Abstract
Stem cells give rise to tissues and organs during development and maintain their integrity during adulthood. They have the potential to self-renew or differentiate at each division. To ensure proper organ growth and homeostasis, self-renewal versus differentiation decisions need to be tightly controlled. Systematic genetic studies in Drosophila melanogaster are revealing extensive regulatory networks that control the switch between stem cell self-renewal and differentiation in the germline. These networks, which are based primarily on mutual translational repression, act via interlocked feedback loops to provide robustness to this important fate decision.
Primordial germ cells (PGCs) form during embryogenesis. In some species, such as fruit flies, nematodes, or anuran frogs, their specification relies on the inheritance of maternally synthesized germ cell determinants, whereas in other species, such as mammals, axolotls, and locusts, they form as a result of specific cell–cell interactions (Extavour and Akam, 2003; Seydoux and Braun, 2006; Chatfield et al., 2014; Donoughe et al., 2014). Irrespective of their exact origin, germ cells express a specific, conserved set of RNA regulatory proteins, such as Vasa, Nanos (Nos), Pumilio (Pum), Dazl, and Tudor (Gao and Arkov, 2013). Furthermore, germ cell–specific small RNA pathways play an important role in regulating gene expression in these cells and in surveillance of the genome against transposable elements and nonself RNAs (Luteijn and Ketting, 2013). Studies in the mammalian testis, the Drosophila melanogaster ovary and testis, and the Caenorhabditis elegans hermaphrodite gonad have revealed many features of adult stem cell systems, such as the importance of the local microenvironment for stem cell maintenance and differentiation, that are applicable to germline stem cells (GSCs) as well as other adult stem cell systems (Spradling et al., 2011). However, given their unique role in generating a new embryo, GSCs appear to be less “programmed” than other stem cell populations. In the mouse testis, stem cells can efficiently be reprogrammed into embryonic stem cell–like cells (Kanatsu-Shinohara et al., 2004), and Drosophila adult ovarian stem cells transplanted back into the embryo performed like PGCs (Niki and Mahowald, 2003). Thus, the analysis of GSC self-renewal, stem cell maintenance, and stem cell differentiation can not only reveal mechanisms shared with other adult stem cell systems that are needed for organ homeostasis but can also provide specific insight into mechanisms that reflect the unique demands on GSCs to generate a completely new organism.
To describe regulatory networks controlling GSC behavior, we chose one of the best-studied systems, the GSCs of the Drosophila ovary. Our emphasis is on highlighting the role of RNA regulatory pathways that control the balance between GSC self-renewal and differentiation. The Drosophila system has many advantages for the analysis of stem cell behavior, as different components of the stem cell compartment can be easily identified and individually manipulated by genetic interference (Xie and Spradling, 1998). Recently, live imaging has been added as a further tool to directly observe the process of stem cell division, signaling, and differentiation (Fichelson et al., 2009). Temporal and spatial aspects of gene function can be addressed by clonal analysis as well as tissue- or stage-specific gene expression or deletion analysis (del Valle Rodríguez et al., 2012). These tools are particularly critical for the analysis of genes that also have other essential functions at earlier stages of development or in the somatic tissues of the adult.
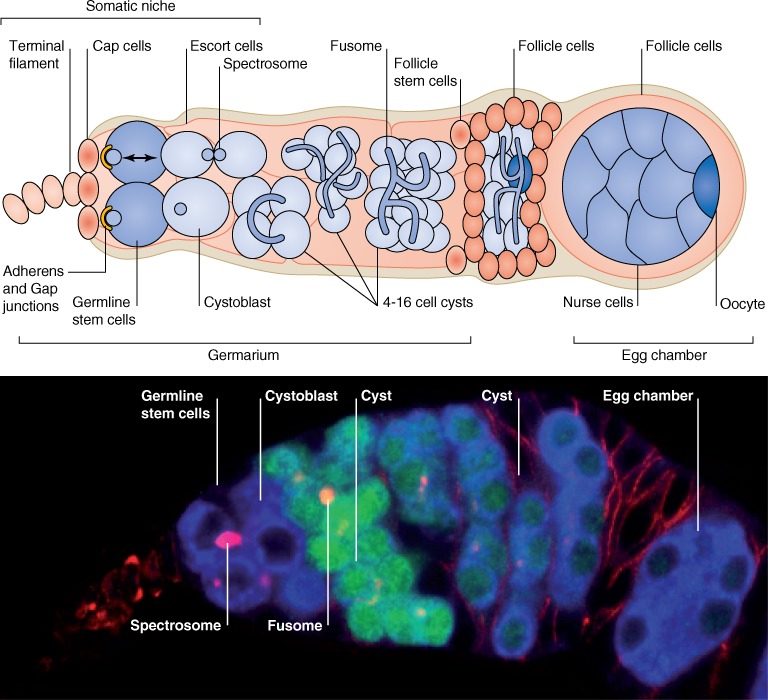
The adult female ovary consists of ∼20 ovarioles, each made of a chain of maturing egg chambers. Sustained egg production is ensured by the division of two to three GSCs at the tip of each ovariole in the germarium (Lin and Spradling, 1993). GSCs and their immediate progeny are surrounded by a somatic gonadal niche consisting of terminal filament, cap, and escort cells (Fig. 1). The cap cells of the niche are in immediate contact with the GSCs through adherens and gap junctions, whereas escort cells form long projections that tightly wrap around the GSCs and their progeny (Song et al., 2002; Tazuke et al., 2002; Kirilly et al., 2011). Generally, each GSC divides perpendicular to the cap cell–GSC interface, producing a new stem cell and a daughter cell that is further away from the niche, called the cystoblast (CB; Hsu et al., 2008). The CB initiates differentiation by undergoing four synchronous divisions with incomplete cytokinesis to form a 16-cell interconnected cyst (Fig. 1). One of the cells in this cyst is specified as an oocyte, and the others become polyploid nurse cells that provide the oocyte with all necessary RNAs and proteins.
Figure 1.
Adult Drosophila ovary. (top) Schematic drawing of a germarium and an egg chamber. Somatic tissues are shown in pink, and germline tissues are shown in blue. (bottom) Immunostaining of a germarium. Blue, anti-Vasa antibody (germ cells); green, anti-GFP antibody showing gfp expression under control of the bam promoter expressed in CB and cysts (note that endogenous Bam protein expression is spatially even more restricted than the GFP expression shown); red, anti-Hts (Hu li tai shao) antibody marks spectrosomes in GSCs and CBs, fusomes in multicellular cysts, and membranes in somatic follicle cells. Anterior is to the left.
Genes required for GSC maintenance, proliferation, and differentiation have been identified largely by unbiased genetic screens for oogenesis-defective mutants. Mutants defective in GSC maintenance or proliferation lose stem cells as the adult ages, whereas mutants defective in early stages of differentiation produce tumors of either single stem cell–like cells or interconnected germline cysts. We will review recent findings on cell-intrinsic regulatory networks that mediate the switch between GSC self-renewal and commitment toward differentiation. We focus on the interactions between specific GSC- and differentiation-promoting factors and discuss how feedback loops, which operate mainly via translational repression mechanisms, provide robustness to the fate decision.
GSC maintenance pathways
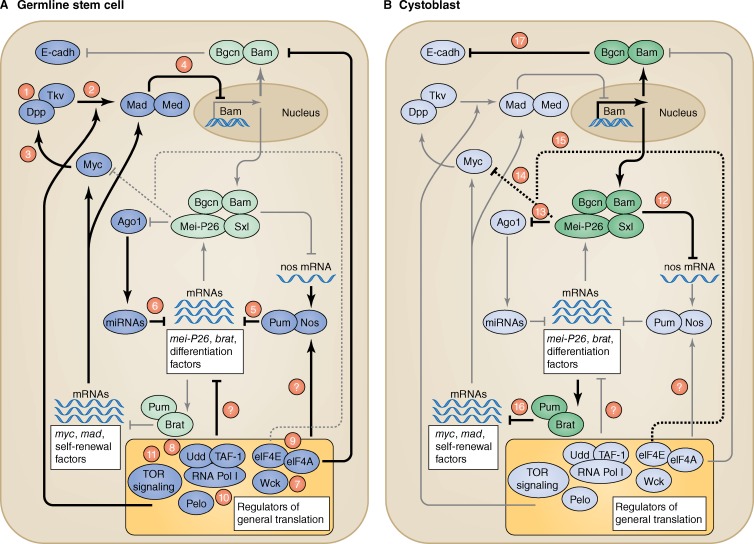
To build networks of interactions important for stem cell maintenance and self-renewal (Fig. 2), we focus first on two genetic pathways that are distinctly associated with GSC maintenance: a bone morphogenetic protein (BMP)–like receptor signaling pathway and the translational repressors Pum and Nos. Both pathways also have essential functions earlier in PGC development. Nos and Pum have early roles in the embryo to specify PGC fate. Both the BMP and Nos–Pum pathways are required during larval stages to prevent PGCs from premature differentiation (Forbes and Lehmann, 1998; Gilboa and Lehmann, 2004; Wang and Lin, 2004). Both pathways also play essential and specific roles in mammalian PGC specification and germline development (Lawson et al., 1999; Tremblay et al., 2001; Tsuda et al., 2003; Miller and Olivas, 2011).
Figure 2.
Gene regulatory networks controlling GSC self-renewal and differentiation. (A) Self-renewal network active in GSC. (B) Differentiation network active in differentiating cells, i.e., cystoblasts (CBs) and cysts. Self-renewal factors (blue) and differentiation factors (green) operating in each cell type are shown in the darker color, and the inactive factors are shown in light blue/green. Arrows illustrate positive and negative regulatory interactions. Numbers on the arrows are described throughout the text. E-cadh, E-cadherin; Pelo, Pelota; Tkv, Thickvein; Udd, Under-developed.
The BMP pathway mediates communication between the niche and GSCs. It is regulated at multiple levels, likely to assure precise calibration of signal reception and response in the germ cells (Fig. 2 A). The BMP ligands, Decapentaplegic (Dpp) and Glass Bottom Boat are secreted by the niche and received in germ cells by the type I and II receptors Thickvein-Saxophone and Punt, respectively (Fig. 2 A, 1 [numbers refer the position of the specific gene in the networks described in Fig. 2]). Mutations either in these ligands or corresponding receptors cause GSC loss (Xie and Spradling, 1998), whereas overexpression of the ligand or constitutive activation of Thickvein leads to the accumulation of single, GSC-like cells. BMP signaling from the niche triggers GSC-restricted phosphorylation of Mothers against Dpp (Mad; pMad) in GSCs (Fig. 2 A, 2; Chen and McKearin, 2003a,b; Song et al., 2004). pMad, with its binding partner Medea (Med), represses the expression of differentiation factors, including Bag of Marbles (Bam; discussed later; Fig. 2 A, 4). The oncoprotein Myc, the small GTPase Rac, and Jun kinase are involved in potentiating Dpp-dependent signaling in GSCs (Fig. 2 A, 3; Neumüller et al., 2008; Rhiner et al., 2009; Lu et al., 2012). The proteoglycan Dally and collagen IV bind to Dpp and restrict ligand diffusion (Wang et al., 2008; Guo and Wang, 2009; Hayashi et al., 2009). This spatial restriction of receptor activation (Fig. 3) specifically at the cap cell/GSC interface directly influences the size of the stem cell compartment.
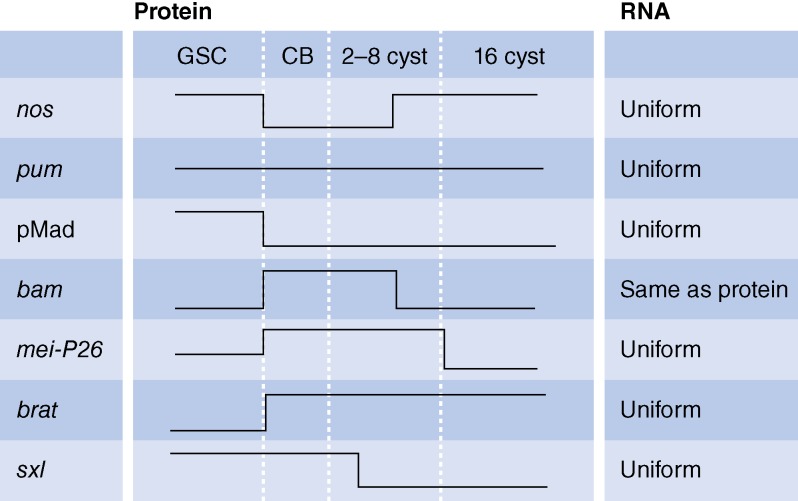
Figure 3.
Expression patterns in germ cells in germaria. (left) For each gene and for the phosphorylated Mad protein (pMad), changes in protein levels (high or low) are depicted during the indicated stages. (right) RNA expression patterns are summarized. Note that protein patterns are dynamic, whereas there is little change in RNA expression with the exception of bam.
In addition to BMP signaling from the niche, GSCs are also physically bound to the niche by high levels of the homotypic adhesion molecule E-cadherin and adherens junctions (Song et al., 2002). This physical attachment ensures reliable asymmetric GSC division by favoring the alignment of the mitotic spindle perpendicular to the niche (Fig. 1). This way, the self-renewing daughter remains attached to the niche, and the other daughter, further away from the niche, loses receptor activation and differentiates. Weakening of this alignment during regeneration and ageing can lead to GSC loss and symmetric divisions (Hsu and Drummond-Barbosa, 2009). For long-term maintenance, GSCs adopt a “modified” cell cycle characterized by a short G1 and a lengthened G2 phase. Mutations in Cyclin B or E, as well as overexpression of a stable form or Cyclin A, lead to GSC loss from the niche (Wang and Lin, 2005; Chen et al., 2009; Ables and Drummond-Barbosa, 2013). The unusual expression of Cyclin E during the G2 phase suggests additional, apparently cell cycle–independent, functions of cyclins in stem cell maintenance.
The conserved translational repressors Nos and Pum control germ cell development in animal species from C. elegans to humans (Tsuda et al., 2003; Miller and Olivas, 2011). In pum or nos mutants, GSCs are lost from the niche and differentiate precociously (Forbes and Lehmann, 1998; Gilboa and Lehmann, 2004). Nos and Pum act in GSCs to repress the translation of differentiation genes, such as the recently characterized targets Mei-P26 and Brain tumor (Brat; Fig. 2 A, 5; Neumüller et al., 2008; Harris et al., 2011; Joly et al., 2013). Nos and Pum inhibit translation of the differentiation-promoting genes by recruiting the CCR4–NOT deadenylase complex to target mRNAs (Joly et al., 2013). The CCR4–NOT complex removes poly(A) from mRNAs, causing a shortening of the poly(A) tail, reduction in the efficiency of translational initiation, and eventually mRNA degradation (Kadyrova et al., 2007).
By a similar mechanism, miRNAs inhibit differentiation genes in GSCs. Mutants in the miRNA-processing machinery, including Ago1 (Argonaute-1), Dicer-1, and Loquacious, lose GSCs as a result of precocious differentiation (Fig. 2 A, 6; Förstemann et al., 2005; Jin and Xie, 2007; Park et al., 2007; Yang et al., 2007). Thus, at least two pathways repress the translation of RNAs encoding differentiation factors. Both Pum and miRNAs bind RNAs in a sequence-specific manner, and translation repression is achieved by similar mechanisms, i.e., shortening of the poly(A) tail (Kadyrova et al., 2007; Piao et al., 2010; Joly et al., 2013). Synergy between Pum and miRNA binding sites on the target RNA may enhance repression and create the sharp transition in protein expression of several differentiation factors, such as Mei-P26 and Brat (discussed later; Figs. 3 and 2 A, 5 and 6).
In addition to these specific regulator pathways, several recent observations suggest that the general translation machinery may play an unexpectedly specific role in GSC maintenance. The expression of several proteins involved in ribosome biosynthesis and translation is increased in GSCs compared with differentiating cells, and manipulating these genes in the germline compartment leads to GSC loss. Specifically, Wicked, a component of the U3 small nucleolar RNP complex, which is required for ribosomal RNA (rRNA) maturation, and Under-developed, a member of the RNA polymerase I regulatory complex, which is required for rRNA synthesis, segregate asymmetrically with the GSC during division and are required for GSC proliferation and maintenance (Fig. 2 A, 7 and 8; Fichelson et al., 2009; Zhang et al., 2014). Consistently, GSCs have an enlarged nucleolus, the site of rRNA transcription (Cmarko et al., 2008; Neumüller et al., 2008). Besides the general translation apparatus, other rate-limiting translation factors, such as components of the translational initiation complex eIF4E and eIF4A (Fig. 2 A, 9) and Pelota, a homologue of eukaryotic translation release factor 1 α Dom34 (Fig. 2 A, 10), are required for GSC maintenance (Gingras et al., 1999; Xi et al., 2005; Shen et al., 2009; Sonenberg and Hinnebusch, 2009; Song and Lu, 2011; Guydosh and Green, 2014). It is unclear whether GSCs require higher general levels of translation or whether the ribosome biosynthesis and translation machinery more specifically targets factors involved in GSC maintenance. In support of a more specific function, modulation of rRNA synthesis specifically affects Mad but not Med or histone H2B levels, and eIF4A regulates E-cadherin but not BMP-signaling components in GSCs (Shen et al., 2009; Zhang et al., 2014). Furthermore, germline-specific translation initiation factors and specific isoforms of ribosomal proteins seem to be enriched in PGCs and GSCs when compared with more differentiated cells (Kai et al., 2005; Shigenobu et al., 2006). These germ cell–specific isoforms may impose a more specific and selective regulation on stem cell factors similar to the proposed transcript selectivity of ribosomal proteins for patterning genes during mammalian embryonic development (Kondrashov et al., 2011).
Adjusting translational activity in germ cells could be used as a fulcrum to maintain GSC–cyst homeostasis under changing nutritional conditions. A likely mediator is the Target of Rapamycin (TOR) signaling pathway, which coordinates growth with nutritional status through translational regulation (Laplante and Sabatini, 2012). TOR plays a conserved role in many stem cell systems, including Drosophila GSCs (Fig. 2 A, 11). Down-regulation of TOR signaling by mutating the central kinase TOR results in defects in GSC maintenance (LaFever et al., 2010). However, up-regulation of TOR signaling by mutating its upstream repressors Tsc1 and Tsc2 similarly results in GSC loss (LaFever et al., 2010; Sun et al., 2010). Thus, derivations from “normal” TOR activity levels may cause precocious differentiation of stem cells. Given the striking up-regulation of the protein biogenesis machinery in GSCs, regulation of protein synthesis is a likely target for TOR signaling. Supporting this, mutations in S6K, a regulator of protein synthesis and direct target of TOR, can rescue GSC maintenance defects caused by TOR hyperactivation in Tsc2 mutants (Sun et al., 2010). Another potent TOR target is Myc. Myc’s ability to promote proliferation, cell growth, and self-renewal has been linked to a direct role in ribosome biogenesis in several stem cell systems as well as cancer (van Riggelen et al., 2010). Myc protein is up-regulated in GSCs in which it promotes GSC maintenance by increasing BMP signaling and the competitiveness of GSCs for niche occupancy; GSCs with reduced Myc activity are lost from the niche (Fig. 2 A, 3; Rhiner et al., 2009). Whether Myc and TOR regulate the activity or expression of the specific GSC maintenance factors described in this review remains unclear. Alternatively, Myc and TOR may primarily regulate proliferation, growth, and quality control in GSCs. These later functions are needed in all stem cells and would be more related to the need of GSCs for sustained growth and less with the GSC differentiation switch.
Collectively, present data suggest that repression of differentiation-promoting genes in the stem cell compartment is the primary target of the known GSC self-renewal and maintenance factors, the BMP receptor pathway, and the translational repressors Nos and Pum (Fig. 2 A). Few, if any, factors have been identified that seem to promote stem cell fate itself. One possibility is that mutations in such factors may cause different phenotypes, such as stem cell death or cessation of stem cell development rather than differentiation, and may be more difficult to be recognized as specific for stem cell function.
GSC differentiation pathways
The first gene identified with a clear role in GSC differentiation was Bam (Fig. 2 B; McKearin and Spradling, 1990). bam mutant ovaries are filled with single, undifferentiated cells, whereas ubiquitous Bam overexpression leads to differentiation of all GSCs into mature eggs (Ohlstein and McKearin, 1997), indicating the important regulatory role of Bam in promoting differentiation. Accordingly, Bam expression is tightly regulated by pMad/Med. As a result, Bam expression is “off” in GSCs, in which Dpp-mediated signaling is high, and “on” in CBs, in which Dpp signaling is attenuated (Figs. 3 and 2 A, 4; Ohlstein and McKearin, 1997; Chen and McKearin, 2003a,b; Song et al., 2004).
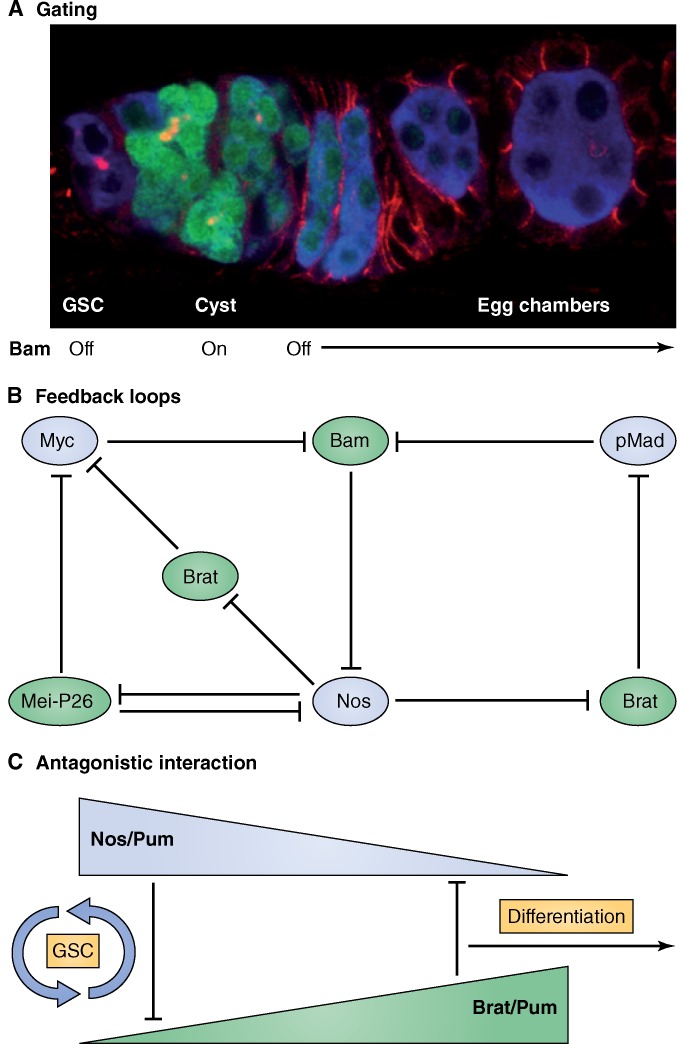
Among the known differentiation genes, Bam regulation and function seem to best exemplify a gating mechanism (Fig. 4 A). This term is used to describe regulatory relationships in which an initial repressor gene is only transiently expressed, nevertheless ensuring repression of target genes even after the initial trigger is no longer present (Davidson, 2010). According to this concept, a short pulse of Bam expression in CBs is sufficient to repress the stem cell program and initiate the differentiation program. The original concept of gating has been mostly used to describe transcription factor–mediated regulation of gene expression; however, most known targets of Bam regulation are translational (see below). Similar to transcriptional gating mechanisms, the precise transcriptional regulation of Bam may only be required to initiate but not instruct the differentiation program. This idea is supported by the finding that reduction in general translation can cause single-celled bam germ cell tumors to differentiate and form multicellular cysts. Thus, in this scenario, general translational repression can overcome the need for Bam to instruct differentiation (Xi et al., 2005; Shen et al., 2009; Sun et al., 2010; Joly et al., 2013; Zhang et al., 2014). Some experiments, however, also suggest that Bam has additional functions during later stages of cyst division (Hawkins et al., 1996; Lilly et al., 2000). Interestingly, in the original characterization of Bam protein, two intracellular localization patterns were identified by different antibodies, one form that was abundant in the cytoplasm and only present during the initiation of differentiation and another form, associated with the fusome, that persisted during later stages of cyst formation (McKearin and Ohlstein, 1995). It is intriguing to speculate that Bam’s roles in gating GSC differentiation and mediating cyst division could be regulated at the intracellular level through putative, location-specific interaction partners. Indeed, Cyclin A associates with the fusome and is involved in controlling cyst division cycles (Lilly et al., 2000).
Figure 4.
Examples of network modules. (A) Transient and spatially highly restricted expression of Bam provides a gate into differentiation. Markers for image shown are same as in Fig. 1. (B) Examples of interlocked double negative gates that result in positive feedback loops when one component in the network increases or decreases. Proteins promoting stem cell fate are shown in blue, and those promoting differentiation fate are shown in green. (C) Nos–Pum and Brat–Pum complexes inhibit each other by competing for Pum binding.
Bam lacks homology to other known proteins but acts in complex with at least three RNA-binding proteins, Benign gonial cell neoplasm (Bgcn), Sex lethal (Sxl), and Mei-P26 (Ohlstein et al., 2000; Li et al., 2013). Together, this complex mediates translation repression of GSC maintenance factors. Bgcn has homology with the DExH box family of ATP-dependent helicases, which are involved in repressing translation of RNAs involved in germline differentiation in other species (Gateff et al., 1996; Insco et al., 2012). Sxl, a sex-specific protein involved in RNA splicing and sequence-specific translational regulation, directs female sex determination in the early embryo and GSC differentiation in the adult ovary. Sxl enables GSC differentiation by down-regulating Nos protein levels during Drosophila oogenesis (Chau et al., 2012). Mei-P26 is a member of the TRIM (tripartite motif containing)-NHL (NCL-1, HT2A, and LIN-41) protein family. These proteins share a single RING (really interesting new gene) domain with E3 ubiquitin ligase activity and a coiled-coil NHL domain, which can bind RNA sequence specifically (Neumüller et al., 2008; Li et al., 2013; Loedige et al., 2014). Association of Bam with these three proteins confers repression on specific target genes, and the ability of Bam to force differentiation relies on each of these three proteins. Bam together with Bgcn represses E-cadherin translation, thereby relieving the tight interaction between GSCs and the niche (Fig. 2 B, 17), and Bam together with Sxl and Mei-P26 represses nos translation (Fig. 2 B, 12; Jin et al., 2008; Li et al., 2009, 2013). Although the relevance of the Bam complex for differentiation is clear, the exact composition of the complex is less well defined. For example, it remains unclear whether the three RNA-binding proteins form one complex with Bam or form separate complexes with a distinct target specificity. In line with a gating function for Bam through its restricted expression, the other components of the complex are expressed more broadly and not restricted to the CB and immediate progeny (Fig. 3). It would be interesting to test whether any other differentiation factors could attain gating function when expressed in a bam pattern.
Attractive candidates for such a test are the two TRIM-NHL proteins Mei-P26 and Brat. Together, these proteins repress most of the known factors required for GSC maintenance and proliferation, and their dual function in protein degradation and RNA translational repression allows regulation at multiple levels. Mei-P26 counteracts GSC self-renewal at different levels. First, as described above, Mei-P26 together with Sxl, Bam, and Bgcn represses nos RNA translation (Fig. 2 B, 12), thus interfering with Nos–Pum-mediated repression of differentiation genes. This provides translational cross-antagonism between Mei-P26 and Nos because Nos represses Mei-P26 translation (Fig. 4 B); thus, akin to a bistable switch, GSC or CB fates are chosen dependent on the balance of Nos and Mei-P26. Second, Mei-P26 binds to Ago1 and thereby lowers the production of miRNAs, which otherwise would block the translation of genes involved in GSC differentiation (Fig. 2, A [6] and B [13]; Neumüller et al., 2008). Third, in Mei-P26 mutants, the levels of Myc (Fig. 2 B, 14; Neumüller et al., 2008) and eIF4E are increased (Fig. 2 B, 15; Song and Lu, 2011). Removal of one copy of Myc or eIF4E partially rescues the Mei-P26 mutant tumor phenotype (Song and Lu, 2011). Thus, down-regulation of general translational efficiency and Myc repression by Mei-P26 provide yet another path into differentiation (Fig. 4 B).
In addition to Mei-P26, another TRIM-NHL protein, Brat, also regulates the GSC-to-CB transition, possibly in parallel to Bam. A specific role for Brat in translational regulation was first shown in the Drosophila embryo, in which Brat represses translation in complex with Nos and Pum (Sonoda and Wharton, 2001). The relationship between Brat, Nos, and Pum apparently differs in ovaries, in which Brat and Nos are in competition for Pum binding. Brat overexpression in germ cells induces precocious GSC differentiation and loss from the niche, whereas overexpression of a Brat mutant that specifically disrupts its binding to Pum has no effect (Harris et al., 2011). Thus, in ovaries, Brat promotes differentiation by forming a complex with Pum, thereby sequestering Pum away from Nos (Fig. 4 C). This antagonistic relationship between a Nos–Pum complex, which represses differentiation genes in GSCs, and the Brat–Pum complex in CBs, which prevents Pum from binding Nos, provides an additional mechanism to repress Nos activity in the CB (Fig. 4 C). The Brat–Pum complex inhibits translation of Mad and Myc (Fig. 3 B, 16), both of which positively regulate BMP signaling. Reduced BMP signaling leads to up-regulation of the transcription of Bam, which together with the other complex components represses Nos translation. This results in a feedback loop toward differentiation (Fig. 4 B; Harris et al., 2011). Contrary to Bam, Bgcn, Sxl, or Mei-P26 mutants, Brat mutant ovaries do not form extensive germ cell tumors; instead, GSC numbers are increased, but overall differentiation is unaffected (Harris et al., 2011). This may suggest overlapping functions with other proteins in the differentiation network or hint at a more supportive role for Brat in promoting the transition toward differentiation. Together, Brat and Mei-P26 seem to coordinate many of the known aspects needed for GSC differentiation: cessation of BMP signaling to promote Bam expression by Brat, translational repression of the GSC factor Nos by the Bam–Mei-P26 complex, regulation of proliferation, BMP signaling and general translation through repression of Myc by Mei-P26 and Brat, and regulation of the miRNA machinery by Mei-P26 (Fig. 4 B). These findings are particularly intriguing given the fact that the function of the TRIM-NHL protein family in stem cell differentiation is conserved. Brat and its human homologue TRIM3 are required for neural stem cell differentiation, and the mouse homologue TRIM32 represses self-renewal of neuronal precursors by a mechanism similar to that of Mei-P26 in GSCs (Bello et al., 2006; Betschinger et al., 2006; Lee et al., 2006; Schwamborn et al., 2009; Chen et al., 2014).
Conclusions
In this review, we have focused on how specific interactions between self-renewal and differentiation factors promote a switch from a GSC program to an oocyte differentiation program. This decision has to occur reliably during each GSC division. The existence of multiple, interlocked feedback loops as outlined in part in Fig. 4 B begins to explain how this switch can be initiated rapidly and then stably maintained. Many aspects of the molecular interplay between the individual components of the network are far from understood. However, it is evident that mutual translational repression plays a major role in promoting and stably maintaining the decision between self-renewal and differentiation. A preponderance of RNA-mediated regulatory mechanisms rather than transcription-centric cross-regulation is a conserved hallmark of germline development. However, in addition to translation, regulation of protein stability and chromatin modifications has also been shown to influence the GSC–CB decision (Casanueva and Ferguson, 2004; Xi and Xie, 2005; Jiang et al., 2008; Xia et al., 2010, 2012; Wang et al., 2011; Barton et al., 2013; Pan et al., 2014). As differentiating cysts can be reprogrammed in vivo either upon GSC loss or during ageing to regain GSC potential (Kai and Spradling, 2004), these mechanisms may reinforce a commitment to differentiation and guarantee a long-lasting, reliable production of progeny. We expect that there are additional factors that regulate general stem cell function, such as stress response, metabolism, and proliferation. Mutations in such regulators might have more complex phenotypes than simply precocious differentiation or tumor formation, making their identification as stem cell or differentiation factors more challenging. Finally, although some players in this network, such as Bam, are fly specific, most are highly conserved, and many are expressed in other stem cell systems. Thus, similar regulatory logics as described here might also influence the stem cell maintenance versus differentiation decision in mammals.
Acknowledgments
We apologize to colleagues whose work could not be cited as a result of space constrains. We thank Carlos Sanchez for providing the images used in Figs. 1 and 4. We are thankful to Dr. Lionel Christiaen for fruitful discussions and comments and appreciated critical reading of the manuscript by Drs. Allison Blum, Thomas Hurd, Felipe Teixeira, and Colin Malone.
We acknowledge grant support from the National Institutes of Health grant R37HD41900. M. Slaidina is a Howard Hughes Medical Institute Fellow of the Life Sciences Research Foundation, and R. Lehmann is a Howard Hughes Medical Institute investigator. Illustrations were provided by Neil Smith, www.neilsmithillustration.co.uk.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- Bam
- Bag of Marbles
- Bgcm
- Benign gonial cell neoplasm
- BMP
- bone morphogenetic protein
- Brat
- Brain tumor
- CB
- cystoblast
- Dpp
- Decapentaplegic
- GSC
- germline stem cell
- Mad
- Mothers against Dpp
- Med
- Medea
- Nos
- Nanos
- PGC
- primordial germ cell
- Pum
- Pumilio
- rRNA
- ribosomal RNA
- Sxl
- Sex lethal
- TOR
- Target of Rapamycin
References
- Ables, E.T., and Drummond-Barbosa D.. 2013. Cyclin E controls Drosophila female germline stem cell maintenance independently of its role in proliferation by modulating responsiveness to niche signals. Development. 140:530–540 10.1242/dev.088583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton, L.J., Pinto B.S., Wallrath L.L., and Geyer P.K.. 2013. The Drosophila nuclear lamina protein otefin is required for germline stem cell survival. Dev. Cell. 25:645–654 10.1016/j.devcel.2013.05.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bello, B., Reichert H., and Hirth F.. 2006. The brain tumor gene negatively regulates neural progenitor cell proliferation in the larval central brain of Drosophila. Development. 133:2639–2648 10.1242/dev.02429 [DOI] [PubMed] [Google Scholar]
- Betschinger, J., Mechtler K., and Knoblich J.A.. 2006. Asymmetric segregation of the tumor suppressor brat regulates self-renewal in Drosophila neural stem cells. Cell. 124:1241–1253 10.1016/j.cell.2006.01.038 [DOI] [PubMed] [Google Scholar]
- Casanueva, M.O., and Ferguson E.L.. 2004. Germline stem cell number in the Drosophila ovary is regulated by redundant mechanisms that control Dpp signaling. Development. 131:1881–1890 10.1242/dev.01076 [DOI] [PubMed] [Google Scholar]
- Chatfield, J., O’Reilly M.-A., Bachvarova R.F., Ferjentsik Z., Redwood C., Walmsley M., Patient R., Loose M., and Johnson A.D.. 2014. Stochastic specification of primordial germ cells from mesoderm precursors in axolotl embryos. Development. 141:2429–2440 10.1242/dev.105346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chau, J., Kulnane L.S., and Salz H.K.. 2012. Sex-lethal enables germline stem cell differentiation by down-regulating Nanos protein levels during Drosophila oogenesis. Proc. Natl. Acad. Sci. USA. 109:9465–9470 10.1073/pnas.1120473109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, D., and McKearin D.. 2003a. Dpp signaling silences bam transcription directly to establish asymmetric divisions of germline stem cells. Curr. Biol. 13:1786–1791 10.1016/j.cub.2003.09.033 [DOI] [PubMed] [Google Scholar]
- Chen, D., and McKearin D.M.. 2003b. A discrete transcriptional silencer in the bam gene determines asymmetric division of the Drosophila germline stem cell. Development. 130:1159–1170 10.1242/dev.00325 [DOI] [PubMed] [Google Scholar]
- Chen, D., Wang Q., Huang H., Xia L., Jiang X., Kan L., Sun Q., and Chen D.. 2009. Effete-mediated degradation of Cyclin A is essential for the maintenance of germline stem cells in Drosophila. Development. 136:4133–4142 10.1242/dev.039032 [DOI] [PubMed] [Google Scholar]
- Chen, G., Kong J., Tucker-Burden C., Anand M., Rong Y., Rahman F., Moreno C.S., Van Meir E.G., Hadjipanayis C.G., and Brat D.J.. 2014. Human Brat ortholog TRIM3 is a tumor suppressor that regulates asymmetric cell division in glioblastoma. Cancer Res. 74:4536–4548 10.1158/0008-5472.CAN-13-3703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cmarko, D., Smigova J., Minichova L., and Popov A.. 2008. Nucleolus: the ribosome factory. Histol. Histopathol. 23:1291–1298 [DOI] [PubMed] [Google Scholar]
- Davidson, E.H.2010. Emerging properties of animal gene regulatory networks. Nature. 468:911–920 10.1038/nature09645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Valle Rodríguez, A., Didiano D., and Desplan C.. 2012. Power tools for gene expression and clonal analysis in Drosophila. Nat. Methods. 9:47–55 10.1038/nmeth.1800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donoughe, S., Nakamura T., Ewen-Campen B., Green D.A. II, Henderson L., and Extavour C.G.. 2014. BMP signaling is required for the generation of primordial germ cells in an insect. Proc. Natl. Acad. Sci. USA. 111:4133–4138 10.1073/pnas.1400525111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Extavour, C.G., and Akam M.. 2003. Mechanisms of germ cell specification across the metazoans: epigenesis and preformation. Development. 130:5869–5884 10.1242/dev.00804 [DOI] [PubMed] [Google Scholar]
- Fichelson, P., Moch C., Ivanovitch K., Martin C., Sidor C.M., Lepesant J.-A., Bellaïche Y., and Huynh J.-R.. 2009. Live-imaging of single stem cells within their niche reveals that a U3snoRNP component segregates asymmetrically and is required for self-renewal in Drosophila. Nat. Cell Biol. 11:685–693 10.1038/ncb1874 [DOI] [PubMed] [Google Scholar]
- Forbes, A., and Lehmann R.. 1998. Nanos and Pumilio have critical roles in the development and function of Drosophila germline stem cells. Development. 125:679–690 [DOI] [PubMed] [Google Scholar]
- Förstemann, K., Tomari Y., Du T., Vagin V.V., Denli A.M., Bratu D.P., Klattenhoff C., Theurkauf W.E., and Zamore P.D.. 2005. Normal microRNA maturation and germ-line stem cell maintenance requires Loquacious, a double-stranded RNA-binding domain protein. PLoS Biol. 3:e236 10.1371/journal.pbio.0030236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, M., and Arkov A.L.. 2013. Next generation organelles: structure and role of germ granules in the germline. Mol. Reprod. Dev. 80:610–623 10.1002/mrd.22115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gateff, E., Kurzik-Dumke U., Wismar J., Löffler T., Habtemichael N., Konrad L., Dreschers S., Kaiser S., and Protin U.. 1996. Drosophila differentiation genes instrumental in tumor suppression. Int. J. Dev. Biol. 40:149–156 [PubMed] [Google Scholar]
- Gilboa, L., and Lehmann R.. 2004. Repression of primordial germ cell differentiation parallels germ line stem cell maintenance. Curr. Biol. 14:981–986 10.1016/j.cub.2004.05.049 [DOI] [PubMed] [Google Scholar]
- Gingras, A.C., Raught B., and Sonenberg N.. 1999. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 68:913–963 10.1146/annurev.biochem.68.1.913 [DOI] [PubMed] [Google Scholar]
- Guo, Z., and Wang Z.. 2009. The glypican Dally is required in the niche for the maintenance of germline stem cells and short-range BMP signaling in the Drosophila ovary. Development. 136:3627–3635 10.1242/dev.036939 [DOI] [PubMed] [Google Scholar]
- Guydosh, N.R., and Green R.. 2014. Dom34 rescues ribosomes in 3′ untranslated regions. Cell. 156:950–962 10.1016/j.cell.2014.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, R.E., Pargett M., Sutcliffe C., Umulis D., and Ashe H.L.. 2011. Brat promotes stem cell differentiation via control of a bistable switch that restricts BMP signaling. Dev. Cell. 20:72–83 10.1016/j.devcel.2010.11.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins, N.C., Thorpe J., and Schüpbach T.. 1996. Encore, a gene required for the regulation of germ line mitosis and oocyte differentiation during Drosophila oogenesis. Development. 122:281–290 [DOI] [PubMed] [Google Scholar]
- Hayashi, Y., Kobayashi S., and Nakato H.. 2009. Drosophila glypicans regulate the germline stem cell niche. J. Cell Biol. 187:473–480 10.1083/jcb.200904118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu, H.J., and Drummond-Barbosa D.. 2009. Insulin levels control female germline stem cell maintenance via the niche in Drosophila. Proc. Natl. Acad. Sci. USA. 106:1117–1121 10.1073/pnas.0809144106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu, H.-J., LaFever L., and Drummond-Barbosa D.. 2008. Diet controls normal and tumorous germline stem cells via insulin-dependent and -independent mechanisms in Drosophila. Dev. Biol. 313:700–712 10.1016/j.ydbio.2007.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insco, M.L., Bailey A.S., Kim J., Olivares G.H., Wapinski O.L., Tam C.H., and Fuller M.T.. 2012. A self-limiting switch based on translational control regulates the transition from proliferation to differentiation in an adult stem cell lineage. Cell Stem Cell. 11:689–700 10.1016/j.stem.2012.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, X., Xia L., Chen D., Yang Y., Huang H., Yang L., Zhao Q., Shen L., Wang J., and Chen D.. 2008. Otefin, a nuclear membrane protein, determines the fate of germline stem cells in Drosophila via interaction with Smad complexes. Dev. Cell. 14:494–506 10.1016/j.devcel.2008.02.018 [DOI] [PubMed] [Google Scholar]
- Jin, Z., and Xie T.. 2007. Dcr-1 maintains Drosophila ovarian stem cells. Curr. Biol. 17:539–544 10.1016/j.cub.2007.01.050 [DOI] [PubMed] [Google Scholar]
- Jin, Z., Kirilly D., Weng C., Kawase E., Song X., Smith S., Schwartz J., and Xie T.. 2008. Differentiation-defective stem cells outcompete normal stem cells for niche occupancy in the Drosophila ovary. Cell Stem Cell. 2:39–49 10.1016/j.stem.2007.10.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joly, W., Chartier A., Rojas-Rios P., Busseau I., and Simonelig M.. 2013. The CCR4 deadenylase acts with Nanos and Pumilio in the fine-tuning of Mei-P26 expression to promote germline stem cell self-renewal. Stem Cell Rev. 1:411–424 10.1016/j.stemcr.2013.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadyrova, L.Y., Habara Y., Lee T.H., and Wharton R.P.. 2007. Translational control of maternal Cyclin B mRNA by Nanos in the Drosophila germline. Development. 134:1519–1527 10.1242/dev.002212 [DOI] [PubMed] [Google Scholar]
- Kai, T., and Spradling A.. 2004. Differentiating germ cells can revert into functional stem cells in Drosophila melanogaster ovaries. Nature. 428:564–569 10.1038/nature02436 [DOI] [PubMed] [Google Scholar]
- Kai, T., Williams D., and Spradling A.C.. 2005. The expression profile of purified Drosophila germline stem cells. Dev. Biol. 283:486–502 10.1016/j.ydbio.2005.04.018 [DOI] [PubMed] [Google Scholar]
- Kanatsu-Shinohara, M., Inoue K., Lee J., Yoshimoto M., Ogonuki N., Miki H., Baba S., Kato T., Kazuki Y., Toyokuni S., et al. 2004. Generation of pluripotent stem cells from neonatal mouse testis. Cell. 119:1001–1012 10.1016/j.cell.2004.11.011 [DOI] [PubMed] [Google Scholar]
- Kirilly, D., Wang S., and Xie T.. 2011. Self-maintained escort cells form a germline stem cell differentiation niche. Development. 138:5087–5097 10.1242/dev.067850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov, N., Pusic A., Stumpf C.R., Shimizu K., Hsieh A.C., Xue S., Ishijima J., Shiroishi T., and Barna M.. 2011. Ribosome-mediated specificity in Hox mRNA translation and vertebrate tissue patterning. Cell. 145:383–397 10.1016/j.cell.2011.03.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaFever, L., Feoktistov A., Hsu H.J., and Drummond-Barbosa D.. 2010. Specific roles of Target of rapamycin in the control of stem cells and their progeny in the Drosophila ovary. Development. 137:2117–2126 10.1242/dev.050351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplante, M., and Sabatini D.M.. 2012. mTOR signaling in growth control and disease. Cell. 149:274–293 10.1016/j.cell.2012.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson, K.A., Dunn N.R., Roelen B.A., Zeinstra L.M., Davis A.M., Wright C.V., Korving J.P., and Hogan B.L.. 1999. Bmp4 is required for the generation of primordial germ cells in the mouse embryo. Genes Dev. 13:424–436 10.1101/gad.13.4.424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, C.-Y., Wilkinson B.D., Siegrist S.E., Wharton R.P., and Doe C.Q.. 2006. Brat is a Miranda cargo protein that promotes neuronal differentiation and inhibits neuroblast self-renewal. Dev. Cell. 10:441–449 10.1016/j.devcel.2006.01.017 [DOI] [PubMed] [Google Scholar]
- Li, Y., Minor N.T., Park J.K., McKearin D.M., and Maines J.Z.. 2009. Bam and Bgcn antagonize Nanos-dependent germ-line stem cell maintenance. Proc. Natl. Acad. Sci. USA. 106:9304–9309 10.1073/pnas.0901452106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y., Zhang Q., Carreira-Rosario A., Maines J.Z., McKearin D.M., and Buszczak M.. 2013. Mei-p26 cooperates with Bam, Bgcn and Sxl to promote early germline development in the Drosophila ovary. PLoS ONE. 8:e58301 10.1371/journal.pone.0058301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilly, M.A., de Cuevas M., and Spradling A.C.. 2000. Cyclin A associates with the fusome during germline cyst formation in the Drosophila ovary. Dev. Biol. 218:53–63 10.1006/dbio.1999.9570 [DOI] [PubMed] [Google Scholar]
- Lin, H., and Spradling A.C.. 1993. Germline stem cell division and egg chamber development in transplanted Drosophila germaria. Dev. Biol. 159:140–152 10.1006/dbio.1993.1228 [DOI] [PubMed] [Google Scholar]
- Loedige, I., Stotz M., Qamar S., Kramer K., Hennig J., Schubert T., Löffler P., Längst G., Merkl R., Urlaub H., and Meister G.. 2014. The NHL domain of BRAT is an RNA-binding domain that directly contacts the hunchback mRNA for regulation. Genes Dev. 28:749–764 10.1101/gad.236513.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, W., Casanueva M.O., Mahowald A.P., Kato M., Lauterbach D., and Ferguson E.L.. 2012. Niche-associated activation of rac promotes the asymmetric division of Drosophila female germline stem cells. PLoS Biol. 10:e1001357 10.1371/journal.pbio.1001357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luteijn, M.J., and Ketting R.F.. 2013. PIWI-interacting RNAs: from generation to transgenerational epigenetics. Nat. Rev. Genet. 14:523–534 10.1038/nrg3495 [DOI] [PubMed] [Google Scholar]
- McKearin, D., and Ohlstein B.. 1995. A role for the Drosophila bag-of-marbles protein in the differentiation of cystoblasts from germline stem cells. Development. 121:2937–2947 [DOI] [PubMed] [Google Scholar]
- McKearin, D.M., and Spradling A.C.. 1990. bag-of-marbles: a Drosophila gene required to initiate both male and female gametogenesis. Genes Dev. 4(12B):2242–2251 10.1101/gad.4.12b.2242 [DOI] [PubMed] [Google Scholar]
- Miller, M.A., and Olivas W.M.. 2011. Roles of Puf proteins in mRNA degradation and translation. Wiley Interdiscip. Rev. RNA. 2:471–492 10.1002/wrna.69 [DOI] [PubMed] [Google Scholar]
- Neumüller, R.A., Betschinger J., Fischer A., Bushati N., Poernbacher I., Mechtler K., Cohen S.M., and Knoblich J.A.. 2008. Mei-P26 regulates microRNAs and cell growth in the Drosophila ovarian stem cell lineage. Nature. 454:241–245 10.1038/nature07014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niki, Y., and Mahowald A.P.. 2003. Ovarian cystocytes can repopulate the embryonic germ line and produce functional gametes. Proc. Natl. Acad. Sci. USA. 100:14042–14045 10.1073/pnas.2235591100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlstein, B., and McKearin D.. 1997. Ectopic expression of the Drosophila Bam protein eliminates oogenic germline stem cells. Development. 124:3651–3662 [DOI] [PubMed] [Google Scholar]
- Ohlstein, B., Lavoie C.A., Vef O., Gateff E., and McKearin D.M.. 2000. The Drosophila cystoblast differentiation factor, benign gonial cell neoplasm, is related to DExH-box proteins and interacts genetically with bag-of-marbles. Genetics. 155:1809–1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, L., Wang S., Lu T., Weng C., Song X., Park J.K., Sun J., Yang Z.-H., Yu J., Tang H., et al. 2014. Protein competition switches the function of COP9 from self-renewal to differentiation. Nature. 10.1038/nature13562 [DOI] [PubMed] [Google Scholar]
- Park, J.K., Liu X., Strauss T.J., McKearin D.M., and Liu Q.. 2007. The miRNA pathway intrinsically controls self-renewal of Drosophila germline stem cells. Curr. Biol. 17:533–538 10.1016/j.cub.2007.01.060 [DOI] [PubMed] [Google Scholar]
- Piao, X., Zhang X., Wu L., and Belasco J.G.. 2010. CCR4-NOT deadenylates mRNA associated with RNA-induced silencing complexes in human cells. Mol. Cell. Biol. 30:1486–1494 10.1128/MCB.01481-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhiner, C., Díaz B., Portela M., Poyatos J.F., Fernández-Ruiz I., López-Gay J.M., Gerlitz O., and Moreno E.. 2009. Persistent competition among stem cells and their daughters in the Drosophila ovary germline niche. Development. 136:995–1006 10.1242/dev.033340 [DOI] [PubMed] [Google Scholar]
- Schwamborn, J.C., Berezikov E., and Knoblich J.A.. 2009. The TRIM-NHL protein TRIM32 activates microRNAs and prevents self-renewal in mouse neural progenitors. Cell. 136:913–925 10.1016/j.cell.2008.12.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seydoux, G., and Braun R.E.. 2006. Pathway to totipotency: lessons from germ cells. Cell. 127:891–904 10.1016/j.cell.2006.11.016 [DOI] [PubMed] [Google Scholar]
- Shen, R., Weng C., Yu J., and Xie T.. 2009. eIF4A controls germline stem cell self-renewal by directly inhibiting BAM function in the Drosophila ovary. Proc. Natl. Acad. Sci. USA. 106:11623–11628 10.1073/pnas.0903325106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigenobu, S., Kitadate Y., Noda C., and Kobayashi S.. 2006. Molecular characterization of embryonic gonads by gene expression profiling in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA. 103:13728–13733 10.1073/pnas.0603767103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg, N., and Hinnebusch A.G.. 2009. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 136:731–745 10.1016/j.cell.2009.01.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, X., Zhu C.-H., Doan C., and Xie T.. 2002. Germline stem cells anchored by adherens junctions in the Drosophila ovary niches. Science. 296:1855–1857 10.1126/science.1069871 [DOI] [PubMed] [Google Scholar]
- Song, X., Wong M.D., Kawase E., Xi R., Ding B.C., McCarthy J.J., and Xie T.. 2004. Bmp signals from niche cells directly repress transcription of a differentiation-promoting gene, bag of marbles, in germline stem cells in the Drosophila ovary. Development. 131:1353–1364 10.1242/dev.01026 [DOI] [PubMed] [Google Scholar]
- Song, Y., and Lu B.. 2011. Regulation of cell growth by Notch signaling and its differential requirement in normal vs. tumor-forming stem cells in Drosophila. Genes Dev. 25:2644–2658 10.1101/gad.171959.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda, J., and Wharton R.P.. 2001. Drosophila Brain Tumor is a translational repressor. Genes Dev. 15:762–773 10.1101/gad.870801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling, A., Fuller M.T., Braun R.E., and Yoshida S.. 2011. Germline stem cells. Cold Spring Harb. Perspect. Biol. 3:a002642 10.1101/cshperspect.a002642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, P., Quan Z., Zhang B., Wu T., and Xi R.. 2010. TSC1/2 tumour suppressor complex maintains Drosophila germline stem cells by preventing differentiation. Development. 137:2461–2469 10.1242/dev.051466 [DOI] [PubMed] [Google Scholar]
- Tazuke, S.I., Schulz C., Gilboa L., Fogarty M., Mahowald A.P., Guichet A., Ephrussi A., Wood C.G., Lehmann R., and Fuller M.T.. 2002. A germline-specific gap junction protein required for survival of differentiating early germ cells. Development. 129:2529–2539 [DOI] [PubMed] [Google Scholar]
- Tremblay, K.D., Dunn N.R., and Robertson E.J.. 2001. Mouse embryos lacking Smad1 signals display defects in extra-embryonic tissues and germ cell formation. Development. 128:3609–3621 [DOI] [PubMed] [Google Scholar]
- Tsuda, M., Sasaoka Y., Kiso M., Abe K., Haraguchi S., Kobayashi S., and Saga Y.. 2003. Conserved role of nanos proteins in germ cell development. Science. 301:1239–1241 10.1126/science.1085222 [DOI] [PubMed] [Google Scholar]
- van Riggelen, J., Yetil A., and Felsher D.W.. 2010. MYC as a regulator of ribosome biogenesis and protein synthesis. Nat. Rev. Cancer. 10:301–309 10.1038/nrc2819 [DOI] [PubMed] [Google Scholar]
- Wang, X., Harris R.E., Bayston L.J., and Ashe H.L.. 2008. Type IV collagens regulate BMP signalling in Drosophila. Nature. 455:72–77 10.1038/nature07214 [DOI] [PubMed] [Google Scholar]
- Wang, X., Pan L., Wang S., Zhou J., McDowell W., Park J., Haug J., Staehling K., Tang H., and Xie T.. 2011. Histone H3K9 trimethylase Eggless controls germline stem cell maintenance and differentiation. PLoS Genet. 7:e1002426 10.1371/journal.pgen.1002426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z., and Lin H.. 2004. Nanos maintains germline stem cell self-renewal by preventing differentiation. Science. 303:2016–2019 10.1126/science.1093983 [DOI] [PubMed] [Google Scholar]
- Wang, Z., and Lin H.. 2005. The division of Drosophila germline stem cells and their precursors requires a specific cyclin. Curr. Biol. 15:328–333 10.1016/j.cub.2005.02.016 [DOI] [PubMed] [Google Scholar]
- Xi, R., and Xie T.. 2005. Stem cell self-renewal controlled by chromatin remodeling factors. Science. 310:1487–1489 10.1126/science.1120140 [DOI] [PubMed] [Google Scholar]
- Xi, R., Doan C., Liu D., and Xie T.. 2005. Pelota controls self-renewal of germline stem cells by repressing a Bam-independent differentiation pathway. Development. 132:5365–5374 10.1242/dev.02151 [DOI] [PubMed] [Google Scholar]
- Xia, L., Jia S., Huang S., Wang H., Zhu Y., Mu Y., Kan L., Zheng W., Wu D., Li X., et al. 2010. The Fused/Smurf complex controls the fate of Drosophila germline stem cells by generating a gradient BMP response. Cell. 143:978–990 10.1016/j.cell.2010.11.022 [DOI] [PubMed] [Google Scholar]
- Xia, L., Zheng X., Zheng W., Zhang G., Wang H., Tao Y., and Chen D.. 2012. The niche-dependent feedback loop generates a BMP activity gradient to determine the germline stem cell fate. Curr. Biol. 22:515–521 10.1016/j.cub.2012.01.056 [DOI] [PubMed] [Google Scholar]
- Xie, T., and Spradling A.C.. 1998. decapentaplegic is essential for the maintenance and division of germline stem cells in the Drosophila ovary. Cell. 94:251–260 10.1016/S0092-8674(00)81424-5 [DOI] [PubMed] [Google Scholar]
- Yang, L., Chen D., Duan R., Xia L., Wang J., Qurashi A., Jin P., and Chen D.. 2007. Argonaute 1 regulates the fate of germline stem cells in Drosophila. Development. 134:4265–4272 10.1242/dev.009159 [DOI] [PubMed] [Google Scholar]
- Zhang, Q., Shalaby N.A., and Buszczak M.. 2014. Changes in rRNA transcription influence proliferation and cell fate within a stem cell lineage. Science. 343:298–301 10.1126/science.1246384 [DOI] [PMC free article] [PubMed] [Google Scholar]