In Duchenne muscular dystrophy, asynchronous regeneration in microenvironments within muscle tissue results in development of fibrosis in lieu of global muscle recovery.
Abstract
We sought to determine the mechanisms underlying failure of muscle regeneration that is observed in dystrophic muscle through hypothesis generation using muscle profiling data (human dystrophy and murine regeneration). We found that transforming growth factor β–centered networks strongly associated with pathological fibrosis and failed regeneration were also induced during normal regeneration but at distinct time points. We hypothesized that asynchronously regenerating microenvironments are an underlying driver of fibrosis and failed regeneration. We validated this hypothesis using an experimental model of focal asynchronous bouts of muscle regeneration in wild-type (WT) mice. A chronic inflammatory state and reduced mitochondrial oxidative capacity are observed in bouts separated by 4 d, whereas a chronic profibrotic state was seen in bouts separated by 10 d. Treatment of asynchronously remodeling WT muscle with either prednisone or VBP15 mitigated the molecular phenotype. Our asynchronous regeneration model for pathological fibrosis and muscle wasting in the muscular dystrophies is likely generalizable to tissue failure in chronic inflammatory states in other regenerative tissues.
Introduction
Duchenne muscular dystrophy (DMD) is a relatively common inborn error, caused by lack of dystrophin in the membrane cytoskeleton of muscle fibers (Hoffman et al., 1987). Myofibers show increased plasma membrane instability and can undergo cell death (necrosis) either individually or in groups. Necrotic myofibers are able to regenerate within the preexisting basal lamina through satellite cell activation, myoblast proliferation, fusion, and maturation—a process that takes ∼2 wk. Robust regeneration is seen in young DMD patient muscle as well as dystrophin-deficient dogs, mice, cats, and pigs (Chen et al., 2005; Kornegay et al., 2012a; Klymiuk et al., 2013). Successful regeneration is associated with retained muscle function at a young age. However, as a DMD patient ages, muscle regeneration gradually fails, with endomysial fibrosis and fatty replacement of the muscle. The fibrofatty infiltration is typically commensurate with muscle wasting and loss of muscle function. Some muscles in both dystrophin-deficient DMD patients and dogs, such as the extraocular muscles and sartorius, seem able to maintain regeneration and are functionally spared (Kornegay et al., 2012b; Nghiem et al., 2013). Most dystrophin-deficient muscles in cats and mice are able to successfully regenerate throughout the normal life span (Gaschen et al., 1992). The failed regeneration seen in the DMD muscle has features that are shared with aging muscle, including chronic inflammatory state and loss of mitochondrial function (Baron et al., 2011). However, the age-, muscle-, and species-specific progressions from successful to failed regeneration are poorly understood, and there has not been a unifying model able to explain the observed transition to muscle wasting and the loss of muscle function (Serrano et al., 2011).
The gradual loss of muscle tissue and disorganization of muscle structure in most muscles in a DMD patient has been attributed, in part, to a progressive exhaustion of satellite cell function in response to the chronic drive to repair degenerating muscle fibers (Blau et al., 1983). However, there are conflicting studies regarding the number and nature of muscle stem cells, and loss of satellite cells with age is questioned as there appear to be higher numbers of satellite cells in DMD muscle than there are in normal muscle (Watkins and Cullen, 1986, 1988; Maier and Bornemann, 1999; Kottlors and Kirschner, 2010). Reports of diminished DMD myoblast proliferation have been associated with low minimal telomere length in DMD whole muscle (Aguennouz et al., 2011) and myoblasts (Decary et al., 2000). However, this remains controversial as another investigation detected no overall reduction in mean telomere length in DMD muscle cells (Oexle et al., 1997).
An alternative hypothesis promoted for failed regeneration is the excessive connective tissue proliferation (fibrosis), a feature in common with many chronic inflammatory states (Serrano et al., 2011). Fibrosis could impair vascularization of muscle tissue, create an insufficient environment for effective satellite cell proliferation and differentiation, and alter cell–cell communication during myofiber regeneration. Histologically, fibrosis is characterized by the exaggerated expression and deposition of extracellular matrix components, particularly collagen I. TGF-β, a key cytokine promoting fibrosis, appears to have a critical role in fibrosis in DMD (Chen et al., 2005; MacDonald and Cohn, 2012) as well as most fibrotic states in most tissues (Akhurst and Hata, 2012; Bowen et al., 2013). Strong induction of TGF-β pathways is seen in both successful and unsuccessful remodeling (Karkampouna et al., 2012). Indeed, TGF-β is important for normal muscle regeneration, and the molecular mechanisms underlying a change in normal versus pathological roles of TGF-β pathways have remained elusive for muscular dystrophy and other chronic inflammatory states (Bhattacharyya et al., 2013). The likely importance of TGF-β pathways in DMD is underscored by the recent finding of two genetic modifiers of disease severity, both of which map to TGF-β pathways (SPP1/osteopontin [Pegoraro et al., 2011; Bello et al., 2012]; LTPB4 [Flanigan et al., 2013]).
Here, we used a data-driven approach involving three large mRNA profiling experiments (two human muscle biopsy and one murine regeneration series) to generate a novel hypothesis regarding the cellular underpinnings of failed regeneration in muscle disease (asynchronous regeneration). The hypothetical model was then tested using an experimental approach of asynchronous bouts of staged degeneration/regeneration in localized microenvironments of wild-type (WT) muscle.
Results
Development of bioinformatics methods for increased sensitivity of biochemical network selection
It is well documented that different analytical methods applied to the same microarray dataset can provide very different interpretations of the data, often with only 30% concordance of lists of differentially regulated genes (Bakay et al., 2002; Seo et al., 2004, 2006; Irizarry et al., 2005). We reasoned that increased sensitivity for selection of differentially regulated genes could be obtained by using multiple probe set algorithms, multiple microarrays (e.g., U133A and U133B arrays on the same samples), and multiple queries of existing literature for biochemical pathways (Fig. S1). Such an iterative approach would be expected to greatly increase sensitivity of gene selection but also increase false positives. However, using an integrative approach with a composite score of gene selection and associated networks, we could decrease false positives by focusing on the top-ranked composite networks (Fig. S1). In effect, as different sources of data are assembled to derive a probability determining the specificity of a network, the most highly scored networks have considerable statistical power and significance.
To our knowledge, this two-tier composite approach has not been previously described. This approach would be expected to be most effective on larger and more complex datasets, and for this reason, we used a human 12-disease group, 117-biopsy dataset, and 234-microarray dataset as a dataset for model generation, with multiple validation and model extension datasets.
Visualization of domain knowledge identifies a potential disease grouping by the histological phenotype of fibrosis and failed regeneration
Seven heat maps with clear patterns of association of disease groups were generated and visually inspected for possible shared pathogenesis between the clustered disease groups. Many of the disease groupings were expected and were not studied further. For example, juvenile dermatomyositis (JDM) is classified as an autoimmune disorder and shows extensive inflammation (immune cells) infiltrating the muscle (Tezak et al., 2002; Chen et al., 2008). Indeed, a strong clustering of JDM distinct from all other disorders was seen with both A and B chips and all probe set algorithms, and the members of this cluster were transcripts involved in immune cell infiltration (unpublished data). Likewise, most muscular dystrophies show some degeneration and regeneration of myofibers, and these gene clusters were evident as a recapitulation of embryonic developmental programs (unpublished data).
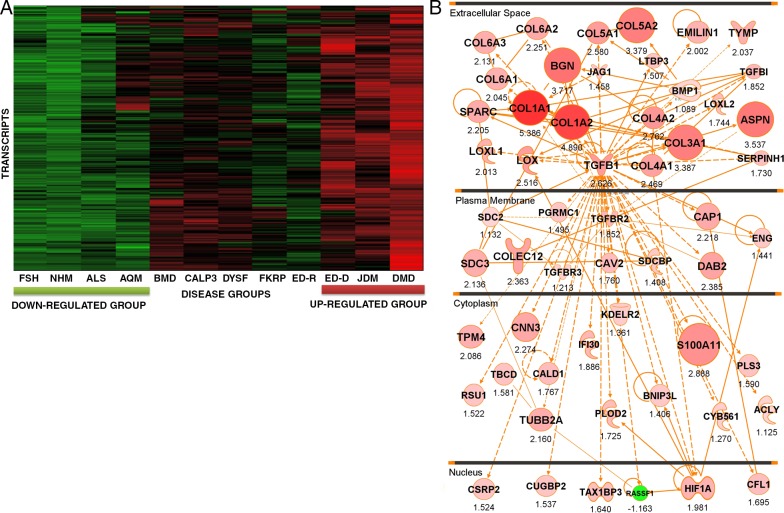
One particular grouping of the seven primary heat maps was believed to be particularly interesting based upon domain knowledge and was studied in some depth (Fig. 1 A). This grouping showed a strong up-regulation of transcripts in three disorders: DMD, JDM, and ED-D (Emery–Dreifuss muscular dystrophy autosomal dominant; LMNA [lamin A/C] mutations). The same cluster was up-regulated, but to a lesser extent, in Becker muscular dystrophy (BMD; dystrophin mutation), LGMD2A (limb girdle muscular dystrophy type 2A; CALP3 [calpain 3] mutation), and LGMD2B (limb girdle muscular dystrophy type 2B; DYSF [dysferlin] mutation). The clustering was seen for the A microarray and PLIER (Probe Logarithmic Intensity Error) probe set algorithm (Fig. 1, heat map) and also all other applied probe set algorithms (not depicted). This grouping could be potentially significant, as these three disorders often show particularly severe histological patterns of fibrosis and failed regeneration early in the disease process.
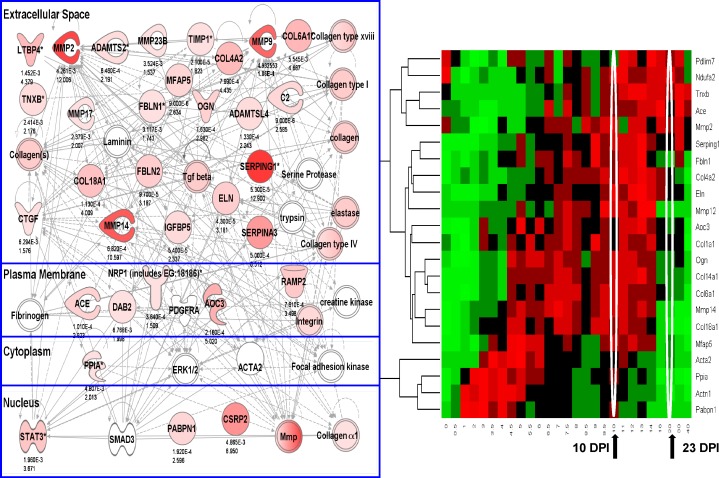
Figure 1.
An iterative composite scoring bioinformatics approach identifies a transcript network associated with progressive muscular dystrophy. (A) Shown is a heat map of 117 muscle biopsies grouped by diagnostic category. The individual transcripts (y axis) defining this disease clustering were used to generate the molecular network in B. Disorders studied were facioscapulohumeral muscular dystrophy (FSH), normal human skeletal muscle (NHM), amyotrophic lateral sclerosis (ALS), acute quadriplegic myopathy (AQM), Becker muscular dystrophy (BMD), LGMD2A (CALP3), LGMD2B (DYSF), LGMD2I (FKRP), Emery–Dreifuss muscular dystrophy X-linked (EMRN [emerin]), Emery–Dreifuss muscular dystrophy autosomal dominant (ED-D; LMNA mutations), juvenile dermatomyositis (JDM), and Duchenne muscular dystrophy (DMD). (B) Shown is the integration of component mRNAs from the clustering in A into functional relationships based upon the literature (IPA). The top-ranked Ingenuity networks were merged into a 56-transcript network. This network is seen to be centered on TGF-β and fibrosis (collagens and other extracellular matrix transcripts), and the network is visualized based on the typical subcellular localization of the encoded proteins of each transcript. Red symbols represent up-regulated transcripts, with the size of the symbol and intensity of color scaled to the observed fold change between groups (DMD, JDM, and ED-D vs. FSH, ALS, and NHM), with fold change with the PLIER algorithm provided under each symbol. Statistical data corresponding to this network are provided in Table 1.
As shown in Fig. S1, this grouping was expanded to both A and B microarrays and both PLIER and dCHIP (DNA chip analyzer) probe set algorithms. Applying IPA (Ingenuity Pathway Analysis) to this clustering, we found the top-ranked networks to share many component members, and these were merged into a 56-member network (Fig. 1 B). Fold changes were displayed using both node size and intensity of color, with the numeric value shown under each node. Statistical support for differential regulation of component members is shown for the PLIER algorithm (Table 1).
Table 1.
List of the 56 transcript genes in the TGF-β network
| Gene | Probe set | P-value | Fold change |
| ACLY | 210337_s_at | 0.051 | 1.125 |
| ASPN | 219087_at | <0.001 | 3.537 |
| BGN | 213905_x_at | <0.001 | 3.717 |
| BMP1 | 207595_s_at | 0.165 | 1.089 |
| BNIP3L | 221478_at | <0.001 | 1.406 |
| CALD1 | 212077_at | <0.001 | 1.767 |
| CAP1 | 200625_s_at | <0.001 | 2.218 |
| CAV2 | 203324_s_at | <0.001 | 1.76 |
| CFL1 | 200021_at | <0.001 | 1.695 |
| CNN3 | 201445_at | <0.001 | 2.274 |
| COL1A1 | 202310_s_at | <0.001 | 5.386 |
| COL1A2 | 202404_s_at | <0.001 | 4.89 |
| COL3A1 | 211161_s_at | <0.001 | 3.387 |
| COL4A1 | 211981_at | <0.001 | 2.469 |
| COL4A2 | 211966_at | <0.001 | 2.762 |
| COL5A1 | 212489_at | <0.001 | 2.58 |
| COL5A2 | 221729_at | <0.001 | 3.379 |
| COL6A1 | 213428_s_at | <0.001 | 2.045 |
| COL6A2 | 209156_s_at | <0.001 | 2.251 |
| COL6A3 | 201438_at | <0.001 | 2.131 |
| COLEC12 | 221019_s_at | <0.001 | 2.363 |
| CSRP2 | 207030_s_at | <0.001 | 1.524 |
| CUGBP2 | 202158_s_at | <0.001 | 1.537 |
| CYB561 | 209163_at | <0.001 | 1.27 |
| DAB2 | 201280_s_at | <0.001 | 2.385 |
| ECGF1 | 204858_s_at | <0.001 | 2.037 |
| EMILIN1 | 204163_at | <0.01 | 2.002 |
| ENG | 201809_s_at | <0.001 | 1.441 |
| HIF1A | 200989_at | <0.001 | 1.981 |
| IFI30 | 201422_at | <0.001 | 1.886 |
| JAG1 | 209098_s_at | <0.001 | 1.458 |
| KDELR2 | 200699_at | <0.001 | 1.361 |
| LOX | 215446_s_at | <0.001 | 2.516 |
| LOXL1 | 203570_at | <0.001 | 2.013 |
| LOXL2 | 202998_s_at | <0.001 | 1.744 |
| LTBP3 | 219922_s_at | <0.001 | 1.507 |
| PGCP | 208454_s_at | <0.01 | 1.237 |
| PGRMC1 | 201121_s_at | <0.001 | 1.495 |
| PLOD2 | 202620_s_at | <0.001 | 1.725 |
| PLS3 | 201215_at | <0.001 | 1.59 |
| RASSF1 | 204346_s_at | <0.05 | −1.163 |
| RSU1 | 201980_s_at | <0.001 | 1.522 |
| S100A11 | 200660_at | <0.001 | 2.888 |
| SDC2 | 212158_at | 0.269 | 1.132 |
| SDC3 | 202898_at | <0.001 | 2.136 |
| SDCBP | 200958_s_at | <0.001 | 1.408 |
| SERPINH1 | 207714_s_at | <0.001 | 1.73 |
| SPARC | 200665_s_at | <0.001 | 2.205 |
| TAX1BP3 | 209154_at | <0.001 | 1.64 |
| TBCD | 211052_s_at | <0.001 | 1.581 |
| TGFB1 | 203085_s_at | <0.001 | 2.626 |
| TGFBI | 201506_at | <0.001 | 1.852 |
| TGFBR2 | 208944_at | <0.001 | 1.852 |
| TGFBR3 | 204731_at | 0.065 | 1.213 |
| TPM4 | 209344_at | <0.001 | 2.086 |
| TUBB2A | 204141_at | <0.001 | 2.16 |
List of the 56 transcript genes in the TGF-β network (Fig. 1 A) and their fold changes and p-values of t test for testing the significance of their changes between two groups: up-regulated group (DMD, JDM, and ED-D) and down-regulated group (FSH, NHM, and ALS). The results shown were from the data processed with PLIER. Those few transcripts showing nonsignificant p-values here are included, as they met significance thresholds with dCHIP mismatch probe set algorithm (not depicted).
This network was centered on TGF-β, a well-studied cytokine associated with pathological fibrosis in many tissues (Akhurst and Hata, 2012; Bowen et al., 2013) and included many of the known components of fibrotic tissue (collagens and lysine oxygenases). Another notable component of the network was HIF1A (hypoxia-inducible factor 1, α subunit), a known transcription factor strongly induced by tissue ischemia (Koh and Powis, 2012). The nature of the network suggested that it was simply reflective of a more severe pathology (greater fibrosis, greater failed remodeling, and regeneration), and DMD, JDM, and LMNA often (but not always) show more severe pathology at the initial diagnosis compared with the other disorders in the dataset.
To test whether the network was more reflective of tissue pathology than clinical and molecular diagnosis, a second validation dataset was generated that was then analyzed by unsupervised clustering. This new dataset contained 49 patient muscle biopsies analyzed on HG-U133 plus 2.0 microarrays. Samples studied were 6 normal volunteers, 17 DMD, 11 BMD, 7 LGMD2I (limb girdle muscular dystrophy type 2I; FKRP [fukutin-related protein deficiency]), and 8 LGMD2B.
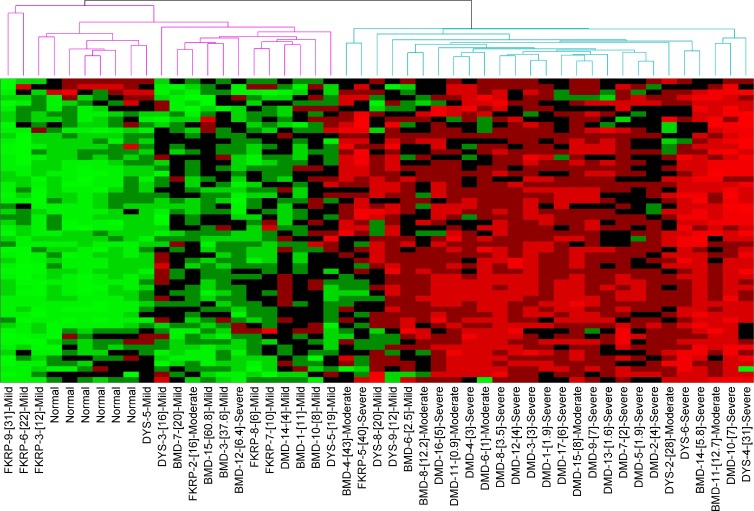
Unsupervised clustering using the 56 network members from Fig. 1 resulted in clustering of the 49 biopsies into two distinct groups indicated by the dendrogram at the top (Fig. 1, purple left group with uniform down-regulation of transcripts and blue right group with uniform up-regulation; and Fig. 2). The two branches did not correspond well to clinical/molecular diagnosis, with Becker, Duchenne, FKRP-deficient and dysferlin-deficient patients seen in both groups. To test whether the segregation of groups was caused by histopathology of the muscle studied, the archived interpretation of the severity of the dystrophy was accessed (all read by the same author, E.P. Hoffman; Fig. 2, normal, mild, moderate, or severe dystrophy groups). Mild was defined by evidence of degeneration and regeneration but little or no fibrosis or inflammation. Moderate showed increased inflammation and fiber size variation but minimal or moderate fibrosis. Severe showed fibrofatty replacement of muscle and evidence of failed regeneration. Normal or mild pathology was seen in 20/22 patients in the left branch but only 3/27 in the right branch. In contrast, moderate or severe pathology was seen in 2/22 patients in the left branch and 24/27 in the right branch. Thus, we concluded that this cluster and associated networks were more reflective of severity of histological pathology than clinical/molecular diagnosis.
Figure 2.
Unsupervised (blinded) clustering of individual muscle biopsies from a 49-biopsy validation mRNA profiling dataset. Shown is a heat map using the same 56-member TGF-β network from Fig. 1 B, with visualization of unsupervised clustering of individual biopsies. The specific mRNAs are on the y axis (not depicted), with individual patient biopsies on the x axis (diagnosis, patient number, [age at biopsy], and blinded classification of severity of the histology [normal, mild, moderate, and severe]). The heat map shows bimodal stratification of biopsies (two branches of dendrogram at the top), with the right branch clustering those biopsies with high levels of all or most of these transcripts, whereas the left branch shows lower levels of these transcripts. This clustering is seen to be driven by the severity of the pathology and not patient diagnosis or age (right branch, moderate or severe dystrophic pathology; left branch, normal or mild dystrophic pathology).
Query of the putative failed regeneration network in normal staged tissue regeneration identifies time-resolved subnetworks
In many respects, the identification of the TGF-β/fibrosis network was quite expected, as fibrosis is associated with end-stage tissue pathology and with failed regeneration in many tissues, including muscle. However, the majority of patient muscle biopsies were studied at initial diagnosis, before end-stage clinical disease. Moreover, there is increasing evidence that TGF-β is important for normal staged muscle regeneration (Philippou et al., 2008; Kollias and McDermott, 2008; MacDonald and Cohn, 2012). To query the relationship of this cluster with normal regeneration, we overlaid these network members over our previously published 27–time point murine in vivo muscle degeneration/regeneration time series (Fig. 3; Zhao et al., 2003, 2006; Zhao and Hoffman, 2004).
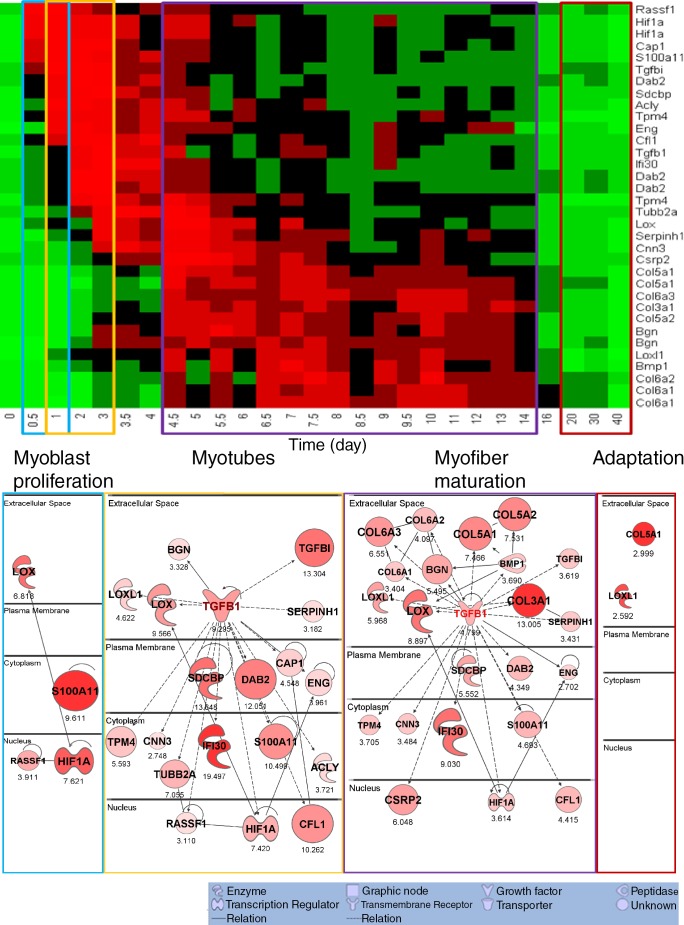
Figure 3.
The TGF-β network superimposed on normal mouse muscle regeneration shows strong induction of subsets of the network at specific time frames during regeneration. The components of the human TGF-β network (Fig. 1 B) were cross-mapped to the mouse genome and queried in a 27–time point muscle regeneration mRNA profiling dataset (Zhao et al., 2003). The top shows heat map visualization, with network members listed on the right y axis, and the time points after regeneration are shown on the x axis (days). Each member of the TGF-β network shows up-regulation within certain stages of normal staged muscle regeneration, although these stages vary depending on the specific network member. The time points are shown grouped (colored boxes) into early myoblast proliferation stage (blue box), myotube stage (yellow box), myofiber maturation stage (purple box), and late myofiber adaptation stage (red box). The transcripts showing up-regulation within each of these stages of regeneration were then used to generate networks (bottom). This shows that the TGF-β network seen in muscular dystrophy muscle (Fig. 1 B) is parsed into subnetworks based upon the stage of normal regeneration that the subset of transcripts is expressed. Numbers indicate fold change. Statistical data associated with this network and heat map visualization are provided in Table 2.
All network members for which we could identify cross-species transcript units and probe sets did indeed show significant regulation of these transcripts in normal regeneration both by heat map (Fig. 3 A) and by statistical comparisons (Table 2). However, we noted from the heat map (Fig. 3 B) that different members of this pathology-associated network were expressed at different time points during normal regeneration. To visualize this temporal-specific pattern, we grouped the 27–time point murine series into four time frames corresponding to relatively distinct periods of muscle regeneration (myoblast proliferation—days 1–3; myotube formation—days 3.5–4.5; myofiber maturation—days 4.5–14; late-stage adaptation—days 20–40; Fig. 3 B). This parsed the TGF-β network in Fig. 1 B into a series of time-resolved subnetworks shown in Fig. 3 B, where the relative expression levels were again coded by both symbol size and color intensity within each subnetwork. The extracellular matrix components commonly associated with pathological fibrosis were seen in the myofiber maturation stage but not other stages.
Table 2.
The subset of genes of TGF-β network significantly up-regulated in each of the four stages in the normal muscle regeneration process and the t test results
| Gene | Probe set | P-value | Fold change |
| 0.5–1 d vs. control | |||
| Hif1a | 98628_f_at | <0.01 | 7.576 |
| Hif1a | 98629_f_at | <0.01 | 7.666 |
| Lox | 160095_at | <0.05 | 6.818 |
| Rassf1 | 102379_at | <0.01 | 3.911 |
| S100a11 | 98600_at | <0.05 | 9.611 |
| 1–3 d vs. control | |||
| Acly | 160207_at | <0.001 | 3.721 |
| Bgn | 162347_f_at | <0.01 | 3.643 |
| Bgn | 96049_at | <0.01 | 3.013 |
| Cap1 | 93721_at | <0.001 | 4.548 |
| Cfl1 | 99119_at | <0.001 | 10.262 |
| Cnn3 | 160150_f_at | <0.05 | 2.748 |
| Dab2 | 104633_at | <0.01 | 9.586 |
| Dab2 | 98044_at | <0.01 | 4.343 |
| Dab2 | 98045_s_at | <0.001 | 22.223 |
| Eng | 100134_at | <0.001 | 3.961 |
| Hif1a | 98628_f_at | <0.001 | 7.293 |
| Hif1a | 98629_f_at | <0.001 | 7.546 |
| Ifi30 | 97444_at | <0.01 | 19.497 |
| Lox | 160095_at | <0.01 | 9.566 |
| Loxl1 | 103850_at | <0.001 | 4.622 |
| Rassf1 | 102379_at | <0.01 | 3.11 |
| S100a11 | 98600_at | <0.001 | 10.499 |
| Sdcbp | 93017_at | <0.001 | 13.648 |
| Serpinh1 | 94817_at | <0.01 | 3.182 |
| Tgfb1 | 101918_at | <0.01 | 9.295 |
| Tgfbi | 92877_at | <0.001 | 13.304 |
| Tpm4 | 95542_at | <0.001 | 5.322 |
| Tpm4 | 95543_at | <0.001 | 5.865 |
| Tubb2a | 94835_f_at | <0.01 | 7.055 |
| 4.5–14 d vs. control | |||
| Bgn | 162347_f_at | <0.001 | 6.38 |
| Bgn | 96049_at | <0.01 | 4.61 |
| Bmp1 | 92701_at | <0.001 | 3.69 |
| Cfl1 | 99119_at | <0.001 | 4.415 |
| Cnn3 | 160150_f_at | <0.001 | 3.484 |
| Col3a1 | 102990_at | <0.001 | 13.005 |
| Col5a1 | 101080_at | <0.001 | 10.476 |
| Col5a1 | 93472_at | <0.001 | 4.456 |
| Col5a2 | 92567_at | <0.001 | 7.531 |
| Col6a1 | 162459_f_at | <0.001 | 3.744 |
| Col6a1 | 95493_at | <0.01 | 3.064 |
| Col6a2 | 93517_at | <0.001 | 4.097 |
| Col6a3 | 101110_at | <0.01 | 6.551 |
| Csrp2 | 93550_at | <0.001 | 6.048 |
| Dab2 | 104633_at | <0.001 | 3.517 |
| Dab2 | 98045_s_at | <0.001 | 5.182 |
| Eng | 100134_at | <0.001 | 2.702 |
| Hif1a | 98628_f_at | <0.001 | 3.57 |
| Hif1a | 98629_f_at | <0.001 | 3.659 |
| Ifi30 | 97444_at | <0.001 | 9.03 |
| Lox | 160095_at | <0.001 | 8.897 |
| Loxl1 | 103850_at | <0.001 | 5.968 |
| S100a11 | 98600_at | <0.001 | 4.693 |
| Sdcbp | 93017_at | <0.001 | 5.552 |
| Serpinh1 | 94817_at | <0.05 | 3.431 |
| Tgfb1 | 101918_at | <0.01 | 4.799 |
| Tgfbi | 92877_at | <0.001 | 3.619 |
| Tpm4 | 95542_at | <0.001 | 3.229 |
| Tpm4 | 95543_at | <0.001 | 4.181 |
| Tubb2a | 94835_f_at | <0.001 | 4.807 |
| 20–40 d vs. control | |||
| Col5a1 | 101080_at | <0.01 | 2.999 |
| Loxl1 | 103850_at | <0.01 | 2.592 |
This analysis evoked a novel hypothesis regarding the distinction between successful regeneration and unsuccessful remodeling (fibrosis). We reasoned that normal regeneration is synchronous, with specific pathways and pathway members having appropriate temporal expression patterns during the 2–3-wk muscle regeneration process. In contrast, dystrophic muscle was an asynchronous remodeling process, with different regions of the small muscle biopsy studied in different stages of regeneration.
This analysis established the hypothetical model in which asynchronous remodeling may be the driver for failed regeneration, muscle wasting, and early death in the severe dystrophies. By this model, the complete TGF-β–centered network shown in Fig. 1 is only possible in dystrophic muscle. In normal muscle, it is parsed into subnetworks resolved by time. We then sought to test this model experimentally.
Establishing a model of asynchronous muscle regeneration
We reasoned that a potential experimental model to test asynchronous regeneration would be to induce localized regions of muscle degeneration/regeneration in WT mice, with repetitive regional injuries in close proximity within the muscle at defined time points. The experimental design is shown in Fig. S2. In brief, low doses of notexin (10 µl notexin at 5 mg/µl) were injected intramuscularly along with a blue tattoo dye. At a defined time point later, a second injection was performed with the same low dose of notexin, using a red tattoo dye. Mice were sacrificed 13 d after the second injection, a time point when regeneration should be largely complete for both areas. We then focused on specific areas of the muscle, where both injection sites were separated by a relatively small area (50–300 myofibers)—the hypothesis being that myofibers in the microenvironments immediately at the injection sites would be synchronously regenerating, whereas the myofibers in between the two injection sites would receive regeneration cues from both sites (cross talk microenvironment).
In an initial experiment, injections were spaced by 0 (two injections at the same time), 1, 2, 4, 5, and 10 d (four muscles at each time point). Cryosections from the belly of each muscle were analyzed for fibrotic content (Fig. S3 A). This analysis showed that increasing time between injections resulted in increasing content of fibrosis of the muscle (Fig. S3 A). This suggested that the timing of injections influenced the amount of fibrotic tissue deposited in the muscle as a whole tissue.
We hypothesized that asynchronous remodeling may result from interference in intercellular signaling when two nearby groups of myofibers are at different stages of regeneration. To test this, we used laser capture microscopy (LCM) to collect groups of myofibers at the sites of injury, as identified by red and blue tattoo dyes, and the myofibers in between the sites of injury. We focused on the mouse groups of 0, 4, and 10 d between injections.
Five regions were excised using LCM from each muscle: (1) blue dye area (injection 1; 17 d after injection for 4-d series and 23 d after injection for 10-d series), (2) red dye area (injection 2; 13 d after injection for both series), (3) muscle fibers in between the two injections sites (called in-between), (4) normal nonregenerating (intact) myofibers (identified by peripheral nuclei), and (5) single regeneration site (single dye area with no evidence of the second dye). These areas were identified by centralized nuclei myofibers located in the external area of one injury site (either blue or red).
LCM-excised regions were used for mRNA isolation and mRNA profiling using Illumina bead arrays. Unsupervised chip clustering showed excellent signal/noise balance, as indicated by the clustering of microarray replicates of LCM from adjacent muscle sections (Fig. S2 C). For data analysis, we first compared region 4 (uninjured) versus region 5 (single regeneration). There were very few differentially regulated transcripts, suggesting that the muscle in region 5 had indeed successfully regenerated to completion. Likewise, there were few statistically significant differences between region 1 (first injury) versus region 5 and region 2 (second injury) versus region 5 (single site), consistent with successful remodeling (unpublished data).
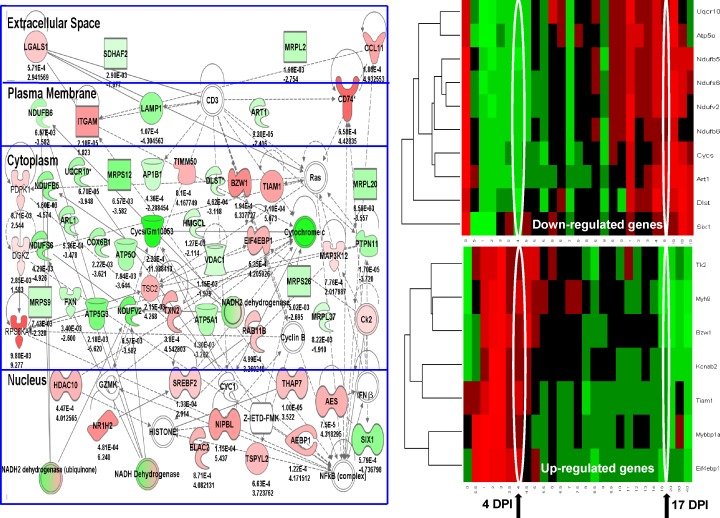
In contrast, comparison of the tissue in-between injuries (area 3) showed mRNA expression profiles that were quite distinct from neighboring regions and also remarkably different for the 4-d series (Fig. 4) versus the 10-d multiinjury series (Fig. 5). Specifically, the 4-d in-between myofiber region showed a relative loss of mitochondrial-related transcripts and an increase in inflammatory transcripts (Fig. 4, left). The 10-d in-between myofiber region showed increase of extracellular matrix components (Fig. 5, left). Statistical comparison of fold changes and p-values between the 4- and 10-d multiple injection sites showed highly significant differences, with the mitochondrial changes specific to the 4-d time series (Table 3) and the extracellular matrix transcripts specific to the 10-d time series (Table 4). Alignment of the subcellular localizations of the components of the top-ranked Ingenuity networks for the 4 and 10 d confirmed the preponderance of intracellular protein changes in the 4 d, in contrast to the extracellular matrix components in the 10 d (Fig. S3, B and C).
Figure 4.
Muscle tissue areas located between asynchronous injuries spaced 4 d apart show chronic inflammation and loss of mitochondrial transcripts. LCM was used to isolate regions of myofibers in between injury sites (cross talk area) spaced 4 d apart, with muscles studied 13 d after the second injection when myofiber regeneration was expected to be largely complete. Shown is the top-ranked Ingenuity network (left) reflective of chronic inflammation (red gene/protein symbols), and loss of mitochondrial components (green symbols). This network was then superimposed on normal murine regeneration time series (right), separated by the mitochondrial-related transcripts (down-regulated genes; top right) and inflammatory transcripts (up-regulated genes; bottom right). The mice that were sacrificed after regeneration should be largely complete (17 d postinjection [DPI]). However, the myofibers between the two injury sites showed expression patterns that were more consistent with the 4-d time point (4 d postinjection). Statistical analyses associated with the heat map are provided in Table 4. These data suggest that the myofibers in between the two injury sites are suspended in a time period of regeneration, defined by the days between the two sequential injuries (cross talk microenvironment). Numbers indicate p-value (top) and fold change (bottom).
Figure 5.
Muscle tissue areas located between asynchronous injuries spaced 10 d apart show a high level expression of profibrotic gene networks. Shown is data for the LCM region of muscle between the two injury sites spaced 10 d apart. The myofibers between the two injury sites show a high level expression of transcripts associated with connective tissue proliferation and fibrosis (top-ranked Ingenuity network, left). Superimposing this network on normal staged murine regeneration shows that the myofibers in the area between the two injury sites show expression patterns consistent with suspension in the 10-d time point of normal regeneration. Statistical analyses associated with the heat map are provided in Table 4. Numbers indicate p-value (top) and fold change (bottom). DPI, day postinjection.
Table 3.
Down-regulated metabolic gene transcripts are shared between 3 and 4 d of synchronous regeneration and 4-d asynchronous remodeling
| Genes | P-value/FC | |||||||
| 27–time point single regeneration series | Double injection LCM series | |||||||
| 4 d between injections | 10 d between injections | |||||||
| 3/4 da | 9/10 da | 13/14 da | 30/40 da | Single siteb | In-betweenb | Single siteb | In-betweenb | |
| ATP5A1 100753_at | 0.06 | NS | NS | NS | NS | 0.004/−3.27 | NS | NS |
| COX6B1 99114_r_at | 0.01 | NS | NS | NS | NS | 0.002/−3.62 | NS | NS |
| CYCS 98132_at | 0.06 | NS | NS | NS | NS | 0.0002/−12 | NS | NS |
| NDUFB5 97307_f_at | 0.002 | 0.05 | 0.05 | 0.04 | NS | 0.001/−4.6 | NS | 0.002/−2.5 |
| UQCRC1 101989_at | 0.07 | NS | NS | NS | NS | 0.009/2.6 | NS | NS |
| ART1 104638_at | 0.02 | NS | NS | NS | NS | 0.00009/−2.4 | NS | NS |
| ARL1 97505_at | 0.02 | NS | NS | NS | NS | 0.0005/−3.5 | NS | NS |
| MRPS12 98547_at | 0.02 | NS | NS | 0.04 | NS | 0.0006/−5.4 | NS | NS |
| MRLP20 94875_at | 0.01 | NS | NS | NS | NS | 0.006/−3.6 | NS | NS |
The nine mRNA transcripts shown were significantly down-regulated in the region between asynchronous injection sites in the double LCM series when injections were separated by 4 d, but not 10 d (8/9). In a separate 27–time point synchronous regeneration time series, these same transcripts were down-regulated at the 3–4-d time point and not later time points. FC, fold change.
Days are versus 0.
Sites are versus nonregenerating.
Table 4.
Up-regulated fibrosis-associated gene transcripts are shared between 9 and 14 d of synchronous regeneration and 10-d asynchronous remodeling
| Genes | P-value/FC | |||||||
| 27–time point single regeneration series | Double injection LCM series | |||||||
| 4 d between injections | 10 d between injections | |||||||
| 3/4 da | 9/10 da | 13/14 da | 30/40 da | Single siteb | In-betweenb | Single siteb | In-betweenb | |
| ACTA2 93100_at | 0.001 | 0.006 | 0.002 | NS | NS | 0.005/8.3 | NS | 0.008/16.8 |
| COL6A1 95493_at | 0.003 | 0.0001 | 0.0001 | 0.02 | NS | 0.002/−3.6 | NS | 0.00004/7.2 |
| COL18A1 101039_at | 0.00001 | 0.0003 | 0.01 | NS | NS | NS | NS | 0.0001/4.0 |
| COL4A2 101039_at | NS | 0.01 | 0.008 | NS | NS | NS | NS | 0.0007/4.5 |
| ELN 92207_at | NS | 0.004 | 0.02 | NS | NS | NS | NS | 0.00004/3.2 |
| MMP14 160118_at | 0.0007 | 0.0002 | 0.006 | NS | NS | NS | NS | 0.0006/10.6 |
| COL14A1 99476_at | 0.007 | 0.004 | 0.002 | NS | NS | NS | NS | 0.0003/1.5 |
| COL15A1 99637_at | 0.02 | NS | 0.006 | NS | NS | NS | NS | 0.004/1.6 |
| C2 103673_at | 0.02 | 0.02 | NS | NS | NS | NS | NS | 0.000009/2.6 |
| STAT3 99099_at | 0.02 | 0.002 | 0.05 | NS | NS | NS | NS | 0.003/4.4 |
| CDKNA1 94881_at | 0.03 | 0.05 | 0.009 | NS | NS | NS | NS | 0.00007/3.25 |
| RAMP2 994444_at | NS | 0.007 | 0.006 | NS | NS | NS | NS | 0.0007/3.5 |
| ACVR2B 994444_at | NS | 0.007 | 0.006 | NS | NS | NS | NS | 0.0004/6.45 |
The 13 gene transcripts shown were significantly up-regulated in the region between asynchronous injection sites in the double LCM series when injections were separated by 10 d, but not 4 d (0/13). In the 27–time point synchronous regeneration time series, these same transcripts showed the highest and consistent up-regulated between 9 and14 d. FC, fold change.
Days are versus 0.
Sites are versus nonregenerating.
We then overlaid the gene networks in Fig. 4 over normal staged muscle regeneration, separating the up-regulated and down-regulated transcripts (Fig. 4, right). This showed that the down-regulated mitochondrial transcripts were highly expressed in uninjured and fully regenerated muscle but were strongly down-regulated both in the 4-d in-between area as well as the 4-d time point during normal staged regeneration. This was consistent with the myofibers in between the two injury sites having suspended regeneration at the 4-d time point. Similarly, the up-regulated inflammatory transcripts in the in-between area separated by 4 d were highly expressed at the 4-d time point of normal staged regeneration but not at the later fully regenerated time point (Fig. 4). Thus, both the down-regulated mitochondrial transcripts and the up-regulated inflammatory transcripts in the in-between area appeared arrested at the time point separating the asynchronous injury times.
The 10-d staged injury series was also consistent with the suspended development model. Connective tissue remodeling transcripts were highly up-regulated in the in-between area separated by 10 d, and this pattern was consistent with the 10-d time point in normal regeneration (Fig. 5, right). Statistical analysis showed that this pattern was largely specific to the 10-d staged remodeling and not seen in the 4-d staged remodeling (Table 4).
The microarray data from the 10-d interval injury model showed that the myofiber region in between asynchronous injuries showed inappropriate persistent expression of connective tissue-remodeling transcripts, and this would be consistent with the pathological findings of localized tissue fibrosis. Although we had previously shown an overall increase in fibrosis that was highest in the day 10 staged series muscle (Fig. 3 A), we refined this analysis to the localized area in between the two injection sites (Fig. 6). Histological visualization of the area between the two injection sites showed dramatic fibrotic replacement of muscle in the 10-d series, which was not seen with the 0- and 4-d series (Fig. 6).
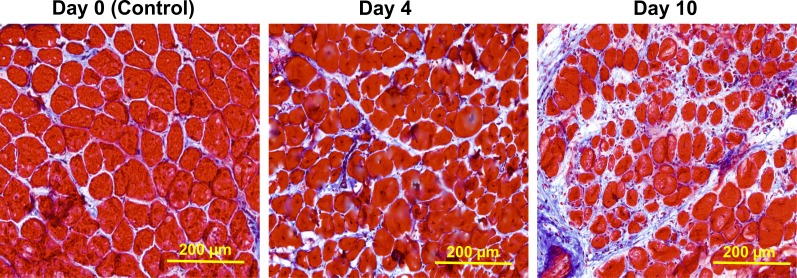
Figure 6.
Histological analysis of connective tissue proliferation (fibrosis). Shown is Masson’s modified trichrome staining of cryosections in the area of muscle between injection sites staged 0, 4, and 10 d apart (n = 3 muscles per group). Endomysial fibrosis is most evident in the area between injections space 10 d apart, consistent with the LCM expression profiling data (Fig. 4 B).
Targeted experiments confirm aberrant expression of fibrotic transcripts (10 d apart) and persistent inflammation and loss of mitochondrial capacity (4 d apart)
To validate the mRNA microarray findings, we selected specific transcripts and proteins to evaluate using an independent series of mice that had been subjected to the same 4- and 10-d asynchronous muscle injuries. Tattoo dyes were used to visualize the first and second injection sites. Tissue for quantitative RT-PCR (qRT-PCR) was obtained by LCM. Immunohistochemistry and enzyme histochemistry were also performed.
The 4-d interval injury model showed an increase in inflammatory transcripts and a relative loss of mitochondrial-associated transcripts (Fig. 4). qRT-PCR confirmed the mRNA profiling data, showing preferential loss of mitochondrial transcripts in the region in between the two injections sites, relative to other regions of the same muscle tissue and the area between two notexin injections made on the same day (Table 5). To confirm the relative loss of mitochondrial oxidative capacity predicted by expression microarray and qRT-PCR microarray data for the 4-d in-between areas, we used standard enzyme histochemistry (NADH, cytochrome c oxidase [COX], succinate dehydrogenase [SDH], and a combination of NADH and COX enzymatic staining). The region between the 4-d interval injection sites showed a relative loss of mitochondrial enzymatic activity stains compared with other injection paradigms, confirming the microarray data (Fig. S4).
Table 5.
RT-PCR validation of loss of mitochondrial transcripts in the area between staged degeneration/regeneration cycles 4 d apart
| Genes | P-value/FC | ||
| Between 4D vs. single injury area | Between 4D vs. intact/uninjured area (same section) | Between 4D vs. between area of the control synchronous mice | |
| Cox6b1 | 0.0005/−2.6 | 0.0004/−4.9 | 0.004/−1.4 |
| Ndufs6 | 0.003/−3.6 | 0.04/−16.0 | 0.001/−6.9 |
| Ndufb5 | 0.01/−4.9 | 0.002/−11.0 | 0.02/−11.0 |
| Ndufb6 | 0.004/−6.5 | 0.009/−7.2 | 0.0002/−20.0 |
FC, fold change.
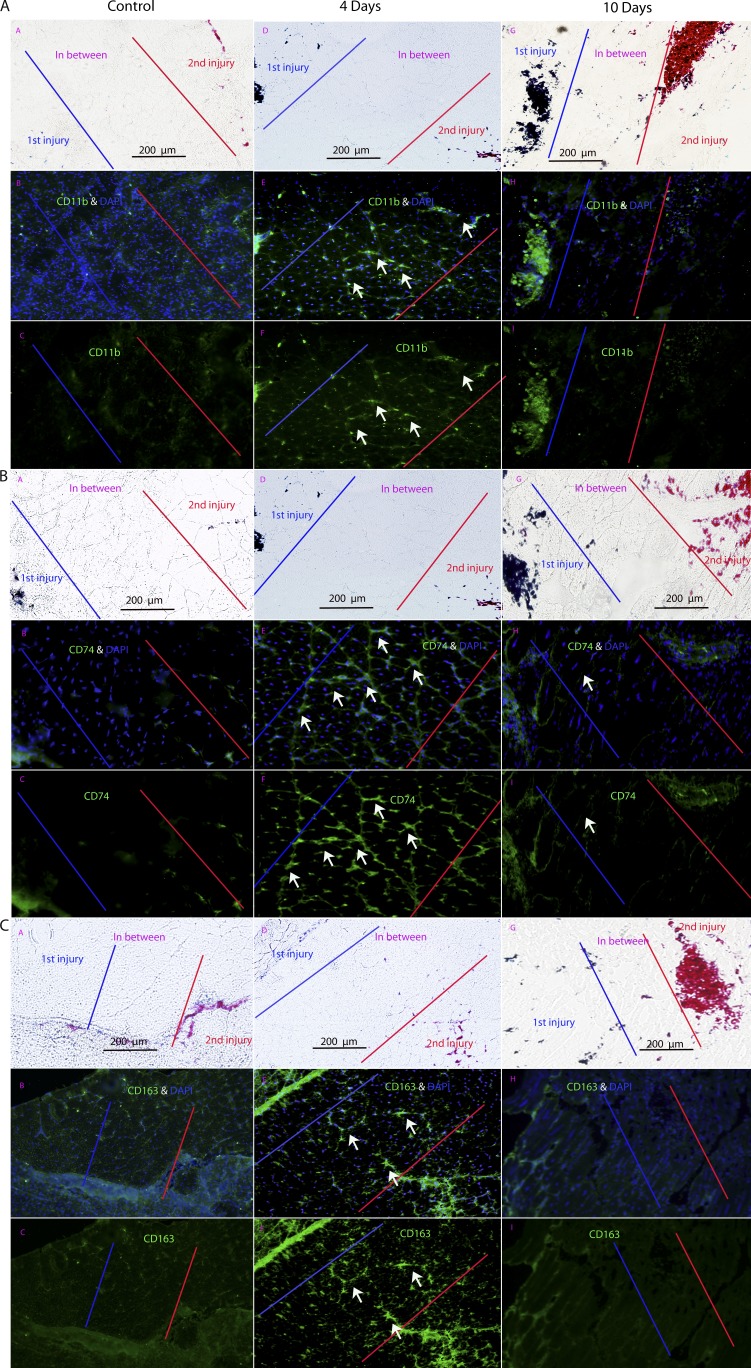
To validate the up-regulation of proinflammatory transcripts, we studied three proteins by immunofluorescence localizations in three double injury mice (injuries spaced 0, 4, and 10 d apart; CD11b, CD74, and CD163; Fig. 7). These proteins were selected based on their role in proinflammatory pathways as well as DMD pathogenesis. CD11b is a leukocyte marker that induces innate immune response resulting in phagocytosis and cell-mediated cytotoxicity (Solovjov et al., 2005). CD11b immunostains showed the highest expression in the in-between region at 4 d but not 10 d or the simultaneous injection control (Fig. 7 A).
Figure 7.
Immunostaining of inflammatory proteins CD11b, CD74, and CD163 shows expression specific to intervening areas of muscle between injuries spaced 4 d apart. (A–C) Shown are immunofluorescence stains for inflammatory protein markers CD11b (A), CD74 (B), and CD163 (C). Each panel shows matched phase contrast (marking injury sites with tattoo dyes), antigen with nuclear stains (DAPI), and antigen alone. All three inflammatory markers show preferential localization in the area between the two injury sites only when injuries are spaced 4 d apart and not 0 or 10 d apart (n = 3 muscles per group). Arrows indicate immunoreactive material in the microenvironment between two neighboring sites of regeneration.
Human leukocyte antigen class II histocompatibility antigen γ chain (CD74) was selected because of its role in regulating adaptive immune response upon injury and its induction of NFK-β pathways (one of the earliest proinflammatory markers of DMD; Cresswell, 1994; Chen et al., 2005). CD74 showed mRNA up-regulation in the 4-d between area and not the 10-d area (Fig. 4 and Table 3). Immunostaining of CD74 showed expression in the region between the 4-d injection sites but not the region in between the 10-d injection sites (Fig. 7 B). CD74 was also not seen in the injection sites themselves.
CD163 is a marker of specific macrophage subtypes (M2c) in dystrophin-deficient muscles, is associated with increased tissue repair (Villalta et al., 2009), and is a known acute phase-regulated receptor (Madsen et al., 2001). The immunostaining patterns for CD163, cell surface markers of monocytes and macrophages class II (M2c), showed similar patterns (Fig. 7 C).
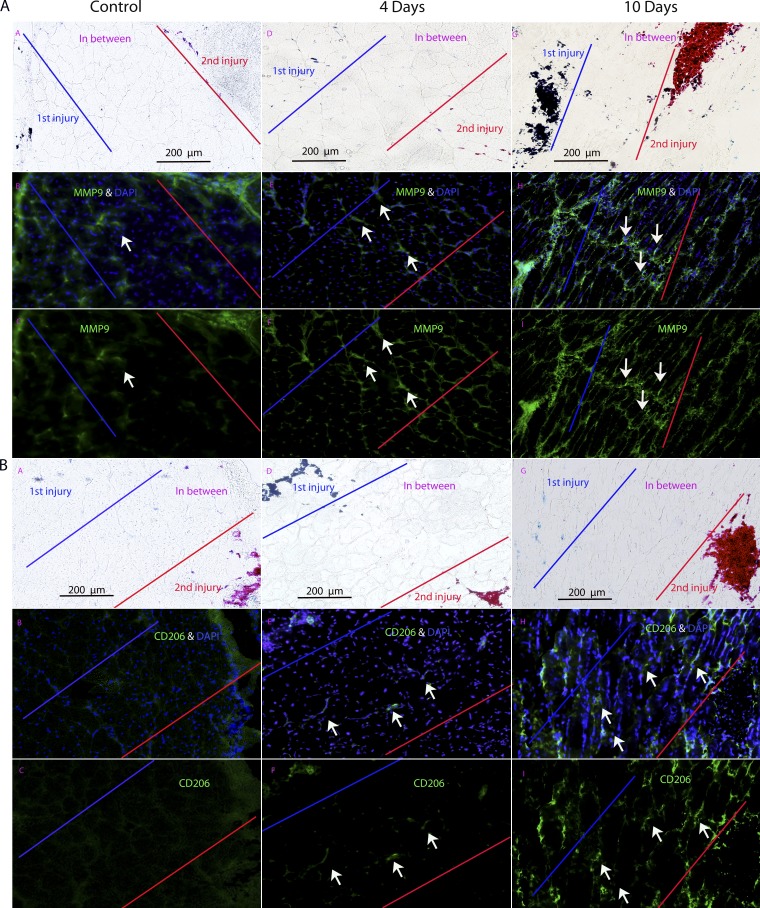
For the 10-d interval injuries, we had shown increases in connective tissue by histological methods (Fig. 6 and Fig. S3 A). To further validate this finding by immunostaining of specific proteins, CD206 (MRC1 [mannose receptor, C type 1]; macrophage marker) and MMP9 (metalloproteinase implicated in dystrophy pathogenesis; Dahiya et al., 2011a,b; Miyazaki et al., 2011) were studied (Fig. 8). Both showed strong expression of the myofiber in-between region of injuries spaced 10 d apart, with lack of immunostaining in 4-d injuries or 0-d control.
Figure 8.
Immunostaining of proteins profibrotic markers MMP9 and CD206 shows preferential expression the area of muscle between injuries spaced 10 d apart. (A and B) Two markers associated with fibrosis, MMP9 (A) and CD206 (B), are shown by immunostaining in muscles from repeated injuries (control = 0 d apart; 4 and 10 d apart). Both markers show preferential expression in the area in between the two injury sites when the injuries are spaced 10 d apart and not 0 (control) or 4 d (n = 3 muscle per group). Arrows indicate immunoreactive material in the microenvironment between two neighboring sites of regeneration.
LCM of human DMD patient muscle shows patterns consistent with asynchronous remodeling
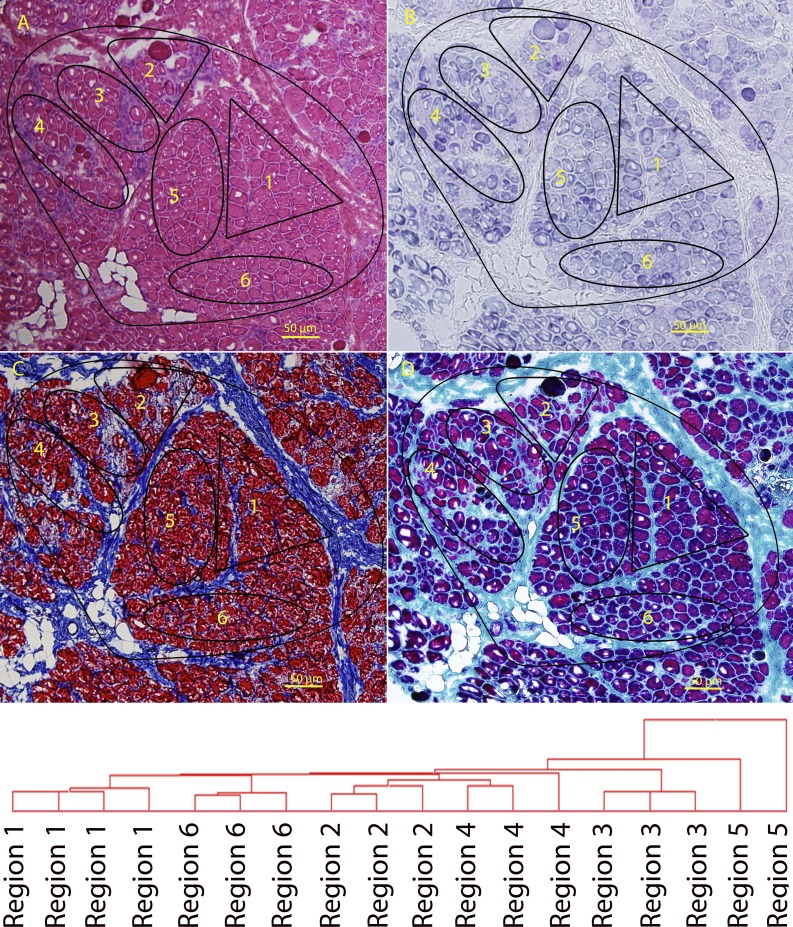
Muscle biopsies from DMD patients have long been recognized to show grouped degeneration and regeneration and focal fibrosis. This histological pattern is consistent with the asynchronous remodeling shown in our murine experimental model. To provide evidence for asynchronous remodeling in human DMD muscle at the molecular level, we studied a frozen diagnostic muscle biopsy from a young child with DMD (3 yr). Frozen sections were stained by hematoxylin and eosin to identify neighboring regions of myofibers both within fascicles and between fascicles (Fig. 9 A), and different subregions (1–6) were identified and then isolated by LCM and subjected to Illumina mRNA profiling. Histochemical staining for mitochondrial enzyme function showed the variable loss of mitochondrial oxidative phosphorylation activity typically seen in DMD biopsies (Fig. 9 B). There were variable degrees of inflammation and fibrosis, as detected by Masson’s trichrome and Gomori’s trichrome (Fig. 9, C and D). Unsupervised clustering of the microarrays from adjacent sections showed clear and consistent differences in expression profiles from the six regions, with replicates of profiles clustering together, yet each region showing a distinct pattern from other regions.
Figure 9.
LCM of regions of a muscle biopsy from a DMD patient. (A–D) A frozen muscle biopsy from a 3-yr-old DMD patient was stained for hematoxylin and eosin (A), mitochondrial enzyme activity (SDH; B), Masson’s modified trichrome (C), and Gomori’s modified trichrome (D). Variable states of degeneration and regeneration can be seen within each fascicle. The different regions indicated (1–6) were isolated by LCM, and individual regions (n ≥ 3 per region) were analyzed by mRNA profiling. The areas were chosen based on visual histopathology, where both fascicles and relatively distinct histological subareas within the fascicles were chosen. Microarray data from replicates of LCM regions microdissected from adjacent cryosections was subjected to unsupervised chip-based clustering. This showed replicates to cluster closely in the dendrogram. This represents heterogeneity within fascicles in DMD muscle, as hypothesized by the asynchronous regeneration model described in the accompanying text.
As we were unable to specifically stage the pathology of the human biopsy, the specific timing of the degeneration and regeneration cycles of the six regions could not be determined. Nonetheless, the profiles were consistent with asynchronous bouts of degeneration/regeneration leading to chronic inflammation and mitochondrial function loss associated with the murine 4-d spaced multiple injuries and the fibrosis associated with the 10-d multiple injuries.
Pharmacogenomics of glucocorticoids on the proposed intercellular signaling networks
Daily glucocorticoids are considered standard of care in DMD (Bushby et al., 2010). The mechanisms of action of glucocorticoids in DMD are thought to be through antiinflammatory mechanisms as in other chronic inflammatory states (Hoffman et al., 2012). However, extensive side effect profiles, such as muscle wasting typically associated with glucocorticoids, and differences in response between human and mouse models have complicated dissection of the molecular mechanism of action (Sali et al., 2012). Recently, we developed a novel dissociative steroid, VBP15, that is similarly antiinflammatory yet lacks most side effects associated with glucocorticoid response element–mediated transactivation subactivities (Heier et al., 2013). We hypothesized that both prednisone and VBP15 might serve to suppress inappropriate intercellular signaling in the regions between asynchronously injured and remodeling areas of muscle, particularly those associated with the proinflammatory 4-d asynchronous cross talk regions.
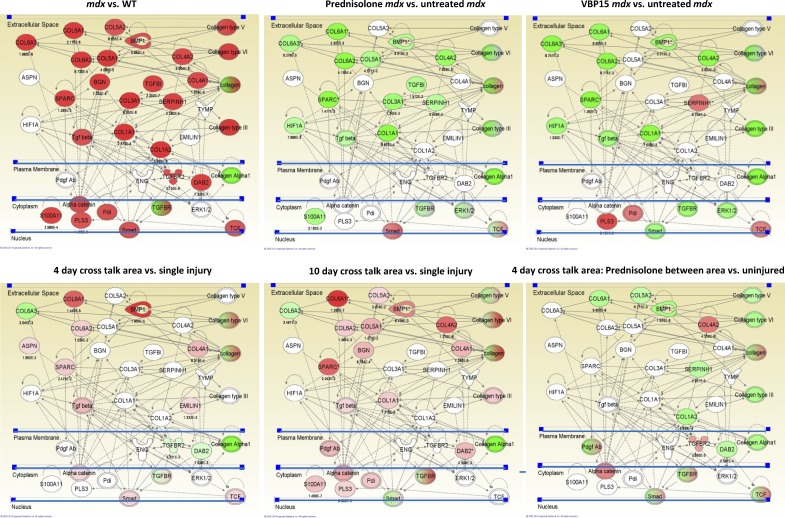
We tested the effects of drug treatment on the TGF-β–based asynchronous remodeling networks both in the mdx mouse model as well as the WT asynchronous remodeling experimental system. For the mdx model, we obtained frozen muscle samples from a previously described preclinical trial of prednisone and VBP15 (Heier et al., 2013). Drug treatments were started at 10 wk of age and continued to 26 wk of age (4 mo of treatment). Skeletal muscles (gastrocnemius) of mice treated with 5 mg/kg/d prednisone and 15 mg/kg/d VBP15 were tested by mRNA profiling (n = 5 per group) using Illumina bead arrays. As a baseline study, the human TGF-β–centered network, including HIF1A and S100A11 nodes, was queried in untreated mdx versus WT mice, and this showed strong up-regulation of the majority of network members (22/29; Fig. 10 A). A marked exception was HIF1A, a key ischemia-responsive gene; however, the rapid expression of HIF1A upon necropsy may lead to postmortem expression in WT animals. Comparing prednisone- and VBP15-treated mdx mice with untreated mdx mice showed both drugs to significantly down-regulate the majority of the pathway members (15/29 prednisone and 12/29 VBP15), including TGF-β ligand and HIF1A. These data suggest that a possible mechanism of action of both prednisone and VBP15 is through suppression of the TGF-β–related networks, leading to more successful regeneration.
Figure 10.
Treatment with prednisone or VBP15 suppresses the TGF-β network in both WT asynchronous remodeling and mdx mice. Top images are mRNA profile data of skeletal muscles from a 4-mo preclinical drug trial of 5 mg/kg/d prednisone and 15 mg/kg/d VBP15. Muscle functional testing and histology on this trial has been previously published (Heier et al., 2013). The TGF-β and fibrosis pathways seen in experimental asynchronous remodeling and DMD biopsies were queried for the relative expression of mRNAs between the indicated samples, with red color indicating up-regulation (p-values underneath network members) and green color indicating down-regulation. Untreated mdx muscle shows strong up-regulation of the network (mdx vs. WT). Treatment with either prednisone (prednisone-treated mdx vs. untreated mdx) or VBP15 (VBP15-treated mdx vs. untreated mdx) shows strong suppression of this network. Bottom images show comparison of expression profiles from LCM samples of asynchronously remodeling regions of WT muscle. The cross talk region between injection sites shows up-regulation of the TGF-β fibrosis network (higher in 10 d compared with 4 d) and prednisone treatment of 4-d asynchronous remodeling mice with prednisone suppresses the network (pretreated, 4-d cross talk area vs. uninjured).
We then used the same data analysis methods for the WT asynchronous remodeling experimental system, similarly determining the effect of drug treatment on expression of the TGF-β network. As a baseline, expression profiles of LCM samples (cross talk area vs. single injury sites) was performed for both 4- and 10-d asynchronous remodeling samples, with both showing up-regulation of the TGF-β network in the cross talk microenvironment (10/29 for 4 d and 16/29 for 10 d; Fig. 10). The 10-d cross talk microenvironment showed more extensive up-regulation of the TGF-β networks compared with 4 d, consistent with the findings presented earlier (Fig. 5). Drug treatment of the WT 4-d asynchronous mouse model was performed 1 d before first injury and then continuously during the 13 d after recovery from the second injection (17 d total), using either 5 mg/kg/d prednisone or 15 mg/kg/d VBP15. LCM and mRNA profiling of the cross talk microenvironment was compared with single injury microenvironment in the same muscle. This showed significant down-regulation by prednisone of TGF-β network members in the cross talk region (Fig. 10).
To validate these findings at the immunohistochemical (protein) level, the WT 4-d asynchronous model drug-treated muscles were immunostained for CD11b, CD74, and CD163 and compared with untreated mice. This analysis showed the expected high expression of each protein marker in the 4-d multiple injury sites (Fig. S5). However, daily administration of either prednisolone or VBP15 markedly reduced the immunopositivity for these antigens (Fig. S5). These data are consistent with both glucocorticoids and VBP15 serving to inhibit inappropriate cross talk in asynchronously remodeling muscle.
Discussion
We used a large-scale data integration approach to query the molecular underpinnings of progressive failure of regeneration in the muscular dystrophies (Fig. S1). Using an initial mRNA profiling dataset of 12 disease groups (117 biopsies and 234 microarrays), we developed an iterative composite approach to identify clusters of transcripts that were associated with the more severe muscle disease classes (DMD, JMD, and LMNA; Fig. 1). Functional clustering of the diagnostic transcripts showed a 56-member TGF-β–centered network consistent with tissue fibrosis. Unsupervised clustering of an independent mRNA profiling dataset of 49 biopsies from four muscle disease groups (DMD, BMD, LGMD2I, and LGMD2B) and controls showed a strong correlation of the TGF-β–centered network with pathological severity rather than diagnostic criteria (Fig. 2). However, all members of this same 56-gene TGF-β–centered network were expressed during specific stages of normal muscle regeneration, using a 27–time point staged murine regeneration dataset (Fig. 3 A). These data analyses established the hypothesis that normal regeneration and progressive dystrophy involved the same proteins; the only distinction was one of timing and degree of expression. The TGF-β network was parsed into stage-specific subnetworks in normal muscle regeneration yet occurred within the same muscle at the same time in severely dystrophic muscle (Fig. 3 B). We hypothesized that asynchronous bouts of degeneration and regeneration in neighboring regions of muscle tissue could result in inappropriate cross talk between the regions.
To test this hypothesis, we developed an experimental model for asynchronous regeneration of muscle in neighboring microenvironments. An initial injury was induced in normal mouse muscle using notexin with the injection site labeled with tattoo dye. A second adjacent injection, labeled with a second tattoo dye, was subsequently made, at defined times (0–10 d). The mice were sacrificed 13 d after the second injection when regeneration would be expected to be largely complete (Fig. S2). Then, regions of the muscle containing both injection sites and regenerating myofibers in between the sites (cross talk microenvironment) were studied by LCM and in situ immunostaining methods. We found that increasing the time between injections led to a greater degree of abnormal collagen deposition (fibrosis; Fig. S3), indicating that the time period between injections was an important determinant of the response of the muscle to the damage. To better define the response of specific subregions of asynchronously remodeling muscle, we used LCM and mRNA profiling as well as immunostaining, histology, and enzyme chemistry experiments. Myofibers in between the two injury sites showed an expression pattern that seemed suspended within the developmental stage that separated the two injections, with the 4- versus 10-d between staged injections sharing little similarity with each other (Fig. 4 and Fig. S3, B and C). The myofibers between the sites of the 10-d repeat injections showed strong expression of transcripts associated with a profibrotic state, with high expression of collagens, and other connective tissue remodeling networks (Fig. 5). This showed considerable overlap of network members, such as TGF-β, COL1A1, COL4A1, COL4A2, and COL6A1, with the human muscle biopsy TGF-β network associated with fibrosis (Fig. 1 B) and was similar to the myotube maturation subgroup of normal staged (synchronous) muscle regeneration (Fig. 3). To assign statistical support to these visualizations of LCM data networks, we took specific network members (mRNAs) and defined expression changes (p-values and fold changes) in specific time points of regeneration, 4-d in-between LCM data and 10-d in-between LCM data (Table 4). The expression patterns were consistent with inappropriately prolonged and extensive connective tissue remodeling, in which signaling led to suspension of progression through the 10-d time window of normal regeneration. In other words, the tissue region between the injection sites staged 10 d apart remained fixed in the 9–10-d regeneration program and appeared unable to successfully mature past this developmental time point.
Our data showed that normal regeneration and failed regeneration involve the same networks, differences largely being caused by the synchrony (episodic) nature of normal regeneration and the asynchronous (repeated injury) nature of failed regeneration. With failed regeneration, regenerating myofibers are likely receiving incorrect temporal cues from their neighbors, and this inappropriate cross talk leads to inability to progress through the normal temporal stages of regeneration.
Glucocorticoids are derivatives of cortisol, the diurnal hormone that sets day/night cycles in most animals and plants. Pharmacological doses of glucocorticoids, typically given on a daily basis, are considered antiinflammatory and remain among the most effective drugs for chronic inflammatory states since their initial use in the 1940s. There is increasing recognition of the role of cortisol and clock genes in establishing cell cycle regulation throughout the body and that disturbances or blunting of diurnal cortisol fluctuations are associated with the proinflammatory state (Niimi et al., 2007; Holderfield and Hughes, 2008). We recently showed that primary bronchial epithelial cells from asthmatic patients are intrinsically dyssynchronous, are proinflammatory, and poorly repair wounds, yet this can be normalized with pulsed glucocorticoids (Freishtat et al., 2011; Alcala et al., 2014).
We reasoned that the effectiveness of glucocorticoids in DMD may be caused by resynchronization of remodeling, thereby rescuing the inappropriate cross talk described in this paper. To test this, we treated WT mice with prednisolone, VBP15 (a dissociative steroid), and vehicle/placebo and then tested for the drug regimens to rescue the inappropriate cross talk in our asynchronous regeneration model. Drug treatments of both mdx mice and the WT asynchronous model were tested (Fig. 10 and Fig. S5). As hypothesized, both prednisolone and VBP15 reduced evidence of cross talk in both mdx and the WT asynchronous models. These data suggest that part of the mechanism of action of both prednisone and VBP15 is through suppression of TGF-β networks induced by asynchronous remodeling.
There is an extensive literature supporting many of the findings in our study, although we believe that this study is the first to synthesize these observations into a single model. Specifically, transient TGF-β activity is essential for maintaining normal tissue repair and regeneration, whereas persistent TGF-β activity results in excessive fibrosis and, ultimately, scarring in skin and internal organs. In dystrophic muscle, TGF-β may affect tissue fibrosis by elevating collagen secretion by fibroblasts and converting myoblasts to fibroblasts. Furthermore, TGF-β can inhibit terminal myogenic differentiation and thus impair muscle repair process (Alexakis et al., 2007).
Our model may provide an explanation for the enigmatic pathology of the mdx mouse. Dystrophin-deficient mouse muscle shows normal histology in early weeks postpartum (0–3 wk) and then a stage of widespread necrosis followed by relatively successful regeneration. The mdx muscle in the range of 2–12 mo shows a mild dystrophy relative to human DMD muscle. Our model suggests that the widespread necrotic phase (2–3 wk) leads to well-synchronized regeneration; the severity of the initial necrotic phase may be beneficial in that it may avoid the poorly synchronized regeneration and pathology-associated signaling cross talk that occurs in human DMD muscle.
The regenerative asynchrony model may be applicable to chronic pathologies in nonmuscle tissues. Asthma involves chronic inflammation of the airway, where pulsed glucocorticoids are standard of care. In other studies, we have shown that normal epithelial wound repair is synchronous, that remodeling of asthmatic cells is asynchronous and proinflammatory, and that pulsed glucocorticoids restores regenerative synchrony and the nonproinflammatory state (Freishtat et al., 2011; Alcala et al., 2014). One could extend these models to chronic liver pathology, poor skin wound healing, or possibly all chronic inflammatory tissue states leading to fibrosis.
Materials and methods
Human patient muscle samples and mRNA profiles
Flash-frozen neuromuscular disease patient muscle biopsies were taken for diagnostic purposes. Normal control biopsies were obtained from baseline samples from exercise experiments in young adult volunteers.
Two datasets were analyzed. The first (test set) has been previously published (Bakay et al., 2006). This set contained Affymetrix mRNA profiles from 117 patient muscle biopsies using both HG-U133A and HG-U133B microarrays (n = 234 microarrays total; GEO accession no. GSE3307). The muscle disease groups were DMD (n = 10), amyotrophic lateral sclerosis (ALS; n = 9), acute quadriplegic myopathy (n = 5), BMD (n = 5), CAL (calpain III gene mutations; LGMD2A; n = 10), DYSF (LGMD2B; n = 10), ED-R (Emery–Dreifuss muscular dystrophy X linked; n = 4), ED-D (n = 4), FKRP (LGMD2I; n = 7), facioscapulohumeral dystrophy (FSH; n = 14), JDM (n = 21), and normal human skeletal muscle (NHM; n = 18). Biopsies were taken generally at the time of diagnosis and were typically from the vastus lateralis. The original published dataset analyzed 13 groups, but we removed the hereditary spastic paraplegia group from analyses reported here because of a high proportion of outlier transcripts and also removed four JDM samples that were not made public. To generate the mRNA profiles, frozen muscle biopsies were solubilized, total RNA was isolated, RNA was converted into biotinylated cRNA probes, hybridized to Affymetrix microarrays, and detected using fluorescent streptavidin, and microarrays were scanned. Quality control metrics used were as previously reported (Tumor Analysis Best Practices Working Group, 2004).
A second set of 49 human patient mRNA profiles was generated using HG-U133 Plus 2.0 microarrays, and this dataset is new to this study. These datasets contained profiles from 6 normal controls, 17 DMD (absence of dystrophin), 11 BMD (present but abnormal dystrophin), 7 LGMD2I (FKRP deficiency, a glycosylation defect), and 8 LGMD2B (DYSF). Patients had a broad range of ages, clinical severity of their disease, and histopathological findings, although all neuromuscular disease patients showed evidence of a dystrophic process (degeneration/regeneration of muscle fibers). Hematoxylin and eosin stains were performed on frozen sections, and the amount of fibrotic replacement (fibrosis) visually approximated for by the same evaluator (E.P. Hoffman) into normal, mild, moderate, or severe fibrosis categories.
Murine muscle regeneration time series
Staged degeneration/regeneration of adult WT mice was induced by intramuscular injection of cardiotoxin into the entire gastrocnemius muscle using a 10-needle manifold (Zhao and Hoffman, 2004). Mice were sacrificed at 27 time points after the injection, and muscles showing homogenous regeneration were selected for mRNA profiling (n = 2 per time point). Frozen muscle samples were solubilized, total RNA was isolated, RNA was converted to biotinylated cRNA, cRNAs were hybridized to MG-U74Av2 microarrays, and fluorescent images were scanned (GEO accession no. GSE469). Analyses and validations of this degeneration/regeneration time series have been previously published (Zhao et al., 2003, 2006; Zhao and Hoffman, 2004).
Biostatistical clustering methods
We developed a novel method of defining gene clusters within a large multigroup dataset, using the 12-group human muscle biopsy dataset (Fig. S1). Mapped image files from microarrays are interpreted using probe set algorithms that provide probe set normalization, chip normalization, and project normalization so that quantitative signals can be assigned to each probe set (transcript) represented on the microarray. We and others have shown that different probe set algorithms often lead to very different interpretations of the same microarray image files (Seo et al., 2004, 2006; Seo and Hoffman, 2006). To avoid loss of sensitivity for gene selection by using a single probe set algorithm, we simultaneously interpreted the muscle biopsy data using three distinct probe set algorithms (dCHIP, MAS 5.0, and PLIER; Fig. S1). Both A and B microarrays were used, leading to two images with distinct probe sets representing each sample.
Two-way clustering on the rows (genes) and columns (muscular disorders) of the data matrix was performed using the VISDA (Visual Statistical Data Analyzer) software to cluster genes (Fig. S1; Wang et al., 2000, 2007; Zhu et al., 2008). Within each gene cluster, the clustering of the muscular disorders was performed using hierarchical clustering with correlation as the similarity measure after a preprocessing step in which all samples within one muscle group were taken as an average.
This led to a series of probe set algorithm-specific gene clusters that grouped the muscular dystrophies into larger groups based upon the specific microarray used (U133A chip or U133B chip). Thus, PA cluster i refers to PLIER algorithm U133A chip dataset, MA cluster i refers to MAS 5.0 algorithm U133A chip dataset, and dA cluster i refers to dCHIP algorithm U133A chip dataset (Fig. S1 and formulas in this section). Each gene cluster was then tested for gene–gene and gene–protein interactions between cluster members using IPA. The top-ranked networks were then retained for each cluster i (PA network i, MA network i, and dA network i; Fig. S1). We then merged networks and assigned cumulative scores. To accomplish this, we implemented programs to query overlaps among networks and then derived a total score for each network based on its overlaps with all other networks of each gene-disorder pattern. The probability for two networks, S1 and S2 with n1 and n2 genes, respectively, having m overlapped genes can be calculated as
given that S1, S2, and the overlapped genes belong to the global network with N genes (N = 26,400, the total number of mammalian genes in Ingenuity Pathway Knowledge base), and C represents a mathematical operation of combination. C of N and m gives the number of subsets of m distinct elements of a set of N elements. Because this probability indicates the likelihood of the overlap of the two networks occurring randomly, −log10(p(m)) may be used as a score for specifying the reliability of the overlap. In addition, the IPA dynamically computes relevant networks by focusing on the genes of interest and delivers a set of scored networks for each gene cluster. A composite score for each network was then calculated by summing up the IPA score and the total score based on network overlaps; the networks were ranked based on their composite scores.
Cross-species query of time series data
Once a gene cluster of interest was selected, probe sets were tested for recognition of canonical transcripts (e.g., 3′ end of well-documented transcript units), and then, these human transcript units were mapped to corresponding rat and mouse transcript units using Affymetrix NetAffx Analysis Center. The Affymetrix probe set IDs corresponding to the transcript units were then queried against the mouse time series data. All mouse image files were analyzed using the PLIER probe set algorithm. Candidate gene clusters were visualized in mouse datasets using heat maps (hierarchical clustering).
All differentially regulated transcripts visualized were tested for statistical significance using t test, without correction for multiple testing. For time series data, adjacent time points were grouped and statistically compared with a relevant control region of the time series.
Asynchronous regeneration protocol
WT BL57 mice of 4–8 wk of age were used for all asynchronous remodeling experiments. To induce asynchronous bouts of regeneration, a 20-guage needle was filled with 10 µl of 5-µg/ml notexin and then dipped in tattoo dye (blue for first injection; red for second injection). Both gastrocnemii were surgically exposed, and the initial injection was made distally to proximally along the axis of the muscle. The wound was closed for the number of days indicated, and then, the second injection was performed after the time annotated in each experiment. The initial injection site could be visualized in the muscle as a result of the tattoo dye, and the second injection was initiated adjacent the first site. Mice were euthanized 13 d after the second injury, and gastrocnemii were collected.
For drug treatments with 5 mg/kg/d prednisolone, 15 mg/kg/d VBP15, or vehicle, the drug was delivered in sugar syrup orally. We had five mice in each group. Drug treatments were started 24 h before the initial notexin injection and then continued daily until sacrifice.
Van Gieson measures of fibrotic tissue
Reinjured muscle sections were stained by the Van Gieson method (n = 4 muscles per group). Images were taken and analyzed with ImageJ (National Institutes of Health) to measure the extent of the connective tissue. We assayed the amount of connective tissue per field of tissue by measuring the number of pixels occupied by red staining and then measuring the number of pixels covered by the muscle tissue. The data are then represented as the percentage of muscle that is collagenous connective tissue.
Laser capture microdissection
Flash-frozen muscle biopsies or specimens were sectioned at 10-µm thickness on RNase-free glass slides (CellCut; Molecular Machines & Industries, Inc.). Slides were immediately placed in 99% ethanol to fix. Rapid hematoxylin and eosin stain using RNase-free solutions (Molecular Machines & Industries, Inc.) was then performed followed by laser capture microdissection (CellCut Plus instrument) to identify and cut the areas of interest. We used tubes with an adhesive cap to collect LCM-isolated regions (Molecular Machines & Industries, Inc.).
In brief, 15 consecutive sections for mice at 4 and 10 d were subjected to LCM to isolate each region on each muscle, and cRNA amplification was performed from the combined sections. 3–4 of the 15 LCM sections (per area per mouse) were successful in mRNA profiling, and these showed excellent chip-based clustering. Although the LCM was performed on only one mouse for 4 d and one mouse for 10 d (in part because of the need to identify closely spaced tattoo dye injection sites), additional mice were used for immunostaining validations.
LCM mRNA profiling
RNA isolation was performed using PicoPure kit (Applied Biosystems), which includes on-column DNase digestion. Two rounds of T7-mediated cRNA amplification were performed (RiboAmp HS kit [Applied Biosystems] and then TotalPrep-96 RNA Amplification kit [Life Technologies]). The mRNA profiling on asynchronous mouse LCM experiments was performed in triplicate on serial sections. Murine WG-6 v2.0 Expression BeadChips (Illumina) containing 45,200 transcript probes or HumanHT-12 v4 Expression BeadChips (Illumina) containing 47,000 transcript probes were used. Results of expression arrays were read using the HiScan SQ System (Illumina). Statistical analyses (P ≤ 0.05 with correction for multiple testing [False Discovery Rate]) and a fold change ≤1.5 or ≥1.5) were performed to identify dysregulated genes in the areas of interest using Partek Genomics Suite software (Partek, Inc.). IPA was used to query protein–protein and gene–protein interactions.
TaqMan gene expression assay
The RiboAmp HS kit was used to perform two rounds of amplification on total RNA isolated from LCM samples (n = 3 muscles per group) and convert amplified RNA to cDNA. Quantitative gene expression analysis was performed by TaqMan-based qRT-PCR on a real-time PCR system (7900 HT Fast; Applied Biosystems). We used TaqMan Gene Expression kits obtained from Applied Biosystems to run qRT-PCR: Cox6b1 (Mm00824357_m1), Ndufs6 (Mm02529639_u1), Ndufb5 (Mm00452592_m1), Ndufb6 (Mm01208591_g1), and HPRT (hypoxanthine guanine phosphoribosyltransferase; Mm01318747_g1). The expression of HPRT was analyzed as endogenous control simultaneously, and the target gene expression in each sample normalized to HPRT. Thermal cycling conditions were 95°C for 10 min, 50 cycles at 95°C for 15 s, and 60°C for 1 min.
Immunostaining
Monoclonal rat anti–mouse CD11b (eBioscience), mouse anti–human CD74 (Abcam), mouse anti–human macrophage surface antigen (AM-3K/CD163), mouse anti–human mannose receptor (CD206; Abcam), and mouse anti–human MMP9 (Santa Cruz Biotechnology, Inc.) monoclonal primary antibodies were used to perform immunofluorescence assays. The Mouse on Mouse kit (Vector Laboratories) was used to reduce background staining when using mouse monoclonal antibodies on mouse tissue sections (n = 3 muscles per group). Goat anti–mouse (Invitrogen) and goat anti–rat (Invitrogen) secondary antibodies conjugated with Cy3 or Alexa Fluor 488 were used. Nuclei were stained with DAPI (OCHEM). The images were acquired using a fluorescence microscope (BX61; Olympus).
Histochemistry
NADH, SDH, COX, costaining of NADH and COX, Masson’s trichrome, and Gomori’s modified trichrome were performed on 10-µm biopsy frozen sections to evaluate histology and mitochondrial enzymatic activities (Dubowitz and Sewry, 2007). Murine gastrocnemius 10-µm sections (n = 3 muscles per group) were stained following a standard Van Gieson protocol (Dubowitz and Sewry, 2007). A minimum of nine nonoverlapping images were analyzed in ImageJ for the percentage of connective tissue per field of muscle tissue.
Drug treatment of mdx and WT asynchronous remodeling mice
Drug-treated mdx mice were from the previously reported preclinical trial comparing 4 mo of 5 mg/kg/d prednisone treatment to 15 mg/kg/d VBP15 treatment (Heier et al., 2013). Frozen archival muscle samples were obtained for WT, mdx untreated, and the two drug-treated groups (prednisone and VBP15). RNA was isolated, and mRNA profiling was performed using Illumina bead arrays, according to the manufacturer’s protocol (n = 5 per group). Bioinformatics analyses were performed using TGF-β pathway candidates. Drug treatment of the WT asynchronous remodeling mice (n = 5 per group) was performed using the same drugs and treatment regimens (5 mg/kg/d prednisone; 15 mg/kg/d VBP15), with treatment starting the day before the first surgery until the day before euthanasia.
Microscopic image acquisition
All images were acquired using a microscope (BX61; Olympus), with U Plan S Apochromat 20×/0.75 NA dry objective, at room temperature. Photography was performed using a camera (DP11; Olympus) with DP controller 3.2.1.276 and DP Manager 3.1.1.208 software.
Online supplemental material
Fig. S1 shows a flow chart of the data integration and biochemical network analysis on the 12-group human muscle transcriptional profiling data. Fig. S2 shows LCM methods and chip clustering of mRNA profiles of LCM regions of multiply injured muscle. Fig. S3 shows quantitative analysis of endomysial fibrosis in asynchronous remodeled muscle samples. Fig. S4 shows histochemical stains for mitochondrial activity showing a relative loss of mitochondrial oxidative phosphorylation in the area between injuries spaced 4 d apart. Fig. S5 shows treatment of asynchronous remodeling mouse muscle with prednisone or VBP15 suppresses the proinflammatory proteins in between injuries spaced by 4 d. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201402079/DC1.
Supplementary Material
Acknowledgments
This work was supported by the US Department of Defense (W81XWH-05-0334), National Institutes of Health (R01 NS029525, U54 HD053177, and R24 HD050846), Children’s National Medical Center Board of Visitors, and the Muscular Dystrophy Association USA. The development of VBP15 is supported by the Congressionally Directed Medical Research Programs of the Department of Defense, Muscular Dystrophy Association Venture Philanthropy program, and the National Institutes of Health National Center for Advancing Translational Sciences, Therapeutics for Rare and Neglected Diseases program.
Competing financial interests: E.P. Hoffman and K. Nagaraju are coinventors of patent WO 2011127048 A3 (Non-hormonal steroid modulators of NF-κB for treatment of disease), and this has been assigned to ReveraGen Biopharma. E.P. Hoffman and K. Nagaraju own founder shares of ReveraGen Biopharma and are members of the Board of Directors. The authors declare no further competing financial interests.
Author contributions: S. Dadgar contributed to the experimental design, data generation and interpretation, and manuscript writing. Z. Wang contributed to the bioinformatics methods development and conduct, figures, and manuscript writing. H. Johnston-Carey contributed to the experimental design, data generation, and writing. A. Kesari contributed to the human muscle biopsy expression profiling design and conduct. K. Nagaraju contributed to the experimental design and data interpretation. Y.-W. Chen contributed to the human muscle biopsy expression profiling design, conduct, and data interpretation. D.A. Hill contributed to the immunohistochemical and histological stains and data interpretation. T. Partridge contributed to the experimental design and data interpretation. M. Giri contributed to the bioinformatics methods and data interpretation. J. Nazarian contributed to the experimental design and data interpretation. R. Freishtat to the contributed to the experimental design and data interpretation. J. Xuan contributed to the bioinformatics methods development. Y. Wang contributed to the bioinformatics methods development. E.P. Hoffman contributed to the experimental design and interpretation and manuscript writing.
Footnotes
Abbreviations used in this paper:
- ALS
- amyotrophic lateral sclerosis
- BMD
- Becker muscular dystrophy
- COX
- cytochrome c oxidase
- DMD
- Duchenne muscular dystrophy
- FSH
- facioscapulohumeral dystrophy
- JDM
- juvenile dermatomyositis
- LCM
- laser capture microscopy
- NHM
- normal human skeletal muscle
- qRT-PCR
- quantitative RT-PCR
- SDH
- succinate dehydrogenase
- WT
- wild type
References
- Aguennouz, M., Vita G.L., Messina S., Cama A., Lanzano N., Ciranni A., Rodolico C., Di Giorgio R.M., and Vita G.. 2011. Telomere shortening is associated to TRF1 and PARP1 overexpression in Duchenne muscular dystrophy. Neurobiol. Aging. 32:2190–2197 10.1016/j.neurobiolaging.2010.01.008 [DOI] [PubMed] [Google Scholar]
- Akhurst, R.J., and Hata A.. 2012. Targeting the TGFβ signalling pathway in disease. Nat. Rev. Drug Discov. 11:790–811 10.1038/nrd3810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcala, S.E., Benton A.S., Watson A.M., Kureshi S., Reeves E.M., Damsker J., Wang Z., Nagaraju K., Anderson J., Williams A.M., et al. . 2014. Mitotic asynchrony induces transforming growth factor-β1 secretion from airway epithelium. Am. J. Respir. Cell Mol. Biol. 51:363–369 10.1165/rcmb.2013-0396OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexakis, C., Partridge T., and Bou-Gharios G.. 2007. Implication of the satellite cell in dystrophic muscle fibrosis: a self-perpetuating mechanism of collagen overproduction. Am. J. Physiol. Cell Physiol. 293:C661–C669 10.1152/ajpcell.00061.2007 [DOI] [PubMed] [Google Scholar]
- Bakay, M., Chen Y.W., Borup R., Zhao P., Nagaraju K., and Hoffman E.P.. 2002. Sources of variability and effect of experimental approach on expression profiling data interpretation. BMC Bioinformatics. 3:4 10.1186/1471-2105-3-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakay, M., Wang Z., Melcon G., Schiltz L., Xuan J., Zhao P., Sartorelli V., Seo J., Pegoraro E., Angelini C., et al. . 2006. Nuclear envelope dystrophies show a transcriptional fingerprint suggesting disruption of Rb-MyoD pathways in muscle regeneration. Brain. 129:996–1013 10.1093/brain/awl023 [DOI] [PubMed] [Google Scholar]
- Baron, D., Magot A., Ramstein G., Steenman M., Fayet G., Chevalier C., Jourdon P., Houlgatte R., Savagner F., and Pereon Y.. 2011. Immune response and mitochondrial metabolism are commonly deregulated in DMD and aging skeletal muscle. PLoS ONE. 6:e26952 10.1371/journal.pone.0026952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bello, L., Piva L., Barp A., Taglia A., Picillo E., Vasco G., Pane M., Previtali S.C., Torrente Y., Gazzerro E., et al. . 2012. Importance of SPP1 genotype as a covariate in clinical trials in Duchenne muscular dystrophy. Neurology. 79:159–162 10.1212/WNL.0b013e31825f04ea [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya, S., Kelley K., Melichian D.S., Tamaki Z., Fang F., Su Y., Feng G., Pope R.M., Budinger G.R., Mutlu G.M., et al. . 2013. Toll-like receptor 4 signaling augments transforming growth factor-β responses: a novel mechanism for maintaining and amplifying fibrosis in scleroderma. Am. J. Pathol. 182:192–205 10.1016/j.ajpath.2012.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blau, H.M., Webster C., and Pavlath G.K.. 1983. Defective myoblasts identified in Duchenne muscular dystrophy. Proc. Natl. Acad. Sci. USA. 80:4856–4860 10.1073/pnas.80.15.4856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen, T., Jenkins R.H., and Fraser D.J.. 2013. MicroRNAs, transforming growth factor beta-1, and tissue fibrosis. J. Pathol. 229:274–285 10.1002/path.4119 [DOI] [PubMed] [Google Scholar]
- Bushby, K., Finkel R., Birnkrant D.J., Case L.E., Clemens P.R., Cripe L., Kaul A., Kinnett K., McDonald C., Pandya S., et al. ; DMD Care Considerations Working Group. 2010. Diagnosis and management of Duchenne muscular dystrophy, part 1: diagnosis, and pharmacological and psychosocial management. Lancet Neurol. 9:77–93 10.1016/S1474-4422(09)70271-6 [DOI] [PubMed] [Google Scholar]
- Chen, Y.W., Nagaraju K., Bakay M., McIntyre O., Rawat R., Shi R., and Hoffman E.P.. 2005. Early onset of inflammation and later involvement of TGFbeta in Duchenne muscular dystrophy. Neurology. 65:826–834 10.1212/01.wnl.0000173836.09176.c4 [DOI] [PubMed] [Google Scholar]
- Chen, Y.W., Shi R., Geraci N., Shrestha S., Gordish-Dressman H., and Pachman L.M.. 2008. Duration of chronic inflammation alters gene expression in muscle from untreated girls with juvenile dermatomyositis. BMC Immunol. 9:43 10.1186/1471-2172-9-43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cresswell, P.1994. Assembly, transport, and function of MHC class II molecules. Annu. Rev. Immunol. 12:259–293 10.1146/annurev.iy.12.040194.001355 [DOI] [PubMed] [Google Scholar]
- Dahiya, S., Bhatnagar S., Hindi S.M., Jiang C., Paul P.K., Kuang S., and Kumar A.. 2011a. Elevated levels of active matrix metalloproteinase-9 cause hypertrophy in skeletal muscle of normal and dystrophin-deficient mdx mice. Hum. Mol. Genet. 20:4345–4359 10.1093/hmg/ddr362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahiya, S., Givvimani S., Bhatnagar S., Qipshidze N., Tyagi S.C., and Kumar A.. 2011b. Osteopontin-stimulated expression of matrix metalloproteinase-9 causes cardiomyopathy in the mdx model of Duchenne muscular dystrophy. J. Immunol. 187:2723–2731 10.4049/jimmunol.1101342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decary, S., Hamida C.B., Mouly V., Barbet J.P., Hentati F., and Butler-Browne G.S.. 2000. Shorter telomeres in dystrophic muscle consistent with extensive regeneration in young children. Neuromuscul. Disord. 10:113–120 10.1016/S0960-8966(99)00093-0 [DOI] [PubMed] [Google Scholar]
- Dubowitz, V., and Sewry C.A.. 2007. Muscle Biopsy: A Practical Approach. Third edition Saunders Elsevier, China: 626 pp [Google Scholar]
- Flanigan, K.M., Ceco E., Lamar K.M., Kaminoh Y., Dunn D.M., Mendell J.R., King W.M., Pestronk A., Florence J.M., Mathews K.D., et al. . United Dystrophinopathy Project. 2013. LTBP4 genotype predicts age of ambulatory loss in Duchenne muscular dystrophy. Ann. Neurol. 73:481–488 10.1002/ana.23819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freishtat, R.J., Watson A.M., Benton A.S., Iqbal S.F., Pillai D.K., Rose M.C., and Hoffman E.P.. 2011. Asthmatic airway epithelium is intrinsically inflammatory and mitotically dyssynchronous. Am. J. Respir. Cell Mol. Biol. 44:863–869 10.1165/rcmb.2010-0029OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaschen, F.P., Hoffman E.P., Gorospe J.R., Uhl E.W., Senior D.F., Cardinet G.H. III, and Pearce L.K.. 1992. Dystrophin deficiency causes lethal muscle hypertrophy in cats. J. Neurol. Sci. 110:149–159 10.1016/0022-510X(92)90022-D [DOI] [PubMed] [Google Scholar]
- Heier, C.R., Damsker J.M., Yu Q., Dillingham B.C., Huynh T., Van der Meulen J.H., Sali A., Miller B.K., Phadke A., Scheffer L., et al. . 2013. VBP15, a novel anti-inflammatory and membrane-stabilizer, improves muscular dystrophy without side effects. EMBO Mol. Med. 5:1569–1585 10.1002/emmm.201302621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman, E.P., Brown R.H. Jr, and Kunkel L.M.. 1987. Dystrophin: the protein product of the Duchenne muscular dystrophy locus. Cell. 51:919–928 10.1016/0092-8674(87)90579-4 [DOI] [PubMed] [Google Scholar]
- Hoffman, E.P., Reeves E., Damsker J., Nagaraju K., McCall J.M., Connor E.M., and Bushby K.. 2012. Novel approaches to corticosteroid treatment in Duchenne muscular dystrophy. Phys. Med. Rehabil. Clin. N. Am. 23:821–828 10.1016/j.pmr.2012.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holderfield, M.T., and Hughes C.C.. 2008. Crosstalk between vascular endothelial growth factor, notch, and transforming growth factor-beta in vascular morphogenesis. Circ. Res. 102:637–652 10.1161/CIRCRESAHA.107.167171 [DOI] [PubMed] [Google Scholar]
- Irizarry, R.A., Warren D., Spencer F., Kim I.F., Biswal S., Frank B.C., Gabrielson E., Garcia J.G., Geoghegan J., Germino G., et al. . 2005. Multiple-laboratory comparison of microarray platforms. Nat. Methods. 2:345–350 10.1038/nmeth756 [DOI] [PubMed] [Google Scholar]
- Karkampouna, S., Ten Dijke P., Dooley S., and Julio M.K.. 2012. TGFβ signaling in liver regeneration. Curr. Pharm. Des. 18:4103–4113 10.2174/138161212802430521 [DOI] [PubMed] [Google Scholar]
- Klymiuk, N., Blutke A., Graf A., Krause S., Burkhardt K., Wuensch A., Krebs S., Kessler B., Zakhartchenko V., Kurome M., et al. . 2013. Dystrophin-deficient pigs provide new insights into the hierarchy of physiological derangements of dystrophic muscle. Hum. Mol. Genet. 22:4368–4382 10.1093/hmg/ddt287 [DOI] [PubMed] [Google Scholar]
- Koh, M.Y., and Powis G.. 2012. Passing the baton: the HIF switch. Trends Biochem. Sci. 37:364–372 10.1016/j.tibs.2012.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollias, H.D., and McDermott J.C.. 2008. Transforming growth factor-beta and myostatin signaling in skeletal muscle. J. Appl. Physiol. 104:579–587 10.1152/japplphysiol.01091.2007 [DOI] [PubMed] [Google Scholar]
- Kornegay, J.N., Bogan J.R., Bogan D.J., Childers M.K., Li J., Nghiem P., Detwiler D.A., Larsen C.A., Grange R.W., Bhavaraju-Sanka R.K., et al. . 2012a. Canine models of Duchenne muscular dystrophy and their use in therapeutic strategies. Mamm. Genome. 23:85–108 10.1007/s00335-011-9382-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornegay, J.N., Childers M.K., Bogan D.J., Bogan J.R., Nghiem P., Wang J., Fan Z., Howard J.F. Jr, Schatzberg S.J., Dow J.L., et al. . 2012b. The paradox of muscle hypertrophy in muscular dystrophy. Phys. Med. Rehabil. Clin. N. Am. 23:149–172 10.1016/j.pmr.2011.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kottlors, M., and Kirschner J.. 2010. Elevated satellite cell number in Duchenne muscular dystrophy. Cell Tissue Res. 340:541–548 10.1007/s00441-010-0976-6 [DOI] [PubMed] [Google Scholar]
- MacDonald, E.M., and Cohn R.D.. 2012. TGFβ signaling: its role in fibrosis formation and myopathies. Curr. Opin. Rheumatol. 24:628–634 10.1097/BOR.0b013e328358df34 [DOI] [PubMed] [Google Scholar]
- Madsen, M., Graversen J.H., and Moestrup S.K.. 2001. Haptoglobin and CD163: captor and receptor gating hemoglobin to macrophage lysosomes. Redox Rep. 6:386–388 10.1179/135100001101536490 [DOI] [PubMed] [Google Scholar]
- Maier, F., and Bornemann A.. 1999. Comparison of the muscle fiber diameter and satellite cell frequency in human muscle biopsies. Muscle Nerve. 22:578–583 [DOI] [PubMed] [Google Scholar]
- Miyazaki, D., Nakamura A., Fukushima K., Yoshida K., Takeda S., and Ikeda S.. 2011. Matrix metalloproteinase-2 ablation in dystrophin-deficient mdx muscles reduces angiogenesis resulting in impaired growth of regenerated muscle fibers. Hum. Mol. Genet. 20:1787–1799 10.1093/hmg/ddr062 [DOI] [PubMed] [Google Scholar]
- Nghiem, P.P., Hoffman E.P., Mittal P., Brown K.J., Schatzberg S.J., Ghimbovschi S., Wang Z., and Kornegay J.N.. 2013. Sparing of the dystrophin-deficient cranial sartorius muscle is associated with classical and novel hypertrophy pathways in GRMD dogs. Am. J. Pathol. 183:1411–1424 10.1016/j.ajpath.2013.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niimi, H., Pardali K., Vanlandewijck M., Heldin C.H., and Moustakas A.. 2007. Notch signaling is necessary for epithelial growth arrest by TGF-β. J. Cell Biol. 176:695–707 10.1083/jcb.200612129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oexle, K., Zwirner A., Freudenberg K., Kohlschütter A., and Speer A.. 1997. Examination of telomere lengths in muscle tissue casts doubt on replicative aging as cause of progression in Duchenne muscular dystrophy. Pediatr. Res. 42:226–231 10.1203/00006450-199708000-00016 [DOI] [PubMed] [Google Scholar]
- Pegoraro, E., Hoffman E.P., Piva L., Gavassini B.F., Cagnin S., Ermani M., Bello L., Soraru G., Pacchioni B., Bonifati M.D., et al. ; Cooperative International Neuromuscular Research Group. 2011. SPP1 genotype is a determinant of disease severity in Duchenne muscular dystrophy. Neurology. 76:219–226 10.1212/WNL.0b013e318207afeb [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippou, A., Maridaki M., and Koutsilieris M.. 2008. The role of urokinase-type plasminogen activator (uPA) and transforming growth factor beta 1 (TGFbeta1) in muscle regeneration. In Vivo. 22:735–750 [PubMed] [Google Scholar]
- Sali, A., Guerron A.D., Gordish-Dressman H., Spurney C.F., Iantorno M., Hoffman E.P., and Nagaraju K.. 2012. Glucocorticoid-treated mice are an inappropriate positive control for long-term preclinical studies in the mdx mouse. PLoS ONE. 7:e34204 10.1371/journal.pone.0034204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo, J., and Hoffman E.P.. 2006. Probe set algorithms: is there a rational best bet? BMC Bioinformatics. 7:395 10.1186/1471-2105-7-395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo, J., Bakay M., Chen Y.W., Hilmer S., Shneiderman B., and Hoffman E.P.. 2004. Interactively optimizing signal-to-noise ratios in expression profiling: project-specific algorithm selection and detection p-value weighting in Affymetrix microarrays. Bioinformatics. 20:2534–2544 10.1093/bioinformatics/bth280 [DOI] [PubMed] [Google Scholar]
- Seo, J., Gordish-Dressman H., and Hoffman E.P.. 2006. An interactive power analysis tool for microarray hypothesis testing and generation. Bioinformatics. 22:808–814 10.1093/bioinformatics/btk052 [DOI] [PubMed] [Google Scholar]
- Serrano, A.L., Mann C.J., Vidal B., Ardite E., Perdiguero E., and Muñoz-Cánoves P.. 2011. Cellular and molecular mechanisms regulating fibrosis in skeletal muscle repair and disease. Curr. Top. Dev. Biol. 96:167–201 10.1016/B978-0-12-385940-2.00007-3 [DOI] [PubMed] [Google Scholar]
- Solovjov, D.A., Pluskota E., and Plow E.F.. 2005. Distinct roles for the α and β subunits in the functions of integrin αMβ2. J. Biol. Chem. 280:1336–1345 10.1074/jbc.M406968200 [DOI] [PubMed] [Google Scholar]
- Tezak, Z., Hoffman E.P., Lutz J.L., Fedczyna T.O., Stephan D., Bremer E.G., Krasnoselska-Riz I., Kumar A., and Pachman L.M.. 2002. Gene expression profiling in DQA1*0501+ children with untreated dermatomyositis: a novel model of pathogenesis. J. Immunol. 168:4154–4163 10.4049/jimmunol.168.8.4154 [DOI] [PubMed] [Google Scholar]
- Tumor Analysis Best Practices Working Group. 2004. Expression profiling—best practices for data generation and interpretation in clinical trials. Nat. Rev. Genet. 5:229–237 10.1038/nrg1297 [DOI] [PubMed] [Google Scholar]
- Villalta, S.A., Nguyen H.X., Deng B., Gotoh T., and Tidball J.G.. 2009. Shifts in macrophage phenotypes and macrophage competition for arginine metabolism affect the severity of muscle pathology in muscular dystrophy. Hum. Mol. Genet. 18:482–496 10.1093/hmg/ddn376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J., Li H., Zhu Y., Yousef M., Nebozhyn M., Showe M., Showe L., Xuan J., Clarke R., and Wang Y.. 2007. VISDA: an open-source caBIG analytical tool for data clustering and beyond. Bioinformatics. 23:2024–2027 10.1093/bioinformatics/btm290 [DOI] [PubMed] [Google Scholar]
- Wang, Y., Luo L., Freedman M.T., and Kung S.Y.. 2000. Probabilistic principal component subspaces: a hierarchical finite mixture model for data visualization. IEEE Trans. Neural Netw. 11:625–636 10.1109/72.846734 [DOI] [PubMed] [Google Scholar]
- Watkins, S.C., and Cullen M.J.. 1986. A quantitative comparison of satellite cell ultrastructure in Duchenne muscular dystrophy, polymyositis, and normal controls. Muscle Nerve. 9:724–730 10.1002/mus.880090808 [DOI] [PubMed] [Google Scholar]
- Watkins, S.C., and Cullen M.J.. 1988. A quantitative study of myonuclear and satellite cell nuclear size in Duchenne’s muscular dystrophy, polymyositis and normal human skeletal muscle. Anat. Rec. 222:6–11 10.1002/ar.1092220103 [DOI] [PubMed] [Google Scholar]
- Zhao, P., and Hoffman E.P.. 2004. Embryonic myogenesis pathways in muscle regeneration. Dev. Dyn. 229:380–392 10.1002/dvdy.10457 [DOI] [PubMed] [Google Scholar]
- Zhao, P., Seo J., Wang Z., Wang Y., Shneiderman B., and Hoffman E.P.. 2003. In vivo filtering of in vitro expression data reveals MyoD targets. C. R. Biol. 326:1049–1065 10.1016/j.crvi.2003.09.035 [DOI] [PubMed] [Google Scholar]
- Zhao, P., Caretti G., Mitchell S., McKeehan W.L., Boskey A.L., Pachman L.M., Sartorelli V., and Hoffman E.P.. 2006. Fgfr4 is required for effective muscle regeneration in vivo. Delineation of a MyoD-Tead2-Fgfr4 transcriptional pathway. J. Biol. Chem. 281:429–438 10.1074/jbc.M507440200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, Y., Li H., Miller D.J., Wang Z., Xuan J., Clarke R., Hoffman E.P., and Wang Y.. 2008. caBIG VISDA: modeling, visualization, and discovery for cluster analysis of genomic data. BMC Bioinformatics. 9:383 10.1186/1471-2105-9-383 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.