishes RGA4 cell death and HR signaling activation, while mutation of the RGA5 P-loop motif has no such effect. This finding suggests that RGA4 activity is likely to be dependent on nucleotide binding and exchange while RGA5 activity is not.
Structurally, the constitutive activity of RGA4 could be caused by its degenerated MHD motif. Modeling studies and functional analysis suggest that the MHD motif located at the C-terminus of the ARC2 subdomain is important for maintaining the NB-ARC domain in a compact inactive state by stabilizing ADP-binding and physical interactions with the NB-subdomain (Riedl et al, 2005; Van Ooijen et al, 2008b). In RGA4, the highly conserved histidine and aspartate are changed to uncharged amino acids, and the normally hydrophobic amino acid preceding the histidine is replaced by a hydrophilic threonine. This degeneration of the MHD motif may trap RGA4 in an active state. This is supported by the finding that introduction of a consensus MHD motif abolishes constitutive activity of RGA4.
RGA5 also carries a modified MHD domain since the conserved aspartate is replaced by a histidine. This altered MHD motif or the introduction of additional mutations usually rendering NB-LRR proteins auto-active does not lead to constitutive activity of RGA5. It may be that a closed inactive conformation of RGA5 is stabilized by additional intramolecular interactions or that RGA5 is unable to interact with downstream signaling partners and is therefore intrinsically inactive in resistance activation. This further supports the view that cell death and HR activation is mediated exclusively by RGA4.
The inspection of other characterized NB-LRR protein pairs from rice did not identify other NB-LRRs with MHD motifs degenerated as in RGA4. However, in several cases, the conserved aspartate is replaced either by an asparagine or a proline, but it remains to be determined if these changes lead to constitutive activity. It may also be that auto-activity is caused by other structural features. It remains to be investigated whether other NB-LRR pairs also rely on auto-active NB-LRRs or interact in another manner.
RGA5 regulates RGA4 in an AVR-Pia-dependent manner
Co-expression of RGA4 and RGA5 in rice and N. benthamiana shows that RGA5 inhibits RGA4-mediated cell death. This suggests that RGA5 regulates RGA4 signaling activity by acting as a repressor in the absence of recognized effectors. This regulation is specific since cell death triggered by the Orin1 CC-NB-LRR auto-active protein (Kawano et al, 2010) or an auto-active allele of L6 (Howles et al, 2005) is not repressed by RGA5. Upon AVR recognition by direct binding, RGA5-mediated repression of RGA4 is relieved and cell death occurs. This is reminiscent of the interaction of other well-studied NB-LRR immune receptors with their corresponding guardees or decoys (Jones & Dangl, 2006; Van der Hoorn & Kamoun, 2008). These host proteins are operational targets of pathogen effectors or their mimic, and binding to effectors or modification by them is recognized by the corresponding NB-LRR immune receptor, generally by direct physical binding. Since there is no indication that RGA5 has other functions in disease or immunity than Avr recognition, RGA5 can be interpreted formally as a decoy surveyed by RGA4. However, AVR-Pia and AVR1-CO39 do not bind the CC, NB, or LRR domains but the uncommon RATX1 domain, and this event is sensed by the immune receptor hetero-pair. It is therefore more appropriate to consider the RATX1 domain as the true decoy integrated into one partner of the NB-LRR hetero-pair and allowing to monitor modification of other RATX1 proteins (Cesari et al, 2013). It will be interesting to test in the future if other hetero-pairs, such as Pik-1/Pik-2 where one of the partners contains a RATX1 domain and RRS1/RPS4 where one of the partners contains a WRKY domain, could also function by such an integrated decoy mechanism (Deslandes et al, 2003; Narusaka et al, 2009; Okuyama et al, 2011; Cesari et al, 2013).
RGA4 and RGA5 form hetero-complexes in planta which appears to be mediated, at least in part, by interaction between their CC domains. Likewise, both proteins individually form homo-complexes in planta which are also apparently mediated via CC domain interactions. Thus, a simple hypothesis is that the RGA4–RGA5 hetero-complex competes with and prevents formation of a signaling active RGA4 homo-complex. Binding of AVR-Pia to RGA5 could disrupt RGA4–RGA5 hetero-complexes resulting in RGA4 homo-complex formation and de-repression. Such homo-complexes are important for activation of the MLA and L6 NB-LRR proteins (Maekawa et al, 2011; Bernoux et al, 2011a,b). However, co-immunoprecipitation of RGA4 by RGA5 is still observed in the presence of AVR-Pia. Therefore, under this hypothesis, it must be assumed that disruption of a portion of the complexes frees sufficient RGA4 to signal cell death. An alternative model is that AVR-Pia binding to the RATX1 domain of RGA5 induces conformational changes within the intact RGA4–RGA5 hetero-complexes leading to their transition from a repressed to an activated state.
The dissection of RGA4 and RGA5 homo- and hetero-complex formation indicates that they are mainly mediated through interactions between the CC domains. This is consistent with the finding that removal of the RGA5 CC domain leads to a loss of RGA4 repression and the previous finding that MLA10 homo-complex formation is mediated by its CC domain (Maekawa et al, 2011). It will be interesting to analyze in the future the impact of mutations in the RGA4 and RGA5 CC domains on homo- and hetero-complex formation and on cell death activation and repression.
Striking similarities in the interaction of the RPS4–RRS1 and RGA4–RGA5 hetero-pairs
The RGA4–RGA5 interaction shows striking similarities to the interaction of RPS4 and RRS1 described in a recent publication (Williams et al, 2014). RPS4 was shown to act as RGA4 as an activator of resistance and cell death, while RRS1 represses RPS4 and binds the Avr proteins PopP2 and AvrRPS4 and resembles in this sense RGA5. RPS4 and RRS1 interact physically, and RPS4–RRS1 hetero-complexes are not or not completely disrupted by Avr protein recognition. The N-terminal TIR domains of RPS4 and RRS1 are important for resistance signaling and repression and form hetero- and homo-dimers like the RGA4–RGA5 CC domains. A P-loop mutation in RPS4 leads to loss of resistance, while an RRS1 P-loop mutant is fully functional (Williams et al, 2014). It therefore looks as if different NB-LRR hetero-pairs possessing different types of N-terminal domains interact molecularly and functionally in a similar manner.
Differences between RPS4 and RGA4 can, however, also be observed. RPS4 has a canonical MHD motif and causes cell death in a temperature-sensitive manner when overexpressed in A. thaliana but not when expressed from its own promoter (Wirthmueller et al, 2007). It will therefore be interesting to investigate in different hetero-pairs in detail the link between effector recognition and activation of signaling.
Similarities exist also with the A. thaliana NB-LRR proteins ADR1 and ADR1-like 1 and 2 (ADR1-L1 and -L2) that interact functionally with NB-LRR immune receptors and are required for their activity (Bonardi et al, 2011). However, on the contrary to RGA5 or RRS1, ADR1 proteins are not involved in Avr recognition, and an MHD mutant of ADR1-L2 shows auto-immune responses. On the contrary to RGA4 and RPS4, ADR1 proteins interact not just with one but with several different NB-LRR immune receptors that are physically not linked to them, they are required for certain but not all downstream responses, and a p-loop mutation in ADR1-L2 does not impair its activity in resistance signaling (Bonardi et al, 2011). ADR1 proteins are therefore considered as helper NB-LRRs and define another type of NB-LRR interactions, different from NB-LRR hetero-pairs such as RAG4–RGA5 and RPS4–RRS1.
Genomic organization: a key feature for proper control and co-evolution of NB-LRR pairs?
The RGA4-RGA5 system which contains a constitutively active cell death activator is potentially dangerous for the plant and requires tight control since loss or modification of RGA5 may lead to inappropriate activation of immune responses and cell death. Other NB-LRR proteins, that are harmless in their native genetic context, were demonstrated to cause constitutive activation of the plant immune system and trigger hybrid necrosis after inter- or intraspecific crosses (Bomblies et al, 2007). The clustered genomic organization of RGA4-RGA5 in an inverted head-to-head orientation in a 7-kb interval where they share the same intergenic region reduces the risk of misregulation and modification or loss of one partner by recombination or genomic rearrangements. However, it is intriguing that the same genomic organization as in RGA4-RGA5 is found in almost all R gene pairs identified, although they are not phylogenetically related. Investigation of other NB-LRR pairs is needed to determine whether clustered organization is a universal feature of NB-LRR gene pairs to favor efficient transmission of resistance after sexual reproduction or whether it is restricted to those that code constitutively active cell death inducers and related to the control of inappropriate activation of immune responses.
Convergent evolution generated NB-LRR pairs in animals and plants
Interestingly, cooperation of two distinct NB-LRR proteins in the recognition of pathogens is not restricted to plants but also occurs in mammals. Activation of immune responses in mice by the inner rod protein of the type III secretion systems (T3SS) of diverse bacterial species (for instance PrgJ of Salmonella typhimurium) requires the cytoplasmic NB-LRR proteins NLRC4 and NAIP2 (NLR family, apoptosis inhibitory protein 2), whereas NLRC4 and NAIP5 are required for recognition of bacterial flagellin in the cytoplasm of mouse macrophages and activation of downstream immune responses (Kofoed & Vance, 2011; Zhao et al, 2011). Biochemical dissection of the NLRC4/NAIP system showed that NAIP2 physically and specifically associates with T3SS inner rod proteins from different bacteria, whereas NAIP5 specifically associates with C-terminal sequences of flagellin. In both cases, ligand binding induces NAIP–NLRC4 association followed by NLRC4 oligomerization and activation of downstream responses such as caspase activation, interleukin maturation and release, and pyroptosis in an NLRC4-dependent manner (Kofoed & Vance, 2011; Zhao et al, 2011; Von Moltke et al, 2013).
The RGA4/RGA5 pair resembles in many instances to the mammalian NLRC4/NAIP system and adds thus one more piece to the striking similarities between plant and animal NB-LRR proteins in domain architectures and molecular and biological functions. These similarities appear even more appealing if one considers that evolutionary analysis strongly suggests that mammalian and plant NB-LRR proteins appeared independently by convergent evolution in plants and animals (Ausubel, 2005; Yue et al, 2012). The frequent observation of NB-LRR pairs in mammals and plants may thus indicate that interaction of two different NB-LRR proteins is a fundamental element of NB-LRR protein function.
Materials and Methods
Growth of plants
For leaf protoplast preparation, etiolated rice plants (Oryza sativa L.) were grown as described (Chen et al, 2006). Faivre-Rampant et al (2008) Nicotiana benthamiana plants were grown in a growth chamber at 22°C with a 16-h light period.
Constructs
All PCR products used for cloning were generated using Phusion High-Fidelity DNA Polymerase (Finnzymes) or PrimeSTAR GLX DNA polymerase (Takara Bio) using primers listed in Supplementary Table S1. Molecular cloning was performed by restriction-ligation, Gateway recombination (Invitrogen) or Quickchange site-directed Mutagenesis (Stratagene) as detailed in Supplementary Table S2. For yeast two-hybrid analysis, pGBKT7-BD and pGADT7-AD (Clontech) or Gateway-compatible vectors derived from pGADT7 and pGBKT7 were used (Bernoux et al, 2011b). For the creation of Gateway entry clones, pDONR207 or pENTR D-TOPO (Invitrogen) were used. For agro infiltration experiments, pBIN19-35S::GTW:3HA, pBIN19-35S::YFP:GTW, pBIN19-35S::GTW:CFP, pBIN19-35S::HA:GTW (Cesari et al, 2013), and pMCD83-35S::GTW:GFP (Curtis & Grossniklaus, 2003) were used. Constructs used for protoplast transfection were generated with pAHC17-pUbi (Yoshida et al, 2009; Okuyama et al, 2011), p35s:GTW (Takahashi et al, 2009), p35s::GTW:GFP (Akamatsu et al, 2013), p35s::Venus:GTW (Kawano et al, 2010), pGII pUbi::GW-T7, and pGII pUbi::10xMyc-GW (Taoka et al, 2011). Constructs used for RNAi in rice protoplasts were generated in pENTR D-TOPO (Invitrogen) and inserted in pPANDA (Miki & Shimamoto, 2014) by LR gateway reaction. Details of constructs and primers are given in Supplementary Tables S1 and S2.
Yeast two-hybrid assays
BD or AD constructs used in Fig6A and Supplementary Fig S10 were transformed into the yeast strain AH109. Co-transformants were plated on synthetic medium lacking leucine, tryptophan, adenine, and histidine (supplemented with 5-bromo-4-chloro-3-indolyl-D-galactopyranoside) and incubated at 28°C for 3 days.
AD or BD fusions of RGA51–133, RGA51–167, RGA51–228, RGA41–130, RGA41–164, RGA41–229, and Sr331–225 constructs used in Fig6B and Supplementary Fig S11 were transformed into HF7c yeast strain as described in the Yeast Protocols Handbook (Clontech).
Transient protein expression and cell death assays in N. benthamiana
For Agrobacterium-mediated N. benthamiana leaf transformations, transformed GV3101-pMP90 strains were grown in Luria-Bertani liquid medium containing 50 mg/ml rifampicin, 15 mg/ml gentamycin, and 25 mg/ml kanamycin at 28°C for 24 h. Bacteria were harvested by centrifugation, resuspended in infiltration medium (10 mM MES pH 5.6, 10 mM MgCl2, 150 μM acetosyringone) to an OD600 of 1, and incubated for 2 h at room temperature before leaf infiltration. The infiltrated plants were incubated in growth chambers under controlled conditions for co-immunoprecipitation experiments and cell death assays, respectively. For documentation of cell death, leaves were photographed or scanned with a Typhoon FLA9000 fluorescence scanner (GE Healthcare) with excitation at 635 nm and use of a long-pass red filter (LPR-665 nm) 3–5 days after infiltration.
Confocal microscopy
Protoplast isolation from rice Oc suspension culture and protoplast transformation using polyethylene glycol (PEG) method were performed as described (Chen et al, 2006). 2.5 μg of RGA4 or 5 μg of RGA5 expression vector with 2 μg of subcellular localization markers was mixed with 0.1 ml of aliquots of 2 × 106 cells. After incubation from 16 to 22 h at 30°C, the protoplasts were observed under a confocal microscope (FV1000; Olympus). Spectral settings for detection of each fluorescence proteins were as follows: CFP; excitation, 458 nm, detection, 475–500 nm, GFP; excitation, 488 nm, detection, 500–525 nm, Venus; excitation, 515 nm, detection, 530–560 nm, mOrange; excitation, 543 nm, detection, 555–600 nm, mCherry; excitation, 543 nm, detection, 610–650 nm.
Protein extraction, Western blot and co-immunoprecipitation
Yeast protein extraction for immunoblotting analysis was performed following a post-alkaline extraction method (Kushnirov, 2000). Protein fusion detection was performed using anti-HA-HRP (Roche, clone 3F10) and anti-Myc (Roche, clone 9E10) antibodies.
Protein extracts of N. benthamiana leaves and related immunoblotting and co-IP experiments were performed as described (Cesari et al, 2013).
For rice protoplast co-immunoprecipitation assays, a large-scale protoplast transformation procedure was used. DNA plasmids (50 μg of pUbi::RGA4-T7 and 250 μg of pUbi::10xMyc-RGA5 or 10 μg of pUbi::10xMyc-GFP) were mixed with 1 ml of aliquots of 5 × 106 cells in each sample (Chen et al, 2006). Protein extract of rice protoplast was prepared in protein extraction buffer (50 mM Tris, pH 7.5, 2 mM EDTA, 150 mM NaCl, 5 mM MgCl2, 10% glycerol, 1% (w/v) Triton X-100, phosphatase inhibitor cocktail 2, phosphatase inhibitor cocktail 3 and 1× cOmplete, EDTA free or 1× cOmplete ULTRA, EDTA free). Cell debris was removed by centrifugation, and extracts were immunoprecipitated by using the μMacs c-Myc isolation kits and μMacs GFP isolation kits (Miltenyi Biotec). Samples were separated by SDS-PAGE and subjected to immunoblotting using anti-c-Myc antibody (Nacalai Tesque, Inc.), anti-T7 antibody (MBL) or anti-GFP antibody (Abcam), and secondary antibody conjugated to horseradish peroxidase (GE Healthcare).
Cell viability assay in rice protoplasts
Rice protoplasts were prepared as described previously from rice-etiolated seedlings of cultivars Himenomochi (Yoshida et al, 2009) or rice Oc cells (Chen et al, 2006). The firefly luciferase gene (LUC) expressed under the control of the maize ubiquitin promoter was used as a reporter to monitor protoplasts viability. Specified combinations of constructs were co-transfected together with the LUC plasmid into rice protoplasts by electroporation or polyethylene glycol (PEG) method (Chen et al, 2006; Yoshida et al, 2009). Luciferase activity was measured 40 h after transfection using a luciferase assay system (Promega), and the reduction in luminescence was compared with a control comprising empty or Venus expression vector-transfected protoplasts.
Acknowledgments
We thank Loic Fontaine for technical assistance. SC was supported by the program ‘contrat jeune scientifique’ of INRA. TF was supported by a fellowship from the Japanese Society for the Promotion of Science. RT was supported by the Program for Promotion of Basic and Applied Researches for Innovations in Bio-oriented Industry, Japan, by Grant-in-aid for Scientific Research from the Ministry of Education, Cultures, Sports and Technology, Japan (Grant-in-Aid for Scientific Research on Innovative Areas, 23113009) and by JSPS KAKENHI (24248004). HK was supported by JSPS KAKENHI (23580065). We acknowledge support by COSTAction SUSTAIN FA1208.
Author contributions
SC, HK, TF, RT, and TK designed the research. SC, HK, TF, MB, VC, and TK performed the research. SC, HK, TF, MB, RT, PD, and TK analyzed the data. SC and TK wrote the paper.
Conflict of interest
The authors declare that they have no conflict of interest.
Supplementary information for this article is available online: http://emboj.embopress.org
References
- Ade J, DeYoung BJ, Golstein C, Innes RW. Indirect activation of a plant nucleotide binding site–leucine-rich repeat protein by a bacterial protease. Proc Natl Acad Sci USA. 2007;104:2531. doi: 10.1073/pnas.0608779104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akamatsu A, Wong HL, Fujiwara M, Okuda J, Nishide K, Uno K, Imai K, Umemura K, Kawasaki T, Kawano Y, Shimamoto K. An OsCEBiP/OsCERK1-OsRacGEF1-OsRac1 module is an essential early component of chitin-induced rice immunity. Cell Host Microbe. 2013;13:465–476. doi: 10.1016/j.chom.2013.03.007. [DOI] [PubMed] [Google Scholar]
- Ashikawa I, Hayashi N, Yamane H, Kanamori H, Wu J, Matsumoto T, Ono K, Yano M. Two adjacent nucleotide-binding site-leucine-rich repeat class genes are required to confer pikm-specific rice blast resistance. Genetics. 2008;180:2267–2276. doi: 10.1534/genetics.108.095034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel FM. Are innate immune signaling pathways in plants and animals conserved? Nat Immunol. 2005;6:973–979. doi: 10.1038/ni1253. [DOI] [PubMed] [Google Scholar]
- Baba A, Hasezawa S, Syōno K. Cultivation of rice protoplasts and their transformation mediated by agrobacterium spheroplasts. Plant Cell Physiol. 1986;27:463–471. [Google Scholar]
- Bai S, Liu J, Chang C, Zhang L, Maekawa T, Wang Q, Xiao W, Liu Y, Chai J, Takken FLW, Schulze-Lefert P, Shen Q-H. Structure-function analysis of barley NLR immune receptor MLA10 reveals its cell compartment specific activity in cell death and disease resistance. PLoS Pathog. 2012;8:e1002752. doi: 10.1371/journal.ppat.1002752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendahmane A, Farnham G, Moffett P, Baulcombe DC. Constitutive gain-of-function mutants in a nucleotide binding site–leucine rich repeat protein encoded at the Rx locus of potato. Plant J. 2002;32:195–204. doi: 10.1046/j.1365-313x.2002.01413.x. [DOI] [PubMed] [Google Scholar]
- Bernoux M, Ellis JG, Dodds PN. New insights in plant immunity signaling activation. Curr Opin Plant Biol. 2011a;14:512–518. doi: 10.1016/j.pbi.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernoux M, Ve T, Williams S, Warren C, Hatters D, Valkov E, Zhang X, Ellis JG, Kobe B, Dodds PN. Structural and functional analysis of a plant resistance protein TIR domain reveals interfaces for self-association, signaling, and autoregulation. Cell Host Microbe. 2011b;9:200–211. doi: 10.1016/j.chom.2011.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomblies K, Lempe J, Epple P, Warthmann N, Lanz C, Dangl JL, Weigel D. Autoimmune response as a mechanism for a dobzhansky-muller-type incompatibility syndrome in plants. PLoS Biol. 2007;5:e236. doi: 10.1371/journal.pbio.0050236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonardi V, Tang S, Stallmann A, Roberts M, Cherkis K, Dangl JL. Expanded functions for a family of plant intracellular immune receptors beyond specific recognition of pathogen effectors. Proc Natl Acad Sci U S A. 2011;108:16463–16468. doi: 10.1073/pnas.1113726108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brotman Y, Normantovich M, Goldenberg Z, Zvirin Z, Kovalski I, Stovbun N, Doniger T, Bolger AM, Troadec C, Bendahmane A, Cohen R, Katzir N, Pitrat M, Dogimont C, Perl-Treves R. Dual resistance of melon to Fusarium oxysporum races 0 and 2 and to papaya ring-spot virus is controlled by a pair of head-to-head-oriented NB-LRR genes of unusual architecture. Mol Plant. 2012;6:235–238. doi: 10.1093/mp/sss121. [DOI] [PubMed] [Google Scholar]
- Cesari S, Thilliez G, Ribot C, Chalvon V, Michel C, Jauneau A, Rivas S, Alaux L, Kanzaki H, Okuyama Y, Morel J-B, Fournier E, Tharreau D, Terauchi R, Kroj T. The rice resistance protein pair RGA4/RGA5 recognizes the Magnaporthe oryzae effectors AVR-Pia and AVR1-CO39 by direct binding. Plant Cell. 2013;25:1463–1481. doi: 10.1105/tpc.112.107201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Tao L, Zeng L, Vega-Sanchez ME, Umemura K, Wang G-L. A highly efficient transient protoplast system for analyzing defence gene expression and protein? Protein interactions in rice. Mol Plant Pathol. 2006;7:417–427. doi: 10.1111/j.1364-3703.2006.00346.x. [DOI] [PubMed] [Google Scholar]
- Collier SM, Moffett P. NB-LRRs work a “bait and switch” on pathogens. Trends Plant Sci. 2009;14:521–529. doi: 10.1016/j.tplants.2009.08.001. [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 2003;133:462–469. doi: 10.1104/pp.103.027979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl JL, Jones JDG. Plant pathogens and integrated defence responses to infection. Nature. 2001;411:826–833. doi: 10.1038/35081161. [DOI] [PubMed] [Google Scholar]
- Dangl JL, Horvath DM, Staskawicz BJ. Pivoting the plant immune system from dissection to deployment. Science. 2013;341:746–751. doi: 10.1126/science.1236011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La Fuente van Bentem S, Vossen JH, Vries KJ, Wees S, Tameling WIL, Dekker HL, Koster CG, Haring MA, Takken FLW, Cornelissen BJC. Heat shock protein 90 and its co-chaperone protein phosphatase 5 interact with distinct regions of the tomato I-2 disease resistance protein. Plant J. 2005;43:284–298. doi: 10.1111/j.1365-313X.2005.02450.x. [DOI] [PubMed] [Google Scholar]
- Dean R, Van Kan JAL, Pretorius ZA, Hammond-Kosack KE, Di Pietro A, Spanu PD, Rudd JJ, Dickman M, Kahmann R, Ellis J, Foster GD. The top 10 fungal pathogens in molecular plant pathology. Mol Plant Pathol. 2012;13:414–430. doi: 10.1111/j.1364-3703.2011.00783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deslandes L, Olivier J, Theulières F, Hirsch J, Feng DX, Bittner-Eddy P, Beynon J, Marco Y. Resistance to Ralstonia solanacearum in Arabidopsis thaliana is conferred by the recessive RRS1-R gene, a member of a novel family of resistance genes. Proc Natl Acad Sci U S A. 2002;99:2404–2409. doi: 10.1073/pnas.032485099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deslandes L, Olivier J, Peeters N, Feng DX, Khounlotham M, Boucher C, Somssich I, Genin S, Marco Y. Physical interaction between RRS1-R, a protein conferring resistance to bacterial wilt, and PopP2, a type III effector targeted to the plant nucleus. Proc Natl Acad Sci U S A. 2003;100:8024–8029. doi: 10.1073/pnas.1230660100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinesh-Kumar SP, Tham WH, Baker BJ. Structure–function analysis of the tobacco mosaic virus resistance gene N. Proc Natl Acad Sci U S A. 2000;97:14789. doi: 10.1073/pnas.97.26.14789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds PN, Rathjen JP. Plant immunity: towards an integrated view of plant–pathogen interactions. Nat Rev Genet. 2010;11:539–548. doi: 10.1038/nrg2812. [DOI] [PubMed] [Google Scholar]
- Eitas TK, Dangl JL. NB-LRR proteins: pairs, pieces, perception, partners, and pathways. Curr Opin Plant Biol. 2010;13:472–477. doi: 10.1016/j.pbi.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis JG, Lawrence GJ, Luck JE, Dodds PN. Identification of regions in alleles of the flax rust resistance gene L that determine differences in gene-for-gene specificity. Plant Cell. 1999;11:495–506. doi: 10.1105/tpc.11.3.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis JG, Dodds PN, Lawrence GJ. Flax rust resistance gene specificity is based on direct resistance-avirulence protein interactions. Annu Rev Phytopathol. 2007;45:289–306. doi: 10.1146/annurev.phyto.45.062806.094331. [DOI] [PubMed] [Google Scholar]
- Faivre-Rampant O, Thomas J, Allègre M, Morel J-B, Tharreau D, Nottéghem J-L, Lebrun M-H, Schaffrath U, Piffanelli P. Characterization of the model system rice-Magnaporthe for the study of nonhost resistance in cereals. New Phytol. 2008;180:899–910. doi: 10.1111/j.1469-8137.2008.02621.x. [DOI] [PubMed] [Google Scholar]
- Frost D, Way H, Howles P, Luck J, Manners J, Hardham A, Finnegan J, Ellis J. Tobacco transgenic for the flax rust resistance gene L expresses allele-specific activation of defense responses. Mol Plant Microbe Interact. 2004;17:224–232. doi: 10.1094/MPMI.2004.17.2.224. [DOI] [PubMed] [Google Scholar]
- Gao Z, Chung E-H, Eitas TK, Dangl JL. Plant intracellular innate immune receptor resistance to Pseudomonas syringae pv. maculicola 1 (RPM1) is activated at, and functions on, the plasma membrane. Proc Natl Acad Sci USA. 2011;108:7619–7624. doi: 10.1073/pnas.1104410108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassmann W, Hinsch ME, Staskawicz BJ. The Arabidopsis RPS4 bacterial-resistance gene is a member of the TIR-NBS-LRR family of disease-resistance genes. Plant J. 1999;20:265–277. doi: 10.1046/j.1365-313x.1999.t01-1-00600.x. [DOI] [PubMed] [Google Scholar]
- Gutierrez JR, Balmuth AL, Ntoukakis V, Mucyn TS, Gimenez-Ibanez S, Jones AME, Rathjen JP. Prf immune complexes of tomato are oligomeric and contain multiple Pto-like kinases that diversify effector recognition. Plant J. 2010;61:507–518. doi: 10.1111/j.1365-313X.2009.04078.x. [DOI] [PubMed] [Google Scholar]
- Howles P, Lawrence G, Finnegan J, McFadden H, Ayliffe M, Dodds P, Ellis J. Autoactive alleles of the flax L6 rust resistance gene induce non-race-specific rust resistance associated with the hypersensitive response. Mol Plant Microbe Interact. 2005;18:570–582. doi: 10.1094/MPMI-18-0570. [DOI] [PubMed] [Google Scholar]
- Jia Y, McAdams SA, Bryan GT, Hershey HP, Valent B. Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J. 2000;19:4004–4014. doi: 10.1093/emboj/19.15.4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JDG, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- Kanzaki H, Yoshida K, Saitoh H, Fujisaki K, Hirabuchi A, Allaux L, Fournier E, Tharreau D, Terauchi R. Arms race co-evolution of Magnaporthe oryzae AVR-Pik and rice Pik genes driven by their physical interactions. Plant J. 2012;72:894–907. doi: 10.1111/j.1365-313X.2012.05110.x. [DOI] [PubMed] [Google Scholar]
- Kawano Y, Akamatsu A, Hayashi K, Housen Y, Okuda J, Yao A, Nakashima A, Takahashi H, Yoshida H, Wong HL, Kawasaki T, Shimamoto K. Activation of a Rac GTPase by the NLR family disease resistance protein pit plays a critical role in rice innate immunity. Cell Host Microbe. 2010;7:362–375. doi: 10.1016/j.chom.2010.04.010. [DOI] [PubMed] [Google Scholar]
- Kofoed EM, Vance RE. Innate immune recognition of bacterial ligands by NAIPs determines inflammasome specificity. Nature. 2011;477:592–595. doi: 10.1038/nature10394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasileva KV, Dahlbeck D, Staskawicz BJ. Activation of an Arabidopsis resistance protein is specified by the in planta association of its leucine-rich repeat domain with the cognate oomycete effector. Plant Cell. 2010;22:2444–2458. doi: 10.1105/tpc.110.075358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushnirov VV. Rapid and reliable protein extraction from yeast. Yeast. 2000;16:857–860. doi: 10.1002/1097-0061(20000630)16:9<857::AID-YEA561>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Lee S-K, Song M-Y, Seo Y-S, Kim H-K, Ko S, Cao P-J, Suh J-P, Yi G, Roh J-H, Lee S, An G, Hahn T-R, Wang G-L, Ronald P, Jeon J-S. Rice Pi5-mediated resistance to Magnaporthe oryzae requires the presence of two coiled-coil-nucleotide-binding-leucine-rich repeat genes. Genetics. 2009;181:1627–1638. doi: 10.1534/genetics.108.099226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leister RT. Molecular genetic evidence for the role of SGT1 in the intramolecular complementation of Bs2 protein activity in Nicotiana benthamiana. Plant Cell. 2005;17:1268–1278. doi: 10.1105/tpc.104.029637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Wang X, Mitchell T, Hu Y, Liu X, Dai L, Wang G-L. Recent progress and understanding of the molecular mechanisms of the rice-Magnaporthe oryzae interaction. Mol Plant Pathol. 2010;11:419–427. doi: 10.1111/j.1364-3703.2009.00607.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loutre C, Wicker T, Travella S, Galli P, Scofield S, Fahima T, Feuillet C, Keller B. Two different CC-NBS-LRR genes are required for Lr10-mediated leaf rust resistance in tetraploid and hexaploid wheat. Plant J. 2009;60:1043–1054. doi: 10.1111/j.1365-313X.2009.04024.x. [DOI] [PubMed] [Google Scholar]
- Maekawa T, Cheng W, Spiridon LN, Töller A, Lukasik E, Saijo Y, Liu P, Shen Q-H, Micluta MA, Somssich IE, Takken FLW, Petrescu A-J, Chai J, Schulze-Lefert P. Coiled-coil domain-dependent homodimerization of intracellular barley immune receptors defines a minimal functional module for triggering cell death. Cell Host Microbe. 2011;9:187–199. doi: 10.1016/j.chom.2011.02.008. [DOI] [PubMed] [Google Scholar]
- Mestre P, Baulcombe DC. Elicitor-mediated oligomerization of the tobacco N disease resistance protein. Plant Cell. 2006;18:491–501. doi: 10.1105/tpc.105.037234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki D, Shimamoto K. Simple RNAi vectors for stable and transient suppression of gene function in rice. Plant Cell Physiol. 2014;45:490–495. doi: 10.1093/pcp/pch048. [DOI] [PubMed] [Google Scholar]
- Monaghan J, Zipfel C. Plant pattern recognition receptor complexes at the plasma membrane. Curr Opin Plant Biol. 2012;15:349–357. doi: 10.1016/j.pbi.2012.05.006. [DOI] [PubMed] [Google Scholar]
- Narusaka M, Shirasu K, Noutoshi Y, Kubo Y, Shiraishi T, Iwabuchi M, Narusaka Y. RRS1 and RPS4 provide a dual resistance-gene system against fungal and bacterial pathogens. Plant J. 2009;60:218–226. doi: 10.1111/j.1365-313X.2009.03949.x. [DOI] [PubMed] [Google Scholar]
- Okuyama Y, Kanzaki H, Abe A, Yoshida K, Tamiru M, Saitoh H, Fujibe T, Matsumura H, Shenton M, Galam DC, Undan J, Ito A, Sone T, Terauchi R. A multifaceted genomics approach allows the isolation of the rice Pia-blast resistance gene consisting of two adjacent NBS-LRR protein genes. Plant J. 2011;66:467–479. doi: 10.1111/j.1365-313X.2011.04502.x. [DOI] [PubMed] [Google Scholar]
- Pennisi E. Armed and dangerous. Science. 2010;327:804–805. doi: 10.1126/science.327.5967.804. [DOI] [PubMed] [Google Scholar]
- Periyannan S, Moore J, Ayliffe M, Bansal U, Wang X, Huang L, Deal K, Luo M, Kong X, Bariana H, Mago R, McIntosh R, Dodds P, Dvorak J, Lagudah E. The gene Sr33, an ortholog of barley Mla genes, encodes resistance to wheat stem rust race Ug99. Science. 2013;341:786–788. doi: 10.1126/science.1239028. [DOI] [PubMed] [Google Scholar]
- Riedl SJ, Li W, Chao Y, Schwarzenbacher R, Shi Y. Structure of the apoptotic protease-activating factor 1 bound to ADP. Nature. 2005;434:926–932. doi: 10.1038/nature03465. [DOI] [PubMed] [Google Scholar]
- Roberts M, Tang S, Stallmann A, Dangl JL, Bonardi V. Genetic requirements for signaling from an autoactive plant NB-LRR intracellular innate immune receptor. PLoS Genet. 2013;9:e1003465. doi: 10.1371/journal.pgen.1003465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinapidou E, Williams K, Nott L, Bahkt S, Tör M, Crute I, Bittner-Eddy P, Beynon J. Two TIR:NB:LRR genes are required to specify resistance to Peronospora parasitica isolate Cala2 in Arabidopsis. Plant J. 2004;38:898–909. doi: 10.1111/j.1365-313X.2004.02099.x. [DOI] [PubMed] [Google Scholar]
- Slootweg EJ, Spiridon LN, Roosien J, Butterbach P, Pomp R, Westerhof L, Wilbers R, Bakker E, Bakker J, Petrescu A-J, Smant G, Goverse A. Structural determinants at the interface of the ARC2 and leucine-rich repeat domains control the activation of the plant immune receptors Rx1 and Gpa2. Plant Physiol. 2013;162:1510–1528. doi: 10.1104/pp.113.218842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiderski MR, Birker D, Jones JDG. The TIR domain of TIR-NB-LRR resistance proteins is a signaling domain involved in cell death induction. Mol Plant Microbe Interact. 2009;22:157–165. doi: 10.1094/MPMI-22-2-0157. [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Teshima KM, Yokoi S, Innan H, Shimamoto K. Expression levels contribute to diversity of flowering time in cultivated rice. Proc Natl Acad Sci U S A. 2009;106:4555–4560. doi: 10.1073/pnas.0812092106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takken FL, Goverse A. How to build a pathogen detector: structural basis of NB-LRR function. Curr Opin Plant Biol. 2012;15:375–384. doi: 10.1016/j.pbi.2012.05.001. [DOI] [PubMed] [Google Scholar]
- Tameling WIL. The tomato R gene products I-2 and Mi-1 are functional ATP binding proteins with ATPase activity. Plant Cell. 2002;14:2929–2939. doi: 10.1105/tpc.005793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y, Yuan F, Leister RT, Ausubel FM, Katagiri F. Mutational analysis of the Arabidopsis nucleotide binding site–leucine-rich repeat resistance gene RPS2. Plant Cell. 2000;12:2541–2554. doi: 10.1105/tpc.12.12.2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taoka K, Ohki I, Tsuji H, Furuita K, Hayashi K, Yanase T, Yamaguchi M, Nakashima C, Purwestri YA, Tamaki S, Ogaki Y, Shimada C, Nakagawa A, Kojima C, Shimamoto K. 14-3-3 proteins act as intracellular receptors for rice Hd3a florigen. Nature. 2011;476:332–335. doi: 10.1038/nature10272. [DOI] [PubMed] [Google Scholar]
- Van der Hoorn RAL, Kamoun S. From guard to decoy: a new model for perception of plant pathogen effectors. Plant Cell. 2008;20:2009–2017. doi: 10.1105/tpc.108.060194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Ooijen G, Mayr G, Albrecht M, Cornelissen BJC, Takken FLW. Transcomplementation, but not physical association of the CC-NB-ARC and LRR domains of tomato R protein Mi-1.2 is altered by mutations in the ARC2 subdomain. Mol Plant. 2008a;1:401–410. doi: 10.1093/mp/ssn009. [DOI] [PubMed] [Google Scholar]
- Van Ooijen G, Mayr G, Kasiem MMA, Albrecht M, Cornelissen BJC, Takken FLW. Structure-function analysis of the NB-ARC domain of plant disease resistance proteins. J Exp Bot. 2008b;59:1383–1397. doi: 10.1093/jxb/ern045. [DOI] [PubMed] [Google Scholar]
- Von Moltke J, Ayres JS, Kofoed EM, Chavarría-Smith J, Vance RE. Recognition of bacteria by inflammasomes. Annu Rev Immunol. 2013;31:73–106. doi: 10.1146/annurev-immunol-032712-095944. [DOI] [PubMed] [Google Scholar]
- Wang C-IA, Guncar G, Forwood JK, Teh T, Catanzariti A-M, Lawrence GJ, Loughlin FE, Mackay JP, Schirra HJ, Anderson PA, Ellis JG, Dodds PN, Kobe B. Crystal structures of flax rust avirulence proteins AvrL567-A and -D reveal details of the structural basis for flax disease resistance specificity. Plant Cell. 2007;19:2898–2912. doi: 10.1105/tpc.107.053611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Richards J, Gross T, Druka A, Kleinhofs A, Steffenson B, Acevedo M, Brueggeman R. The rpg4-mediated resistance to wheat stem rust (Puccinia graminis) in barley (Hordeum vulgare) requires Rpg5, a second NBS-LRR gene, and an actin depolymerization factor. Mol Plant Microbe Interact. 2013;26:407–418. doi: 10.1094/MPMI-06-12-0146-R. [DOI] [PubMed] [Google Scholar]
- Williams SJ, Sornaraj P, deCourcy-Ireland E, Menz RI, Kobe B, Ellis JG, Dodds PN, Anderson PA. An autoactive mutant of the M flax rust resistance protein has a preference for binding ATP, whereas wild-type M protein binds ADP. Mol Plant Microbe Interact. 2011;24:897–906. doi: 10.1094/MPMI-03-11-0052. [DOI] [PubMed] [Google Scholar]
- Williams SJ, Sohn KH, Wan L, Bernoux M, Sarris PF, Segonzac C, Ve T, Ma Y, Saucet SB, Ericsson DJ, Casey LW, Lonhienne T, Winzor DJ, Zhang X, Coerdt A, Parker JE, Dodds PN, Kobe B, Jones JDG. Structural basis for assembly and function of a heterodimeric plant immune receptor. Science. 2014;344:299–303. doi: 10.1126/science.1247357. [DOI] [PubMed] [Google Scholar]
- Wirthmueller L, Zhang Y, Jones JDG, Parker JE. Nuclear accumulation of the Arabidopsis immune receptor RPS4 is necessary for triggering EDS1-dependent defense. Curr Biol. 2007;17:2023–2029. doi: 10.1016/j.cub.2007.10.042. [DOI] [PubMed] [Google Scholar]
- Yoshida K, Saitoh H, Fujisawa S, Kanzaki H, Matsumura H, Yoshida K, Tosa Y, Chuma I, Takano Y, Win J, Kamoun S, Terauchi R. Association genetics reveals three novel avirulence genes from the rice blast fungal pathogen Magnaporthe oryzae. Plant Cell. 2009;21:1573–1591. doi: 10.1105/tpc.109.066324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan B, Zhai C, Wang W, Zeng X, Xu X, Hu H, Lin F, Wang L, Pan Q. The Pik-p resistance to Magnaporthe oryzae in rice is mediated by a pair of closely linked CC-NBS-LRR genes. Theor Appl Genet. 2011;122:1017–1028. doi: 10.1007/s00122-010-1506-3. [DOI] [PubMed] [Google Scholar]
- Yue J-X, Meyers BC, Chen J-Q, Tian D, Yang S. Tracing the origin and evolutionary history of plant nucleotide-binding site-leucine-rich repeat (NBS-LRR) genes. New Phytol. 2012;193:1049–1063. doi: 10.1111/j.1469-8137.2011.04006.x. [DOI] [PubMed] [Google Scholar]
- Zhai C, Lin F, Dong Z, He X, Yuan B, Zeng X, Wang L, Pan Q. The isolation and characterization of Pik, a rice blast resistance gene which emerged after rice domestication. New Phytol. 2011;189:321–334. doi: 10.1111/j.1469-8137.2010.03462.x. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Wu Y, Gao M, Zhang J, Kong Q, Liu Y, Ba H, Zhou J, Zhang Y. Disruption of PAMP-induced MAP kinase cascade by a Pseudomonas syringae effector activates plant immunity mediated by the NB-LRR protein SUMM2. Cell Host Microbe. 2012;11:253–263. doi: 10.1016/j.chom.2012.01.015. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Yang J, Shi J, Gong Y-N, Lu Q, Xu H, Liu L, Shao F. The NLRC4 inflammasome receptors for bacterial flagellin and type III secretion apparatus. Nature. 2011;477:596–600. doi: 10.1038/nature10510. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.