Valdivia studies how the pathogen Chlamydia trachomatis interacts with its host cell.
Valdivia studies how the pathogen Chlamydia trachomatis interacts with its host cell.
Intracellular pathogens are capable of profoundly altering the biology of their host cell. For example, the bacterium Chlamydia trachomatis (the causative agent of a common sexually transmitted disease in humans) secretes proteins that transform a host cell vacuole into a protected enclave for bacterial reproduction.

Raphael Valdivia
PHOTO COURTESY OF DUKE UNIVERSITY PHOTOGRAPHY
Raphael Valdivia is fascinated by how Chlamydia manages this makeover (1) and how it alters host cell behavior to support bacterial reproduction (2). His lab has already identified some of the tricks the bacterium uses to modulate host cell lipid trafficking (3, 4) and cytoskeletal architecture (5) and has constructed a new approach to systematically interrogate the roles of its as-yet-uncharacterized genes. We called him at his office at Duke University to learn about his work and to hear about some recent transformations in his career.
CHOOSING A PATH
When did you first consider a career in the sciences?
I was born and raised in Lima, Peru, but I left before finishing high school and came to an international school in the United States, part of a school system called the United World Colleges. I got to meet people from all over the world, so that was a pretty transformative event for me.
“It’s like a candy store for the trainees in my lab right now.”
I was always good at science and numbers came easily to me, so I thought I would become a physician or agricultural engineer and then return to Peru at some point and live there. However, it was a time of civil unrest in Peru, so instead I ended up going to Cornell, where I fell in love with microbiology. I had a great laboratory research experience with Steve Winans studying the genetics of the plant pathogen Agrobacterium tumefaciens. But college was an age of confusion for me because, while science was easier for me than other subjects, I had also become very involved in experiential education and outdoor activities.
I was very torn about what path to take, so I took a year off to teach outdoor education. At the end of the year I was still undecided, so I applied to graduate schools just in case. I chose to go to Stanford. Yosemite was two-and-a-half hours away, so I figured I would work in the lab Monday through Friday and then go to Yosemite and climb during the weekend. That lasted three months before I realized it was not a sustainable path.
So you set your sights on the lab…
By that time, I had met my scientific hero, Stanley Falkow. Stanley stated that he was no longer taking new students, but after I did a rotation in his lab he agreed to make an exception and let me join. I later found out, of course, that I was not his last student and he made exceptions all the time. [Laughs]
GFP had just been discovered as a live reporter for gene expression, and I worked to adapt it as a meaningful way to measure bacterial gene expression within infected cells and tissues. In collaboration with Brendan Cormack, we retooled GFP to make it fold better and make it brighter so that it could work well in pathogenic bacteria like Salmonella. By my second or third year of graduate school we had this new bright GFP, which Clontech bought and is now sold as EGFP.
A GIFT
You probably had your pick of postdocs after that…
Pathogenic bacteria have elaborate secretion systems to deliver proteins directly into the cytoplasm of their host cell, and we had some inkling that, for Salmonella, these proteins were involved in modulating endosomal dynamics. I wanted to change direction a little and go to a place where I could learn all about endosome function.
By this time I had also become engaged to my wife, whom I had originally met in high school and had re-encountered in California. We wanted to stay in the area, so I went to Randy Schekman’s lab at Berkeley. I’d had a productive graduate career and received some prestigious postdoctoral fellowships, and I really thought I was God’s gift to science. But during my first month in Randy’s lab, I wrecked the ultracentrifuge.
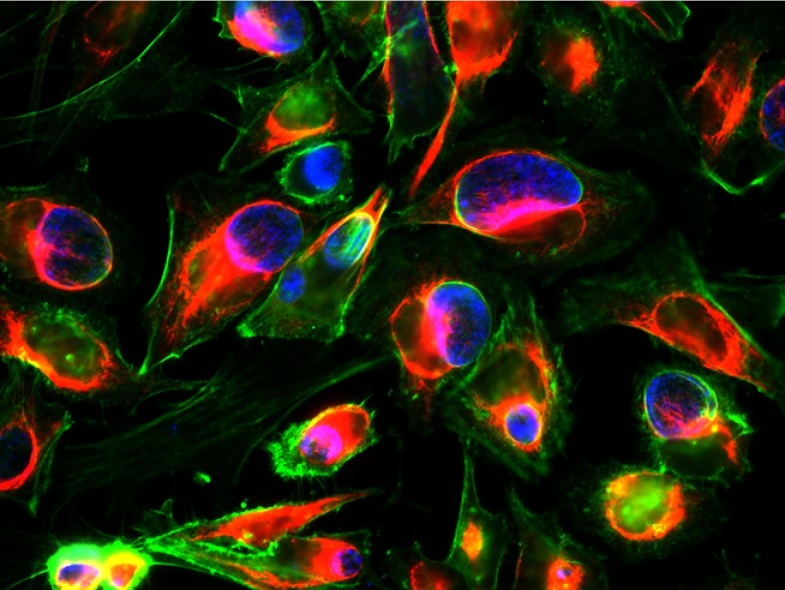
Chlamydia reorganizes the host cell cytoskeleton. Chlamydia inclusions (blue) are encased in a network of actin (green) and intermediate filaments (red).
IMAGE COURTESY OF MARCELA KOKES
How did you do that?
To this day, I have no idea. But in dealing with the aftermath of that, I took a very close inward look and assessed whether my failure to listen or my pride had led to such a disaster. It was a humbling experience, and it reset me in a much better frame of mind. I told myself, “I know nothing. I’m here to learn.”
Was your aim always to return to studying pathogenic bacteria?
Yes. Towards the end of my postdoc, I looked at the list of understudied pathogens, and the one that caught my eye was Chlamydia. It can only live in the context of a host cell, where it creates a unique membrane-bound compartment called an inclusion. Inclusions lack classical endomembrane markers, yet somehow the bacterium efficiently acquires host lipids. This seemed like a fascinating cell biological problem in a medically relevant biological system. It had been understudied because the pathogen lacked a system for molecular genetic manipulation, so the repertoire of tools we could use to study it was very limited.
I also had another problem in that all my postdoctoral training was in yeast. How was I going to land a job proposing to do things that I had not worked on? I proposed to use an expression system to essentially express individual Chlamydia proteins in yeast and ask whether this led to phenotypes consistent with disruption of membrane transport. At some of the places I interviewed I just got laughed off. Others were more open to the risk and to taking a gamble on me—that’s how I ended up at Duke.
AN INTERESTING PROBLEM
Your approach worked…
To our surprise, we found that some Chlamydia proteins targeted yeast lipid droplets, which are unusual organelles that were traditionally thought of as an inert cellular fat store. So the first thing we did was to look in mammalian cells to see whether these proteins target lipid droplets—and they do. We saw that during infection the lipid droplets proliferated in an infected cell, and in fact the bacteria were able to translocate some of those droplets into the lumen of the inclusion, presumably to fuel the creation of new membranes in dividing bacteria.
Frustrated by the inability to generate mutants defective for this process, we took a step back and tried to develop a system for genetics in Chlamydia. Despite the lack of molecular genetic tools, we could still create mutants the old-fashioned way with chemicals and then map mutations by whole-genome sequencing. We made a large library of mutants, sequenced them all, and in the process generated a platform for both forward and reverse genetics. With this approach we are examining the function of a cohort of proteins that the bacterium translocates after invasion as it begins to form an inclusion. More excitingly, we are performing multiple forward genetic screens to identify mutations that alter how the bacterium interacts with its host cell. It’s like a candy store for the trainees in my lab right now.
“We scientists do a very poor job communicating our work to the public.”
Finally, we are taking the lessons we learned from what was a “genetically intractable” organism to other bacteria that have been difficult to study, in particular components of the microbiome. We have a collaboration with an investigator here at Duke, John Rawls, who studies the microbial communities of the fish gut to try to tackle the genetic basis of how bacteria interact with their host and with each other.
Bouldering in Hanging Rock, North Carolina
PHOTO COURTESY OF RAPHAEL VALDIVIA
You were also recently appointed Vice Dean for Basic Science at Duke…
Two or three years ago, I got the opportunity to start the Duke Center for the Genomics of Microbial Systems. We brought together people from all over to build a new enterprise, and I had a lot of fun doing that. Around the same time I was becoming more preoccupied with questions beyond my lab’s research, finding myself increasingly concerned with the declining state of funding for research and science education. And it dawned on me that we scientists do a very poor job communicating our work to the public and especially to lawmakers. We think that the value of what we do is self-evident, but it is not. We need to be more effective in making the argument as to why the taxpayer should invest money in science. So in addition to helping implement new research initiatives and shaping biomedical training, I view this transition to administration as a chance to help change the culture and make us more effective advocates for research and science education.
Are you still an outdoors enthusiast?
Well, with a family and my new position, it’s harder to justify taking off for a long weekend or a week to go sleep on a cliff. Not only does it take away time from my family, but it’s also a risky activity. I still go climbing on plastic walls indoors, which I enjoy.
Climbing is like science. It’s a puzzle. You have to weave and find a route through, follow your instincts, and, if you make mistakes, you will fall. Then you correct them, try again, and learn along the way.
References
- 1.Spaeth, K.E., Chen Y.-S., and Valdivia R.H.. 2009. PLoS Pathog. 5:e1000579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen, Y.-S., et al. 2014. PLoS Pathog. 10:e1003954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kumar, Y., Cocchiaro J., and Valdivia R.H.. 2006. Curr. Biol. 16:1646–1651. [DOI] [PubMed] [Google Scholar]
- 4.Cocchiaro, J.L., et al. 2008. Proc. Natl. Acad. Sci. USA. 105:9379–9384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kumar, Y., and Valdivia R.H.. 2008. Cell Host Microbe. 4:159–169. [DOI] [PMC free article] [PubMed] [Google Scholar]