In both Xenopus laevis egg extract and human cells, an increase in cytosolic calcium activates the endogenous ribonuclease XendoU/hEndoU, which localizes to the ER, promotes RNA cleavage and RNP removal, and induces ER network assembly.
Abstract
How cells shape and remodel organelles in response to cellular signals is a poorly understood process. Using Xenopus laevis egg extract, we found that increases in cytosolic calcium lead to the activation of an endogenous ribonuclease, XendoU. A fraction of XendoU localizes to the endoplasmic reticulum (ER) and is required for nuclear envelope assembly and ER network formation in a catalysis-dependent manner. Using a purified vesicle fusion assay, we show that XendoU functions on the surface of ER membranes to promote RNA cleavage and ribonucleoprotein (RNP) removal. Additionally, RNA removal from the surface of vesicles by RNase treatment leads to increased ER network formation. Using human tissue culture cells, we found that hEndoU localizes to the ER, where it promotes the formation of ER tubules in a catalysis-dependent manner. Together, these results demonstrate that calcium-activated removal of RNA from membranes by XendoU promotes and refines ER remodeling and the formation of tubular ER.
Introduction
Developmental differentiation results in changes to intracellular organization and organelle structure as cells adapt to fulfill more specialized roles. The transition from oocyte to embryo requires several coordinated cellular events, including release from meiotic arrest, exocytosis of cortical granules, fusion of male and female pronuclei, cytoskeletal rearrangements, and changes in protein expression (for review see Vacquier, 1981; Whitaker, 2006a; Horner and Wolfner, 2008; Perry and Verlhac, 2008). The developmental program and signaling cascade is initiated by a transient intracellular calcium wave released through IP3 receptors that reside on the surface of the ER, in addition to an influx of extracellular calcium (Ridgway et al., 1977; Steinhardt et al., 1977; Gilkey et al., 1978; Busa and Nuccitelli, 1985; Miyazaki et al., 1992). Work in many other cell types has shown that calcium is one of the most widely used signaling molecules regulating such diverse processes as neuronal signaling (Berridge, 1998), transcription (Ikura et al., 2002), mitochondrial function (Brookes et al., 2004), and apoptosis (Nicotera and Orrenius, 1998). Although it is known that calcium plays a role in numerous cellular events, it is unknown if there are additional roles for calcium during fertilization and early development.
One of the major internal stores for calcium within a cell is the ER (Jaffe, 1983; Eisen and Reynolds, 1985). The ER itself is a complex, continuous membrane structure with a common lumen and includes the nuclear envelope and peripheral ER, which is comprised of sheets (rough ER) and tubules (smooth ER; Baumann and Walz, 2001; Shibata et al., 2006). ER tubules are connected by three-way junctions and include regions of high membrane curvature (Voeltz et al., 2006). In contrast, ER sheets are flat membrane structures with very little curvature except at the edges (Shibata et al., 2010). During interphase, the ER maintains its basic structure, though changes in interactions with the microtubule cytoskeleton (Waterman-Storer and Salmon, 1998; Grigoriev et al., 2008; Friedman et al., 2010; Wang et al., 2013) and membrane-bound organelles (English and Voeltz, 2013a), as well as continual structural rearrangements (Du et al., 2004), render this organelle quite dynamic. During mitosis, the ER changes morphology and the nuclear envelope fragments and becomes part of the ER (Poteryaev et al., 2005; Puhka et al., 2007; Lu et al., 2009, 2011; Wang et al., 2013). ER tubules and sheets are present in all eukaryotic cells, though the organization and ratios of sheets to tubules vary by cell type (Shibata et al., 2006). In general, cells with a higher ratio of sheets, which usually contain more ribosomes, are specialized for synthesizing secretory and membrane proteins (Hu et al., 2011). In turn, tubules contain very few ribosomes and usually serve as sites of lipid synthesis, vesicle budding, and points of contact with other organelles and the cytoskeleton (Hu et al., 2011).
Recent advances have provided insight into how these different structures are formed and maintained. The reticulons, and related DP1/Yop1 family, localize to the tubular ER (Voeltz et al., 2006) and to the highly curved edges of sheets (Shibata et al., 2010). These proteins form a unique hairpin configuration within the outer membrane leaflet, leading to the highly curved nature of tubules. Depletion of DP1 and reticulons in yeast cells leads to an overabundance of sheets and loss of tubules (Voeltz et al., 2006). In addition, purified reticulon and DP1/Yop1 family members were sufficient to induce tubule formation from vesicles (Hu et al., 2008), which indicates that these proteins play a role in promoting tubule formation. Homotypic fusion of ER tubules leads to network formation and is mediated by members of the Atlastin family (Hu et al., 2009; Wang et al., 2013), a class of dynamin-like GTPases, and the Rab10 GTPase (English and Voeltz, 2013b). When Atlastins are depleted in cells, long unbranched tubules are observed, which results from the lack of fusion events (Hu et al., 2009). Alternatively, the maintenance and stabilization of ER sheets is mediated by the sheet-promoting factors Climp-63, kinectin, and p180 (Shibata et al., 2010).
In addition to proteins, a large fraction of the transcriptome localizes to the surface of the ER where these mRNAs are translated on membrane-bound ribosomes (Reid and Nicchitta, 2012). Several proteins that promote ER sheet formation have been shown to interact with mRNAs and ribosomes (Shibata et al., 2010), and changes in the translational status of the cell can alter ER morphology and protein distribution (Puhka et al., 2007; Shibata et al., 2010). However, it is not clear how the regulation of stability and localization of mRNAs and ribosomes to the ER affects ER structure. RNA degradation may significantly influence the localization and function of an mRNA. Cellular nucleases are the key enzymes mediating RNA turnover, and their enzymatic activity is generally dependent on divalent cations. Here we used Xenopus laevis egg extracts to explore the hypothesis that an increase in intracellular calcium could regulate the RNA content and morphology of the ER. We show that the calcium-activated ribonuclease XendoU (Caffarelli et al., 1994; Laneve et al., 2003) localizes to the surface of the ER, where it functions in local RNA degradation and eviction of ribosomes, RNPs, and RNA from the membrane surface to ensure proper ER morphology. Additionally, we show that the localization and function of XendoU are maintained in human cells, demonstrating a conserved role for calcium-regulated RNA degradation in control of ER morphology. Thus, we propose that the EndoU family of proteins regulate ER structure and function by mediating local RNA degradation in response to changes in intracellular calcium levels, thereby contributing to the spatial reorganization of the ER network throughout the cell cycle.
Results
Identification of a calcium-dependent nuclease activity
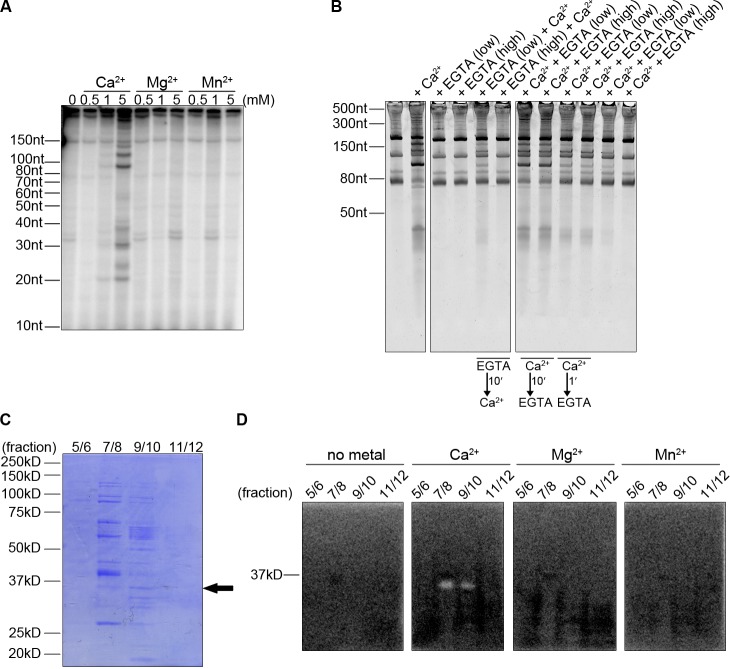
Metaphase-arrested egg extracts from X. laevis are routinely used to study various aspects of the cell cycle in vitro (Murray, 1991; Desai et al., 1999). “Cytostatic factor (CSF)-arrested” extracts are prepared with the addition of the calcium chelator EGTA in order to maintain the CSF arrest during extract preparation. The addition of ∼0.6 mM calcium chloride to CSF extracts leads to an increase in the free calcium concentration to ∼1.5 µM, which mimics the calcium release from intra- and extracellular stores that occurs at fertilization (Busa and Nuccitelli, 1985; Lorca et al., 1991), leading to activation of the calcium/calmodulin-dependent protein kinase II (CamKII) and exit from CSF-mediated meiotic arrest. X. laevis egg extracts have previously been used to identify several important calcium-regulated factors that play important roles during fertilization, demonstrating the fidelity of this cell-free system (Lorca et al., 1993; Mochida and Hunt, 2007). To determine if calcium regulated RNA metabolism, increasing concentrations of calcium chloride were added to egg extract and total RNA was isolated to determine if there was a change in RNA stability. Surprisingly, specific RNA cleavage products were identified at physiologically relevant calcium concentrations (Fig. 1 A). Addition of magnesium chloride or manganese chloride, common metal cofactors for nucleases, at the same concentrations did not lead to RNA cleavage, which indicates that the nuclease activity in egg extract was dependent on calcium. Treating the extract with EGTA, a known calcium chelator, prevented RNA degradation when added before, and in excess over calcium (Fig. 1 B). When calcium was added before EGTA, cleavage still occurred, indicating that the nuclease acts rapidly and efficiently.
Figure 1.
A calcium-dependent nuclease activity in X. laevis egg extract. (A) CaCl2, MgCl2, or MnCl2 were added to extract at increasing concentrations for 60 min. RNA was isolated from extract, 5′ end-labeled, and run on a denaturing acrylamide gel. (B) Total RNA was isolated from reactions supplemented with no metal, CaCl2, EGTA alone, or EGTA and CaCl2 in the same reaction. RNAs were run on denaturing acrylamide gels and stained with SYBR Green II. (C) Precleared egg extract was run over a Heparin HiTrap affinity column, and flow-through was collected. CaCl2 was added to the flow-through and run over a Heparin HiTrap column preequilibrated with CaCl2. Proteins were eluted with a salt gradient from 0.1 M to 1 M KCl. Peak protein–containing fractions were pooled and run on a 10% SDS-PAGE gel and Coomassie stained. A band running just under 37 kD in fractions 7 + 8 and 9 + 10 (arrow) was submitted for mass spec analysis and identified XendoU as the putative nuclease. (D) Peak protein fractions from C were run on 10% SDS-PAGE gels containing 32P-labeled RNA in the resolving portion of the gel. Gels were soaked in buffer to remove SDS and renature proteins, then soaked in buffer containing 1 mM of CaCl2, MgCl2, MnCl2, or no metal overnight at 4°C and imaged by PhosphorImager.
Purification of the calcium-dependent ribonuclease
A previous report described purification of a calcium-dependent ribonuclease activity from X. laevis oocyte extract (Seidel and Peck, 1994), and we used this purification technique to search for similar activity in egg extract. Precleared egg extract was applied to a heparin column equilibrated in calcium-free buffer, and nuclease activity was found in the flow-through. The heparin flow-through was adjusted to 1 mM CaCl2 and reapplied to the heparin column, which retained the nuclease activity in the presence of CaCl2. Bound proteins were eluted with a linear gradient of KCl and analyzed for protein content and nuclease activity (Fig. 1, C and D).
Using these partially purified fractions, we used an in-gel nuclease activity assay in which proteins are electrophoresed through an SDS-PAGE gel containing radiolabeled RNA in the resolving portion of the gel (Rosenthal and Lacks, 1977; Seidel and Peck, 1994). After electrophoresis, the gels were soaked in a buffer to remove SDS and renature the proteins followed by incubation in a buffer containing either no metal, CaCl2, MgCl2, or MnCl2. Ribonuclease activity was detected by a clearing in the gel after exposure to a phosphor image plate. Activity was only detected in the presence of CaCl2 and only in fractions 7–10 (Fig. 1 D). Examination of Coomassie blue–stained fractions revealed a protein at the expected size of the nuclease activity that also corresponded to the size of the protein isolated in oocytes (Fig. 1 C, black arrow). Mass spectrometry of this band resulted in the identification of XendoU, a nuclease previously studied in the maturation of specific small nucleolar RNAs (snoRNAs; Laneve et al., 2003). These results collectively suggest that XendoU is the protein responsible for the calcium-dependent nuclease activity observed in egg extract.
Characterization of XendoU activity
To examine the catalytic activity of XendoU, we cloned the cDNAs from X. laevis egg extract and purified recombinantly expressed His-tagged versions of XendoU from bacteria (Fig. S1 E). Two paralogues of XendoU exist in X. laevis: A and B, which differ by 15 amino acids throughout the protein. We expressed both paralogues and found that XendoU B purified as a single band, whereas XendoU A purified with additional bands, potentially heat shock proteins (unpublished data).
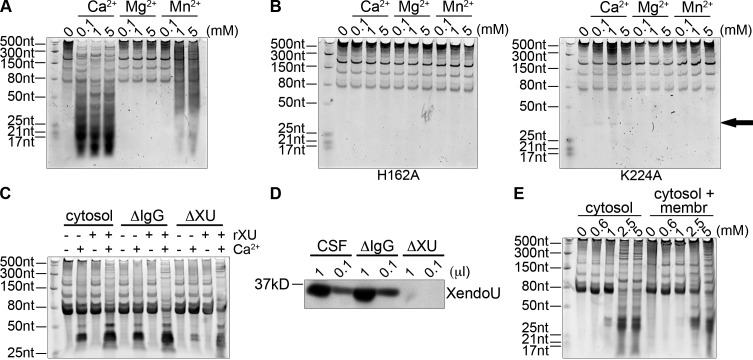
Next we incubated the recombinant, wild-type proteins with increasing concentrations of CaCl2, MgCl2, and MnCl2 in the presence of total RNA purified from egg extract (Fig. 2 A, Fig. S1 B [XendoU B], and Fig. S1 D [XendoU A]). Both wild-type XendoU protein isoforms exhibited activity in the presence of CaCl2 and lacked activity in the presence of MgCl2. XendoU B exhibited RNase activity with MnCl2, but this activity required 20-fold higher levels of MnCl2 and exhibited weaker nuclease activity, which suggests that XendoU B prefers CaCl2 for nuclease activity.
Figure 2.
XendoU is required for calcium-mediated RNA degradation in vitro. (A) Recombinant wild-type XendoU was incubated with increasing concentrations of CaCl2, MgCl2, MnCl2, and total RNA from egg extract. RNA was visualized on a denaturing acrylamide gel stained with SYBR Green II. (B) Recombinant H162A and K224A were incubated with metal and RNA and visualized as in A. A very faint band (arrow) appears in the presence of calcium and K224A. (C) XendoU or mock-depleted (IgG) cytosol was incubated with 1 mM CaCl2. Recombinant wild-type XendoU (rXU) was added to XendoU-depleted, mock-depleted (IgG), or undepleted cytosol and analyzed as in A. (D) Western blot of XendoU demonstrating the antibody-mediated depletion in CSF extract as compared with undepleted (CSF) or mock-depleted (IgG) extracts. (E) Cytosol or cytosol mixed with light membranes were incubated with increasing concentrations of CaCl2, and RNA was visualized as in A.
To further test the metal-dependent catalytic capacity of XendoU, the wild-type recombinant protein was subjected to the in-gel nuclease assay. In the presence of CaCl2, MgCl2, or MnCl2, a clearing was only detected in the presence of CaCl2 (Fig. S1 A [XendoU B] and Fig. S1 C [XendoU A]) as seen with the endogenous protein (Fig. 1 D). It has previously been shown that XendoU is active in oocyte nuclear extract in the presence of MnCl2 on a specific snoRNA target, though it has also been shown that the protein is generally very active in the presence of CaCl2 (Caffarelli et al., 1994, 1997). Here we show that recombinant XendoU B is an extremely robust, nonspecific ribonuclease active at all calcium concentrations tested. Subsequent assays were performed using recombinant XendoU B protein because it showed greater activity and purified more homogenously than XendoU A.
The catalytic site of XendoU is similar to RNase A and T1, and is composed of a triad of residues important for snoRNA catalysis (Gioia et al., 2005). To determine if mutations to catalytic and substrate-binding residues would reduce calcium-dependent RNase activity, we mutated residues H162 (predicted catalytic residue) and K224 (positively charged residue predicted to stabilize substrate binding and previously demonstrated to be important for snoRNA processing) to alanine and subjected the mutant proteins to the same in vitro cleavage assay as the wild-type protein. XendoU B H162A showed no catalytic activity against total RNA in the presence of any metal at any of the concentrations tested (Fig. 2 B). XendoU B K224A showed dramatically reduced activity in the presence of CaCl2 (Fig. 2 B, black arrow) and no activity with any other metal. Previous work demonstrated that both recombinant proteins were capable of binding RNA (Gioia et al., 2005), demonstrating that these specific residues play an important role in metal-dependent RNA catalysis, but not in RNA binding.
To determine if XendoU is responsible for the specific calcium-dependent cleavage products in egg extracts, we immunodepleted XendoU from cleared cytosolic extracts using custom antibodies (Fig. 2 D). Depletion of XendoU from the cytosol resulted in a near-complete loss of cleavage activity in the presence of calcium (Fig. 2 C). Cleavage activity was restored after the addition of recombinant wild-type XendoU to the previously depleted extract. RNA cleavage products of the same size and pattern as seen in the IgG control were detected after addition of the recombinant protein, demonstrating that XendoU is the protein responsible for the calcium-dependent RNA catalysis. Interestingly, activity in membrane-free cytosolic extract was more robust than in total egg extract at the same concentration of exogenously added calcium. When comparing cytosolic extract alone and cytosolic extract with the ER and membrane fraction added back, we found that more calcium was required for nuclease activity when membranes were present. This indicates that although the ER is a main source of calcium in the extract, it can also reabsorb cellular calcium (Fig. 2 E) and reduce global RNase activity.
One possible model for a calcium-activated nuclease is that activity would be coupled to calcium release to completely degrade a specific set of transcripts important for meiosis upon fertilization. To determine if specific transcripts were degraded in a XendoU-dependent manner after CaCl2 addition to egg extracts, we prepared mRNA-seq libraries from control and XendoU-depleted extracts arrested in meiosis and after calcium treatment. Because the poly-A tails of many mRNAs change at fertilization (Horner and Wolfner, 2008), we isolated mRNAs using CAP-capture (Blower et al., 2013). We found that there were several mRNAs that appeared to be reduced in control-depleted extracts upon calcium addition but were stable in XendoU-depleted extracts (Fig. S2 A). We hypothesized that mRNAs that are completely degraded after calcium addition would be absent from both CAP-capture and oligo-dT selected mRNA libraries. However, when we prepared mRNA-seq libraries from an additional extract using oligo-dT selection, we did not find any mRNAs that appeared to be degraded upon CaCl2 addition using both mRNA purification methods (Fig. S2 B), which suggests that mRNAs that change in dT or CAP-capture libraries are likely to be the result of deadenylation or decapping, but not RNA degradation. Additionally, RT-PCR analysis failed to confirm consistent degradation of any mRNAs upon addition of CaCl2 (unpublished data). We conclude that XendoU does not function to completely degrade a specific subset of mRNAs upon fertilization (Fig. S2 C), but may function in a local RNA degradation pathway.
XendoU regulates membrane dynamics after meiotic exit
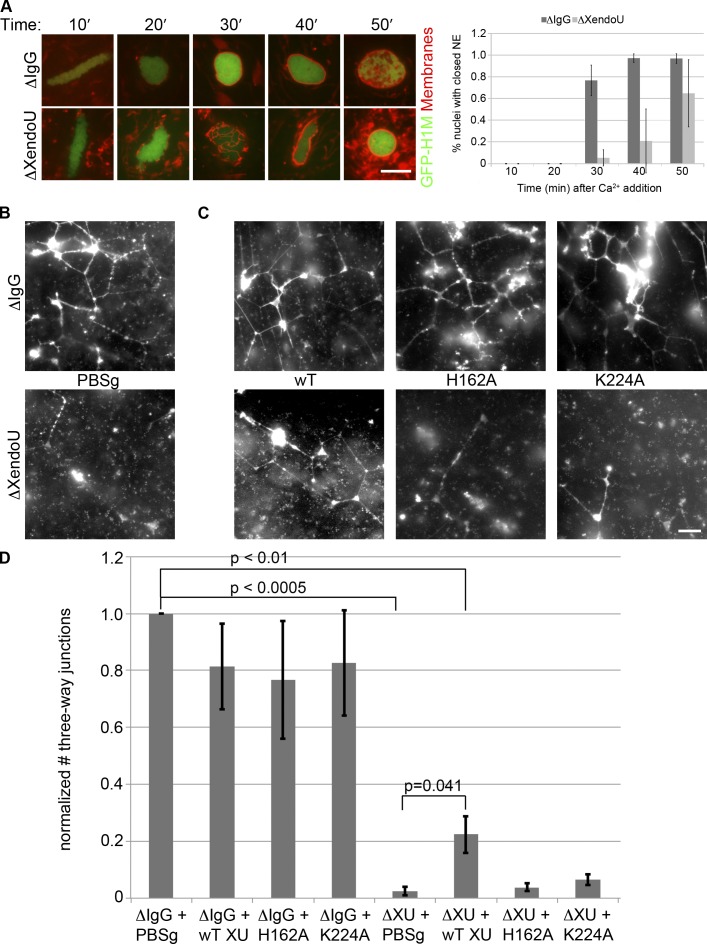
The calcium-mediated activation of XendoU suggested that XendoU could regulate aspects of the cell cycle or cell morphology in response to calcium signaling. To determine if XendoU regulated the cell cycle after meiotic exit, we incubated sperm nuclei, CaCl2, GFP-Histone H1 (to identify chromatin in live images), and Vybrant DiI (to image the nuclear envelope/membranes) in control and XendoU-depleted extracts. We found that XendoU depletion resulted in a significant delay in nuclear envelope formation (Fig. 3 A) and onset of DNA replication (Fig. S3, A and B), common hallmarks of interphase. Importantly, the timing of cyclin B degradation was not affected (Fig. S3 C) after XendoU depletion (Fig. S3 D), which suggests that the observed effects were not the result of changes in cell cycle timing. Taking into consideration the defect seen in nuclear envelope closure, we investigated general membrane dynamics in the extract, as the nuclear envelope and ER comprise a continuous membranous structure sharing a common lumen. Examination of bulk membranes in the XendoU-depleted extract using octadecyl rhodamine revealed a lack of network formation and characteristic three-way junctions between tubules (Fig. 3 B). Instead, only a few long tubules were observed compared with IgG control-depleted extracts, where a complete network of ER tubules joined by three-way junctions was detected. Previous studies have found that antibody-mediated protein depletion can cause physical damage to extracts, resulting in suboptimal assembly of cellular structures in depleted extracts (Desai et al., 1999). We found this to be especially true for the formation of the ER network, as treatment of extracts with IgG or XendoU antibody beads resulted in a reduction in ER network formation (Fig. 3 B) compared with network formation assays performed in extract that was never incubated with beads (Fig. S4 A). However, XendoU-depleted extracts consistently exhibited ER network assembly defects that far exceeded those caused by control antibody depletion alone (Fig. 3, C and D). Collectively, these results demonstrate that XendoU plays an important role in regulating membrane dynamics during meiotic exit.
Figure 3.
XendoU plays a role in nuclear envelope formation and proper ER morphology. (A) Demembranated X. laevis sperm nuclei were incubated in mock-depleted (IgG) or XendoU-depleted CSF in the presence of GFP-Histone H1 and Vybrant DiI membrane dye. 10–15 random fields were taken from live images every 10 min and the percent nuclei with closed nuclear envelopes was counted. n = 3. (B) IgG mock-depleted (top) or XendoU-depleted (bottom) CSF was incubated at room temperature in the presence of 1 mM CaCl2 and 1× PBS + 15% glycerol (PBSg, protein storage buffer) for 60 min followed by staining of membranes by octadecyl rhodamine and imaged live. (C) Mock-depleted (top) or XendoU-depleted (bottom) extracts were incubated with recombinant proteins (wild-type [left], H162A [middle], or K224A [right]) on ice for 30 min followed by addition of 1 mM CaCl2 and incubated for 60 min at room temperature. Membranes were stained and imaged as above. (D) 10–12 randomly selected fields were taken for each condition in B and C in three separate egg extracts. Three-way junctions between ER tubules were counted for each field to assess network formation. Statistical comparison of IgG + PBSg to experimental samples was performed using a one-sample t test. Comparison of XendoU-depleted to rescued extract was performed using an unpaired Student’s t test. Error bars indicate SD. Bars, 10 µm.
To determine if the nuclease activity of XendoU is required for the regulation of membrane dynamics, we performed “rescue” experiments of XendoU immunodepletions using both the wild-type (Fig. 2 A) and the catalytically dead point mutants (Fig. 2 B). The addition of the recombinant proteins (wild-type or catalytically dead) did not affect network formation in the IgG mock-depleted extract (Fig. 3 C, top; and Fig. 3 D). The wild-type recombinant protein was able to partially rescue the defect in ER network formation while the catalytically dead point mutants were unable to rescue the network formation phenotype (Fig. 3 C, bottom; and Fig. 3 D). This indicates that RNA cleavage mediated by XendoU is important for proper ER network formation.
XendoU localizes and functions on ER membranes to promote network formation
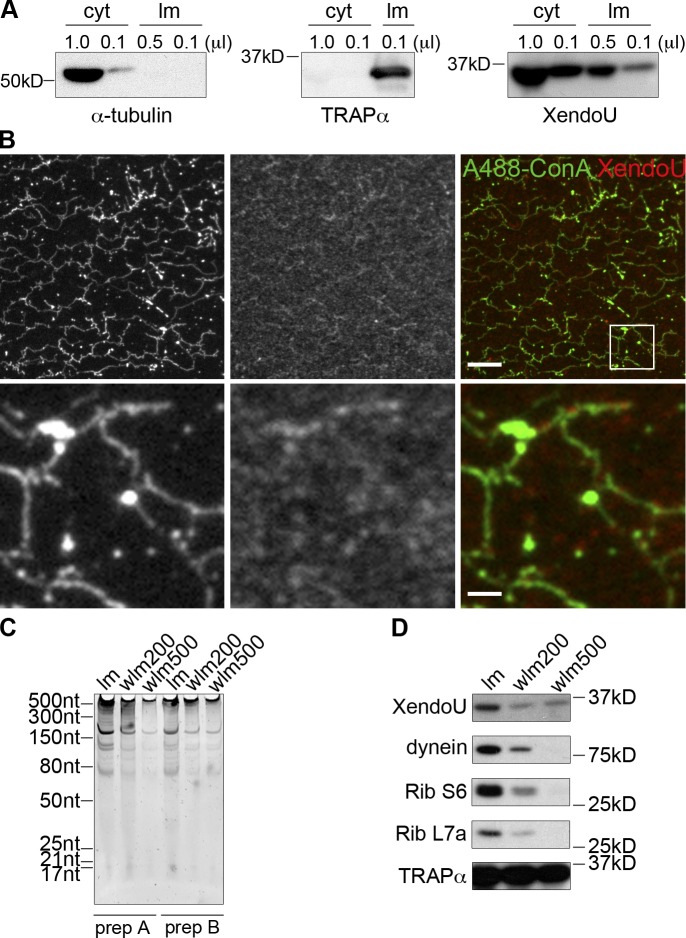
The finding that XendoU regulates ER dynamics suggested that the nuclease may be performing a novel role on the membrane to control ER structure and function. To determine if XendoU associated with the ER, we fractionated egg extract into cytosol and light membrane fractions, which are enriched in ER membranes (Powers et al., 2001). Membrane proteins such as TRAPα were only found in the light membrane fractions, whereas highly abundant proteins, such as α-tubulin, were only found in the cytosol (Fig. 4 A). XendoU was present in both fractions. We found that endogenous XendoU consistently colocalized with ER networks and exhibited a punctate localization to the ER (Fig. 4 B). Interestingly, both XendoU and RNA were present on membranes, and subpopulations of both remained stably bound to membranes washed in moderate (200 mM KCl, washed light membrane [wlm] 200) or high salt (500 mM KCl, wlm 500; Fig. 4, C and D). Conversely, dynein and ribosomal proteins S6 and L7a were largely dissociated from light membranes at 500 mM KCl, whereas the association of TRAPα with light membranes and salt-washed membranes remained unchanged. Collectively, these results demonstrate that a subpopulation of XendoU is tightly bound to membranes, where it may directly regulate ER dynamics.
Figure 4.
A subpopulation of XendoU localizes to ER membranes. (A) Cytosol (cyt) and light membrane (lm) preps (∼20×) were isolated as described in the Materials and methods. Western blots were performed on each fraction for α-tubulin, TRAPα, and XendoU. (B) ER networks were formed in flow cells and fixed with paraformaldehyde and glutaraldehyde. Immunofluorescence for XendoU was performed with αXendoU antibody and α-rabbit Cy3 secondary antibody (red). Alexa fluor 488 Concanavalin A (conA) was added in with secondary antibodies to detect glycoproteins and the ER (green). The boxed region is magnified 5× in the lower panels. Bars: (top) 10 µm; (bottom) 2 µm. (C) Light membranes (lm) were washed in buffers containing 200 mM (wlm 200) or 500 mM (wlm 500) KCl. Membranes were washed twice and resuspended in the same original volume. RNA was isolated from each membrane prep (prep A and B), run on denaturing acrylamide gels, and stained with SYBR Green II. (D) Western blots for XendoU, dynein, ribosomal protein S6 (Rib S6), ribosomal protein L7a (Rib L7a), and TRAPα were performed on light membranes (lm) and membranes washed in 200 mM KCl (wlm 200) or 500 mM KCl (wlm 500). TRAPα serves as a loading and wash control.
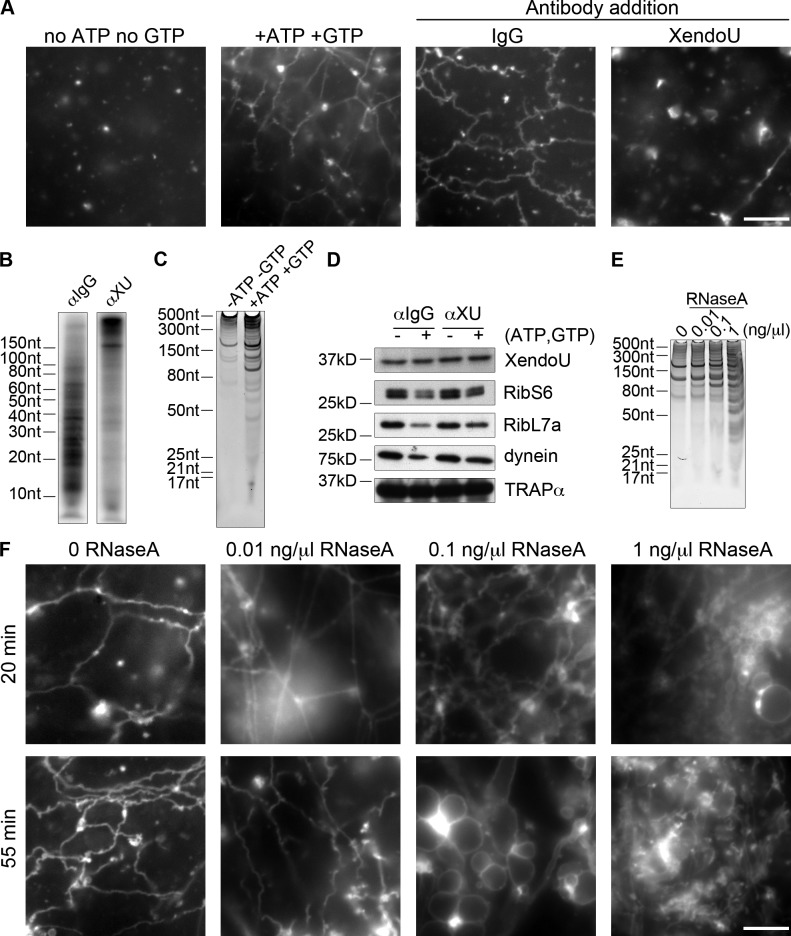
To determine if membrane-bound XendoU acts on membranes to regulate ER network formation, we performed vesicle fusion reactions in the absence of cytosol. These vesicle fusion reactions have been used to demonstrate a direct role for the reticulons (Voeltz et al., 2006), Atlastin (Hu et al., 2009), and Rab10 (English and Voeltz, 2013b) in mediating homotypic ER fusion events. Salt-washed vesicles (200 mM KCl) devoid of cytosolic and peripheral membrane proteins were incubated at 25°C in the presence of ATP and GTP, resulting in the formation of a network (Fig. 5 A). No networks formed in the absence of ATP and GTP (Fig. 5 A). Networks formed in the presence of IgG (Fig. 5 A). Conversely, when XendoU antibody was included in the reaction, networks failed to form and only small vesicles were detected (Fig. 5 A). It is important to note that vesicle fusion in this system does not rely on the addition of exogenous calcium, but the vesicle fusion reaction itself releases calcium from the lumen of the ER (Voeltz et al., 2006). Consistent with our results, addition of 5′5′-dibromo BAPTA, a calcium chelator, blocks vesicle fusion in this system (Dreier and Rapoport, 2000), and BAPTA blocks nuclear envelope formation in X. laevis egg extracts (Sullivan et al., 1993). These results demonstrate that the membrane-bound subpopulation of XendoU plays an integral role in vesicle fusion and network formation.
Figure 5.
XendoU plays a role in vesicle fusion and controls local RNA degradation on membranes. (A) Light membranes were washed one time in buffer containing 200 mM KCl (wlm 200) and incubated in the absence of ATP and GTP (no fusion) or in the presence of ATP and GTP (vesicle fusion). Alternatively, wlms were incubated for 30 min at RT with 5 µM control (IgG) or XendoU antibody followed by the addition of ATP and GTP. Aliquots were mixed with octadecyl rhodamine at 15 min, incubated for an additional 15 min at RT, and imaged live. Representative images of each condition are shown. (B) RNA was isolated from membrane pellets from the end points of reactions containing IgG or XendoU antibodies in A, 5′ end-labeled with [32P-γ]ATP as described, and run on a denaturing gel. (C) Membranes were pelleted from standard vesicle fusion reactions (+ATP +GTP) or control reactions (−ATP –GTP), and RNA was isolated from the supernatant, run on a denaturing gel, and imaged with SYBR Green II. (D) Western blots of XendoU, ribosomal protein S6 (RibS6), ribosomal protein L7a (RibL7a), dynein, and TRAPα on membranes after vesicle fusion (+ATP +GTP) or in the absence of fusion (−ATP −GTP) in the presence of IgG or XendoU antibody. (E) Wlms were mock-treated or RNaseA treated (0.01 ng/µl, 0.1 ng/µl, or 1 ng/µl), then washed once more, and RNA was isolated and imaged as in C. (F) RNaseA-treated vesicles from E were incubated and imaged as in A at 20 min and 50 min after addition of ATP and GTP. Bars, 10 µm.
ER membranes exhibit localized nuclease activity
To determine if the nuclease function of XendoU was active on membranes, we isolated RNA from membrane pellets of vesicle fusion reactions in the presence of IgG control or XendoU antibody and labeled the 5′ end of cleavage products with 32P (Fig. 5 B). Many cleavage products (between 10 and 100 nt) were present in IgG reactions and absent in reactions containing XendoU antibodies, demonstrating that the XendoU antibody blocks the catalytic activity of XendoU on membranes. These data, combined with the ability of the XendoU antibody to block network formation (Fig. 5 A), suggest that the nuclease function of XendoU on membranes is required for the generation of the ER network.
To determine if RNAs were released from membranes over the course of vesicle fusion, we repeated the reaction in the absence of ATP and GTP (no vesicle fusion) or in the presence of ATP and GTP (vesicle fusion). When the supernatant was isolated after membrane pelleting, we found that RNA was released from the membranes after vesicle fusion (Fig. 5 C), indicating that RNA cleavage and release from membranes are important steps for efficient vesicle fusion.
Because we observed that XendoU persisted on membranes before and after vesicle fusion (Fig. 5 D), we wondered if the abundance of other proteins on membranes was affected during vesicle fusion. Interestingly, we detected that a subpopulation of ribosomal proteins L7a and S6 as well as the microtubule motor dynein are removed from membranes after vesicle fusion (Fig. 5 D), as seen by a reduction in protein levels on membranes isolated after fusion. In addition, when vesicle fusion is blocked by the presence of XendoU antibody, this subpopulation of L7a and S6 and dynein remains on the membranes, which indicates that RNA degradation induced by XendoU results in the ejection of a subpopulation of ribosomal proteins from membranes (Fig. 5 D).
To determine if general RNase activity on membranes was sufficient to promote vesicle fusion, we RNAaseA treated salt-washed vesicles, which is a technique previously used to strip ribosomes and mRNAs from isolated ER membranes (Lee et al., 1971; Rosbash and Penman, 1971). Using a very conservative RNase treatment at 0.01 ng/µl, we observed that only a small fraction of the RNA present on membranes was degraded (as compared with 0.1 and 1 ng/µl), as seen by comparing the accumulation of specific RNA fragments over increasing concentrations to untreated wlms (Fig. 5 E). When these treated membranes were used in the vesicle fusion reaction in the presence of ATP and GTP, a dense ER network formed more rapidly, with a far greater number of tubules as compared with standard wlms (Fig. 5 F). In the absence of ATP and GTP, neither membrane preparation formed a network (Fig. S4, B and C). Additionally, in the presence of high RNase concentrations (0.1–1 ng/µl), extremely dense, aberrant networks formed, exhibiting large round vesicles and an absence of tubular ER network (Fig. 5 F). These results indicate that low-level nonspecific RNA cleavage on ER membranes promotes network formation, but that excess RNA removal results in aberrant ER morphology, indicating that controlled RNA removal from the ER is important for normal ER morphology.
EndoU activity and localization in human cells
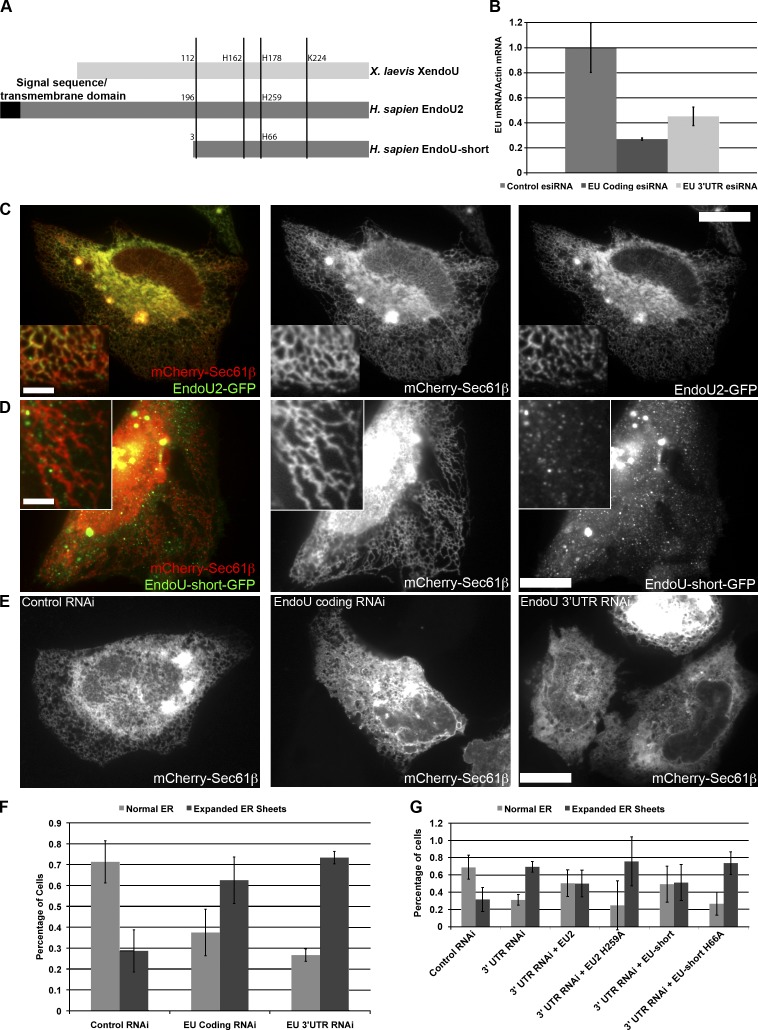
To determine if the localization and function of XendoU are evolutionarily conserved, we examined human EndoU in cultured cells. The human EndoU gene encodes splice variants that encode at least four different polypeptides, three of which contain a predicted N-terminal signal sequence or transmembrane domain (Poe et al., 2014). We examined two EndoU isoforms in HeLa cells: EndoU2, which contains a putative signal sequence/transmembrane domain; and EndoU-short (Fig. 6 A), which contains the catalytic domain and is similar to X. laevis EndoU. To determine the localization of the two human EndoU isoforms, we created GFP fusion constructs and cotransfected these into HeLa cells with an mCherry-Sec61β plasmid to identify ER (Fig. 6, C and D). EndoU2 localizes throughout the ER sheets and tubules (Fig. 6 C), whereas EndoU-short localizes throughout the cytoplasm and to puncta along, or directly abutting, the ER tubules (Fig. 6 D), similar to the localization observed for XendoU.
Figure 6.
Human EndoU localizes to the ER and controls ER morphology. (A) Schematic of the structures of XendoU, human EndoU2, and EndoU-short. The locations of conserved catalytic residues and putative signal sequence/transmembrane domains are indicated. Residue 112 in XendoU is where the homology between the X. laevis and human proteins begins; the N termini are relatively unconserved. (B) Quantitative RT-PCR showing effective knockdown of EndoU with esiRNAs directed against the coding region and 3′ UTR. (C) HeLa cells were cotransfected with EndoU2-GFP and mCherry-Sec61β, and analyzed by fluorescence microscopy. (D) HeLa cells were cotransfected with EndoU-short-GFP and mCherry-Sec61β and analyzed as in C. (E) Cells treated with control esiRNAs (left) or esiRNAs against the coding region (middle) or 3′ UTR (right) of EndoU were transfected with mCherry-Sec61β and analyzed by fluorescent microscopy. (F) Quantification of the percentage of cells exhibiting expanded ER sheets in control, EndoU coding, and EndoU 3′ UTR RNAi. Error bars indicate standard deviation. At least 200 cells were scored and the experiment was performed in triplicate. (G) HeLa cells were treated with control or EndoU 3′ UTR esiRNAs, then transfected with wild-type or catalytically dead EndoU2-GFP or EndoU-short-GFP (resistant to RNAi, lacking the 3′ UTR) and mCherry Sec61β. The percentage of cells exhibiting expanded ER sheets is quantified. Error bars indicate standard deviation. At least 200 cells from three replicates of each condition were scored. Inset panels in C and D are magnified 3×. Bars: (main panels) 10 µm; (insets) 2 µm.
To determine if EndoU also regulated ER morphology in human cells, we used endoribonuclease-prepared siRNAs (esiRNAs; Yang et al., 2002, 2004) to knock down EndoU in HeLa cells. esiRNAs were designed for a portion of the coding region or the 3′ UTR of EndoU (see Materials and methods), and therefore targeted all EndoU isoforms. Quantitative RT-PCR demonstrates knockdown of EndoU (Fig. 6 B). Knockdown of EndoU, either by esiRNAs directed against the coding region or 3′ UTR, resulted in loss of the tubular ER and expansion ER sheets (Fig. 6, E and F), similar to the phenotype observed upon loss of Rab10 function (English and Voeltz, 2013b). We observed an identical expansion of ER sheets upon knockdown of EndoU with a pool of siRNAs directed against the coding region and 3′ UTR (Fig. S5, B–E).
To determine if EndoU can rescue this phenotype, we transfected an RNAi-resistant plasmid (lacking the 3′ UTR) after treatment of the cells with the 3′ UTR–directed esiRNAs (Fig. 6 G). Catalytically active, but not catalytically inactive, EndoU2 and EndoU-short are able to partially rescue the ER defect phenotype (Fig. 6 G and Fig. S5 A), similar to the rescue observed using the recombinant XendoU protein in X. laevis extract (Fig. 3, C and D). Partial rescue of the ER morphology phenotype by both ER-localized EndoU2 and largely cytoplasmic EndoU-short demonstrates that EndoU can regulate ER morphology whether it is statically localized to the ER (EndoU2) or recruited to the ER from the cytoplasm (EndoU-short), similar to the situation observed with XendoU. Collectively, these results demonstrate that EndoU exhibits a conserved localization and function at the ER in humans and X. laevis in a catalysis-dependent manner.
Discussion
Our study has uncovered a novel role of calcium signaling in the regulation of ER morphology. Here we show that XendoU is a calcium-activated nuclease that localizes to the ER and affects ER morphology and nuclear envelope formation. Our results support the hypothesis that localized RNA degradation on the surface of the ER is important for promoting and refining ER network formation.
Using X. laevis egg extracts, we identified a ribonuclease activity that is activated by physiologically relevant levels of calcium (Busa and Nuccitelli, 1985; Lorca et al., 1991). We purified the protein responsible for calcium-dependent catalysis and found that it was XendoU, a protein previously described to play a role in snoRNA processing in X. laevis oocyte nuclear extract (Caffarelli et al., 1994, 1997). Although it has been shown that XendoU activity on a snoRNA target in nuclear extract and with recombinant protein relies on manganese, we also note that the previously described XendoU was dramatically more active in calcium (Caffarelli et al., 1994, 1997), which is consistent with our results. The XendoU crystal structure does not contain Mn2+ ions even when the crystals are soaked in MnCl2 (Renzi et al., 2006), which suggests that XendoU likely has a higher affinity for calcium than for manganese. Activity of XendoU in the presence of manganese occurs at 5–7 mM (Caffarelli et al., 1994), which is significantly higher than what occurs in the cell (Tholey et al., 1988; Zhang and Ellis, 1989), whereas XendoU activation by calcium in extracts occurs at the same concentration that mimics fertilization (Busa and Nuccitelli, 1985; Lorca et al., 1991). Collectively, these data suggest that calcium is likely the relevant cofactor for the majority of XendoU functions, whereas manganese may play a more limited role in activating XendoU for snoRNA processing.
Structures of both XendoU and the viral homologue NendoU have demonstrated that both of these ribonucleases possess a catalytic site similar to RNase A (Renzi et al., 2006; Ricagno et al., 2006), which is consistent with production of cleavage products containing 2′,3′ cyclic phosphates (Renzi et al., 2006). RNase A, which also produces cleavage products with ends identical to those produced by XendoU, does not require a divalent cation for catalysis. In turn, XendoU lacks an obvious metal binding site (Renzi et al., 2006), yet requires a metal cofactor for catalysis. These observations are consistent with XendoU belonging to a novel class of ribonucleases.
In this study, we used X. laevis egg extracts to mimic elevation of cytosolic calcium levels that occur upon calcium release from the ER at fertilization. It is known that many changes in organelle structure and distribution take place at fertilization, including the mitochondria and ER (Terasaki et al., 2001; Sun et al., 2011). In addition to fertilization, there is evidence from many different systems that calcium transients occur during mitosis (for review see Whitaker, 2006b) and may play a role in various aspects of cell cycle control. A recent study has demonstrated that X. laevis egg extracts faithfully recapitulate changes in ER morphology that occur during the cell cycle (Wang et al., 2013). Previous work has demonstrated that the mitotic ER is composed almost entirely of sheets, which reorganize to form the nuclear envelope, and an ER composed of a mixture of sheets and tubules at mitotic exit (Lu et al., 2009; Lu et al., 2011; Wang et al., 2013). The dramatic morphological changes of the ER during mitosis highlight the dynamic nature of this organelle (Voeltz et al., 2002) and demonstrate that large changes in ER structure are well correlated with calcium transients. Interestingly, we found that immunodepletion of XendoU in interphase extracts blocked formation of the nuclear envelope and ER network assembly. The requirement for XendoU in ER network assembly was strictly dependent on the catalytic activity of XendoU, which is identical to what we observed for human EndoU in cultured cells. These results are consistent with XendoU playing a role in ER remodeling at fertilization, although further studies will be required to directly test this hypothesis.
Previous studies of X. laevis, human, and viral XendoU homologues have demonstrated that these nucleases have very limited sequence specificity, cleaving RNAs after UU dinucleotides or a single U in vitro (Caffarelli et al., 1997; Bhardwaj et al., 2006; Laneve et al., 2008), although no target RNAs have been identified in vivo. In an attempt to determine if specific mRNAs were degraded in a XendoU-dependent manner, we sequenced mRNAs before and after calcium addition to egg extracts. We found that there were no mRNAs that were dramatically degraded upon calcium addition, demonstrating that XendoU does not function to completely degrade any specific set of mRNAs. Because we have observed that XendoU is active on the surface of ER membranes, and is likely to have very little sequence specificity, we speculate that the RNAs targeted by XendoU will likely be determined by proximity to the nuclease upon activation by calcium release from the ER. We hypothesize that these target RNAs will include both ribosomal RNAs (rRNAs) and mRNAs localized to the surface of the ER that are in the process of being translated into secreted and membrane-bound proteins. Rapid uptake of free calcium by the ER will likely limit the RNase activity of XendoU, which is consistent with our observation that the addition of purified membranes to clarified cytosol dampens XendoU activity as compared with the same concentration of calcium added to cytosol alone. This mechanism would limit RNase activity on the surface of the ER, which is important for proper ER morphology, as we found that high RNase concentrations result in aberrant ER morphology.
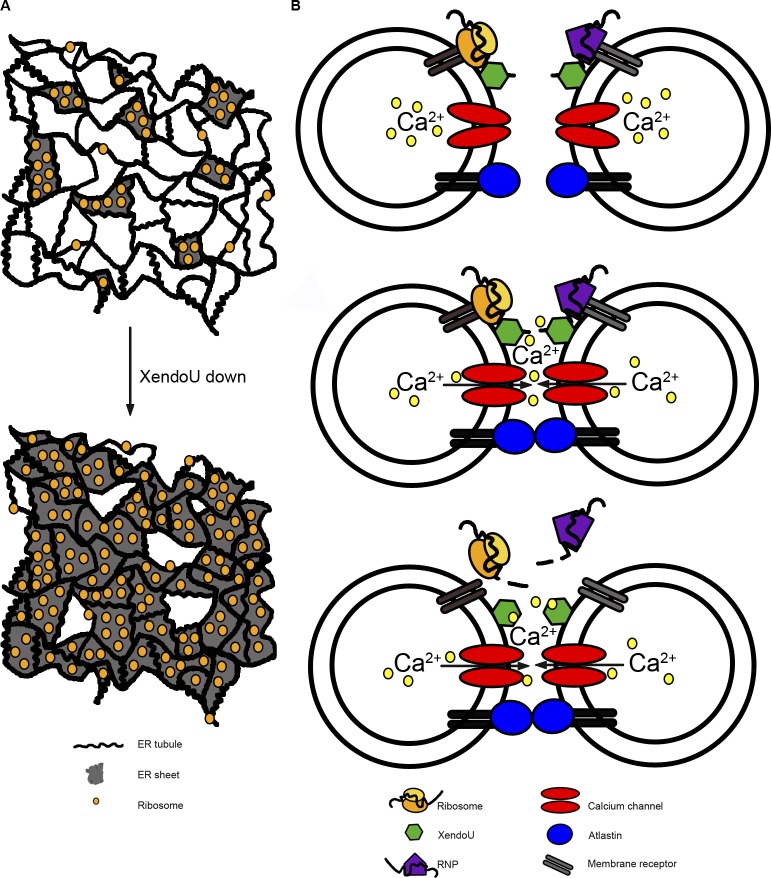
Our data, in particular the use of purified vesicles, support the hypothesis that XendoU acts by locally cleaving membrane-bound mRNAs and rRNAs followed by release of a subpopulation of ribosomes and other RNA-associated proteins from the ER. We speculate that RNA cleavage functions to clear a patch of ER membrane, possibly to remove complexes that could sterically hinder the vesicle fusion machinery, allowing for tubule growth and fine-tuning of network remodeling (Fig. 7 A). In X. laevis egg extracts, the loss of XendoU leads to a failure of vesicle fusion and a lack of tubular ER network. Loss of EndoU in HeLa cells leads to a loss of tubular ER and an expansion of ER sheets. This is consistent with the observation that ER tubules generally contain fewer ribosomes than ER sheets (Puhka et al., 2007; West et al., 2011), though it is unknown how the difference in ribosome occupancy is achieved by the cell. Our results support the model that XendoU is part of the molecular mechanism that regulates ribosome occupancy on the surface of the ER, and therefore affects ER morphology.
Figure 7.
Model of XendoU nuclease activity on membranes. (A) The ER network exists as a mixture of tubules and sheets. A decrease in XendoU results in the expansion of sheets. (B) Ribosomes, ribonucleoproteins (RNPs), XendoU, Atlastin, and (closed) calcium channels are localized to membrane vesicles containing Ca2+. Dimerization of Atlastin leads to eventual membrane fusion and calcium release through calcium channels on the membrane. XendoU binds calcium and mediates local RNA degradation (mRNAs, rRNAs, other RNAs), resulting in the release of ribosomes, RNPs, and RNA from the surface of membranes.
Recent work has indicated that several proteins play a key role in regulating the shape of the ER. Among the most intriguing is the membrane-bound GTPase Atlastin, which is required for ER formation in several different systems (Hu et al., 2009; Orso et al., 2009; Wang et al., 2013) and is sufficient to promote vesicle fusion in a purified system (Orso et al., 2009; Bian et al., 2011). Structural and biochemical studies have demonstrated that Atlastin can form two different types of dimers that are likely to be important intermediates in the vesicle fusion reaction (Bian et al., 2011). Because XendoU is not homologous to other proteins that are important in vesicle fusion, it seems likely that XendoU facilitates vesicle fusion at a step after Atlastin functions (Fig. 7 B). Perhaps Atlastin-mediated vesicle dimerization promotes calcium release from the ER lumen that activates XendoU activity to promote efficient vesicle fusion.
Our original identification of XendoU used the addition of CaCl2 to egg extracts to mimic fertilization-induced calcium increases. However, vesicle fusion experiments performed in the absence of cytosol did not require exogenous calcium, demonstrating that XendoU is activated by calcium released from the lumen of the ER. Calcium plays a well-documented role in the fusion of vesicles with the plasma membrane during exocytosis (for review see Jahn and Fasshauer, 2012). In addition, several studies have shown that release of calcium from the lumen of fusing vesicles is required for SNARE-mediated vesicle fusion (Peters and Mayer, 1998; Schekman, 1998; Ungermann et al., 1998), although it is unclear how vesicle fusion triggers calcium release or what calcium channel is used. Work using X. laevis extracts has demonstrated a role for both calcium and the IP3 receptor in mediating nuclear envelope formation (Sullivan et al., 1993, 1995), which suggests that the IP3 receptor could be a potential calcium channel activated during vesicle fusion.
Consistent with a role for luminal calcium in ER structure, we also observe the EndoU-mediated effects on tubule formation in human cells, where knockdown of EndoU results in an expansion of ER sheets, a phenotype also seen in loss-of-function Rab10 (English and Voeltz, 2013b). In human cells, all changes in intracellular calcium arise as a consequence of release from intracellular stores, primarily the ER. Recent work has shown that calcium signaling in neuronal cells leads to a dramatic rearrangement of the ER (Cui-Wang et al., 2012). In addition, a recent study demonstrated that down-regulation of mouse EndoU suppressed activation-induced cell death in B cells (Poe et al., 2014). B cell activation is accompanied by increased calcium signaling and an enormous expansion of the ER, which is consistent with the observation that changes in EndoU levels modulate ER structure. Collectively these results suggest that EndoU proteins are likely to function in general ER structure, but that they may play an increased role in response to calcium signaling.
Materials and methods
Extract preparation
X. laevis metaphase-arrested egg extract was prepared fresh as described previously (Hannak and Heald, 2006). In brief, female frogs were primed with 100 U pregnant mare serum gonadotropin (PMSG; EMD Millipore) followed by injection of 500 U human chorionic gonadotropin (HCG; Sigma-Aldrich) at least 48 h later. Laid eggs (16–18 h) were collected, dejellied, washed (with buffer containing 5 mM EGTA), packed (200 g for 1 min followed by 800 g for 30 s), and crushed (17,000 g for 15 min). Cytoplasm was collected via needle puncture through the side of the tube, protease inhibitors and energy mix were added, and extract was kept at 12°C until use.
Fractionated interphase extract was prepared from X. laevis unfertilized eggs, as described previously, into cytosolic and light membrane fractions (Powers et al., 2001). In brief, frogs were injected as described for CSF preparation, eggs were collected (100 mM NaCl), dejellied, washed (without EGTA), and packed (400 g for 15 s). Protease inhibitors and cycloheximide (50 µg/µl final) were added on top of the packed eggs that were then lysed (12,000 g for 15 min). Crude interphase extract was collected via needle puncture on the side of the tube and centrifuged (250,000 g for 70 min at 4°C). A clear cytosolic fraction was collected and spun again (250,000 g for 25 min at 4°C) while the light membrane fraction was isolated, diluted, and spun over a sucrose gradient (26,000 g for 20 min at 4°C). Light membranes were collected and along with the cytosolic fraction were aliquoted, snap frozen in liquid nitrogen, and stored at −80°C.
RNA isolation and electrophoresis
Aliquots of egg extract reactions (in the presence of metal at the concentrations indicated) were added to 100 µl 2× PK buffer (200 mM Tris-Cl, pH 7.5, 25 mM EDTA, pH 8.0, 300 mM NaCl, and 2% wt/vol SDS) and deproteinized with 10 µl of 20 mg/ml proteinase K (Roche) at 65°C for 15 min. Samples were phenol extracted using equal volume of phenol:chloroform and precipitated with 1 µl of 20 mg/ml glycogen (USB) and 3 vol absolute ethanol (Haley et al., 2003). Alternatively, RNA was isolated using TRIzol (Invitrogen) according to manufacturer’s directions. RNAs were resuspended in 2× urea dye (8 M urea, 25 mM EDTA, pH 8.0, 0.025% wt/vol xylene cyanol, and 0.025% wt/vol bromophenol blue) and separated on 10% 8 M urea acrylamide gels. RNA was detected by staining with SYBR Green II (Invitrogen) according to the manufacturer’s directions and visualized using a Typhoon TRIO (GE Healthcare). Alternatively, RNAs were 5′ end labeled with 50 µCi ATP [γ-32P] (6000 Ci/mmol; PerkinElmer) and T4 polynucleotide kinase (New England Biolabs, Inc.) according to the manufacturer’s directions. RNAs were electrophoresed on 10% 8 M urea acrylamide gels, then gels were exposed to a PhosphorImager plate (GE Healthcare) and imaged on a Typhoon TRIO (GE Healthcare).
To test the effects of chelators on RNA cleavage, reactions were assembled in the presence of EGTA (either 2 or 10 mM over exogenous CaCl2). EGTA was either added alone (no CaCl2 present), at the same time as CaCl2, or at the times indicated before, or after, the addition of CaCl2. RNA was isolated and processed as described for isolation of RNA from CSF extract.
Purification of nuclease activity from X. laevis egg extract
X. laevis egg extract (200 µl) was precleared (200,000 g for 2 h) and run over a 1-ml HiTrap Heparin HP column (GE Healthcare) on an AKTA Purifier 10 (GE Healthcare) equilibrated in 1× XB (without CaCl2): 100 mM KCl, 1 mM MgCl2, 50 mM sucrose, and 10 mM Hepes-KOH, pH 7.7. Flow-through fractions were collected and bound proteins were eluted using a linear salt gradient to 1 M KCl. Flow-through (2.5 ml) was adjusted to 1 mM CaCl2 and run over the same Heparin column preequilibrated in 1× XB + 1 mM CaCl2. Bound proteins were eluted using a linear salt gradient to 1 M KCl (Seidel and Peck, 1994). Fractions (0.5 ml) containing protein were analyzed by SDS-PAGE and Coomassie Imperial staining (Thermo Fisher Scientific). Bands were excised and submitted for mass spectrometry (LC/MS/MS) analysis (Taplin Mass Spectrometry Facility, Harvard Medical School).
Cloning, expression, and purification of recombinant proteins
Full-length XendoU A and B were cloned from total egg extract RNA. First strand synthesis was performed with random hexamers (Promega) and reverse transcription was performed with Superscript III RT (Invitrogen). PCR was performed using the Fast Start High Fidelity system (Roche) using primers oMB 929 (5′-TATTGACTCGAGTCAGTACAGATCCGGGTT-3′, XhoI site underlined) and oMB 910 (5′-TTACTAGGATCCGCGAGTAACAGGGGGCAG-3′, BamHI site underlined) for XendoU A and oMB930 (5′-TATTGACTCGAGTCAGTACAGATCTGGGTT-3′, XhoI site underlined) and oMB912 (5′-TTACTAGGATCCGAGGCCAACAGGGGGCAG-3′, BamHI site underlined) for XendoU B. PCR products were digested with BamHI (New England Biolabs, Inc.) and XhoI (New England Biolabs, Inc.), and gel purified (QIAGEN). pET30a plasmid was digested with BamHI (New England Biolabs, Inc.) and XhoI (New England Biolabs, Inc.), treated with calf alkaline phosphatase (New England Biolabs, Inc.), and gel purified (QIAGEN). Linearized plasmid and digested PCR products were ligated with T4 DNA (Quick) ligase (New England Biolabs, Inc.) and transformed into DH5α competent cells (Invitrogen). Sequence-verified clones (XendoU A [pMB523] and XendoU B [pMB524]) were transformed into BL21 Rosetta-gami competent cells (EMD Millipore). Single colonies were used to inoculate overnight cultures. 10 ml of overnight culture was used to inoculate 1 liter of LB + kanamycin with 0.1% glucose. Cells were grown at 37°C until OD600 = 0.6. Cells were induced with 0.5 mM IPTG for 6 h at 21°C and collected by centrifugation at 4,000 g for 30 min and snap frozen in liquid nitrogen. Cell pellets were thawed on ice and resuspended in 1× PBS with 150 mM NaCl and 20 mM imidazole and lysed with a French press. Native purification was performed on NiNTA beads (QIAGEN) according to manufacturer’s directions and dialyzed into 1× PBS + 15% glycerol. Aliquots were snap frozen in liquid nitrogen and stored at −80°C.
Oligos used for site-directed mutagenesis (H162A, K224A) were as follows: H162A (oMB 1619, 5′-GGAGAATCTAAGAGAGGGAAGGAG-3′; and oMB 1614, 5′-CACAAACACGGCCTCAAAGCCGCAC-3′, where underlined nucleotides represent amino acid change) and K224A (oMB 1617, 5′-AAGGAGATGGTGGCACCCGTCGGC-3′; and oMB 1618, 5′-CCAACTGAACTGCAGGTTCAACACC-3′, where underlined nucleotides represent amino acid change). Oligos were phosphorylated using T4 PNK (New England Biolabs, Inc.) according to manufacturer’s directions. PCR was performed using Phusion polymerase (New England Biolabs, Inc.) according to the manufacturer’s directions based on the Tm of the oligos. PCR products were purified using the QIAquick PCR purification kit (QIAGEN), ligated using T4 DNA (Quick) ligase (New England Biolabs, Inc.) according to manufacturer’s directions, and transformed into chemically competent cells. Sequence-verified clones (XendoU B H162A [pMB837] and XendoU B K224A [pMB838]) were transformed and proteins were expressed and purified as described for wild-type XendoU.
In vitro cleavage assays using recombinant XendoU
Purified recombinant XendoU was used at 0.05 µM final concentration in reaction buffer (50 mM NaCl, 25 mM Hepes, pH 7.4, 1 mM DTT, 0.1% glycerol, and 20 U RNasin) in a 25-µl reaction containing 2 µg of total RNA isolated from egg extract (as described for isolation of RNA from CSF extract). CaCl2, MgCl2, or MnCl2 were used at the concentrations indicated (0–5 mM) and incubated at 25°C for 30 min, and RNA was isolated and electrophoresed on 10% 8 M urea acrylamide gels.
In-gel nuclease assay
In-gel nuclease assays (Rosenthal and Lacks, 1977; Seidel and Peck, 1994) were performed essentially as described previously. In brief, heparin column fractions or purified recombinant XendoU (at 0.4 or 1 µg) were run on a 10% SDS-PAGE gel containing 32P-labeled full-length Xpat RNA incorporated into the resolving portion of the gel. Gels were washed four times for 30 min each at room temperature in 50 mM Tris-Cl, pH 7.5, 1 mM EDTA, and 0.5 mM DTT to renature proteins. Identical gels were soaked in buffer containing either no metal, 1 mM CaCl2, 1 mM MgCl2, or 1 mM MnCl2 overnight at 4°C and imaged using a Typhoon PhosphorImager.
In vitro transcription of 32P RNAs
T7 transcription reactions were performed as described previously (Haley et al., 2003). RNA labeling with UTP [α-32P] was performed using 50 µCi (3,000 Ci/mmol; PerkinElmer) per 20 µl reaction. RNAs were treated with Turbo DNase (Ambion) and purified over G-25 microspin columns (GE Healthcare) according to manufacturer’s directions. RNAs were stored in milliQ H2O and concentrations were determined by NanoDrop (Thermo Fisher Scientific).
Antibodies
Rabbit α-XendoU B antibodies were generated by Covance against full-length native protein and affinity purified over Affi-Gel 10 agarose beads (Bio-Rad Laboratories). In brief, 5 mg of purified recombinant protein was incubated overnight at 4°C with 1 ml packed agarose beads, proceeded by a 1-h incubation at RT with 100 mM Tris-Cl, pH 7.8, 100 mM KCl, and 200 mM NaCl to neutralize unreacted sites. 1 ml of serum was incubated with washed beads overnight at 4°C, beads were washed with 1× PBS, and affinity purified antibody was collected after 2 × 1.2 ml addition of 200 mM glycine and 300 mM NaCl. Fractions were collected into 1/10 volume of 1.5 M Tris-Cl, pH 8.8, to neutralize, then dialyzed into 1× PBS + 50% glycerol. Antibodies were used at dilutions indicated. Rabbit α-TRAP alpha was raised against the C-terminal domain (CTD) by Covance and affinity purified from serum. Gifted antibodies used in this study include α-Importin β (rabbit; K. Weis laboratory, ETH Zurich, Zurich, Switzerland) and α-Cyclin B2 (mouse; T. Hunt laboratory, London Research Institute, London, England, UK). Commercial antibodies used in this study include α-Dynein (74 kD intermediate chain MAB1618, mouse; EMD Millipore), α-α tubulin (DM1A, mouse; Sigma-Aldrich), α-ribosomal protein S6 (5G10, rabbit; Cell Signaling Technology), α-ribosomal protein L7a (E109, rabbit; Cell Signaling Technology), α-kinectin-C (rabbit, K1644; Sigma-Aldrich), and ChromPure Rabbit IgG (whole molecule; Jackson ImmunoResearch Laboratories, Inc.).
Depletions of XendoU in egg extract
40 µg of affinity purified antibody or rabbit IgG was incubated with 160 µl washed protein A Dynabeads (Invitrogen) for 1 h at room temperature. 50 µl of extract was incubated with coupled and washed beads on ice for 30 min, flicking every 10 min. A second round of depletion was performed identically to the first round of depletion.
Rescue of depleted extract
Recombinant proteins (or protein storage buffer, 1× PBS + 15% glycerol) were added to depleted or mock-depleted extracts at 10 µM for 30 min on ice. CaCl2 was added at a 0.8-mM final concentration, and reactions were incubated at 25°C and monitored by squash (2 µl reaction mixed with 1 µl 0.025% octadecyl rhodamine [Molecular Probes] placed on a glass slide and covered with an 18-mm glass coverslip).
Visualization of ER membranes
Calcium chloride was added to a final concentration of 0.8 mM in metaphase-arrested extracts to send them into interphase. Reactions were performed at 25°C for 60 min, and 2 µl of the reaction was mixed with 1 µl of 0.025% octadecyl rhodamine in buffer (10 mM Hepes, pH 7.4, 250 mM sucrose, 50 mM KCl, and 2.5 mM MgCl2). Images were acquired with a 60× oil objective lens (NA 1.42) on a microscope (BX61; Olympus) equipped with a charge-coupled device camera (ORCA-ER; C4742-80; Hamamatsu Photonics) at ∼20°C. Images were acquired and analyzed using the MetaMorph software package (Molecular Devices), and three-way junctions were counted to assess network formation.
Monitoring rate of replication and nuclear envelope formation in CSF extract
CSF extracts were depleted of XendoU or control IgG, and Cy5-dCTP (GE Healthcare) was added to extracts at a concentration of 1 µM. Samples were extracted every 10 min and examined by squash using the Olympus microscope setup described for visualization of ER networks. Nuclear intensity of Cy5-dCTP was calculated at each time point using MetaMorph. To monitor nuclear envelope formation in depleted extracts, Vybrant DiL (Invitrogen) was diluted 1:500 in extract that contained sperm nuclei and GFP Histone H1. Aliquots were monitored by squash every 10 min with 10–15 random fields imaged live.
Immunofluorescence on fixed ER networks
Light membranes (∼20×) were mixed with cytosol in a 10-µl reaction including 1 mM ATP and loaded into a flow cell that had been preblocked for 2 min at RT with cytosol/light membranes diluted 1:20 in 1× XB (100 mM KCl, 0.1 mM CaCl2, 1 mM MgCl2, 50 mM sucrose, and 10 mM Hepes, pH 7.7). Flow cells consisted of a standard glass slide, two rows of double-sided tape with an 18 × 18 × 1-mm coverslip (Corning) inverted on top of the tape, creating a channel ∼3–4 mm in width. All glass slides and coverslips were treated with 1 M HCl at 55°C overnight, cooled to RT, then washed several times with milliQ H2O. Cleaned slides were rinsed with ethanol and allowed to dry on Whatman paper. Additionally, coverslips were treated with poly-lysine (Sigma-Aldrich) diluted 1:10 in milliQ H2O for 15 min RT. Coverslips were rinsed several times with milliQ H2O and ethanol and allowed to dry.
Reactions in flow cells were kept in a humid chamber for 60 min at RT. Chambers were gently washed three times with 1× XB, leaving the chamber filled with 1× XB. Fix (4% PFA and 0.1% glutaraldehyde [Sigma-Aldrich] in 1× XB) was slowly flowed into the chamber. Flow cells were kept at RT for 20 min in a humid chamber then washed with 1× XB. Coverslips were removed and post-fixed for 5 min in −20°C methanol. Unreacted glutaraldehyde was reduced with 10 mM sodium borohydride in 1× PBS for 4 min, repeated twice, and washed three times with 1× PBS (Allan, 1995). Coverslips were blocked for 30 min at RT with 0.2% fish skin gelatin in 1× PBS and incubated with primary antibodies by inverting coverslips over antibody solution overnight at 4°C. Coverslips were washed and incubated with secondary antibody (α-rabbit Cy3; Jackson ImmunoResearch Laboratories, Inc.) and Alexa Fluor 488 Concanavalin A (A488 conA; 1:250; Life Technologies) for 1 h at RT. Washed coverslips were mounted in Vectashield (1:2,000; Vector Laboratories) and visualized as described previously (Allan, 1995; Wollert et al., 2002).
Vesicle fusion assay
Light membranes were washed twice with BufferA200 (50 mM Hepes-KOH, pH 7.5, 2.5 mM MgCl2, 250 mM sucrose, and 200 mM KCl). Network formation was performed by incubating wlms (0.5 mg/ml final concentration) in BufferA200 with 1 mM ATP and 0.5 mM GTP at 25°C for 60 min. Images for vesicle fusion assays were acquired on an inverted microscope (Eclipse TE2000-E; Nikon) equipped with a charge-coupled device camera (ORCA-ER; C4742-80; Hamamatsu Photonics) and a 100× oil objective lens (NA 1.4) using the Openlab software package (PerkinElmer). Antibody addition (5 µM) experiments included a 30-min preincubation of antibody at RT with wlms followed by addition of ATP and GTP as described for visualization of membranes in CSF extract. Networks were visualized by taking aliquots of the reactions at 15 min onto a passivated coverslip and mixed with octadecyl rhodamine as described for CSF extracts. Coverslips were attached with double-sided tape to the bottom of a glass slide with a 6-mm hole drilled into it and sealed with another coverslip on top. Coverslips were passivated essentially as described previously (Field et al., 2014) with minor modifications. Coverslips (gold seal; BD) were etched for 1 min in an oxygen plasma etcher (March Instruments) and silanized using 10% wt/vol methoxy-PEG-silane (5,000 MW; JenKem Technology) for 2 h at 70°C, rinsed with 100% ethanol and milliQ H20, air dried, and sealed until use.
RNAs were isolated from membranes (pellet) or supernatants after a 10-min pelleting spin at 20,000 g at 4°C. Membrane RNAs were labeled with ATP [γ-32P] as described for labeling of total extract RNA after treatment with calf intestinal phosphatase (CIP; New England Biolabs, Inc.) according to the manufacturer’s directions to remove 5′ phosphates and enable more efficient labeling with PNK.
RNaseA treatment of light membranes
Light membranes were washed twice in 15 volumes of BufferA200 and resuspended in the original volume. Wlms were mock-treated (H2O) or treated with RNaseA (Type X IIA; Sigma-Aldrich) at 0.01 ng/µl, 0.1 ng/µl, or 1 ng/µl final concentration for 20 min at RT. Treated membranes were washed one more time in BufferA200 and reactions were set up for each condition with or without ATP and GTP for 15 min at RT. Aliquots of each sample were placed on a passivated coverslip and mixed with octadecyl rhodamine and imaged at 20 min and 60 min from the start of the reaction.
DNA constructs
Human EndoU-2 was PCR amplified from cDNA corresponding to BC074763 (Refseq EndoU-2) using primers (F, 5′-AGACTTGTACTCGGCACCCA-3′; R, 5′-GGAAGACACTATGTAGGC-3′) and subcloned into pEGFP-N1 using SalI and SacII (pMB651). A short version of EndoU (corresponding to cDNA AK295963) was PCR amplified from the same cDNA using the forward primer (5′-AGACTTGTACTCGGCACCCA-3′) and the reverse primer (R, 5′-GGAAGACACTATGTAGGC-3′) and subcloned into pEGFP-N1 (pMB836). Sec61β was amplified from HeLa cell cDNA and subcloned into pEGFP-C1 using BglII and EcoRI (pMB648). mCherry Sec61β (pMB680) was generated by replacing GFP with mCherry using NheI and BglII. Catalytically dead, GFP-tagged point mutants of EndoU2 H259A (pMB678) and EndoU-short H66A (pMB679) were created using site directed mutagenesis.
esiRNA production
A portion of the coding region of EndoU (bp 172–704 of EndoU2) was amplified by PCR from the cDNA BC074763 using primers (F, 5′-ATCTCCCTGGTATTGGCCGT-3′; R, 5′-TCCTGGGCACTGAAGTGCT-3′) or 1,000 bp of the 3′ UTR were amplified by PCR from HeLa cDNA using primers (F, 5′-TAGAACTTCGAGCCAGAAAGG-3′; R, 5′-CTTCAATGACTTTAATGGAAAC-3′). PCR products were TOPO cloned into pCR2.1 (Invitrogen) and verified by sequencing. Control esiRNA sequences came from the 3′ UTR of X. laevis MenF (pMB463 and pMB464). One clone in each orientation with respect to the T7 promoter sequence in pCR2.1 was amplified by PCR using M13 primers. Each PCR product was used as the template for in vitro transcription using T7 RNA polymerase. Complementary RNAs were annealed by heating to 90°C for 3 min followed by cooling at 0.1°C/s to 70°C (holding for 3 min), cooling at 0.1°C/s to 50°C (holding for 3 min), and cooling at 0.1°C/s to 25°C (holding 3 min), cooling to 4°C. Double-stranded RNA (dsRNA) was processed into esiRNA using the ShortCut RNaseIII system (New England Biolabs, Inc.) and purified using the Purelink miRNA isolation kit (Invitrogen). Digested esiRNAs were run on denaturing PAGE gels to ensure that RNAs were of the correct size. siRNAs against human EndoU were purchased from Thermo Fisher Scientific (siGENOME SMART pool). The sequences of these siRNAs are: 5′-GCAGUGAUGCCAUAACAAA-3′, 5′-CAACUGGGACGGCUACUAU-3′, 5′-GAGGAUAUCCCUUAGCUGU-3′, and 5′-GGUUUGGGCUCUAUAUUCAAG-3′. Verification of knockdown was confirmed by quantitative RT-PCR as described previously (Sharp et al., 2011) and normalized to endogenous actin levels.
Cell culture, transfection, and RNAi
HeLa cells were grown in DMEM (Invitrogen) supplemented with 10% FBS. To examine localization of GFP- or mCherry tagged proteins, cells were plated on glass coverslips in a 24-well plate at ∼80% confluency and transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Each transfection contained 400 ng of DNA. Transfected cells were examined 24 h after transfection. For esiRNA experiments, cells were seeded at 50% confluency and transfected using 10 nM of each esiRNA and Lipofectamine RNAiMAX (Invitrogen). Cells were examined 48–72 h after transfection. For RNAi rescue experiments, cells were transfected with esiRNA, incubated for ∼36 h until cells had reached ∼80% confluency, then transfected with mCherry-Sec61β and various GFP-tagged EndoU constructs. For siRNA experiments, cells were seeded at 50% confluency and transfected with 5 nM siRNA using Lipofectamine RNAiMAX (Invitrogen). Cells were examined 72 h after transfection.
Fixation and immunofluorescence
Cells were washed twice with 37°C 1× PBS then fixed with 37°C 1× PBS + 4% paraformaldehyde (Electron Microscopy Sciences) + 0.1% glutaraldehyde (Electron Microscopy Sciences) for 10 min at 37°C. Cells were washed twice with 1× PBS, incubated with 10 mM NaBH4 for 10 min, washed three times with 1× PBS, then examined by microscopy or processed for immunofluorescence. Cells were mounted in Vectashield and examined using the Olympus microscope setup described for visualization of ER networks at 20°C, with the exception that all images were acquired using an Olympus spinning disc confocal attachment (DSU) and all images were acquired as 3D stacks. For immunofluorescence with kinectin antibodies, anti–rabbit Cy3 was used as the secondary antibody. All images were acquired and processed using MetaMorph software.
RNA-seq and analysis
RNA was purified from CSF-arrested or interphase (30 min after addition of 0.6 mM CaCl2) extracts using TRIzol (Invitrogen). Extracts were untreated, immunodepleted using control IgG, or immunodepleted using XendoU antibodies. mRNA was purified by CAP-capture (Blower et al., 2013) for IgG and XendoU-depleted extracts or oligo-dT (untreated extracts) and used as input for the TruSeq mRNA library (Illumina) preparation kit. Libraries were multiplexed (four samples per lane) and sequenced on an Illumina HiSeq as single-end 50 mers.
The resulting reads were reduced to unique reads (we assumed that all perfectly matching reads were the result of PCR duplication) using a custom Perl script. Reads were the aligned to the X. laevis rRNA precursor (gi|65056, gi|64487, gi|65094, gi|65095) using Bowtie (Langmead et al., 2009). Reads aligning to X. laevis rRNA were subtracted using a custom Perl script. The remaining reads were aligned to a draft assembly of the X. laevis genome (7.0 downloaded from Xenbase, June 2013) using tophat2. Reads aligning to transcripts were counted using Cufflinks.
Online supplemental material
Fig. S1 presents additional characterization of recombinant XendoU. Fig. S2 presents analysis of mRNA stability in X. laevis CSF-arrested and calcium-treated egg extracts. Fig. S3 presents the effects of XendoU depletion on DNA replication timing and cyclin B degradation. Fig. S4 presents additional information on ER network formation in untreated CSF extract and in purified light membranes. Fig. S5 presents images of the rescue of EndoU RNAi phenotype and the phenotypes of HeLa cells treated with EndoU siRNAs. Supplemental script S1, uniquefastq.pl, is a Perl script that collapses all the reads in a fastq file into unique reads. Supplemental script S2, removerrna.pl is a Perl script that uses the SAM alignment file of reads aligned against rRNA to remove these reads from a fastq file. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201406037/DC1.
Supplementary Material
Acknowledgments
We thank members of the Blower laboratory, members of the Zamore laboratory, Nelson Lau, Songyu Wang, and Tom Rapoport for helpful discussions and comments on the manuscript. We thank Phuong Nguyen for advice on passivation of coverslips, Aleksandar Marinkovic for help with the oxygen plasma etcher, and Derek Lessing for assistance with the inverted microscope. We thank Tim Hunt and Karsten Weis for the kind gift of antibodies and the MGH Molecular Biology Next Generation Sequencing Core for deep sequencing samples.
This work was supported by a Jane Coffin Childs Memorial Fund for Medical Research postdoctoral fellowship to D.S. Schwarz, a Massachusetts General Hospital Executive Committee on Research (ECOR) Fund for Medical Discovery award to D.S. Schwarz, and a Career Award in the Biomedical Sciences from the Burroughs Wellcome Fund to M.D. Blower.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- CSF
- cytostatic factor
- esiRNA
- endoribonuclease-prepared siRNA
- rRNA
- ribosomal RNA
- snoRNA
- small nucleolar RNA
- wlm
- washed light membrane
References
- Allan, V.1995. Protein phosphatase 1 regulates the cytoplasmic dynein-driven formation of endoplasmic reticulum networks in vitro. J. Cell Biol. 128:879–891 10.1083/jcb.128.5.879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann, O., and Walz B.. 2001. Endoplasmic reticulum of animal cells and its organization into structural and functional domains. Int. Rev. Cytol. 205:149–214 10.1016/S0074-7696(01)05004-5 [DOI] [PubMed] [Google Scholar]
- Berridge, M.J.1998. Neuronal calcium signaling. Neuron. 21:13–26 10.1016/S0896-6273(00)80510-3 [DOI] [PubMed] [Google Scholar]
- Bhardwaj, K., Sun J., Holzenburg A., Guarino L.A., and Kao C.C.. 2006. RNA recognition and cleavage by the SARS coronavirus endoribonuclease. J. Mol. Biol. 361:243–256 10.1016/j.jmb.2006.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian, X., Klemm R.W., Liu T.Y., Zhang M., Sun S., Sui X., Liu X., Rapoport T.A., and Hu J.. 2011. Structures of the atlastin GTPase provide insight into homotypic fusion of endoplasmic reticulum membranes. Proc. Natl. Acad. Sci. USA. 108:3976–3981 10.1073/pnas.1101643108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blower, M.D., Jambhekar A., Schwarz D.S., and Toombs J.A.. 2013. Combining different mRNA capture methods to analyze the transcriptome: analysis of the Xenopus laevis transcriptome. PLoS ONE. 8:e77700 10.1371/journal.pone.0077700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brookes, P.S., Yoon Y., Robotham J.L., Anders M.W., and Sheu S.S.. 2004. Calcium, ATP, and ROS: a mitochondrial love-hate triangle. Am. J. Physiol. Cell Physiol. 287:C817–C833 10.1152/ajpcell.00139.2004 [DOI] [PubMed] [Google Scholar]
- Busa, W.B., and Nuccitelli R.. 1985. An elevated free cytosolic Ca2+ wave follows fertilization in eggs of the frog, Xenopus laevis. J. Cell Biol. 100:1325–1329 10.1083/jcb.100.4.1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caffarelli, E., Arese M., Santoro B., Fragapane P., and Bozzoni I.. 1994. In vitro study of processing of the intron-encoded U16 small nucleolar RNA in Xenopus laevis. Mol. Cell. Biol. 14:2966–2974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caffarelli, E., Maggi L., Fatica A., Jiricny J., and Bozzoni I.. 1997. A novel Mn++-dependent ribonuclease that functions in U16 SnoRNA processing in X. laevis. Biochem. Biophys. Res. Commun. 233:514–517 10.1006/bbrc.1997.6487 [DOI] [PubMed] [Google Scholar]
- Cui-Wang, T., Hanus C., Cui T., Helton T., Bourne J., Watson D., Harris K.M., and Ehlers M.D.. 2012. Local zones of endoplasmic reticulum complexity confine cargo in neuronal dendrites. Cell. 148:309–321 10.1016/j.cell.2011.11.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai, A., Murray A., Mitchison T.J., and Walczak C.E.. 1999. The use of Xenopus egg extracts to study mitotic spindle assembly and function in vitro. Methods Cell Biol. 61:385–412 10.1016/S0091-679X(08)61991-3 [DOI] [PubMed] [Google Scholar]
- Dreier, L., and Rapoport T.A.. 2000. In vitro formation of the endoplasmic reticulum occurs independently of microtubules by a controlled fusion reaction. J. Cell Biol. 148:883–898 10.1083/jcb.148.5.883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, Y., Ferro-Novick S., and Novick P.. 2004. Dynamics and inheritance of the endoplasmic reticulum. J. Cell Sci. 117:2871–2878 10.1242/jcs.01286 [DOI] [PubMed] [Google Scholar]
- Eisen, A., and Reynolds G.T.. 1985. Source and sinks for the calcium released during fertilization of single sea urchin eggs. J. Cell Biol. 100:1522–1527 10.1083/jcb.100.5.1522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- English, A.R., and Voeltz G.K.. 2013a. Endoplasmic reticulum structure and interconnections with other organelles. Cold Spring Harb. Perspect. Biol. 5:a013227 10.1101/cshperspect.a013227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- English, A.R., and Voeltz G.K.. 2013b. Rab10 GTPase regulates ER dynamics and morphology. Nat. Cell Biol. 15:169–178 10.1038/ncb2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field, C.M., Nguyen P.A., Ishihara K., Groen A.C., and Mitchison T.J.. 2014. Xenopus egg cytoplasm with intact actin. Methods Enzymol. 540:399–415 10.1016/B978-0-12-397924-7.00022-4 [DOI] [PubMed] [Google Scholar]
- Friedman, J.R., Webster B.M., Mastronarde D.N., Verhey K.J., and Voeltz G.K.. 2010. ER sliding dynamics and ER-mitochondrial contacts occur on acetylated microtubules. J. Cell Biol. 190:363–375 10.1083/jcb.200911024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilkey, J.C., Jaffe L.F., Ridgway E.B., and Reynolds G.T.. 1978. A free calcium wave traverses the activating egg of the medaka, Oryzias latipes. J. Cell Biol. 76:448–466 10.1083/jcb.76.2.448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gioia, U., Laneve P., Dlakic M., Arceci M., Bozzoni I., and Caffarelli E.. 2005. Functional characterization of XendoU, the endoribonuclease involved in small nucleolar RNA biosynthesis. J. Biol. Chem. 280:18996–19002 10.1074/jbc.M501160200 [DOI] [PubMed] [Google Scholar]
- Grigoriev, I., Gouveia S.M., van der Vaart B., Demmers J., Smyth J.T., Honnappa S., Splinter D., Steinmetz M.O., Putney J.W. Jr, Hoogenraad C.C., and Akhmanova A.. 2008. STIM1 is a MT-plus-end-tracking protein involved in remodeling of the ER. Curr. Biol. 18:177–182 10.1016/j.cub.2007.12.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley, B., Tang G., and Zamore P.D.. 2003. In vitro analysis of RNA interference in Drosophila melanogaster. Methods. 30:330–336 10.1016/S1046-2023(03)00052-5 [DOI] [PubMed] [Google Scholar]
- Hannak, E., and Heald R.. 2006. Investigating mitotic spindle assembly and function in vitro using Xenopus laevis egg extracts. Nat. Protoc. 1:2305–2314 10.1038/nprot.2006.396 [DOI] [PubMed] [Google Scholar]
- Horner, V.L., and Wolfner M.F.. 2008. Transitioning from egg to embryo: triggers and mechanisms of egg activation. Dev. Dyn. 237:527–544 10.1002/dvdy.21454 [DOI] [PubMed] [Google Scholar]
- Hu, J., Shibata Y., Voss C., Shemesh T., Li Z., Coughlin M., Kozlov M.M., Rapoport T.A., and Prinz W.A.. 2008. Membrane proteins of the endoplasmic reticulum induce high-curvature tubules. Science. 319:1247–1250 10.1126/science.1153634 [DOI] [PubMed] [Google Scholar]
- Hu, J., Shibata Y., Zhu P.P., Voss C., Rismanchi N., Prinz W.A., Rapoport T.A., and Blackstone C.. 2009. A class of dynamin-like GTPases involved in the generation of the tubular ER network. Cell. 138:549–561 10.1016/j.cell.2009.05.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, J., Prinz W.A., and Rapoport T.A.. 2011. Weaving the web of ER tubules. Cell. 147:1226–1231 10.1016/j.cell.2011.11.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikura, M., Osawa M., and Ames J.B.. 2002. The role of calcium-binding proteins in the control of transcription: structure to function. BioEssays. 24:625–636 10.1002/bies.10105 [DOI] [PubMed] [Google Scholar]
- Jaffe, L.F.1983. Sources of calcium in egg activation: a review and hypothesis. Dev. Biol. 99:265–276 10.1016/0012-1606(83)90276-2 [DOI] [PubMed] [Google Scholar]
- Jahn, R., and Fasshauer D.. 2012. Molecular machines governing exocytosis of synaptic vesicles. Nature. 490:201–207 10.1038/nature11320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laneve, P., Altieri F., Fiori M.E., Scaloni A., Bozzoni I., and Caffarelli E.. 2003. Purification, cloning, and characterization of XendoU, a novel endoribonuclease involved in processing of intron-encoded small nucleolar RNAs in Xenopus laevis. J. Biol. Chem. 278:13026–13032 10.1074/jbc.M211937200 [DOI] [PubMed] [Google Scholar]
- Laneve, P., Gioia U., Ragno R., Altieri F., Di Franco C., Santini T., Arceci M., Bozzoni I., and Caffarelli E.. 2008. The tumor marker human placental protein 11 is an endoribonuclease. J. Biol. Chem. 283:34712–34719 10.1074/jbc.M805759200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead, B., Trapnell C., Pop M., and Salzberg S.L.. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S.Y., Krsmanovic V., and Brawerman G.. 1971. Attachment of ribosomes to membranes during polysome formation in mouse sarcoma 180 cells. J. Cell Biol. 49:683–691 10.1083/jcb.49.3.683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorca, T., Galas S., Fesquet D., Devault A., Cavadore J.C., and Dorée M.. 1991. Degradation of the proto-oncogene product p39mos is not necessary for cyclin proteolysis and exit from meiotic metaphase: requirement for a Ca(2+)-calmodulin dependent event. EMBO J. 10:2087–2093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorca, T., Cruzalegui F.H., Fesquet D., Cavadore J.C., Méry J., Means A., and Dorée M.. 1993. Calmodulin-dependent protein kinase II mediates inactivation of MPF and CSF upon fertilization of Xenopus eggs. Nature. 366:270–273 10.1038/366270a0 [DOI] [PubMed] [Google Scholar]
- Lu, L., Ladinsky M.S., and Kirchhausen T.. 2009. Cisternal organization of the endoplasmic reticulum during mitosis. Mol. Biol. Cell. 20:3471–3480 10.1091/mbc.E09-04-0327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, L., Ladinsky M.S., and Kirchhausen T.. 2011. Formation of the postmitotic nuclear envelope from extended ER cisternae precedes nuclear pore assembly. J. Cell Biol. 194:425–440 10.1083/jcb.201012063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki, S., Yuzaki M., Nakada K., Shirakawa H., Nakanishi S., Nakade S., and Mikoshiba K.. 1992. Block of Ca2+ wave and Ca2+ oscillation by antibody to the inositol 1,4,5-trisphosphate receptor in fertilized hamster eggs. Science. 257:251–255 10.1126/science.1321497 [DOI] [PubMed] [Google Scholar]
- Mochida, S., and Hunt T.. 2007. Calcineurin is required to release Xenopus egg extracts from meiotic M phase. Nature. 449:336–340 10.1038/nature06121 [DOI] [PubMed] [Google Scholar]
- Murray, A.W.1991. Cell cycle extracts. Methods Cell Biol. 36:581–605 10.1016/S0091-679X(08)60298-8 [DOI] [PubMed] [Google Scholar]
- Nicotera, P., and Orrenius S.. 1998. The role of calcium in apoptosis. Cell Calcium. 23:173–180 10.1016/S0143-4160(98)90116-6 [DOI] [PubMed] [Google Scholar]
- Orso, G., Pendin D., Liu S., Tosetto J., Moss T.J., Faust J.E., Micaroni M., Egorova A., Martinuzzi A., McNew J.A., and Daga A.. 2009. Homotypic fusion of ER membranes requires the dynamin-like GTPase atlastin. Nature. 460:978–983 10.1038/nature08280 [DOI] [PubMed] [Google Scholar]
- Perry, A.C., and Verlhac M.H.. 2008. Second meiotic arrest and exit in frogs and mice. EMBO Rep. 9:246–251 10.1038/embor.2008.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters, C., and Mayer A.. 1998. Ca2+/calmodulin signals the completion of docking and triggers a late step of vacuole fusion. Nature. 396:575–580 10.1038/25133 [DOI] [PubMed] [Google Scholar]
- Poe, J.C., Kountikov E.I., Lykken J.M., Natarajan A., Marchuk D.A., and Tedder T.F.. 2014. EndoU is a novel regulator of AICD during peripheral B cell selection. J. Exp. Med. 211:57–69 10.1084/jem.20130648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poteryaev, D., Squirrell J.M., Campbell J.M., White J.G., and Spang A.. 2005. Involvement of the actin cytoskeleton and homotypic membrane fusion in ER dynamics in Caenorhabditis elegans. Mol. Biol. Cell. 16:2139–2153 10.1091/mbc.E04-08-0726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers, M., Evans E.K., Yang J., and Kornbluth S.. 2001. Preparation and use of interphase Xenopus egg extracts. Curr. Protoc. Cell Biol. Chapter 11:10. [DOI] [PubMed] [Google Scholar]
- Puhka, M., Vihinen H., Joensuu M., and Jokitalo E.. 2007. Endoplasmic reticulum remains continuous and undergoes sheet-to-tubule transformation during cell division in mammalian cells. J. Cell Biol. 179:895–909 10.1083/jcb.200705112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid, D.W., and Nicchitta C.V.. 2012. Primary role for endoplasmic reticulum-bound ribosomes in cellular translation identified by ribosome profiling. J. Biol. Chem. 287:5518–5527 10.1074/jbc.M111.312280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renzi, F., Caffarelli E., Laneve P., Bozzoni I., Brunori M., and Vallone B.. 2006. The structure of the endoribonuclease XendoU: From small nucleolar RNA processing to severe acute respiratory syndrome coronavirus replication. Proc. Natl. Acad. Sci. USA. 103:12365–12370 10.1073/pnas.0602426103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricagno, S., Egloff M.P., Ulferts R., Coutard B., Nurizzo D., Campanacci V., Cambillau C., Ziebuhr J., and Canard B.. 2006. Crystal structure and mechanistic determinants of SARS coronavirus nonstructural protein 15 define an endoribonuclease family. Proc. Natl. Acad. Sci. USA. 103:11892–11897 10.1073/pnas.0601708103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridgway, E.B., Gilkey J.C., and Jaffe L.F.. 1977. Free calcium increases explosively in activating medaka eggs. Proc. Natl. Acad. Sci. USA. 74:623–627 10.1073/pnas.74.2.623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosbash, M., and Penman S.. 1971. Membrane-associated protein synthesis of mammalian cells. I. The two classes of membrane-associated ribosomes. J. Mol. Biol. 59:227–241 10.1016/0022-2836(71)90048-9 [DOI] [PubMed] [Google Scholar]
- Rosenthal, A.L., and Lacks S.A.. 1977. Nuclease detection in SDS-polyacrylamide gel electrophoresis. Anal. Biochem. 80:76–90 10.1016/0003-2697(77)90627-3 [DOI] [PubMed] [Google Scholar]
- Schekman, R.1998. Membrane fusion. Ready … aim … fire! Nature. 396:514–515 10.1038/24986 [DOI] [PubMed] [Google Scholar]
- Seidel, C.W., and Peck L.J.. 1994. Purification of a calcium dependent ribonuclease from Xenopus laevis. Nucleic Acids Res. 22:1456–1462 10.1093/nar/22.8.1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp, J.A., Plant J.J., Ohsumi T.K., Borowsky M., and Blower M.D.. 2011. Functional analysis of the microtubule-interacting transcriptome. Mol. Biol. Cell. 22:4312–4323 10.1091/mbc.E11-07-0629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata, Y., Voeltz G.K., and Rapoport T.A.. 2006. Rough sheets and smooth tubules. Cell. 126:435–439 10.1016/j.cell.2006.07.019 [DOI] [PubMed] [Google Scholar]
- Shibata, Y., Shemesh T., Prinz W.A., Palazzo A.F., Kozlov M.M., and Rapoport T.A.. 2010. Mechanisms determining the morphology of the peripheral ER. Cell. 143:774–788 10.1016/j.cell.2010.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhardt, R., Zucker R., and Schatten G.. 1977. Intracellular calcium release at fertilization in the sea urchin egg. Dev. Biol. 58:185–196 10.1016/0012-1606(77)90084-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan, K.M., Busa W.B., and Wilson K.L.. 1993. Calcium mobilization is required for nuclear vesicle fusion in vitro: implications for membrane traffic and IP3 receptor function. Cell. 73:1411–1422 10.1016/0092-8674(93)90366-X [DOI] [PubMed] [Google Scholar]
- Sullivan, K.M., Lin D.D., Agnew W., and Wilson K.L.. 1995. Inhibition of nuclear vesicle fusion by antibodies that block activation of inositol 1,4,5-trisphosphate receptors. Proc. Natl. Acad. Sci. USA. 92:8611–8615 10.1073/pnas.92.19.8611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, L., Yu F., Ullah A., Hubrack S., Daalis A., Jung P., and Machaca K.. 2011. Endoplasmic reticulum remodeling tunes IP3-dependent Ca²+ release sensitivity. PLoS ONE. 6:e27928 10.1371/journal.pone.0027928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terasaki, M., Runft L.L., and Hand A.R.. 2001. Changes in organization of the endoplasmic reticulum during Xenopus oocyte maturation and activation. Mol. Biol. Cell. 12:1103–1116 10.1091/mbc.12.4.1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tholey, G., Ledig M., Mandel P., Sargentini L., Frivold A.H., Leroy M., Grippo A.A., and Wedler F.C.. 1988. Concentrations of physiologically important metal ions in glial cells cultured from chick cerebral cortex. Neurochem. Res. 13:45–50 10.1007/BF00971853 [DOI] [PubMed] [Google Scholar]
- Ungermann, C., Sato K., and Wickner W.. 1998. Defining the functions of trans-SNARE pairs. Nature. 396:543–548 10.1038/25069 [DOI] [PubMed] [Google Scholar]
- Vacquier, V.D.1981. Dynamic changes of the egg cortex. Dev. Biol. 84:1–26 10.1016/0012-1606(81)90366-3 [DOI] [PubMed] [Google Scholar]
- Voeltz, G.K., Rolls M.M., and Rapoport T.A.. 2002. Structural organization of the endoplasmic reticulum. EMBO Rep. 3:944–950 10.1093/embo-reports/kvf202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voeltz, G.K., Prinz W.A., Shibata Y., Rist J.M., and Rapoport T.A.. 2006. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell. 124:573–586 10.1016/j.cell.2005.11.047 [DOI] [PubMed] [Google Scholar]
- Wang, S., Romano F.B., Field C.M., Mitchison T.J., and Rapoport T.A.. 2013. Multiple mechanisms determine ER network morphology during the cell cycle in Xenopus egg extracts. J. Cell Biol. 203:801–814 10.1083/jcb.201308001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterman-Storer, C.M., and Salmon E.D.. 1998. Endoplasmic reticulum membrane tubules are distributed by microtubules in living cells using three distinct mechanisms. Curr. Biol. 8:798–807 10.1016/S0960-9822(98)70321-5 [DOI] [PubMed] [Google Scholar]
- West, M., Zurek N., Hoenger A., and Voeltz G.K.. 2011. A 3D analysis of yeast ER structure reveals how ER domains are organized by membrane curvature. J. Cell Biol. 193:333–346 10.1083/jcb.201011039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitaker, M.2006a. Calcium at fertilization and in early development. Physiol. Rev. 86:25–88 10.1152/physrev.00023.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitaker, M.2006b. Calcium microdomains and cell cycle control. Cell Calcium. 40:585–592 10.1016/j.ceca.2006.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollert, T., Weiss D.G., Gerdes H.H., and Kuznetsov S.A.. 2002. Activation of myosin V-based motility and F-actin-dependent network formation of endoplasmic reticulum during mitosis. J. Cell Biol. 159:571–577 10.1083/jcb.200204065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, D., Buchholz F., Huang Z., Goga A., Chen C.Y., Brodsky F.M., and Bishop J.M.. 2002. Short RNA duplexes produced by hydrolysis with Escherichia coli RNase III mediate effective RNA interference in mammalian cells. Proc. Natl. Acad. Sci. USA. 99:9942–9947 10.1073/pnas.152327299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, D., Goga A., and Bishop J.M.. 2004. RNA interference (RNAi) with RNase III-prepared siRNAs. Methods Mol. Biol. 252:471–482 [DOI] [PubMed] [Google Scholar]
- Zhang, R.Q., and Ellis K.J.. 1989. In vivo measurement of total body magnesium and manganese in rats. Am. J. Physiol. 257:R1136–R1140 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.