Abstract
Background
By encoding a group of small secretory peptides, the members of the CLAVATA3/EMBRYO-SURROUNDING REGION (CLE) family play important roles in cell-to-cell communication to control the balance between stem cell proliferation and differentiation in plant development. Despite recent identification and characterization of members of this gene family in several plant species, little is known about its functional role in plants with fleshy fruits.
Results
In total, fifteen CLE genes (SlCLE1-15) were identified from tomato (Solanum lycopersicum cv. ‘Heinz-1706’) genome and their multiple characters including phylogeny, gene structures, chromosome locations, conserved motifs and cis-elements in the promoter sequences, were analyzed. Real-time PCR analysis showed that 13 out of 15 identified SlCLE genes are transcribed and exhibit remarkably unique expression patterns among tissues and organs. In particular, SlCLE12, the homologue of Arabidopsis CLE41/44 gene, appears to be the dominant CLE gene in most of tested tissues with its maximum expression found in vascular tissues. Meanwhile, SlCLE1, 10, 13 exhibit specific but distinct expression in flower bud, root and shoot apex, respectively. More notably, several SlCLEs are dramatically regulated in their transcriptional levels during fruit development and ripening, indicating significant role these genes may potentially play in the critical physiological process. Upon the treatment with synthetic peptides corresponding to the 12-aa CLE domains of SlCLE 10, 12 and 13, tomato seedlings exhibit a clear reduction in root length to varying degrees.
Conclusions
This study provides a comprehensive genomic analysis of CLE gene family in tomato, a crop species with fleshy fruit. Differential expression patterns of various SlCLEs provide important insights into the functional divergence of CLE signaling cascade in Solanaceae species, especially their potential involvements in the regulation of fruit development and ripening.
Electronic supplementary material
The online version of this article (doi:10.1186/1471-2164-15-827) contains supplementary material, which is available to authorized users.
Keywords: Solanum lycopersicum, CLE gene family, Phylogenetic analysis, Tissue expression pattern, Peptide application
Background
For multicellular organisms, intercellular communication is essential for coordinating their well orchestrated growth and development. In higher plants, besides the conventional phytohormones such as auxin and cytokinin, a group of small secretory peptides, namely the CLAVATA3/EMBRYO-SURROUNDING REGION-RELATED (CLV3/ESR, CLE) peptides, has been recognized as important players that are responsible for mediating cell-to-cell signaling.
CLE proteins usually include a signal peptide at the N-terminus followed by a variable domain and a conserved CLE domain composed of 12–14 amino acids at the C-terminus [1–3]. When exogenously applied, synthetic peptides corresponding to the CLE domains could reduce the size of the shoot apex meristem (SAM) or the root length, or inhibit tracheary element differentiation [1, 2, 4]. With 32 CLE members in Arabidopsis thaliana genome, synthetic peptides encoded by at least 19 of them have been shown to be functional [1, 2, 4–6]. Recent work indicated that CLE peptides are involved in many aspects of plant growth and development, including plant-pathogen interaction, cell division, anther-stigma interaction and stem cell maintenance [7]. Representing a founding member of the CLE family, CLAVATA3 (CLV3) has been identified as a key regulator of stem cell homeostasis at the SAM through suppressing the homeodomain transcription factor, WUSCHEL (WUS) [8–11]. In roots, CLE40 peptide functions to maintain the quiescent center (QC) and columella stem cell identity by repressing the WUS-related gene, WOX5 [12]. In vascular tissues, TRACHEARY ELEMENT DIFFERENTIATION INHIBITORY FACTOR (TDIF) encoded by CLE41 and CLE44 promotes the accumulation of undifferentiated procambial cells and inhibits their differentiation into xylem [2]. CLE peptides also take additional biological roles beyond stem cell regulation. For example, CLE8 has recently been identified as a regulator of seed development [13]. Moreover, CLEs have been identified as signaling ligands in plant responses to environmental cues. Two CLE peptides, Lotus japonicus CLE ROOT SIGNAL1 and 2 (LjCLE-RS1 and LjCLE-RS2) have been identified as root-derived infection signals that are transmitted from roots to shoots and systemically repress excess nodulation [14]. Although that CLE signaling plays an important role during plant development and in responses to various environmental stimuli, its role in fruit formation and ripening process has yet to be investigated.
Genes encoding CLE peptides and their receptors have been found in a diverse range of plant species [15–17] including monocots (Oryza sativa and Zea mays), dicots (Glycine max, Populus trichocarpa, Medicago truncatula) and even the moss Physcomitrella patens and the alga Chlamydomonas reinhardtii [18], thus suggesting that CLE signaling pathway has been conserved during the course of plant evolution [19]. However, the organization and roles of CLEs in crops, especially in fruit-bearing plants has fallen far behind Arabidopsis. Therefore, we chose tomato (S. lycopersicum), as a model crop with very high economic impact to understand the similarity and diversity of CLE cascade among various plants. Taking advantage of the available online database, we first carried out a genome-wide search for the CLE family members in tomato and then investigated their tissue-specific expression patterns. We also investigated the roles of CLE signaling in tomato by synthesizing the 12-aa CLE motif peptides of three SlCLEs specifically expressed in vegetative tissues and observed the effects of their exogenous application on tomato seedlings. Data presented here shed light on the evolutionary conservation of CLE gene families across various plant species as well as the various signaling components of CLE pathway in fleshy fruit-bearing crops.
Results and discussion
Members and phylogenetic analysis of CLEgene family in tomato
We performed a systematic analysis of the tomato CLE small signaling polypeptide family to expand our insights into the developmental processes that are potentially regulated by CLE peptide-mediated signal transduction pathways. Many small proteins including potential ligand peptides, are frequently not detectable by automated annotation programs due to their sizes [16]. To determine the CLE family members in tomato, all previously reported CLE proteins from various species were used as query sequences to perform full-length BLASTP searches against tomato proteome database at Phytozome v9.1 [20, 21]. As results, 15 apparent SlCLE genes were identified (Additional file 1). Considering the bigger size of Arabidopsis CLE gene family (32 members), this low yield could be due to the poor sequence conservation outside the CLE domains and there remained a chance to identify few more CLE family members in tomato genome.
We then examined the phylogenetic relationship of the putative SlCLE family members to Arabidopsis CLEs and certain well-known CLE proteins from other species. Because the majority of SlCLE family members have very divergent amino acid sequences outside the CLE motifs, their phylogenetic relationship was not well supported (Figure 1A). However, some branches in the tree were supported by high bootstrap values and thus indicated the most closely related CLE proteins. We are confident of the close relationship of SlCLE15 (Solyc11g071380) to FLORAL ORGAN NUMBER2 and 4 (OsFON2/FON4), which have been reported to regulate floral meristem size in rice [22, 23]. SlCLE10 (Solyc07g053370) and SlCLE11 (Solyc07g062670) are clustered together with AtCLE1-7, which genes form a single subclade among Arabidopsis 32 CLE members. The overexpression of AtCLE1-7 is known to cause a premature termination of SAM and longer roots as well [24, 25]. Interestingly, the functional forms of the above-mentioned three SlCLEs (SlCLE15, SlCLE10 and SlCLE11) are also the SlCLEs sharing the highest sequence similarity with the well-known AtCLV3 and ZmESR1-3. CLV3 encodes a stem cell-specific protein that plays a key role in stem cell fate determination through mediating the intercellular communication during Arabidopsis development [26, 27]. ZmESRs are endosperm-specific genes in maize and are supposed to play roles in the nutrition of the developing embryo and in the establishment of a physical barrier between embryo and endosperm [28]. When compared to the 26 predicted active CLE peptides encoded by Arabidopsis CLE genes, a perfect match in tomato was only found for SLCLE12 (Solyc09g061410) with the TDIF peptide encoded by AtCLE41/44. TDIF plays a specific role in xylem differentiation [2] and very recently, its perfect conservation in amino acid sequences has been reported across eight conifer species [29] .
Figure 1.
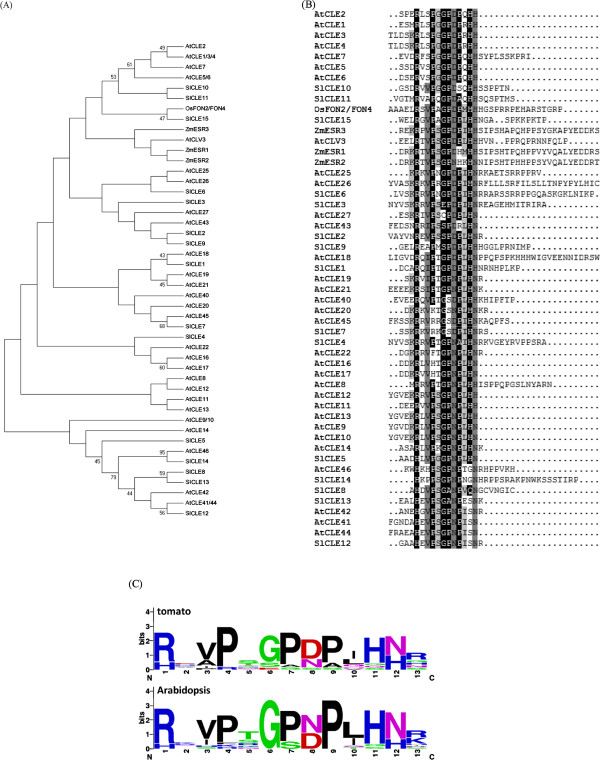
Phylogenetic tree and sequence analysis of tomato and Arabidopsis CLE gene families. (A) Maximum likelihood bootstrap tree phylogeny based on the CLE motif sequences of SlCLE genes in tomato. The unrooted tree was constructed using MEGA 5.05. Numbers at nodes indicate the percentage bootstrap scores and only bootstrap values higher than 40% from 1,000 replicates are shown. Sl, S. lycopersicum; At, A. thaliana; Zm, Z. Maize. (B) Predicted protein sequences of SlCLEs were aligned with AtCLEs with flanking sequences using ClustalX and the output was displayed with Box Shade. (C) Sequence logos for the CLE motifs of tomato and Arabidopsis CLE gene family members. The height of the bars indicates the number of identical residues per position.
Detailed description of all SlCLE polypeptides is provided in Additional file 1 and summarized in Table 1. Briefly, SlCLE genes encode polypeptides of 66–110 aa, with a predicted molecular mass range of 7.1–12.6 kDa. Though the vast majority of known CLE sequences have basic pI values [16], the theoretical pI values of SlCLEs range from 4.78 to 11.06 (Table 1). CLE proteins are synthesized as inactive protein precursors and the active 12 or 13-aa peptide ligands are liberated by proteolysis. Unlike the sequence degeneracy among the AtCLEs, all active SlCLE peptides are different in their sequences. When the sequence logo was built for tomato CLE motifs to visualize the conserved residues, the consensus sequence was slightly different from the one generated for AtCLEs (Figure 1B). Four residues including P(4), P(7), P(9) and H(11) are almost invariant over the whole SlCLE family (Additional file 2). Five predicted SlCLEs (SlCLE5, 8, 12, 13 and 14) diverge from the consensus at the otherwise perfectly conserved Arg residue at position 1 with a His residue. A similar occurrence of V/A at position 3, D/N at position 8 and N/H at position 12 are also observed (Figure 1B). Other positions in the CLE domain are highly variable, such as positions 2, 5, 6, 10 and 13 (Figure 1B). In a previous report, 174 predicted CLE motifs from various plant species were classified into 13 distinct groups based on their CLE domain sequences [4]. The signature amino acids of each group in that work could also be identified for some SlCLEs in this study. For instance, the His at position 1 and the Asn at position 11 of SlCLE12 and 13 were both identified as the group V-specific residues [4]. Four residues upstream of the CLE domain have been recently shown to be required for CLE peptide endoproteolytic processing [30]. In the case of SlCLEs, the upstream short regions adjacent to the CLE motif are highly variable and no similarity as previously reported for Arabidopsis CLEs [16] has been detected (Additional file 2). At the C terminus of the CLE domain, addition of an arginine has been shown to cause a decrease of peptide activity [2, 31]. In tomato, five out of 15 CLEs include this arginine residue at the C terminus of their respective CLE domains (Additional file 2).
Table 1.
A complete list of 15 SlCLEs identified in the present study
| Gene name | Gene locus | Unigene ID | Length (bp) | Protein (aa) | MW (kDa) | PI |
|---|---|---|---|---|---|---|
| SlCLE1 | Solyc01g014100.2.1 | 676 | 75 | 8.65 | 6.39 | |
| SlCLE2 | Solyc01g098890.1.1 | 246 | 81 | 8.91 | 4.78 | |
| SlCLE3 | Solyc02g067550.1.1 | 672 | 96 | 10.47 | 9.61 | |
| SlCLE4 | Solyc02g087470.2.1 | 516 | 69 | 7.62 | 11.06 | |
| SlCLE5 | Solyc03g025960.1.1 | SGN-U566466 | 201 | 66 | 7.13 | 10.02 |
| SlCLE6 | Solyc05g006610.2.1 | SGN-U599368 | 2858 | 110 | 12.60 | 10.87 |
| SlCLE7 | Solyc05g007650.1.1 | 252 | 83 | 9.25 | 10.14 | |
| SlCLE8 | Solyc05g053630.1.1 | SGN-U575421 | 270 | 89 | 9.72 | 9.57 |
| SlCLE9 | Solyc06g074060.1.1 | 403 | 74 | 8.40 | 9.51 | |
| SlCLE10 | Solyc07g053370.1.1 | 264 | 87 | 9.83 | 6.90 | |
| SlCLE11 | Solyc07g062670.1.1 | SGN-U581442 | 267 | 88 | 9.94 | 6.89 |
| SlCLE12 | Solyc09g061410.1.1 | SGN-U574832 | 318 | 105 | 11.67 | 8.86 |
| SlCLE13 | Solyc09g091810.1.1 | SGN-U566187 | 264 | 87 | 9.48 | 8.89 |
| SlCLE14 | Solyc11g066120.1.1 | SGN-U600211 | 568 | 95 | 10.71 | 10.45 |
| SlCLE15 | Solyc11g071380.1.1 | 555 | 93 | 10.60 | 9.63 |
Genomic DNA sequences are obtained from Phytozome v9.1 (http://www.phytozome.net) (Additional file 1). Amino acid sequences are deduced from the corresponding coding sequences (Additional file 1).
Gene structures, chromosomal locations, motif analysis and cis-elements in promoter regions of SlCLEs
Analysis of the predicted SlCLEs suggests that they are all secreted proteins, with signal peptides as predicted by SignalP. The presence of an intron in the sequence upstream of the CLE domain has been reported in Arabidopsis for CLE40 and CLV3 [9, 32] and in rice, 9 out of 44 OsCLE genes include introns [16]. In tomato, 7 out of 15 SlCLE coding sequences were interrupted by a single or two introns (Figure 2). By contrast, most Arabidopsis CLE genes consist of one exon with a single open reading frame [20]. As to the chromosome distribution, 15 SlCLE genes are distributed over 8 out of 12 tomato chromosomes with 1–3 genes per chromosome (Additional file 3). Although neighboring SlCLE genes could be found in pairs on chromosome 5, 7 and 11, they only share 9.7-26.1% identity within pairs and thus could not be taken as tandem segmental duplication event (Additional file 4). By contrast, whole genome duplication and reshuffling as well as localized gene duplication followed by gene retention have been reported to all contribute to the expansion of CLE gene family in Arabidopsis during evolution [33], which might explain the fairly large size of Arabidopsis CLE gene family.
Figure 2.
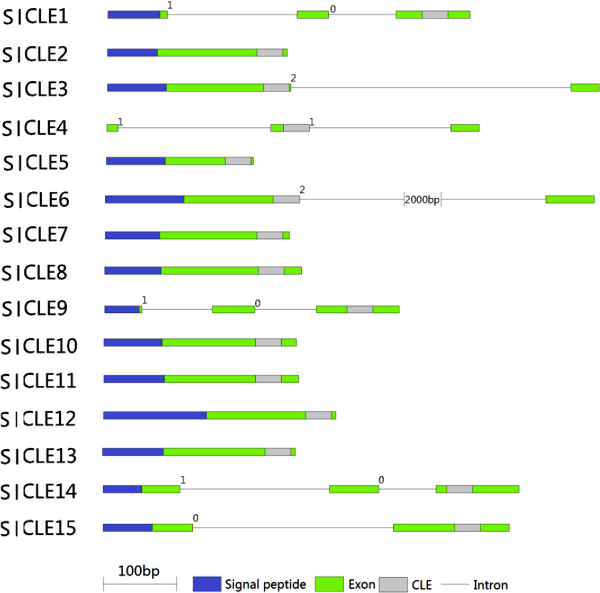
Schematic diagram of genomic structures of tomato CLE family members. Exons are shown as green boxes and introns as grey lines. The blue and grey boxes mark the signal peptides and CLE domains, respectively.
The motif distribution in tomato and Arabidopsis CLE proteins were analyzed using Multiple Expectation Maximization for Motif Elicitation (MEME) tool. Despite that multiple CLE domains have been reported for two OsCLEs [17], which implied a possibility of generation of multiple CLE peptides from one polypeptide precursor, no more than one CLE motif was observed within any of the SlCLEs. Besides the CLE motif, another one extremely conserved motif enriched with hydrophobic amino acids is found in the signal peptide regions of SlCLEs (Additional file 5). Except for these two motifs, no other consensus motif was found to be shared by the majority of SlCLEs. However, a close motif comparison between tomato and Arabidopsis CLEs revealed that individual SlCLE does share specific motifs with certain AtCLE, such as SolycCLE1 and CLV3 (Additional file 5).
The search for putative cis elements within the 2 kb genomic sequences upstream of the 5’-UTR of SlCLEs was performed in the PLACE database (http://www.dna.affrc.go.jp/PLACE/signalscan.html). A total of 200 putative cis elements were identified and only 29 out of them were able to be found in all promoter regions, suggesting the very complex regulatory behaviors of the SlCLE family genes (Additional file 6). Most putative cis elements could be classified into four major types. The first type include the elements that are regulated by one of the most important developmental signals, hormones, including abscisic acid (ABA), gibberellins (GA), auxin, jasmonic acid (JA) and ethylene (ETH). For example, the GADOWNAT sequence similar to ABRE was found to be present in 24 GA down-regulated genes involved in Arabidopsis seed germination [34]. Diverse cis-elements responsive to hormones identified in the promoters of SlCLE genes indicate that CLE peptide signaling transduction pathway interacts dynamically with other phytohormone signaling pathways. Cis-elements of the second type identified are either related to organgenesis or required for tissue-specific gene expression patterns, which make them very attractive considering the important roles played by CLEs in plant meristem maintenance and organogenesis. For example, the core AACA motifs found in rice glutelin genes are involved in controlling their endosperm-specific expression [35] and the TGA1a motif is likely to contribute to the root tip meristem-specific expression of GST isoenzymes [36]. Cis-elements of the third type are light-related ones, including the GT1 CONSENSUS binding site present in many light-regulated genes [37]. Cis-elements of the fourth type are generally related to environmental stimulus including both abiotic and biotic stresses and this type of elements outnumbered other element categories except the light regulation-related type, which indicates the involvement of SlCLEs in stress responses. In addition, two very interesting elements, WUSATAg and XYLAT could be found in 5 (SlCLE11, 2, 7, 13 and 12) and 2 (SLCLE3 and 6) genes, respectively, with 1 or 2 copy numbers. The WUSATAg has been identified as a target sequence of WUS in the intron of AGAMOUS gene in Arabidopsis [38]. In both SAM and RAM, the interplay between CLE genes and different WUS gene lays the foundation of meristem cell population regulation. The XYLAT element is a cis-element characterized among the promoters of the “core xylem gene set”, reminding us of the TDIF, which is highly relevant to the xylem cell differentiation [39].
SlCLEexpression patterns indicate their involvement in many developmental processes including fruit ripening
In Arabidopsis, the specificity of CLE function has been proposed to be mainly achieved by differential expression patterns of CLE genes. In order to gain insights into the potential biological roles of SlCLEs, we analyzed their temporal and spatial expression profiles in various tissues and fruits at different developmental stages using qRT-PCR (Additional file 7 and Figure 3). In Arabidopsis, all but one of the 32 AtCLE genes are transcribed [33]. In tomato, thirteen out of fifteen SlCLEs were able to be cloned from a pooled cDNA derived from an array of tomato tissues. All tissues examined express multiple SlCLE genes and the overlap among different CLE gene expression patterns was conspicuous (Additional file 7 and Figure 3). In brief, the expression level of all SlCLEs is low in small green fruit and increases after the breaker stage (Additional file 6 and Figure 4). SlCLE2, 3, 4, 6, 7, and 14 exhibit a relatively uniform expression among the examined tissues; however, the remaining SlCLEs exhibit a quite differential expression pattern among tissues and organs (Additional file 7 and Figure 3). Notably, SlCLE12 appears to be the most abundantly expressed SlCLE in almost all tested tissues except for the green fruit of 1 cm size (Additional file 7 and Figure 3). In Arabidopsis, an exactly same CLE peptide (TDIF) as SlCLE12p and a very similar one are produced by CLE41/44 and CLE42, respectively [2, 40]. Using the GUS (β-glucuronidase) reporter gene assay, CLE41/44 are found to be expressed preferentially in vascular bundles, while CLE42 is expressed strongly in SAM and axillary meristems. Therefore, CLE41/44 and CLE42 have been suggested to play roles in regulating wood development [41–43] and the meristematic activity [44], respectively. Considering its vascular-specific expression, SlCLE12 could be most likely denoted as the orthologue of CLE41/44 in tomato; however, its broad and significant expression in a range of tissues and organs imply an even higher functional diversity or a basic function of TDIF in regulating biological processes. As an evidence, a novel function in enhancing axillary bud formation has been recently disclosed for Arabidopsis TDIF peptide [44].
Figure 3.

Expression patterns of SlCLE genes in selected tissues and fruits at different developmental stages. (A) Photos of tomato fruits at seven developmental stages assessed. (B) Transcription levels of SlCLEs investigated by RT-qPCR shown as heat-map for better illustration. Relative gene expression levels were normalized to have mean zero and variance one with actin transcript values before producing the heat maps. The RT-qPCR analyses were repeated at least three times in three independent experiments. Color scale at the top of each dendrogram represents log2 values. Total RNA isolated from apex (A), root (R), young, medium and old stems (YS, MS, and OS), young and mature leaf (YL and ML), flower (FL), flower buds (FB), and 1-cm, 2-cm and 3-cm sized green fruits (F1, F2 and F3), mature green fruit (MG), fruit at breaker stage (BF), fruit turning red (TF) and mature fruit in red(RF).
Figure 4.
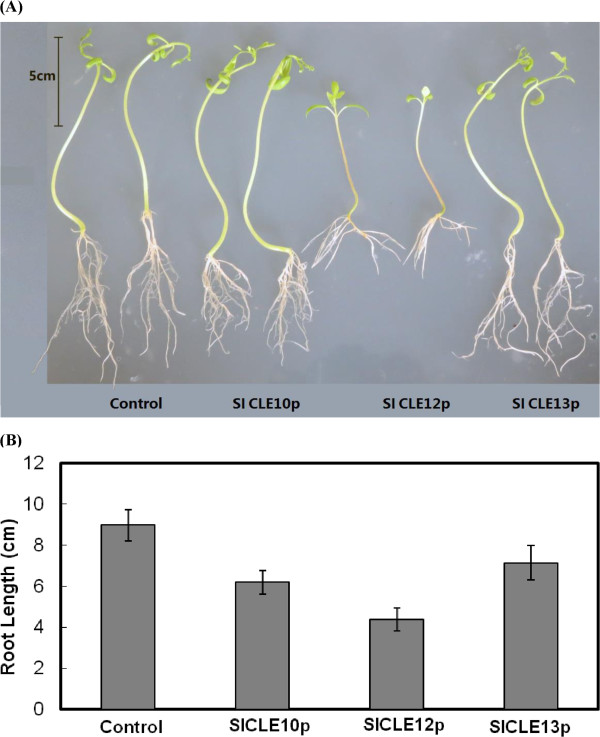
Phenotypes of 10-d tomato seedlings upon the treatment of SlCLE motif peptides. (A) Effect of 100 μM concentration of SlCLE10p, 12p and 13p on 10-day old tomato seedling development. (B) Effects of different SlCLE motif peptides on the root length of tomato seedlings. The lengths of the main roots were measured after 10 d of growth on peptide-containing media (n = 25 for each treatment). Data and error bars represent mean ± SD.
SlCLE1, SlCLE10, SlCLE12 and SlCLE13 are specifically expressed at a high transcript level in flower bud, root, stem and shoot apex, respectively, suggesting that they might play particular roles during the development of these organs (Additional file 7 and Figure 3). The Arabidopsis counterpart of SlCLE1 is AtCLE18 (At1g66145) and the synthetic 12-aa peptide derived from the CLE18 motif has been shown to suppress root growth and cause a short-root phenotype when applied exogenously [2]. SlCLE13 is highly and specifically expressed in tomato shoot apex, where Arabidopsis CLV3 plays a critical role in the maintenance of stem cell reservation. However, since they only share low sequence similarity, they might not be the functional equivalence. SlCLE5, 8 and 11 reveal their expression peaks in fruit indicating their potential involvements in fruit development and ripening process. In particular, the transcript level of SlCLE8 sharply increases in green fruit at 1 cm stage and continuously increases during early fruit development and dramatically declines when green fruit reaches maturation (Additional file 7 and Figure 3). It is noteworthy that despite of its attracting green-fruit expression specificity, the transcript level of SlCLE8 is lower than that of SlCLE11, 12 and 13, which are the dominant SlCLEs in green fruit (Additional file 7 and Figure 3). With its maximum expression found in stem tissue, SlCLE12 is still the most highly expressed SlCLE gene in green fruit until its transcript level is surpassed by SlCLE5 and SlCLE11 in orange and red ripe fruits, respectively (Additional file 7 and Figure 3). SlCLE5 reaches its maximum transcription level at the turning stage and keeps a high expression level together with SlCLE11 in red ripe fruit, indicating their roles in later process of fruit and/or embryo/seed development. Based on these results, we propose a hypothesis that SlCLEs play important and sequential roles in the fruit ripening process. It has been reported that receptors of CLE peptide including CLV1, CLV2, and CRN function together in meristems and gynoecia to regulate Arabidopsis fruit organ number and the authors also suggested that CLE ligand(s) other than CLV3 is acting in the CLV pathway in gynoecia [45]. The results shown here further support the importance of the CLE-mediated signaling network in regulating organogenesis throughout plant life cycle and may provide information about key players in the regulation of fruit quality in tomato and other crop plants.
In the original tomato genome sequencing report, the relative expression of all tomato genes was determined by replicated strand-specific Illumina RNA-Seq using tissues including root, leaf, flower (2 stages) and fruit (6 stages) [46]. Based on their data, we performed an in silico gene expression analysis of tomato CLE gene family members. Similar expression patterns as shown by the qPCR results were identified for some SlCLE genes (Additional file 8). For example, CLE5 and CLE11 are highly expressed in orange and red fruit at maturation, while SlCLE7 and 13 are preferentially expressed with abundance in flower buds (Additional file 8). However, there are also some discrepancies between the RNA-sequencing work and the results we obtained by qPCR method. For example, we did not confirm the reported high expression level of SlCLE2, 4, 6, 14 in root (Additional file 8); instead, SlCLE10 appear to be the most specifically and highly expressed CLE gene in root (Additional file 7 and Figure 3).
Effects of synthetic SlCLEs application on tomato seedlings
In literature, the active structure of AtCLV3 has been determined as a 12-aa hydroxylated peptide through matrix-assisted laser desorption/ionization–time-of-flight mass spectrometry (MALDI-TOF MS) [1] and the TDIF-peptide isolated from Zinnia elegans mesophyll cell culture is also a dodecapeptide with two hydroxyproline (HyP) residues [2]. Thus, in this work we synthesized the 12–amino acid peptides representing the CLE domains of SlCLE10, 12, 13 with hydroxyzed pralines and used them for in vitro application assays since these three SlCLEs are most specifically expressed genes in vegetative tissues (root, stem and apex, respectively). Tomato seeds were germinated on 1/2 MS media plates containing individual peptide at 10, 50 or 100 μm and 10 days after germination, a dose-dependent inhibitory effect on root growth was observed. Treatments with SlCLE10, 12 and 13 CLE motif peptides dramatically inhibited the root growth by 31%, 51% and 20%, respectively, when applied at the 100 μM concentrations (Figure 4). These results basically resembled the short root phenotype generated by in vitro application of AtCLE peptides on Arabidopsis seedlings and gave us the first indication of the functional role played by the motif peptides encoded by SlCLEs. In Arabidopsis, application of 19 different synthetic AtCLE peptides has been reported to arrest root meristem growth [2, 24, 47], whereas the application of CLE41/44 (TDIF), CLE42, or CLE43 peptides only caused suppressed xylem differentiation [2, 47]. In a previous work, after growing on CLV3p, CLE19p, and CLE40p-containing media for 7 days, a similar inhibition was found for the lengths of the Arabidopsis main roots [6]. In contrast, three synthetic SlCLE peptides in this work produced varied short root phenotypes (Figure 4). Unexpectedly, the root-specific SlCLE10p inhibited the root length to a slighter degree than the TDIF encoded by SlCLE12. In phylogenetic tree, SlCLE10p is positioned in a single clade together with AtCLE1-7 peptides by sharing 10 identical amino acids over the 12-aa CLE motif sequence. Overexpression of AtCLE1-7 in Arabidopsis result in the developmental timing delay and wus-like phenotypes (premature SAM termination, misshapen leaves, flower abnormalities and long root accompanied with anthocyanin accumulation) [24], but no phenotypes has been reported for roots treated with synthetic peptides corresponding to the CLE2 and CLE4-7 CLE motifs [2]. In contrast, we did not observe any of the above-mentioned Arabidopsis phenotypes in the current work and instead, an inhibition on root length was detected upon the application of SlCLE10p, which might indicate the functional variance of SlCLE10p and AtCLE1-7p in two different plant species.
SlCLE12 produces an active CLE motif exactly as same as TDIF, a dodecapeptide corresponding to the CLE motif of Arabidopsis CLE41/44. In vitro, TDIF could independently suppress the differentiation of procambial cells to treacheary elements and promote cell division, but is unable to terminate the RAM development and cause root development retardation [42]. Overexpression of TDIF has been reported to cause dwarf growth and shrub-like phenotype due to a lack of apical dominance without affecting the growth from the SAM or root elongation [24]. Different from these data, in this work SLCLE12p dramatically reduced the growth rate of primary root when applied to tomato seedlings accompanied with an accumulation of anthocynin in the hypocotyl part (Figure 4). Seven AtCLE genes not including CLE42 or CLE44 have been reported to cause apparent anthocyanin overproduction when overexpressed in Arabidopsis plants and this phenotype promoted the authors to suggest possible roles for CLE family members in the regulation of plant stress responses [24]. Similar to SlCLE 10p and 12p, the application of SlCLE13p on tomato seedlings also suppressed the root length elongation but to a slightest degree (Figure 4). Despite of its apex-predominant expression, no misfunctioning of the SAM and any arrest of organogenesis from the shoot tip was observed upon the application of SlCLE13p. Since SlCLE13p is very similar to SlCLE12p in the amino acid composition except that Pro7 and Ile10 in SlCLE12 are substituted by Ala7 and Glu10 in SlCLE13, respectively, it is intriguing to speculate why they inhibit the root growth differentially. In a previous work, when Pro4 and Pro7 of CLV3 was substituted with sarcosine, the simplest imino acid (N-methylglycine), its inhibition activity on Arabidopsis roots was significantly decreased accompanied with a weaker receptor binding activity [48]. In addition, Pro7 is also the posttranslational arabinosylation location of several CLE peptides including CLV3 and this sugar modification strongly enhances its binding to corresponding receptor CLV1 and its biological effect [49]. Therefore, tomato CLE peptides probably also act in a sequence-specific manner as previously reported [6] and the substitution of Pro at position 7 to Ala in SlCLE13p might lead to its decreased activity compared to SlCLE12p.
It is noteworthy that the reduction in root length observed for tomato seedlings treated by SlCLE10p, 12p, or 13p could only be taken as the first sign of their potential physiological roles and by no means limit their role in additional processes or to rule out other SlCLEs as key regulators of growth and development in various parts of plant. In order to uncover the potentially diverse functions and activities of SlCLE peptides, functional analysis by ectopic expression or gene silencing techniques is undoubtedly needed to provide further insights into the molecular properties of CLE peptides and their modes of action in the tomato life cycle, in particular in the events of fruit formation and maturation.
Conclusions
Characterization of CLE genes in a fleshy fruit-bearing crop species would facilitate a better understanding of the functions of CLE signaling cascade. Using tomato as a model, a comprehensive overview of SlCLE gene family is presented, including the gene structures, phylogeny, chromosome locations, conserved motifs and cis-elements in promoter sequences. qRT-PCR analysis showed that 13 of the 15 SlCLE genes are transcribed and multiple SlCLE genes are actively expressed in a overlapping manner within a given tissue. Several SlCLE genes are tissue-specific expressed and developmentally regulated during fruit organogenesis and maturation. SlCLE12, the TDIF homologue in tomato, exhibits an exceptionally high expression level in almost all investigated tissues and organs including fruits at various developmental stages. Synthetic peptides corresponding to the CLE motif of SlCLE10, 12 and 13 could significantly and variably inhibit the elongation of tomato primary root and SlCLE12p also leads to an arrest of shoot growth and anthocynin accumulation in hypocotyl, suggesting that such CLE peptides are likely to be the functional CLE gene products. Data shown in this study would provide a very useful reference for future functional analysis of member of CLE genes in Solanum crops, one of the largest angiosperm genera.
Methods
Gene identification and sequence analysis
To search the tomato CLE genes, the amino acid sequences of Arabidopsis, rice, poplar and other known CLEs were used to perform BLASTP searches against the Phytozome v9.1 (http://www.phytozome.net/) and the tomato database on the SOL Genomics Network (http://solgenomics.net). The retrieved tomato sequences were used as queries to repeat the step in an iterative manner. The positive hits were characterized with a conserved 12-aa CLE domain at the C-terminus and a hydrophobic signal peptide at the N-terminus. Full-length amino acid sequences (60–120 amino acids) or sequences corresponding to the CLE domains were aligned using ClustalX2.1. Phylogenetic analysis was performed using MEGA5.05 program by the maximum likelihood (ML) method with 1000 bootstrap replicates. MEME utility (http://meme.ebi.edu.au/meme/cgi-bin/meme.cgi) was used to display motifs of SlCLE proteins with default parameters except that the motif width was set from 10 to 300 amino acids and the maximum number of motifs to find was five [50]. To determine the locations of SlCLE genes on tomato chromosomes, each sequence was further used as query sequence for the BLASTN search against SGN tomato whole genome scaffolds data (2.30) (http://www.sgn.cornell.edu/tools/blast/). To investigate cis-elements in promoter sequences of tomato CLE genes, 2000 bp genomic sequences upstream of the initiation codons were analyzed for cis regulatory elements at the PLACE website (http://www.dna.affrc. go.jp/PLACE/) [51, 52].
Plant growth and organ-specific expression analysis
Tomato seeds were sown and grown in soil for 5 weeks with a day length of 16 h at a constant temperature of 25°C. The different organs were cut from the plants and total RNA was isolated from fresh tissue using the EasyPureTM Plant RNA Kit (TransGen, China) according to the manufacturer’s instructions and genomic DNA was removed with DNase I treatment. The concentration and quality of the RNA samples were examined using Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific, Finland). One μg of total RNA was used to synthesize cDNA with the oligo-(dT)18 primer using the EasyScript® First-Strand cDNA Synthesis SuperMix (TransGen, China). Quantitative Real-Time PCR (qRT-PCR) analysis of cDNA was performed on a PikoReal 96 Real-time Thermal Cycler and PikoReal Software (V2.2) (Thermo Fisher Scientific, Finland) using SYBR® Premix Ex TaqTM (TaKaRa, Japan) and specific primers shown in Additional file 9. The following thermal cycle conditions were used: 95°C for 2 min, followed by 45 cycles of 95°C for 20 s and 58°C for 20 s, 72°C for 30 s. All reactions were performed in triplicate from three independent pooled samples (4 plants per sample). Relative quantification of specific mRNA levels was analyzed using the cycle threshold (Ct) 2-ΔΔCt method. Relative expression levels are normalized using the housekeeping gene actin and shown in folds of root expression value. Student’s t test (P < 0.05) was used to determine the significant difference of relative expression of individual genes among different samples. A gene in a given tissue was defined as preferentially expressed only if the expression value of the gene in this tissue was more than 2-fold and had a P value less than 0.05 compared to other tissues. Based on the data, the heat map was generated using the MeV4.9.0 software (http://www.tm4.org/) following the instruction.
Synthetic SlCLE motif peptide application on tomato seedlings
For peptide treatments, the sterilized seeds were placed on half strength Murashige and Skoog media (Duchefa, Netherlands) containing 10, 50 or 100 μM SlCLE peptides, 1% (w/v) sucrose, with 1.5% (w/v) Daishin agar (Brunschwig Chemie, Netherlands). Plates were first incubated at 4°C in the dark for 2 d and then transferred to a room with a temperature of 22°C, 16 h light per day. Three peptides including SlCLE10p (RVVHypGGHypDSQHH), SlCLE12p (HEVHypSGHypNPISN), SlCLE13p (HEVHypSGANPESN), were ordered from Scilight Biotechnology (Beijing, China) with a purity of >90% and dissolved in a filter-sterilized sodium phosphate buffer (50 mM, pH 6).
Availability of supporting data
The data sets supporting the results of this article are included within the article and its additional files.
Electronic supplementary material
Additional file 1: Nucleic acid sequences and amino acid sequences of all 15 tomato CLE gene family members. (XLS 40 KB)
Additional file 2: Multiple alignments of the full-length amino acid sequences of SlCLE gene family members using DNAMAN alignment program. Black and light gray shading indicates identical and conversed amino acid residues, respectively. (TIFF 140 KB)
Additional file 3: Locations of SlCLE genes on the tomato chromosomes. Physical distances are in megabases (Mb) and gene transcription orientations are marked by solid arrows. (DOCX 34 KB)
Additional file 4: Homology, similarity and identity of SlCLE family members over their full-length sequences. (DOC 110 KB)
Additional file 5: Schematic illustrations of the types and distributions of motifs in tomato and Arabidopsis CLE family. The motif characterization was based on full-length proteins by MEME web server. (XLS 1 MB)
Additional file 6: The variety and numbers of cis-acting elements in the 2 kb regions upstream of SlCLE genes searched in the PLACE database (http://www.dna.affrc.go.jp/PLACE/signalscan.html). (XLS 132 KB)
Additional file 7: Tissue-specific transcription levels of 13 SlCLEs using qRT-PCR method. The expression levels of individual SlCLE gene among various tissues/organs as well as multiple SlCLE genes within one individual tissue/organ were presented. Relative gene expression levels are normalized with actin transcript values. The RT-qPCR analyses were repeated at least three times in three independent experiments. Total RNA isolated from apex (A), root (R), young, medium and old stems (YS, MS, and OS), young and mature leaf (YL and ML), flower (FL), flower buds (FB), and 1-cm, 2-cm and 3-cm sized green fruits (F1, F2 and F3), mature green fruit (MG), fruit at breaker stage (BF), fruit turning red (TF) and mature fruit in red (RF). (DOC 86 KB)
Additional file 8: Normalized expression of tomato SlCLE genes based on RNA-seq data downloaded from previously published data [46]. Based on the data, the heat map was generated using the MeV4.9.0 software (http://www.tm4.org/) and gene-wise normalized and hierarchical clustered based on Pearson correlation. Color scale at the top of each dendrogram represents fold change values. Bud: unopened flower buds; flower: fully opened flowers; MG fruit: mature green fruit; B_fruit: fruits at breaker; B + 10_fruit: fruits at breaker + 10 days stage. (ZIP 15 KB)
Additional file 9: qRT-PCR primers used in the current study. (XLS 27 KB)
Acknowledgements
We thank Dr. Shuzhen Men, Nankai University, China, for kindly providing us the tomato seeds (S. lycopersicum cv. ‘Heinz-1706’). This study was supported by National Natural Science Foundation of China (31270644 and 31170564).
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
YZ performed most of the data mining, data analysis and experimental work. YS participated in the illustrations of the figures and tables. SY helped to retrieve data from GEO database and draw the heat-map. JW designed and coordinated the work and wrote the manuscript. All authors read and approved the final manuscript.
Contributor Information
Yu Zhang, Email: zy_mafia@hotmail.com.
Shaohui Yang, Email: shaohuiyang77@tju.edu.cn.
Yingjin Song, Email: songyingjin@yahoo.com.cn.
Jiehua Wang, Email: jiehuawang@tju.edu.cn.
References
- 1.Kondo T, Sawa S, Kinoshita A, Mizuno S, Kakimoto T, Fukuda H, Sakagami Y. A plant peptide encoded by CLV3 identified by in situ MALDI-TOF MS analysis. Science. 2006;313(5788):845–848. doi: 10.1126/science.1128439. [DOI] [PubMed] [Google Scholar]
- 2.Ito Y, Nakanomyo I, Motose H, Iwamoto K, Sawa S, Dohmae N, Fukuda H. Dodeca-CLE peptides as suppressors of plant stem cell differentiation. Science. 2006;313(5788):842–845. doi: 10.1126/science.1128436. [DOI] [PubMed] [Google Scholar]
- 3.Suzaki T, Yoshida A, Hirano HY. Functional diversification of CLAVATA3-related CLE proteins in meristem maintenance in rice. Plant Cell. 2008;20(8):2049–2058. doi: 10.1105/tpc.107.057257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kinoshita A, Nakamura Y, Sasaki E, Kyozuka J, Fukuda H, Sawa S. Gain-of-function phenotypes of chemically synthetic CLAVATA3/ESR-related (CLE) peptides in Arabidopsis thaliana and Oryza sativa. Plant Cell Physiol. 2007;48(12):1821–1825. doi: 10.1093/pcp/pcm154. [DOI] [PubMed] [Google Scholar]
- 5.Fiers M, Golemiec E, van der Schors R, van der Geest L, Li KW, Stiekema WJ, Liu CM. The CLAVATA3/ESR motif of CLAVATA3 is functionally independent from the nonconserved flanking sequences. Plant Physiol. 2006;141(4):1284–1292. doi: 10.1104/pp.106.080671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fiers M, Golemiec E, Xu J, van der Geest L, Heidstra R, Stiekema W, Liu CM. The 14-amino acid CLV3, CLE19, and CLE40 peptides trigger consumption of the root meristem in Arabidopsis through a CLAVATA2-dependent pathway. Plant Cell. 2005;17(9):2542–2553. doi: 10.1105/tpc.105.034009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miyawaki K, Tabata R, Sawa S. Evolutionarily conserved CLE peptide signaling in plant development, symbiosis, and parasitism. Curr Opin Plant Biol. 2013;16(5):598–606. doi: 10.1016/j.pbi.2013.08.008. [DOI] [PubMed] [Google Scholar]
- 8.Clark SE, Running MP, Meyerowitz EM. CLAVATA3 is a specific regulator of shoot and floral meristem development affecting the same processes as CLAVATA1. Development. 1995;121:2057–2067. [Google Scholar]
- 9.Fletcher LC, Brand U, Running MP, Simon R, Meyerowitz EM. Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science. 1999;283(5409):1911–1914. doi: 10.1126/science.283.5409.1911. [DOI] [PubMed] [Google Scholar]
- 10.Mayer KF, Schoof X, Haecker H, Lenhard A, Jurgens M, Laux GT. Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell. 1998;95(6):805–815. doi: 10.1016/S0092-8674(00)81703-1. [DOI] [PubMed] [Google Scholar]
- 11.Schoof H, Lenhard M, Haecker A, Mayer KFX, Jurgens G, Laux T. The stem cell population of Arabidopsis shoot meristems is maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell. 2000;100(6):635–644. doi: 10.1016/S0092-8674(00)80700-X. [DOI] [PubMed] [Google Scholar]
- 12.Stahl Y, Wink RH, Ingram GC, Simon R. A signaling module controlling the stem cell niche in Arabidopsis root meristems. Curr Biol. 2009;19(11):909–914. doi: 10.1016/j.cub.2009.03.060. [DOI] [PubMed] [Google Scholar]
- 13.Fiume E, Fletcher JC. Regulation of Arabidopsis embryo and endosperm development by the polypeptide signaling molecule CLE8. Plant Cell. 2012;24(3):1000–1012. doi: 10.1105/tpc.111.094839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Okamoto S, Shinohara H, Mori T, Matsubayashi Y, Kawaguchi M. Root-derived CLE glycopeptides control nodulation by direct binding to HAR1 receptor kinase. Nat Commun. 2013;4:2191. doi: 10.1038/ncomms3191. [DOI] [PubMed] [Google Scholar]
- 15.Yamamoto E, Karakaya H,C, Knap HT. Molecular characterization of two soybean homologs of Arabidopsis thaliana CLAVATA1 from the wild type and fasciation mutant. Biochim Biophys Acta. 2000;1491(1–3):333–340. doi: 10.1016/S0167-4781(00)00061-0. [DOI] [PubMed] [Google Scholar]
- 16.Oelkers K, Goffard N, Weiller GF, Gresshoff PM, Mathesius U, Frickey T. Bioinformatic analysis of the CLE signaling peptide family. BMC Plant Biol. 2008;8:1. doi: 10.1186/1471-2229-8-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sawa S, Kinoshita A, Betsuyaku S, Fukuda H. A large family of genes that share homology with CLE domain in Arabidopsis and rice. Plant Signal Behav. 2008;3(5):337–339. doi: 10.4161/psb.3.5.5344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miwa H, Kinoshita A, Fukuda H, Sawa S. Plant meristems: CLAVATA3/ESR-related signalling in the shoot apical meristem and the root apical meristem. J Plant Res. 2009;122:31–39. doi: 10.1007/s10265-008-0207-3. [DOI] [PubMed] [Google Scholar]
- 19.Kiyohara S, Sawa S. CLE signaling systems during plant development and nematode infection. Plant Cell Physiol. 2012;53(12):1989–1999. doi: 10.1093/pcp/pcs136. [DOI] [PubMed] [Google Scholar]
- 20.Cock JM, McCormick S. A large family of genes that share homology with CLAVATA3. Plant Physiol. 2001;126(3):939–942. doi: 10.1104/pp.126.3.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suzaki T, Ohneda M, Toriba T, Yoshida A, Hirano HY. FON2 SPARE1 redundantly regulates floral meristem maintenance with FLORAL ORGAN NUMBER2 in rice. PLoS Genet. 2009;5(10):e1000693. doi: 10.1371/journal.pgen.1000693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chu H, Zhang D. The Shoot Apical Meristem Size Regulated by FON4 in Rice. Plant Signal Behav. 2007;2(2):115–116. doi: 10.4161/psb.2.2.3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Strabala TJ, O’Donnell PJ, Smit AM, Ampomah-Dwamena C, Martin EJ, Netzler N, Nieuwenhuizen NJ, Quinn BD, Foote HC, Hudson KR. Gain-of-function phenotypes of many CLAVATA3/ESR genes, including four new family members, correlate with tandem variations in the conserved CLAVATA3/ESR domain. Plant Physiol. 2006;140(4):1331–1344. doi: 10.1104/pp.105.075515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meng L, Ruth KC, Fletcher JC, Feldman L. The roles of different CLE domains in Arabidopsis CLE polypeptide activity and functional specificity. Mol Plant. 2010;3(4):760–772. doi: 10.1093/mp/ssq021. [DOI] [PubMed] [Google Scholar]
- 26.Brand U, Fletcher JC, Hobe M, Meyerowitz EM, Simon R. Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science. 2000;289(5479):617–619. doi: 10.1126/science.289.5479.617. [DOI] [PubMed] [Google Scholar]
- 27.Rojo E, Sharma VK, Kovaleva V, Raikhel NV, Fletcher JC. CLV3 is localized to the extracellular space, where it activates the Arabidopsis CLAVATA stem cell signaling pathway. Plant Cell. 2002;14(5):969–977. doi: 10.1105/tpc.002196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Opsahl-Ferstad HG, Le Deunff E, Dumas C, Rogowsky PM. ZmEsr, a novel endosperm-specific gene expressed in a restricted region around the maize embryo. Plant J. 1997;12(1):235–246. doi: 10.1046/j.1365-313X.1997.12010235.x. [DOI] [PubMed] [Google Scholar]
- 29.Strabala TJ, Phillips L, West M, Stanbra L. Bioinformatic and phylogenetic analysis of the CLAVATA3/EMBRYO-SURROUNDING REGION (CLE) and the CLE-LIKE signal peptide genes in the Pinophyta. BMC Plant Biol. 2014;14(1):47. doi: 10.1186/1471-2229-14-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ni J, Guo Y, Jin H, Hartsell J, Clark SE. Characterization of a CLE processing activity. Plant Mol Biol. 2011;75(1–2):67–75. doi: 10.1007/s11103-010-9708-2. [DOI] [PubMed] [Google Scholar]
- 31.Sawa S, Kinoshita A, Nakanomyo I, Fukuda H. CLV3/ESR-related (CLE) peptides as intercellular signaling molecules in plants. Chem Rec. 2006;6(6):303–310. doi: 10.1002/tcr.20091. [DOI] [PubMed] [Google Scholar]
- 32.Hobe M, Muller R, Grunewald M, Brand U, Simon R. Loss of CLE40, a protein functionally equivalent to the stem cell restricting signal CLV3, enhances root waving in Arabidopsis. Dev Genes Evol. 2003;213(8):371–381. doi: 10.1007/s00427-003-0329-5. [DOI] [PubMed] [Google Scholar]
- 33.Sharma VK, Ramirez J, Fletcher JC. The Arabidopsis CLV3-like (CLE) genes are expressed in diverse tissues and encode secreted proteins. Plant Mol Biol. 2003;51(3):415–425. doi: 10.1023/A:1022038932376. [DOI] [PubMed] [Google Scholar]
- 34.Busk PK, Pages M. Regulation of abscisic acid-induced transcription. Plant Mol Biol. 1998;37(3):425–435. doi: 10.1023/A:1006058700720. [DOI] [PubMed] [Google Scholar]
- 35.Wu C, Washida H, Onodera Y, Harada K, Takaiwa F. Quantitative nature of the Prolamin-box. ACGT and AACA motifs in a rice glutelin gene promoter: minimal cis-element requirements for endosperm-specific gene expression. Plant J. 2000;23(3):415–421. doi: 10.1046/j.1365-313x.2000.00797.x. [DOI] [PubMed] [Google Scholar]
- 36.Johnson C, Glover G, Arias J. Regulation of DNA binding and trans-activation by a xenobiotic stress-activated plant transcription factor. J Biol Chem. 2001;276(1):172–178. doi: 10.1074/jbc.M005143200. [DOI] [PubMed] [Google Scholar]
- 37.Villain P, Mache R, Zhou DX. The mechanism of GT element-mediated cell type-specific transcriptional control. J Biol Chem. 1996;271(51):32593–32598. doi: 10.1074/jbc.271.51.32593. [DOI] [PubMed] [Google Scholar]
- 38.Lohmann JU, Hong RL, Hobe M, Busch MA, Parcy F, Simon R, Weigel R. A Molecular Link between Stem Cell Regulation and Floral Patterning in Arabidopsis. Cell. 2003;105:793–803. doi: 10.1016/S0092-8674(01)00384-1. [DOI] [PubMed] [Google Scholar]
- 39.Ko JH, Beers EP, Han KH. Global comparative transcriptome analysis identifies gene network regulating secondary xylem development in Arabidopsis thaliana. Mol Genet Genomics. 2006;276(6):517–531. doi: 10.1007/s00438-006-0157-1. [DOI] [PubMed] [Google Scholar]
- 40.Fukuda H, Hirakawa Y, Sawa S. Peptide signaling in vascular development. Curr Opin Plant Biol. 2007;10(5):477–482. doi: 10.1016/j.pbi.2007.08.013. [DOI] [PubMed] [Google Scholar]
- 41.Hirakawa Y, Kondo Y, Fukuda H. TDIF peptide signaling regulates vascular stem cell proliferation via the WOX4 homeobox gene in Arabidopsis. Plant Cell. 2010;22(8):2618–2629. doi: 10.1105/tpc.110.076083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hirakawa Y, Shinohara H, Kondo Y, Inoue A, Nakanomyo I, Ogawa M, Sawa S, Ohashi-Ito K, Matsubayashi Y, Fukuda H. Non-cell-autonomous control of vascular stem cell fate by a CLE peptide/receptor system. Proc Natl Acad Sci U S A. 2008;105(39):15208–15213. doi: 10.1073/pnas.0808444105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hirakawa Y, Kondo Y, Fukuda H. Regulation of vascular development by CLE peptide-receptor systems. J Integr Plant Biol. 2010;52(1):8–16. doi: 10.1111/j.1744-7909.2010.00904.x. [DOI] [PubMed] [Google Scholar]
- 44.Yaginuma H, Hirakawa Y, Kondo Y, Ohashi-Ito K, Fukuda H. A novel function of TDIF-related peptides: promotion of axillary bud formation. Plant Cell Physiol. 2011;52(8):1354–1364. doi: 10.1093/pcp/pcr081. [DOI] [PubMed] [Google Scholar]
- 45.Durbak AR, Tax FE. CLAVATA signaling pathway receptors of Arabidopsis regulate cell proliferation in fruit organ formation as well as in meristems. Genetics. 2011;189(1):177–194. doi: 10.1534/genetics.111.130930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.International Tomato Genome Sequencing Project The tomato genome sequence provides insights into fleshy fruit evolution. Nature. 2012;485(7400):635–641. doi: 10.1038/nature11119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Whitford R, Fernandez A, De Groodt R, Ortega E, Hilson P. Plant CLE peptides from two distinct functional classes synergistically induce division of vascular cells. Proc Natl Acad Sci U S A. 2008;105(47):18625–18630. doi: 10.1073/pnas.0809395105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kondo T, Yokomine K, Nakagawa A, Sakagami Y. Analogs of the CLV3 peptide: synthesis and structure-activity relationships focused on proline residues. Plant Cell Physiol. 2011;52(1):30–36. doi: 10.1093/pcp/pcq146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ohyama K, Shinohara H, Ogawa-Ohnishi M, Matsubayashi Y. A glycopeptide regulating stem cell fate in Arabidopsis thaliana. Nat Chem Biol. 2009;5(8):578–580. doi: 10.1038/nchembio.182. [DOI] [PubMed] [Google Scholar]
- 50.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37(Web Server issue):W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Higo K, Ugawa Y, Iwamoto M, Korenaga T. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 1999;27(1):297–300. doi: 10.1093/nar/27.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Higo K, Ugawa Y, Iwamoto M, Higo H. PLACE: a database of plant cis-acting regulatory DNA elements. Nucleic Acids Res. 1998;26(1):358–359. doi: 10.1093/nar/26.1.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Nucleic acid sequences and amino acid sequences of all 15 tomato CLE gene family members. (XLS 40 KB)
Additional file 2: Multiple alignments of the full-length amino acid sequences of SlCLE gene family members using DNAMAN alignment program. Black and light gray shading indicates identical and conversed amino acid residues, respectively. (TIFF 140 KB)
Additional file 3: Locations of SlCLE genes on the tomato chromosomes. Physical distances are in megabases (Mb) and gene transcription orientations are marked by solid arrows. (DOCX 34 KB)
Additional file 4: Homology, similarity and identity of SlCLE family members over their full-length sequences. (DOC 110 KB)
Additional file 5: Schematic illustrations of the types and distributions of motifs in tomato and Arabidopsis CLE family. The motif characterization was based on full-length proteins by MEME web server. (XLS 1 MB)
Additional file 6: The variety and numbers of cis-acting elements in the 2 kb regions upstream of SlCLE genes searched in the PLACE database (http://www.dna.affrc.go.jp/PLACE/signalscan.html). (XLS 132 KB)
Additional file 7: Tissue-specific transcription levels of 13 SlCLEs using qRT-PCR method. The expression levels of individual SlCLE gene among various tissues/organs as well as multiple SlCLE genes within one individual tissue/organ were presented. Relative gene expression levels are normalized with actin transcript values. The RT-qPCR analyses were repeated at least three times in three independent experiments. Total RNA isolated from apex (A), root (R), young, medium and old stems (YS, MS, and OS), young and mature leaf (YL and ML), flower (FL), flower buds (FB), and 1-cm, 2-cm and 3-cm sized green fruits (F1, F2 and F3), mature green fruit (MG), fruit at breaker stage (BF), fruit turning red (TF) and mature fruit in red (RF). (DOC 86 KB)
Additional file 8: Normalized expression of tomato SlCLE genes based on RNA-seq data downloaded from previously published data [46]. Based on the data, the heat map was generated using the MeV4.9.0 software (http://www.tm4.org/) and gene-wise normalized and hierarchical clustered based on Pearson correlation. Color scale at the top of each dendrogram represents fold change values. Bud: unopened flower buds; flower: fully opened flowers; MG fruit: mature green fruit; B_fruit: fruits at breaker; B + 10_fruit: fruits at breaker + 10 days stage. (ZIP 15 KB)
Additional file 9: qRT-PCR primers used in the current study. (XLS 27 KB)


