Abstract
N-methyl-d-aspartate (NMDA) receptor function is modulated by several endogenous molecules, including zinc, polyamines, protons, and sulfated neurosteroids. Zinc, polyamines, and phenylethanolamines exert their respective modulatory effects by exacerbating or relieving tonic proton inhibition. Here, we report that pregnenolone sulfate (PS) uses a unique mechanism for enhancement of NMDA receptor function that is independent of the proton sensor. We identify a steroid modulatory domain, SMD1, on the NMDA receptor NR2B subunit that is critical for both PS enhancement and proton sensitivity. This domain includes the J/K helices in the S2 region of the glutamate recognition site and the fourth membrane transmembrane region (M4). A molecular model based on α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptor structure suggests that steroid modulatory domain 1 contributes residues to a hydrophobic pocket that is capable of accommodating PS. The results demonstrate that the J/K helices and the fourth membrane transmembrane region participate in transducing allosteric interactions induced by steroid and proton binding to their respective sites.
N-methyl-d-aspartate (NMDA) receptors mediate fast glutamatergic synaptic transmission, a core element of nervous system function, and are key loci for control of synaptic plasticity, learning and memory, and neuronal development. In particular, memory consolidation involves NMDA receptor-dependent synaptic reinforcement (1) and augmentation of N-methyl-d-aspartate receptor subtype 2B (NR2B) subunit expression leads to enhancement of learning and memory in mice (2). Abnormal activation of NMDA receptors may, however, be associated with certain acute and chronic neurological disorders, including neuropathic pain, stroke, and neurodegenerative diseases.
NMDA receptor function is known to be regulated pharmacologically, and in some cases physiologically, by endogenous molecules such as zinc, polyamines, protons, arachidonic acid, and sulfated neurosteroids, but the mechanism(s) by which modulators function to control transmitter-induced gating of ionotropic glutamate receptors (iGluRs) is unresolved. Moreover, the development of small molecule modulators provides a basis for the use of the NMDA receptor as a key target for future drug discovery (3, 4).
Pregnenolone sulfate (PS) is one of the most abundant neurosteroids synthesized de novo in the nervous system (5–7). In vivo administration of PS promotes the release of acetylcholine in the cerebral cortex and hippocampus (8, 9), and dopamine in the nucleus accumbens (10), whereas exogenous and endogenous alterations of PS correlates with changes in spatial recognition and cognitive functions (11–16). At the molecular level, PS enhances NMDA receptor function while inhibiting several other ligand gated ion channels, including γ-aminobutyric acid type A and α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptors (17–20).
The proton sensor of the NMDA receptor has been demonstrated to be a point of convergence for the actions of different modulators, including spermine, zinc, and redox agents (21–23). However, PS seems to use a distinct molecular mechanism to modulate receptor activity. PS, spermine, and arachidonic acid have been shown to regulate native NMDA receptors via distinct recognition sites; PS modulation is also not dependent on the redox state of the receptor (24). To define the mode of action of PS and to determine structural components at the NMDA receptor that are critical for PS modulation, we took advantage of our previous finding that PS differentially modulates activity of recombinant receptors containing different NR2 subunits (25). PS potentiates the response of recombinant NMDA receptors containing NR2A or NR2B subunits, while inhibiting the response of receptors containing NR2C or NR2D subunits. We constructed chimeric NR2 subunits assembled from fragments of NR2B and NR2D subunits and coexpressed them with NR1 subunits in Xenopus oocytes to explore the molecular basis for this subunit-dependent phenomenon of PS modulation.
In this report, we define the first steroid modulatory domain (SMD1) on the NMDA receptor NR2B subunit that mediates the selective potentiating effects of PS (20, 26). SMD1, which contains the intact J/K helices and contiguous fourth membrane (M4) transmembrane domain, also participates in controlling both proton sensitivity and spermine modulation demonstrating that SMD1 participates in the core mechanism of NMDA receptor modulation. SMD1 is conserved within the iGluR family, and the corresponding amino acid sequence is involved in regulating the gating kinetics and binding of a positive modulator, cyclothiazide, to the AMPA receptor (27). Our results strongly suggest that the mechanism of regulating channel gating is conserved in the iGluR family and provides an answer to the long standing question of whether neurosteroids exert their modulatory effects through a specific domain on the NMDA receptor.
Materials and Methods
Construction of Chimeric Subunits and Point Mutants. Chimeric subunits were generated by exchanging restriction fragments between the cDNAs of NR2B and NR2D subunits by using appropriate restriction endonucleases followed by subcloning and sequencing to confirm identity. Restriction sites were generated in both the NR2B and NR2D cDNAs by PCR-based bridging, by using Turbo-pfu DNA ploymerase (Stratagene) (28, 29).
Electrophysiology. Xenopus laevis (Nasco, Fort Atkinson, WI) oocytes were microinjected with mRNAs transcribed from cDNAs by using mMESSAGE mMACHINE High Yield Capped RNA Transcription kits (Ambion, Austin, TX). Each oocyte was injected with 0.5 ng of NR1-1a or NR1-1b in combination with 5 ng of WT or chimeric NR2 subunits. Oocytes were maintained in Barth's solution (25) at 18°C for 2–4 days before recording. Recordings were at 22–24°C by using an OC-725C two-electrode voltage clamp amplifier (Warner Instruments, Hamden, CT) at a holding potential (Vh) of –70 mV, except for spermine experiments (–20 mV) and ifenprodil experiments (–40 mV). Membrane currents were filtered at 1 kHz, digitized, and sampled with MacADIOS II at 100 Hz (GW Instruments, Somerville, MA). Data acquisition and perfusion was controlled via SuperScopeII (GW Instruments). Microelectrodes were fabricated with a programmed puller (Sutter Instruments, Novato, CA) from borosilicate glass capillaries and filled with 3 M KCl solution (electrode resistance = 1–3 MΩ). Oocytes were perfused continuously with Mg2+-free Ba2+-Ringer's solution (96 mM NaCl/2 mM KCl/1.8 mM BaCl2/5 mM Hepes/0.5% DMSO, pH 7.5), unless otherwise indicated. Drug application lasted 10 s, and wash lasted 60 s. NMDA (300 μM), glycine (50 μM), and steroids (100 μM) were used at near saturating concentrations. Application of NMDA and glycine produced a nondesensitizing current. Absolute currents elicited by agonists alone from receptors containing χ2, χ3, NR2BΔ*, χ4, and all of the NR2B mutants are comparable to those from the WT NR2B (>1 μA). Currents from χ1, χ5, χ6, and χ7 are similar to those from the WT NR2D (≈100–400 nA). Currents elicited from oocytes injected with only NR1-1a mRNA are significantly smaller (<10 nA). Differences in NMDA/glycine currents in the presence and the absence of the modulator are expressed as percentage changes {[(INMDA + glycine + modulator/INMDA + glycine) – 1] × 100%, where INMDA + glycine + modulator is the current elicited by NMDA, glycine and the modulator; INMDA + glycine is the current elicited by NMDA and glycine}. All data are presented as mean ± SEM.
Molecular Modeling. The model of a dimer of the extracellular S1/S2 domain of NR1-1a and NR2B were constructed by threading the sequences of the NMDA receptor subunits onto the crystal structure of the homodimer of the GluR2 subunit of the AMPA receptor complexed with cyclothiazide (Protein Data Bank ID 1LBC). Coordinates of the 1LBC were obtained from the Protein Data Bank. The sequences of NR1-1a and NR2B were aligned with the GluR2 template, and the backbone atoms of the sequence were assigned the corresponding coordinates from the crystal structure of the GluR2 by using the Homology Module of insight 2000 (Accelrys, San Diego). Loops to fill in the gaps between the NMDA receptor sequences and the template sequence were built and refined by using the autorotomer feature in the Biopolymer Module of insight 2000. The backbone atoms (C, Ca, N) of each NMDA receptor residue were tethered to the coordinates of corresponding residues in the GluR2 template with a force constant of 5 kcal Å–2, and the structure was optimized with the discover 2000 module of insight 2000. The PS molecule was placed in the general area where cyclothiazide was on the template. The model was then again optimized with discover 2000.
Data Analysis. Peak or steady-state current measurements were normalized and expressed as a fraction of the peak or steadystate control current measurements, which were performed before and after application of every single concentration of agonists or steroids. IC50s and Hill coefficients for proton and ifenprodil inhibition were derived from experimental dose–response data fitted by logistic equations. Student's t test was used as the statistical analysis to determine statistical significance.
Materials. The cDNAs of NR1 were provided by S. Nakanishi and the cDNAs of NR2B and NR2D were gifts from P. H. Seeburg (Max Planck Institute). All chemicals except 3α5βS were purchased from Sigma. The 3α5βS were purchased from Steraloids (Newport, RI). Oligonucleotides for PCR reactions were purchased from Oligos Etc. (Wilsonville, OR). Turbo-pfu DNA polymerase was purchased from Stratagene. Restriction enzymes were purchased from New England Biolabs.
Results
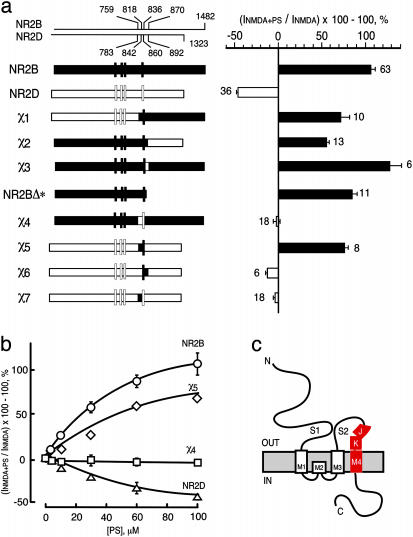
Identification of SMD1: Critical Residues for PS Potentiation Reside in the J/K Helices and/or M4 Domain of NR2B. To identify domains in NR2B that are responsible for differential responses of NMDA receptors to PS, a series of chimeric receptors were constructed by using NR2B and NR2D subunits. Replacing either the region C-terminal to residue S870 (χ1), N-terminal to residue S759 (χ2), or I836–S870 (χ3), of NR2B with the corresponding regions of NR2D does not alter polarity of modulation (Fig. 1a). These regions, therefore, do not determine the NR2B-specific response to PS. To test for posttranslational modification of the C-terminal domain, NR2B was truncated at residue 850 and alanine substituted for Y843, yielding NR2BΔ* (which lacks phosphorylation sites). The response of NR2BΔ*-containing receptors to PS is similar to that of WT, demonstrating that potentiation does not rely on NR2B phosphorylation. However, replacing the J/K helices and M4 domain (S759–I836) of NR2B with the equivalent region of NR2D (χ4) abolishes potentiation, indicating that residues necessary for PS potentiation reside in this 78-aa segment (Fig. 1). The EC50s of NMDA or glycine for receptors containing NR1-1a/χ4 are not significantly different from those of WT, showing that the reduction in PS potentiation is not due to a change in agonist potency. Transplanting (S759–I836) of NR2B into NR2D reverses the polarity of PS modulation from inhibition to potentiation. These “χ5”-containing receptors are similar to NR2B-containing receptors in terms of EC50 and Emax for PS modulation, whereas similar to NR2D in terms of EC50 and Emax for NMDA and glycine action. Therefore, the segment (S759–I836) of NR2B is not only necessary but also sufficient to confer potentiation by PS upon NMDA receptors, and we have termed this SMD1.
Fig. 1.
Identification of a steroid modulatory domain (SMD1) in NR2B. (a Left) A schematic representation of the WT NR2B, NR2D, and the NR2B/NR2D chimeras is displayed. Vertical bars correspond to the four hydrophobic domains. The contribution of NR2B and NR2D to chimeras is depicted in black and white, respectively. The scales at the top indicate the residue numbers in the WT subunits at junctions. Vertical bars represent the four hydrophobic membrane domains. (a Right) The percentage increase in the NMDA/glycine response (elicited by 300 μM NMDA and 50 μM glycine in oocytes expressing NR1-1a and NR2 subunits) in the presence of 100 μM PS is indicated. Error bars are SEMs. Numbers adjacent to the error bars indicate the number of oocytes used in the study. (b) Concentration–response curves of PS modulation for receptors containing NR2B (○), NR2D (▵), χ4(□), and χ5(⋄), were determined in the presence of saturating concentrations of NMDA (300 μM) and glycine (50 μM). The EC50 (NMDA) for NR1-1a/χ4 and NR1-1a/NR2B are both 22 ± 1 μM, and EC50 (glycine) is 0.3 ± 0.02 μM and 0.1 ± 0.02 μM, respectively. (c) The topological representation of the NR2B subunit and the location of the identified segment (colored in red) are depicted. Membrane domains are denoted as M1–M4. The amino terminus (N) is located on the extracellular side and the carboxyl terminus (C) on the intracellular side of the plasma membrane.
Dividing SMD1 into two parts at M818 (the first residue of M4) generated a χ6 (818–870) that contains NR2B M4 but not J/K, and a χ7 (759–818) that contains NR2B J/K but not M4. PS failed to potentiate the NMDA response of receptors containing either chimeric subunit, demonstrating that residues in both M4 and the 59-aa region containing J/K are required to confer PS potentiation.
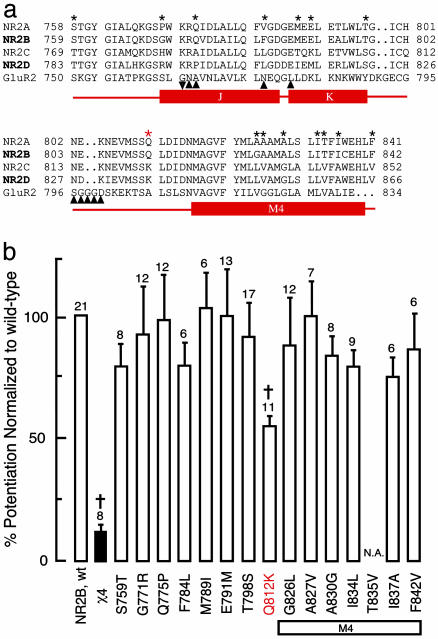
To identify the residues necessary for steroid modulation, the amino acid sequences for SMD1 in the four NR2 subunits were compared (Fig. 2a). SMD1 is highly conserved between subunits but significant differences exist. We clustered NR2 subunits into two groups (2A/2B and 2C/2D) according to their differential responses to PS. Ten residues are identical within groups but different between the two groups. Three residues are identical within one group but different in the other group. One residue is different in all subunits. Each NR2B residue was replaced by the equivalent residue from NR2D and evaluated for response to agonists (NMDA and glycine) and neurosteroid modulator (PS). Of the 15 site-directed single mutants tested, one mutant did not respond to agonists and 14 functional receptors responded to PS (Fig. 2c). Substituting NR2D lysine for NR2B glutamine at residue 812 (Q812K) significantly reduces the effect of PS (P < 0.01, Fig. 2c). However, the single residue substitution does not reproduce the phenotype of χ4-containing receptors. Thus, glutamine 812 in the J/K to M4 linker participates in PS potentiation in conjunction, either synergistically or additively, with at least one additional amino acid. This conclusion is further strengthened by the observation that substituting the corresponding residue of Q812 on χ6, K836, with glutamine could not convey PS potentiation as seen in χ5 chimeric receptors (data not shown).
Fig. 2.
Multiple residues in SMD1 participate in modulation of NMDA receptor function by PS. (a) Multiple sequence alignment of the identified segment in NR2A, NR2B, NR2C, and NR2D of the NMDA receptor and GluR2 of the AMPA receptor. Locations of J/K helices and the M4 domain are indicated by rectangular bars. Residues of GluR2 at RNA editing (▾) and the alternative splicing (▴) sites are marked underneath the GluR2 sequence. Residues subjected to site-directed mutagenesis are indicated by black stars (★). (b) The residues of NR2B (★) were replaced by the corresponding residues of NR2D. Responses of receptors containing the WT NR2B, χ4 and the single mutants to PS are presented as percentage increases in NMDA/glycine currents in the presence of 100 μM PS. All NR2 subunits were coexpressed with NR1-1a. Error bars are SEMs. Numbers adjacent to the error bars are the number of oocytes used in this study. †, Statistically significant reduction in PS potentiation (P < 0.05). Red star, position of Q812 that reduces PS potentiation by 45 ± 5%. N.A., No currents were recorded from oocytes injected with mRNA of NR1-1a and T835V NR2B mutant.
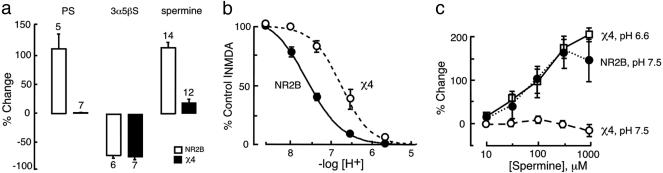
SMD1 Is Critical for Proton Sensitivity. To investigate whether SMD1 is a general control point for NMDA receptor modulation, we next compared modulatory effects of protons, spermine, and pregnanolone sulfate (3α5βS, an inhibitory neurosteroid) on chimeric NR1-1a/χ4 and WT NR1-1a/NR2B receptors.
The χ4 mutation reduces glycine-independent spermine modulation by 80%, which is comparable to its effect on PS, but 3α5βS inhibition remains unaltered (Fig. 3a), and the pKa for proton inhibition of mutant (χ4 containing) receptor is shifted 0.6 unit to the left (more acidic) (Fig. 3b). To ask whether the loss of spermine potentiation might be a consequence of reduced proton sensitivity, the effect of spermine at pH ≈ pKa for the proton sensor (pH 6.6 for NR1-1a/χ4 and pH 7.5 for NR1-1a/NR2B) was determined. At pH = 7.5, NR1-1a/χ4 receptors lack spermine modulation but modulation is entirely restored at pH 6.6 and is indistinguishable from WT receptor at pH 7.5 (Fig. 3c). Thus, mutating SMD1 of NR2B results in decreased PS potentiation, proton inhibition, and spermine potentiation. The apparent complete loss of spermine potentiation at neutral pH results from the diminished proton sensitivity of NR1-1a/χ4.
Fig. 3.
SMD1 is essential for positive and negative modulation by spermine and protons but not for negative modulation by 3α5βS. (a) Percentage change of NMDA/glycine current elicited by application of 300 μM NMDA plus 50 μM glycine in the presence of 100 μM PS, 100 μM3α5βS, or 100 μM spermine. (b) Proton inhibition concentration–response relationship for NMDA/glycine responses of receptors containing NR1-1a and NR2B (•, n = 10; pKa = 7.4 ± 0.1) or χ4(○, n = 10; pKa = 6.8 ± 0.2); P < 0.01. Responses are expressed as a percentage of the value at pH 9. (c) Spermine potentiation concentration–response curves for receptors containing NR1-1a and NR2B at pH 7.5 (•, n = 7), χ4atpH7.5 (○, n = 8), or χ4atpH6.6 (□, n = 8). Vh =–70 mV except in spermine modulation experiments where Vh = –20 mV. Error bars are SEMs. Numbers adjacent to the error bars are the number of oocytes used in this study.
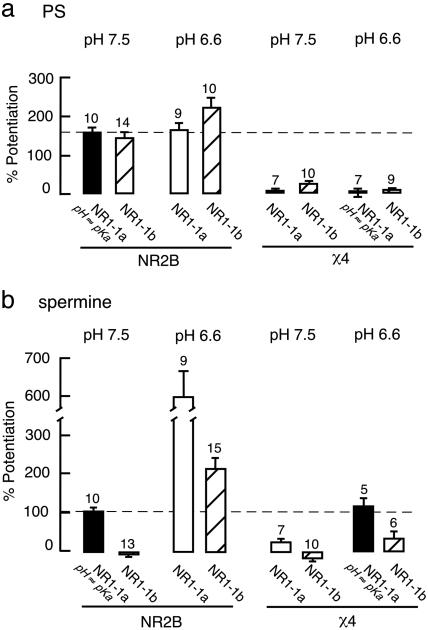
PS Potentiation and Proton Inhibition Are Functionally Independent. We next investigated whether PS potentiation is also protonsensor dependent as the glycine-independent form of spermine potentiation. The effect of PS on receptors containing different NR1 splice variants under different pH conditions was then compared. It has previously been shown that the spermine potentiation is reduced by the inclusion of the α-exon (exon 5) on the NR1 subunit, which acts as a “tethered” modulator by binding near the spermine recognition site (22, 30). The NR2B subunit was coexpressed with NR1-1a or NR1-1b (the isoform of NR1-1a containing the α-exon) to assess the influence of pH and the α-exon on the potentiating effect of PS. In contrast to the effect of spermine, the potentiating effect of PS showed little or no pH or α-exon dependency at WT NR2B-containing receptors (Fig. 4). Furthermore, we compared the ratio of the NMDA/glycine response of NR1-1a/NR2B at pH 6.6 vs. pH 7.5 in the absence and presence of 100 μM PS to determine whether PS would alter the sensitivity of the receptor to protons. The ratio serves as an indicator for the proton sensitivity. Any change in the proton sensitivity would be reflected by a change in the ratio. There is no difference between ratios measured in the absence (0.3 ± 0.05, n = 10) and presence of PS (0.3 ± 0.06, n = 6, P > 0.9), consistent with little or no functional interaction between PS and proton modulation.
Fig. 4.
The proton sensor does not control steroid modulation. Percentage change of NMDA/glycine currents elicited by 300 μM NMDA + 50 μM glycine to NR1-1a or NR1-1b with NR2B (Left) or χ4(Right) in the presence of 100 μM PS (a;Vh = –70 mV) or 100 μM spermine (b;Vh = –20 mV) at pH 7.5 or 6.6.
We then coexpressed χ4, the PS-insensitive NR2B chimera, with the NR1-1a and NR1-1b splice-variants and assessed the influence of α-exon and pH on the potentiating effects of spermine and PS. The prominent charge shielding effect (22) of the α-exon can be seen at pH 7.5 (pH = pKa for NR1-1a/NR2B) and 6.6 by virtue of the reduction in spermine enhancement of the NMDA response in NR1-1b/NR2B- vs. NR1-1a/NR2B-containing receptors. At pH 6.6 (pH = pKa for NR1-1a/χ4), spermine potentiation of NR1-1a/χ4 receptors equals that of NR1-1a/NR2B receptors at pH 7.5 (pH = pKa for NR1-1a/NR2B). Under these conditions, the α-exon reduces spermine potentiation with χ4 as well as with NR2B-containing receptors. Similarly, at pH 6.6, little or no effect of the χ4 mutation on inhibition of spermine potentiation by the α-exon is observed. In contrast to the results with spermine, the loss of PS potentiation with χ4 is pH and α-exon independent. The results demonstrate that PS and spermine modulate NMDA receptor activity via two different routes: Whereas PS uses a mechanism that is independent of the level of receptor protonation, spermine binding is coupled to gating through the proton sensor. Lastly, the results show that the tethered modulator α-exon does not interact with the PS site.
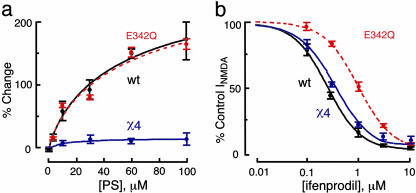
Several residues in the NR1 subunit are important structural components of the proton sensor contributing electronegativity either directly or indirectly to the sensor (21). A receptor with a defective proton sensor was constructed to test whether decreasing proton sensitivity through the NR1 subunit would eliminate PS modulation. One such mutant, NR1-1a(E342Q) that contains a single amino acid substitution at glutamate 342 of the NR1-1a subunit was constructed and the pKa for NR1-1a(E342Q)/NR2B-containing receptors is shifted to a more acidic value as compared to WT (pKa = 6.9 ± 0.1; Fig. 5b). However, the E342Q mutation has little effect on PS modulation (Fig. 5a).
Fig. 5.
Reducing proton sensitivity through mutation of the NR1-1a subunit at E342 does not reduce PS modulation. (a) NR1-1a(E342Q)/NR2B shows a WT response to PS. Concentration–response curves for PS positive modulation of NMDA receptor activity for NR1-1a/NR2B (black), NR1-1a/χ4 (purple), and NR1-1a(E342Q)/NR2B (red). Percentage increases in NMDA/glycine-elicited currents by PS are plotted as a function of the concentration of PS (n = 12–18). (b) NR1-1a/χ4 shows a WT response to ifenprodil. Concentration–response curves for ifenprodil inhibition of NMDA receptor activity for NR1-1a/NR2B (black), NR1-1a/χ4 (purple), and NR1-1a(E342Q)/NR2B (red). All points are normalized to NMDA/glycine currents in the absence of ifenprodil (n = 10–15). Oocytes were clamped at Vh = –40 mV to eliminate the voltage-dependent blockade by ifenprodil.
Similarly, ifenprodil, a noncompetitive NMDA receptor antagonist, selectively inhibits the activity of receptors containing the NR2B subunit by enhancing the sensitivity of the receptors to protons. Parallel changes in the pKa of the proton sensor and the IC50 of ifenprodil inhibition occur at recombinant receptors that contain mutations in the putative proton sensor on NR1-1a, suggesting that ifenprodil and proton inhibition share similar structural elements on the NR1 subunit (21).
Whereas the NR1-1a E342Q mutation reduces receptor sensitivity to ifenprodil, NR1-1a(E342Q)/NR2B has reduced sensitivity to ifenprodil (IC50 = 1 ± 0.1 μM, n = 4, Fig. 5c), the NR2B χ4 mutation is without effect on ifenprodil [IC50 (NR1-1a/χ4) = 0.4 ± 0.03 μM, n = 6; IC50 (NR1-1a/NR2B) = 0.3 ± 0.03 μM, n = 5; Fig. 5c]. The results demonstrate that whereas χ4, NR1 α-exon, and NR1 E342Q all reduce proton sensitivity, only NR2B χ4 eliminates PS modulation.
Discussion
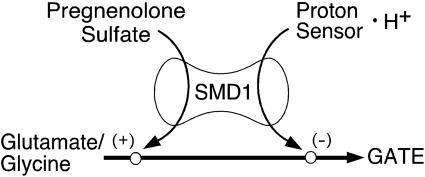
Fine-tuning of chemical synaptic transmission can be achieved in a number of ways, including allosteric modulation of neurotransmitter receptors. This is illustrated by the NMDA receptor, where the neurostransmitter glutamate and its coagonist glycine interact to provide synergistic activation of channel gating. In a parallel fashion, protons and spermine bind to distinct sites on the NMDA receptor and comodulate neurotransmitter induced gating through a mechanism of negative allosteric coupling wherein spermine enhances receptor activity by relieving proton inhibition. This “proton sensor” is a convergent point for the actions of additional modulators, such as zinc and ifenprodil, that inhibit channel activity by enhancing proton inhibition (21–23). In this study, we demonstrate that PS modulates NMDA receptor activity via an unique route that involves a steroid modulatory domain is independent of the level of inhibition by the proton sensor. The loss of both spermine and PS-positive modulation in receptors containing NR1-1a/χ4 (which lacks SMD1), however, suggests that PS potentiation and proton inhibition share a common downstream structural element(s) (Fig. 6). These elements must funnel the free energy of modulator binding to the channel gate.
Fig. 6.
Proposed model for SMD1-mediated allosteric modulation of agonist-induced gating of NR2B containing NMDA receptors. Diagram shows convergence of independent pathways for positive allosteric modulation by PS (+) and negative allosteric modulation by H+ (–) of neurotransmitter (glutamate and glycine)-induced gating of the NMDA receptor through SMD1. SMD1 spans the J/K helices and M4 regions of NR2B and is located near the glutamate-binding site.
The demonstration that SMD1 participates, whether directly or indirectly, in two functionally independent modes of modulation, PS potentiation and proton inhibition, suggests that this domain represents at least part of an allosteric modulatory module within the NR2B subunit. The identified segment on the NR2B subunit includes the J/K helices and M4 domains (Fig. 2b). The functional roles of this segment on the NMDA receptor have not been previously assigned, but its amino acid sequence is fairly conserved among the iGluRs. The same region on the AMPA receptor has been shown to be involved in regulating receptor gating kinetics. It also contains an RNA-editing site and all of the alternatively spliced (flip/flop) sites (32–36) (Fig. 2a). The crystal structure of the region, along with the glutamatebinding core (S1/S2 domain) of the GluR2 subunit of the AMPA receptor has been solved (37). The region contains two α-helices, J and K, which are located at the back of the bilobate structure of the glutamate binding site. The S1/S2 domain of the GluR2 subunit forms a dimer in a unit cell when crystallized. J/K helices reside at the dimerization interface between two subunits (27). The rearrangement of structures at this interface has been shown to be a mechanism for desensitization of the response of AMPA receptors (27), and an allosteric modulator, cyclothiazide, that binds to this interface promotes subunit dimer formation and alleviates desensitization (27).
In addition to playing an important role in modulating gating, J/K helices, along with membrane domains and an N-terminal domain of AMPA and kainate receptors, have been shown to be critical in determining the compatibility of subunits from different iGluR families in functional receptor assembly (38–40). Our results suggest that NMDA receptors may have adopted a similar strategy to regulate channel gating. The free energy of binding of protons and PS between NR1-1a and NR2B might be translated into a reorientation of the interface between subunits in the tetrameric receptor. Together with M4, the energy is then transferred to the channel gate. The recent identification of a prokaryotic iGluR (GluR0), which lacks M4, provides evidence that M4 is an evolutionally younger module in eukaryotic iGluRs and may play a modulatory role in regulating channel activity (41). M4 is not essential for gating the channel but couples the modulatory functions of the intracellular C-terminal domain to channel gating. Our finding that both J/K helices and M4 of the NR2B subunit are required to confer PS potentiation indicates that M4 is also critical in coupling allosteric modulation from extracelluar binding regions to the gating mechanism.
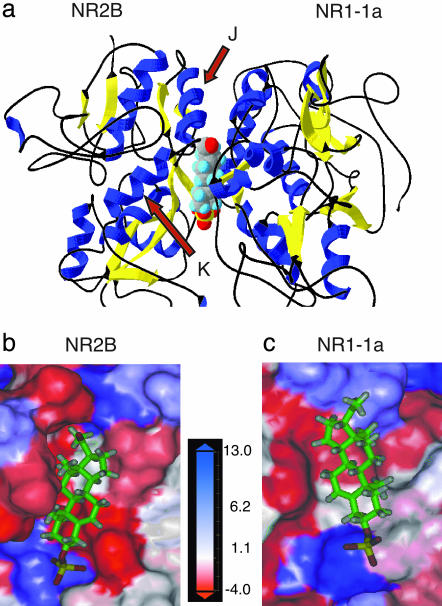
Perturbation of the dimer interface between subunits either by mutating residues at the interface or binding of cyclothiazide has been demonstrated to have marked effects on channel kinetics of GluR2 containing AMPA receptors (27). We have shown that modifying the amino acid sequence near the dimer interface on NR2B to that of NR2D eliminates positive modulation by PS and negative modulation by protons. To investigate whether a pocket also could be found at the interface between NMDA receptor subunits that could accommodate small molecules, molecular-modeling experiments were conducted by using the structure of the cyclothiazide-sensitive S1S2J construct as the template (27). Homology models were generated by threading the sequences of the NR1 and the NR2B subunits through the template (Fig. 7a).
Fig. 7.
Molecular modeling of a dimer comprising the S1/S2 domains of NR2B and NR1-1a reveals a potential binding pocket for PS. (a) The 3D model is depicted in a ribbon configuration with α helices colored in blue and β sheets colored in yellow. PS is docked at the interface between two subunits. Detailed views of the potential binding pocket for PS on NR2B (b) and NR1-1a (c) are illustrated. NR1-1a or NR2B is removed from the model to show the hydrophobic pocket on NR2B (b) or NR1-1a (c), respectively. The receptor surface is colored according to a hydrophobicity scale. Hydrophobic residues are colored in red and charged residues in blue. PS is depicted in a stick configuration and colored by the atom type with hydrogen in white, carbon in green, oxygen in red, and sulfur in yellow.
A hydrophobic pocket was defined between the NR1 and the NR2B subunit, which could complement the hydrophobic steroid core of PS. Docking the PS molecule into the pocket revealed residues that could contribute side chains to stabilize the binding of PS: V527, F529, T532, S759, I780, F784, and M789 from NR2B (Fig. 7b) and I519, F529, G757, R755, and F758 from NR1-1a (Fig. 7c). T532 and S759 from NR2B and R755 from NR1-1a are located within 5 Å next to the sulfate anion of PS and might form hydrogen bonds or salt bridges with the sulfate group, which is essential for activity (31). High-resolution crystal structure data and further mutagenesis studies are needed to validate the computer simulation of the proposed PS-binding site that we have generated; however, the model of a binding pocket at the subunit interface and its potential cognate ligand presents an opportunity to define an unique modulatory site on the receptor and to provide a new molecular target for the development of potential therapeutics that regulate NMDA receptor function.
Acknowledgments
We thank Dr. Terrell Gibbs for fruitful discussions and helpful comments and Dr. Stella Martin for advice. This work was supported by the National Institute of Mental Health Grant MH-49469.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: NMDA, N-methyl-d-aspartate; PS, pregnenolone sulfate; SMD1, first steroid modulatory domain; AMPA, α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid; iGluR, ionotropic glutamate receptor; M4, fourth membrane; NR2B, N-methyl-d-aspartate receptor subtype 2B.
References
- 1.Shimizu, E., Tang, Y. P., Rampon, C. & Tsien, J. Z. (2000) Science 290, 1170–1174. [DOI] [PubMed] [Google Scholar]
- 2.Tang, Y. P., Shimizu, E., Dube, G. R., Rampon, C., Kerchner, G. A., Zhuo, M., Liu, G. & Tsien, J. Z. (1999) Nature 401, 63–69. [DOI] [PubMed] [Google Scholar]
- 3.Weaver, C. E., Jr., Marek, P., Park-Chung, M., Tam, S. W. & Farb, D. H. (1997) Proc. Natl. Acad. Sci. USA 94, 10450–10454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kemp, J. A. & McKernan, R. M. (2002) Nat. Neurosci. 5, Suppl., 1039–1042. [DOI] [PubMed] [Google Scholar]
- 5.Baulieu, E. E., Robel, P. & Schumacher, M. (2001) Int. Rev. Neurobiol. 46, 1–32. [DOI] [PubMed] [Google Scholar]
- 6.Robel, P., Young, J., Corpechot, C., Mayo, W., Perche, F., Haug, M., Simon, H. & Baulieu, E. E. (1995) J. Steroid Biochem. Mol. Biol. 53, 355–360. [DOI] [PubMed] [Google Scholar]
- 7.Corpechot, C., Synguelakis, M., Talha, S., Axelson, M., Sjovall, J., Vihko, R., Baulieu, E. E. & Robel, P. (1983) Brain Res. 270, 119–125. [DOI] [PubMed] [Google Scholar]
- 8.Darnaudery, M., Koehl, M., Pallares, M., Le Moal, M. & Mayo, W. (1998) J. Neurochem. 71, 2018–2022. [DOI] [PubMed] [Google Scholar]
- 9.Darnaudery, M., Koehl, M., Piazza, P. V., Le Moal, M. & Mayo, W. (2000) Brain Res. 852, 173–179. [DOI] [PubMed] [Google Scholar]
- 10.Barrot, M., Vallee, M., Gingras, M. A., Le Moal, M., Mayo, W. & Piazza, P. V. (1999) Eur. J. Neurosci. 11, 3757–3760. [DOI] [PubMed] [Google Scholar]
- 11.Flood, J. F., Morley, J. E. & Roberts, E. (1992) Proc. Natl. Acad. Sci. USA 89, 1567–1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flood, J. F., Morley, J. E. & Roberts, E. (1995) Proc. Natl. Acad. Sci. USA 92, 10806–10810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mathis, C., Paul, S. M. & Crawley, J. N. (1994) Psychopharmacology 116, 201–206. [DOI] [PubMed] [Google Scholar]
- 14.Mathis, C., Vogel, E., Cagniard, B., Criscuolo, F. & Ungerer, A. (1996) Neuropharmacology 35, 1057–1064. [DOI] [PubMed] [Google Scholar]
- 15.Mathis, C., Meziane, H. & Ungerer, A. (1999) J. Soc. Biol. 193, 299–306. [PubMed] [Google Scholar]
- 16.Vallee, M., Mayo, W., Darnaudery, M., Corpechot, C., Young, J., Koehl, M., Le Moal, M., Baulieu, E. E., Robel, P. & Simon, H. (1997) Proc. Natl. Acad. Sci. USA 94, 14865–14870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bowlby, M. R. (1993) Mol. Pharmacol. 43, 813–819. [PubMed] [Google Scholar]
- 18.Yaghoubi, N., Malayev, A., Russek, S. J., Gibbs, T. T. & Farb, D. H. (1998) Brain Res. 803, 153–160. [DOI] [PubMed] [Google Scholar]
- 19.Park-Chung, M., Malayev, A., Purdy, R. H., Gibbs, T. T. & Farb, D. H. (1999) Brain Res. 830, 72–87. [DOI] [PubMed] [Google Scholar]
- 20.Wu, F. S., Gibbs, T. T. & Farb, D. H. (1991) Mol. Pharmacol. 40, 333–336. [PubMed] [Google Scholar]
- 21.Mott, D. D., Doherty, J. J., Zhang, S., Washburn, M. S., Fendley, M. J., Lyuboslavsky, P., Traynelis, S. F. & Dingledine, R. (1998) Nat. Neurosci. 1, 659–667. [DOI] [PubMed] [Google Scholar]
- 22.Traynelis, S. F., Hartley, M. & Heinemann, S. F. (1995) Science 268, 873–876. [DOI] [PubMed] [Google Scholar]
- 23.Traynelis, S. F., Burgess, M. F., Zheng, F., Lyuboslavsky, P. & Powers, J. L. (1998) J. Neurosci. 18, 6163–6175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park-Chung, M., Wu, F. S., Purdy, R. H., Malayev, A. A., Gibbs, T. T. & Farb, D. H. (1997) Mol. Pharmacol. 52, 1113–1123. [DOI] [PubMed] [Google Scholar]
- 25.Malayev, A., Gibbs, T. T. & Farb, D. H. (2002) Br. J. Pharmacol. 135, 901–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gibbs, T. T. & Farb, D. H. (2000) Sci. STKE 60, PE1. [DOI] [PubMed] [Google Scholar]
- 27.Sun, Y., Olson, R., Horning, M., Armstrong, N., Mayer, M. & Gouaux, E. (2002) Nature 417, 245–253. [DOI] [PubMed] [Google Scholar]
- 28.Ho, S. N., Hunt, H. D., Horton, R. M., Pullen, J. K. & Pease, L. R. (1989) Gene 77, 51–59. [DOI] [PubMed] [Google Scholar]
- 29.Williams, K. (1997) Biochem. J. 325, 289–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Durand, G. M., Bennett, M. V. & Zukin, R. S. (1993) Proc. Natl. Acad. Sci. USA 90, 6731–6735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weaver, C. E., Land, M. B., Purdy, R. H., Richards, K. G., Gibbs, T. T. & Farb, D. H. (2000) J. Pharmacol. Exp. Ther. 293, 747–754. [PubMed] [Google Scholar]
- 32.Borges, K. & Dingledine, R. (1998) Prog. Brain Res. 116, 153–170. [DOI] [PubMed] [Google Scholar]
- 33.Lomeli, H., Mosbacher, J., Melcher, T., Hoger, T., Geiger, J. R., Kuner, T., Monyer, H., Higuchi, M., Bach, A. & Seeburg, P. H. (1994) Science 266, 1709–1713. [DOI] [PubMed] [Google Scholar]
- 34.Sommer, B., Keinanen, K., Verdoorn, T. A., Wisden, W., Burnashev, N., Herb, A., Kohler, M., Takagi, T., Sakmann, B. & Seeburg, P. H. (1990) Science 249, 1580–1585. [DOI] [PubMed] [Google Scholar]
- 35.Partin, K. M., Bowie, D. & Mayer, M. L. (1995) Neuron 14, 833–843. [DOI] [PubMed] [Google Scholar]
- 36.Partin, K. M., Fleck, M. W. & Mayer, M. L. (1996) J. Neurosci. 16, 6634–6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Armstrong, N., Sun, Y., Chen, G. Q. & Gouaux, E. (1998) Nature 395, 913–917. [DOI] [PubMed] [Google Scholar]
- 38.Leuschner, W. D. & Hoch, W. (1999) J. Biol. Chem. 274, 16907–16916. [DOI] [PubMed] [Google Scholar]
- 39.Ayalon, G. & Stern-Bach, Y. (2001) Neuron 31, 103–113. [DOI] [PubMed] [Google Scholar]
- 40.Madden, D. R. (2002) Nat. Rev. Neurosci. 3, 91–101. [DOI] [PubMed] [Google Scholar]
- 41.Chen, G. Q., Cui, C., Mayer, M. L. & Gouaux, E. (1999) Nature 402, 817–821. [DOI] [PubMed] [Google Scholar]