Abstract Abstract
In order to resolve better the deep relationships among salticid spiders, we compiled and analyzed a molecular dataset of 169 salticid taxa (and 7 outgroups) and 8 gene regions. This dataset adds many new taxa to previous analyses, especially among the non-salticoid salticids, as well as two new genes – wingless and myosin heavy chain. Both of these genes, and especially the better sampled wingless, confirm many of the relationships indicated by other genes. The cocalodines are placed as sister to lapsiines, in a broader clade with the spartaeines. Cocalodines, lapsiines, and spartaeines are each supported as monophyletic, though the first two have no known morphological synapomorphies. The lyssomanines appear to be non-monophyletic, of three separate groups: (1) Lyssomanes plus Chinoscopus, (2) Onomastus, and (3) the remainder of Old World species. Several previously-inferred relationships continue to be supported: hisponines as sister to the Salticoida, Amycoida as sister to the remaining Salticoida, and Saltafresia as monophyletic. The relationship of Salticus with Philaeus and relatives is now considered well enough corroborated to move the latter into the subfamily Salticinae. A new clade consisting of the Plexippoida + Aelurilloida + Leptorchesteae + Salticinae is recognized. Nungia is found to be an astioid, and Echeclus, Gedea and Diplocanthopoda to be hasariines. The euophryines are corroborated as monophyletic. The agoriines Agorius and Synagelides are salticoids, within the sister group to amycoids, but their further placement is problematical, perhaps because of their nuclear ribosomal genes’ high GC bias, as also seen in the similarly problematic Eupoa.
Keywords: Jumping spiders, Salticidae, phylogeny, systematics
Introduction
Salticid spiders, remarkable for their excellent vision (Land 1969, Blest et al. 1990), include more than 5000 species (Platnick 2014) with a great diversity of body forms and behaviours. While this diversity has long resisted phylogenetic organization, recent molecular studies (Maddison and Hedin 2003, Su et al. 2007, Maddison et al. 2008, Bodner and Maddison 2012, Zhang and Maddison 2013), aided by compilations of morphological taxonomic knowledge (Prószyński 2013) have resolved much of the phylogenetic structure of the family. One of the best-supported clades is the Salticoida, recognized by both morphological and molecular data (Maddison 1996, Maddison and Hedin 2003) and containing about 95% of the known species in the family. Within the Salticoida, large groups such as the Amycoida, Astioida, Marpissoida and Plexippoida are well-corroborated (Maddison and Hedin 2003, Maddison et al. 2008). However, many of the deeper relationships of salticoids have been poorly resolved. Outside the Salticoida are the spartaeines, lyssomanines, and hisponines, showing ancestral features like limited tracheal systems, complex palpi, and the retention of a tarsal claw on the female palp. These non-salticoids (often called “basal salticids”) have been studied phylogenetically (Su et al. 2007), but with limited taxon sampling.
In this work we attempt to resolve more firmly the basic structure of the family by increasing the taxon sampling, especially among non-salticoid salticids, and by using additional genes. Two of the genes, wingless and myosin heavy chain, are new to salticid molecular phylogenetics. By building a dataset that has a greater number of genes among selected species, we hoped to obtain a phylogenetic resolution with stronger confidence.
Methods
Taxon sampling
Taxa included in the analysis are 169 species of salticids and representatives of four dionychan families as outgroups (Table 1, Suppl. material 1). Based on previous phylogenetic work (Maddison and Hedin 2003, Maddison et al. 2008, Bodner and Maddison 2012, Zhang and Maddison 2013, in press), about 70 species of salticids from the major clade Salticoida were selected because they would represent most known major lineages, and because several genes are available for each (Table 1, Suppl. material 1). In addition, a few salticoids were added because their placement was unclear: Agorius, Diplocanthopoda, Echeclus, Gedea, Nungia, Phaulostylus, and Synagelides.
Table 1.
Specimens and sequences used in phylogenetic analyses, with GenBank numbers indicated. * marks previously published sequences. Specimen localities given in Suppl. material 1.
| Reference | 28s | 18s | wingless | myosin HC | actin 5c | histone 3 | CO1 | 16sND1 | |
|---|---|---|---|---|---|---|---|---|---|
| Outgroups | |||||||||
| Anyphaenidae: Hibana sp. | s318 | AY297295* | KM033091 | KM032961 | KM032929 | AY297422* | AY297295 / AY297358* | ||
| Gnaphosidae: Cesonia sp. | s319 | AY297293; EF201663* | KM032996 | EU522700* | DQ665720* | AY297420* | AY296711 / AY297356* | ||
| Miturgidae: Cheiracanthium sp. | s321 | AY297294; EF201664* | KM032997 | KM032928 | AY297421* | AY296712 / AY297357* | |||
| Oxyopidae: Oxyopes birmanicus Thorell, 1887 | Su et al. 2007 | EF419032 / EF419065* | EF418998* | EF419126* | EF419097* | EF418969 / EF419150* | |||
| Philodromidae: Philodromus alascensis Keyserling, 1884 | GR011 | KM033130 | KM033092 | KM032998 | KM032962 | ||||
| Thomisidae: Misumenops nepenthicola (Pocock, 1898) | Su et al. 2007 | EF419029 / EF419062* | EF418996* | EF419123* | EF419094* | EF418967 / EF419148* | |||
| Thomisidae: Xysticus sp. | s316 | AY297296; EF201665* | KM033093 | EU522701* | DQ665704* | AY297296* | AY296714 / AY297359* | ||
| Lyssomanines | |||||||||
| Asemonea sichuanensis Song & Chai, 1992 | SC-03-0055 | EF418986* | EF419082* | ||||||
| Asemonea sichuanensis Song & Chai, 1992 | MRB084 | KM033131 | KM032931 | ||||||
| Asemonea cf. stella Wanless, 1980 | MRB083 | JX145767* | KM033094 | KM032930 | JX145686* | ||||
| Asemonea tenuipes (O. P.-Cambridge, 1869) | d186 | KM033132 | KM033095 | KM032999 | KM032963 | KM032932 | |||
| Chinoscopus cf. flavus (Peckham, Peckham & Wheeler, 1889) | d273 | KM033133 | KM033096 | KM032888 | |||||
| Goleba lyra Maddison & Zhang, 2006 | d051 | DQ665768* | KM033097 | KM033000 | EU522709* | DQ665707* | DQ665755* | ||
| Lyssomanes amazonicus Peckham & Wheeler, 1889 | ECU11-6112 | KM033134 | KM032889 | ||||||
| Lyssomanes antillanus Peckham & Wheeler, 1889 | d298 | KM033135 | KM033001 | ||||||
| Lyssomanes cf. benderi Logunov, 2002 | ECU11-5402 | KM033136 | KM032890 | ||||||
| Lyssomanes cf. jemineus Peckham & Wheeler, 1889 | ECU11-5682 | KM033137 | KM032891 | ||||||
| Lyssomanes longipes (Taczanowski, 1871) | MRB086 | KM033138 | KM032933 | KM033208 | KM032892 | ||||
| Lyssomanes pauper Mello-Leitão, 1945 | d297 | KM033139 | KM033002 | ||||||
| Lyssomanes taczanowskii Galiano, 1980 | ECU11-4193 | KM033141 | KM032894 | ||||||
| Lyssomanes tenuis Peckham & Wheeler, 1889 | ECU11-4869 | KM033142 | KM032895 | ||||||
| Lyssomanes viridis (Walckenaer, 1837) | s160 | AY297231* | AY297360* | AY296652 / AY297297* | |||||
| Lyssomanes viridis (Walckenaer, 1837) | d129 | KM033098 | KM033003 | EU522715* | DQ665715* | ||||
| Lyssomanes sp. [Esmeraldas] | d408 | KM033140 | KM032893 | ||||||
| Onomastus nigrimaculatus Zhang & Li, 2005 | Su et al. 2007 | EF419031 / EF419064* | EF418997* | EF419125* | EF419096* | EF418968 / EF419149* | |||
| Onomastus sp. [Guangxi] | MRB085 | JX145768* | KM033099 | KM033004 | KM032964 | KM032934 | JX145687* | JX145910* | |
| Pandisus cf. decorus Wanless, 1980 | d303 | KM033143 | KM033005 | ||||||
| Cocalodines | |||||||||
| Allococalodes madidus Maddison, 2009 | d236 | KM033144 | KM033006 | KM032896 | |||||
| Cocalodes longicornis Wanless, 1982 | d291 | KM033145 | KM033007 | KM032935 | KM032897 | ||||
| Cocalodes macellus (Thorell, 1878) | d230 | KM033146 | KM033100 | KM033008 | KM032936 | KM033209 | |||
| Cucudeta gahavisuka Maddison, 2009 | d234 | KM033147 | KM033009 | KM032898 | |||||
| Cucudeta zabkai Maddison, 2009 | d235 | KM033148 | KM033010 | KM032965 | KM032899 | ||||
| Tabuina aff. baiteta Maddison, 2009 | d313 | KM033149 | KM033011 | ||||||
| Tabuina rufa Maddison, 2009 | d232 | KM033151 | KM033013 | KM032900 | |||||
| Tabuina aff. rufa Maddison, 2009 | d312 | KM033150 | KM033012 | ||||||
| Tabuina varirata Maddison, 2009 | d233 | KM033152 | KM033014 | KM032901 | |||||
| Yamangalea frewana Maddison, 2009 | d231 | KM033153 | KM033015 | KM032902 | |||||
| Spartaeines | |||||||||
| Brettus cf. adonis Simon, 1900 | SWK12-4323 | KM033154 | |||||||
| Brettus sp. [Yunnan] | LiD-026-053-05 | KM033155S | KM033101S | KM033195S | |||||
| cf. Phaeacius sp. [Sarawak] | SWK12-3728 | KM033156 | |||||||
| Cocalus murinus Simon, 1899 | LiD-013-027-05 | EF419019 / EF419053* | EF418988* | EF419116* | EF419084* | EF418959 / EF419140* | |||
| Cyrba algerina (Lucas, 1846) | Su et al. 2007 | EF419021 / EF419054* | EF418989* | EF419086* | EF418961 / EF419142* | ||||
| Cyrba lineata Wanless, 1984 | MRB106 | JX145792* | KM033016 | KM032966 | KM032937 | JX145704* | |||
| Cyrba ocellata (Kroneberg, 1875) | Su et al. 2007 | EF418990* | EF419087* | EF418962 / EF419143* | |||||
| Cyrba ocellata (Kroneberg, 1875) | MRB104 | KM033157 | |||||||
| Cyrba sp. [Kenya] | Su et al. 2007 | EF419023 / EF419056* | EF418991* | EF419088* | |||||
| Gelotia cf. bimaculata Thorell, 1890 | d250 | KM033158 | KM033017 | KM032938 | |||||
| Gelotia syringopalpis Wanless, 1984 | Su et al. 2007 | EF419024 / EF419057* | EF419118* | ||||||
| Gelotia syringopalpis Wanless, 1984 | MRB105 | KM033019 | KM033212 | KM032903 | |||||
| Gelotia sp. [Guangxi] | MRB199 | KM033018 | KM032939 | KM033210 | |||||
| Gelotia sp. [Yunnan] | LiD002-053-05 | KM033102S | KM033196S | KM033211S | |||||
| Holcolaetis vellerea Simon, 1910 | Su et al. 2007 | EF419025 / EF419058* | EF418992* | EF419119* | EF419090* | EF418963 / EF419144* | |||
| Holcolaetis cf. zuluensis Lawrence, 1937 | d036 | DQ665770* | KM033103 | EU522711* | DQ665721* | DQ665757* | |||
| Meleon aff. kenti (Lessert, 1925) | d287 | KM033159 | KM032940 | ||||||
| Mintonia mackiei Wanless, 1984 | SWK12-4202 | KM033161 | |||||||
| Mintonia cf. melinauensis Wanless, 1984 | d441 | KM033160 | |||||||
| Mintonia ramipalpis (Thorell, 1890) | SWK12-1442 | KM033162 | |||||||
| Mintonia silvicola Wanless, 1987 | d104 | KM033020 | KM032904 | ||||||
| Mintonia silvicola Wanless, 1987 | SWK12-1653 | KM033163 | |||||||
| Mintonia silvicola Wanless, 1987 | Su et al. 2007 | EF418995* | EF419122* | EF419093* | |||||
| Mintonia tauricornis Wanless, 1984 | d249 | KM033164 | KM033021 | KM032941 | KM032905 | ||||
| Neobrettus tibialis (Prószyński, 1978) | LiD-001-055-05 | EF419030 / EF419063* | EF419124* | EF419095* | |||||
| Neobrettus sp. [Sarawak] | SWK12-1040 | KM033165 | |||||||
| Paracyrba wanlessi Zabka & Kovac, 1996 | Su et al. 2007 | EF419033 / EF419066* | EF418999* | EF419098* | |||||
| Phaeacius lancearius (Thorell, 1895) | d111 | DQ665775* | KM033022 | DQ665759* | |||||
| Phaeacius malayensis Wanless, 1981 | Su et al. 2007 | EF419034 / EF419067* | EF419000* | EF419099* | EF418970 / EF419151* | ||||
| Phaeacius sp. [Guangxi] | LQ-24-06 | KM033166S | KM033104S | KM033213S | KM032906S | ||||
| Phaeacius sp. [Hainan] | Su et al. 2007 | EF419035 / EF419068* | EF419001* | EF418971 / EF419152* | |||||
| Phaeacius sp. [Sarawak] | SWK12-4541 | KM033167 | |||||||
| Portia africana (Simon, 1886) | Su et al. 2007 | EF419037 / EF419069* | EF419003* | EF419128* | EF419101* | ||||
| Portia crassipalpis (Peckham & Peckham, 1907) | SWK12-2354 | KM033168 | |||||||
| Portia fimbriata (Doleschall, 1859) | LiD-001-04 | EF419038 / EF419070* | EF419004* | EF419129* | EF419102* | EF418973 / EF419154* | |||
| Portia heteroidea Xie & Yin, 1991 | Su et al. 2007 | EF419039 / EF419071* | EF419005* | EF419130* | EF419103* | EF418974 / EF419155* | |||
| Portia jianfeng Song & Zhu, 1998 | Su et al. 2007 | EF419040 / EF419072* | EF419006* | EF419104* | EF418975 / EF419156* | ||||
| Portia labiata (Thorell, 1887) | S206 | AY297232* | AY297361* | AY296653 / AY297298* | |||||
| Portia cf. schultzi Karsch, 1878 | d131 | DQ665776* | KM033105 | KM033023 | KM032967 | EU522718* | DQ665708* | ||
| Portia quei Zabka, 1985 | Su et al. 2007 | EF419042 / EF419074* | EF419008* | EF419132* | EF419106* | EF418977 / EF419158* | |||
| Portia taiwanica Zhang & Li, 2005 | MRB103 | KM033169 | KM032942 | KM033214 | KM032907 | ||||
| Portia sp. [Sichuan] | SC-03-0011 | EF419043 / EF419075* | EF419009* | EF419133* | EF418978 / EF419159* | ||||
| Sonoita lightfooti Peckham & Peckham, 1903 | d226 | KM033170 | KM033215 | ||||||
| Sonoita aff. lightfooti Peckham & Peckham, 1903 | MRB200 | JX145791* | JX145705* | JX145927* | |||||
| Sparbambus gombakensis Zhang, Woon & Li, 2006 | d251 | KM033171 | KM033024 | KM032943 | |||||
| Spartaeus jianfengensis Song & Chai, 1991 | Su et al. 2007 | EF419045 / EF419076* | EF419011* | EF419109* | EF418980 / EF419161* | ||||
| Spartaeus platnicki Song, Chen & Gong, 1991 | SC-03-069 | EF419046 / EF419077* | EF419012* | EF419135* | EF419110* | EF418981 / EF419162* | |||
| Spartaeus spinimanus (Thorell, 1878) | S199 | KM033216 | KM032908 | ||||||
| Spartaeus thailandicus Wanless, 1984 | BV-004 | EF419047 / EF419078* | EF419013* | EF419136* | EF419111* | EF418982 / EF419163* | |||
| Spartaeus uplandicus Barrion & Litsinger, 1995 | S185/S186 | AY297233* | AY297363* | AY296655* | |||||
| Spartaeus wildtrackii Wanless, 1987 | Su et al. 2007 | EF419048 / EF419079* | EF419014* | EF419137* | EF419112* | EF418983 / EF419164* | |||
| Taraxella sp. [Johor] | d246 | KM033172 | KM032944 | KM032909 | |||||
| Taraxella sp. [Pahang] | d248 | KM033173 | KM032945 | KM033197 | |||||
| Taraxella sp. [Pahang] | LiD-001-003-06 | KM033106S | KM033217S | KM032910S | |||||
| Yaginumanis wanlessi Zhang & Li, 2005 | Su et al. 2007 | EF419050 / EF419081* | EF419016* | EF419139* | EF419114* | EF418985 / EF419166* | |||
| Lapsiines | |||||||||
| Galianora bryicola Maddison, 2006 | d124 | DQ665771* | DQ665741* | KM033025 | EU522706* | DQ665717* | DQ665758* | DQ665727* | |
| Galianora sacha Maddison, 2006 | d116 | DQ665766* | DQ665734* | KM033026 | KM032968 | EU522707* | DQ665716* | DQ665754* | |
| Lapsias canandea Maddison, 2012 | d442 | KM033174 | |||||||
| Lapsias guamani Maddison, 2012 | UBC-SEM AR00191 | KM033175 | KM033027 | ||||||
| Lapsias lorax Maddison, 2012 | UBC-SEM AR00194 | KM033176 | KM033028 | ||||||
| Soesiladeepakius lyra Ruiz & Maddison, 2012 | GR130 | JQ312077 | KM033029 | JQ312074* | JQ312079* | ||||
| Thrandina bellavista Maddison, 2012 | d396 | KM033177 | KM033030 | ||||||
| Thrandina cosanga Maddison, 2012 | d395 | KM033178 | |||||||
| Thrandina parocula Maddison, 2006 | d123 | DQ665779* | KM033107 | EU522720* | DQ665718* | DQ665761* | DQ665726* | ||
| Thrandina parocula Maddison, 2006 | d394 | KM033031 | KM032969 | ||||||
| Eupoa | |||||||||
| Eupoa nezha Maddison & Zhang, 2007 | d220/MRB102 | EF201648* | EF201666* | KM033032 | EF201668* | EF201667* | |||
| Hisponines | |||||||||
| cf. Tomocyrba sp. [Madagascar] | d305 | KM032881* | |||||||
| Hispo macfarlanei Wanless, 1981 | d404 | KM032882* | KM032970 | ||||||
| Hispo sp. [Madagascar] | d309 | KM032883* | |||||||
| Jerzego cf. alboguttatus Simon, 1903 | SWK12-4787 | KM032884* | |||||||
| Jerzego corticicola Maddison, 2014 | SWK12-2900 | KM032885* | KM032887* | ||||||
| Massagris contortuplicata Wesolowska & Haddad, 2013 | d082 | DQ665772* | KM033108 | KM033033 | DQ665705* | DQ665722* | |||
| Massagris schisma Maddison & Zhang, 2006 | d081 | DQ665762* | KM033109 | KM033034 | DQ665728* | ||||
| Tomobella andasibe (Maddison & Zhang, 2006) | d127 | DQ665780* | DQ665752* | KM033035 | KM033198 | DQ665725* | |||
| Tomocyrba sp. [Madagascar] | d306 | KM032886* | |||||||
| Tomomingi sp. [Gabon] | MRB243 | JX145764* | KM033110 | KM033036 | KM032971 | JX145850* | JX145684* | ||
| Salticoida | |||||||||
| Agoriines | |||||||||
| Agorius constrictus Simon, 1901 | d172 | KM032953 | |||||||
| Agorius constrictus Simon, 1901 | d213 | KM033119 | KM033072 | KM032921 | |||||
| Agorius sp. [Selangor] | d299 | KM033189 | KM033073 | ||||||
| Synagelides cf. lushanensis Xie & Yin, 1990 | d214 | KM033074 | |||||||
| Synagelides cf. palpalis Zabka, 1985 | MRB050 | KM032922 | |||||||
| Synagelides cf. palpalis Zabka, 1985 | d225 | KM033190 | KM033226 | ||||||
| Amycoids | |||||||||
| Cotinusa sp. [Ecuador] | MRB024 | JX145746* | KM033120 | KM033075 | KM032987 | JX145832* | JX145671* | JX145896* | |
| Hurius vulpinus Simon, 1901 | S213 | AY297239* | AY297368* | AY296662 / AY297306* | |||||
| Hurius cf. vulpinus Simon, 1901 | d156 | KM033076 | EU522712* | KM033203 | |||||
| Hypaeus aff. miles Simon, 1900 [Ecuador] | d130 | EU815499* | KM033121 | KM033077 | KM032988 | EU522702* | KM032923 | ||
| Sarinda cutleri (Richman, 1965) | MRB193 | JX145744* | KM033078 | KM032954 | JX145669* | JX145895* | |||
| Sitticus floricola palustris (Peckham & Peckham, 1883) | d030 | DQ665778* | KM033122 | KM033079 | KM032989 | KM033204 | DQ665760* | DQ665729* | |
| Astioids | |||||||||
| Arasia mollicoma (L. Koch, 1880) | d046 | EU815483* | EU815532* | KM032990 | JX145834* | KM033205 | EU815598* | EU815550* | |
| Helpis minitabunda (L. Koch, 1880) | d265 | KM033123 | KM033080 | KM032991 | KM032955 | KM033227 | |||
| Ligurra latidens (Doleschall, 1859) | d175 | JX145749* | KM033081 | JX145835* | JX145898* | ||||
| Ligurra latidens (Doleschall, 1859) | LiD-001-027-05 | EF418993* | EF419120* | EF419091* | |||||
| Mopsus mormon Karsch, 1878 | d018 | EU815470* | EU815529* | KM033082 | JX145836* | KM033206 | EU815586* | ||
| Myrmarachne sp. [Pahang] | d162 | EU815507* | KM033124 | KM033083 | KM032992 | JX145837* | EU815616* | EU815565* | |
| Neon reticulatus (Blackwall, 1853) | d283 | KM033191 | KM033125 | KM033084 | KM032993 | KM032956 | |||
| Nungia epigynalis Zabka, 1985 | d221 | KM033192 | KM032924 | ||||||
| Simaetha sp. | d027 | EU815477* | KM033126 | KM033085 | JX145839* | EU815592* | EU815546* | ||
| Trite pennata Simon, 1885 | d035 | EU815478* | KM033086 | KM032957 | KM033207 | EU815593* | EU815547* | ||
| Baviines | |||||||||
| Bavia aff. aericeps Simon, 1877 [Sabah] | d079 | EU815490* | KM033127 | KM032958 | EU815603* | KM032925 | |||
| Stagetilus sp. [Selangor] | MRB079 | KM033193 | KM033087 | KM032959 | KM032926 | ||||
| Marpissoids | |||||||||
| Afromarengo sp. [Gabon] | MRB262 | JX145758* | KM033128 | KM033088 | KM032994 | JX145842* | JX145682* | JX145905* | |
| Dendryphantes hastatus (Clerck, 1757) | d043 | EF201646* | KM033129 | KM033089 | KM033228 | KM032927 | |||
| Platycryptus californicus (Peckham & Peckham, 1888) | d316 | KM033194 | KM033090 | KM032995 | KM032960 | KM033229 | |||
| Rhene sp. [Pahang] | LiD-001-021-05 | EF419044* | EF419010* | EF419134* | EF419108* | EF418979 / EF419160* | |||
| Tisaniba mulu Zhang & Maddison, 2014 | SWK12-1244 | KM032876* | KM032880* | ||||||
| Saltafresians | |||||||||
| Aelurillus cf. ater (Kroneberg, 1875) | d140 | EU815504* | EU815536* | KM033037 | KM032972 | JX145831* | KM033199 | EU815615* | EU815564* |
| Amphidraus complexus Zhang & Maddison, 2012 | JXZ035 | KC615380* | KM033038 | KC616069* | KC615640* | KC615806* | |||
| Athamas cf. whitmeei O. P.-Cambridge, 1877 | JXZ345 | KC616286* | KC615649* | KC615822* | |||||
| Bacelarella pavida Szüts & Jocqué, 2001 | d195 | EU815511* | EU815538* | KM033039 | KM032973 | KM032946 | EU815618* | EU815569* | |
| Bathippus macrognathus (Thorell, 1881) | JXZ372 | KC615407* | KM033040 | KC616305* | KC615835* | ||||
| Bianor maculatus (Keyserling, 1883) | d017 | EU815469* | KM033041 | KM033200 | EU815585* | EU815542* | |||
| Bristowia afra Szüts, 2004 | JXZ363 | KC615409* | KC616301* | ||||||
| Bristowia afra Szüts, 2004 | MRB230 | KM033042 | KM033218 | ||||||
| Cheliceroides longipalpis Zabka, 1985 | d222 | KM033111 | KM033043 | JX145830* | KM033219 | EU815579* | |||
| Cheliceroides cf. longipalpis Zabka, 1985 | d415 | KM033179 | |||||||
| Chinattus parvulus (Banks, 1895) | d009 | EU815464* | EU815525* | KM033044 | JX145848* | KM033201 | EU815581* | ||
| Chinophrys pengi Zhang & Maddison, 2012 | JXZ145 | KC615416* | KM033045 | KC616146* | KC615843* | ||||
| Corythalia locuples (Simon, 1888) | JXZ315 | KC615390* | KM033046 | KC616260* | KC615645* | KC615816* | |||
| Cosmophasis umbratica Simon, 1903 | Su et al. 2007 | EF419020* | EF419117* | EF419085* | EF418960 / EF419141* | ||||
| Cytaea nimbata (Thorell, 1881) | JXZ229 | KC615474* | KM033047 | KC616197* | KC615693* | KC615899* | |||
| Diolenius varicus Gardzińska & Zabka, 2006 | JXZ349 | KC615480* | KM033048 | KC616290* | KC615695* | KC615905* | |||
| Diplocanthopoda marina Abraham, 1925 | d209 | KM033180 | KM032947 | KM033220 | KM032911 | ||||
| Eburneana sp. [Gabon] | MRB231 | KM033181 | KM033049 | JX145858* | KM033221 | KM032912 | |||
| Echeclus sp. [Selangor] | MRB089 | KM033182 | KM032948 | KM033222 | KM032913 | ||||
| Euophrys frontalis (Walckenaer, 1802) | JXZ137 | KC615536* | KM033050 | KC616139* | KC615960* | ||||
| Evarcha proszynskii Marusik & Logunov, 1998 | d096 | DQ665765* | KM033112 | EU522704* | DQ665723* | ||||
| Evarcha proszynskii Marusik & Logunov, 1998 | d323 | KM033051 | KM032974 | ||||||
| Freya decorata (C. L. Koch, 1846) | d211 | EU815521* | EU815539* | KM032975 | EU522705* | JX145908* | |||
| Gedea cf. tibialis Zabka, 1985 | MRB090 | KM033183 | KM032949 | KM033223 | KM032914 | ||||
| Habrocestum cf. albimanum Simon, 1901 | d132 | EU815500* | EU815611* | EU815562* | |||||
| Habronattus borealis (Banks, 1895) | d207 | KM033184 | KM033052 | KM032976 | KM032950 | KM033224 | KM032915 | ||
| Hasarius adansoni (Audouin, 1826) | d295 | KM033113 | KM033053 | KM032977 | |||||
| Hasarius adansoni (Audouin, 1826) | S130/S131/S324 | AY297281* | AY297409* | ||||||
| Heliophanus cupreus (Walckenaer, 1802) | d044 | DQ665769* | KM033114 | EU522710* | DQ665710* | DQ665756* | KM032916 | ||
| Idastrandia cf. orientalis (Szombathy, 1915) | d108 | EU815535; EU815496* | EU815535* | JX145852* | EU815608* | EU815560* | |||
| Langerra aff. longicymbium Song & Chai, 1991 | d182 | KM033185 | KM033054 | KM032917 | |||||
| Leptorchestes berolinensis (C. L. Koch, 1846) | d086 | EU815491* | EU815534* | KM033055 | EU815604* | EU815556* | |||
| Longarenus brachycephalus Simon, 1903 | MRB258 | JX145798* | KM033056 | KM032978 | KM032951 | JX145707* | KM032918 | ||
| Nannenus sp. [Pahang] | d105 | EU815493* | KM033057 | KM032979 | JX145853* | EU815558* | |||
| Naphrys pulex (Hentz, 1846) | JXZ081 | JX145760* | KM033115 | KM032980 | JX145844* | KC615749* | JX145907* | ||
| Omoedus orbiculatus (Keyserling, 1881) | d008 | KC615792* | |||||||
| Omoedus orbiculatus (Keyserling, 1881) | JXZ136 | JX145762* | KM033116 | KM033058 | JX145846* | KM033202 | |||
| Omoedus papuanus Zhang & Maddison, 2012 | JXZ286 | KC615619* | KM033059 | KC616234* | KC615790* | KC616042* | |||
| Pellenes peninsularis Emerton, 1925 | d057 | DQ665774* | KM033117 | KM033060 | JX145864* | DQ665712* | |||
| Pellenes peninsularis Emerton, 1925 | d400 | KM032981 | |||||||
| Phaulostylus grammicus Simon, 1902 | d304 | KM033186 | KM033061 | ||||||
| Philaeus chrysops (Poda, 1761) | d025 | EU815475* | EU815530* | KM033062 | JX145855* | EU815590* | EU815545* | ||
| Phintella sp. [Gabon] | d402 | KM033187 | KM033063 | KM032982 | |||||
| Plexippus paykulli (Audouin, 1826) | LiD-001-029-05 | EF419002* | EF419127* | ||||||
| Plexippus paykulli (Audouin, 1826) | MRB016 | JX145784* | KM033064 | EU522713* | |||||
| Plexippus paykulli (Audouin, 1826) | S73 | AY297384* | AY296674 / AY297317* | ||||||
| Pochyta cf. pannosa Simon, 1903 | MRB257 | JX145806* | KM033065 | KM032983 | KM032952 | JX145715* | KM032919 | ||
| Saitis barbipes (Simon, 1868) | JXZ147 | KC615589* | KM033066 | KC616147* | KC615767* | KC616011* | |||
| Salticus scenicus (Clerck, 1757) | d003 | DQ665777* | KM033118 | KM033067 | KM032984 | EU522719* | DQ665713* | JX145663* | AY296707 / AY297352* |
| Thiania bhamoensis Thorell, 1887 | LiD-001-028-05 | EF419049 / EF419080* | EF419015* | EF419138* | EF419113* | EF418984 / EF419165* | |||
| Trydarssus cf. nobilitatus (Nicolet, 1849) | MRB270 | KM033188 | KM033068 | KM032985 | JX145847* | KM033225 | KM032920 | ||
| Tusitala lyrata (Simon, 1903) | MRB226 | JX145771* | KM033069 | JX145856* | JX145689* | JX145912* | |||
| Yllenus arenarius Menge, 1868 | d013 | EU815527* | EU815583* | EU815541* | |||||
| Yllenus arenarius Menge, 1868 | JXZ173 | JX145766* | KM033070 | KM032986 | JX145851* | ||||
| Zabkattus furcatus Zhang & Maddison, 2012 | JXZ218 | KC615503* | KM033071 | KC616190* | KC615928* | ||||
Our sample targeted especially the non-salticoid salticids, those that lie outside the major clade of familiar salticids (Maddison and Needham 2006). We included most available data from non-salticoid salticids, both new data and data previously published by Su et al. (2007) and others (Maddison and Hedin 2003, Maddison and Needham 2006, Maddison et al. 2007, Bodner and Maddison 2012, Ruiz and Maddison 2012, Zhang and Maddison 2013, Maddison and Piascik, in press). Included for the first time in a molecular phylogeny are the cocalodines (Wanless 1982, Maddison 2009), which are Australasian non-salticoid salticids. Also analyzed for the first time are the lyssomanine genera Chinoscopus and Pandisus, the lapsiine Lapsias, and the spartaeines Brettus, Meleon, Sparbambus, and Taraxella.
Some previously-published data from non-salticoid salticids was either excluded or represented under a different species name here. Excluded are sequences of Hispo cf. frenata, because its limited data made it unstable in the analyses (see Maddison and Piascik 2014), “Portia labiata” from Su et al. (2007), because its identification is in doubt and no voucher specimen is available, and the actin 5C sequence of Tomomingi sp. voucher d243, which we discovered to have been a contaminant from the euophryine Ilargus. The species labeled as Phaeacius yixin by Su et al. (2007) is included here as “Phaeacius sp. [Hainan]”, because the specimen was a juvenile female and thus identified with doubt; by its DNA we suspect it is Phaeacius lancearius. The specimen labeled as Mintonia ramipalpis by Su et al. (2007) is actually a female Mintonia silvicola. This misidentification arose because of an error in male-female matching by Wanless (1984), whose female “Mintonia ramipalpis” is actually the female of Mintonia silvicola. The correct match of male and female Mintonia silvicola is evident by intimate co-collecting in a recent expedition to Sarawak (Maddison and Piascik, unpublished) and in DNA sequence comparison. We have therefore blended data from Su et al.’s female with that from our males to represent Mintonia silvicola.
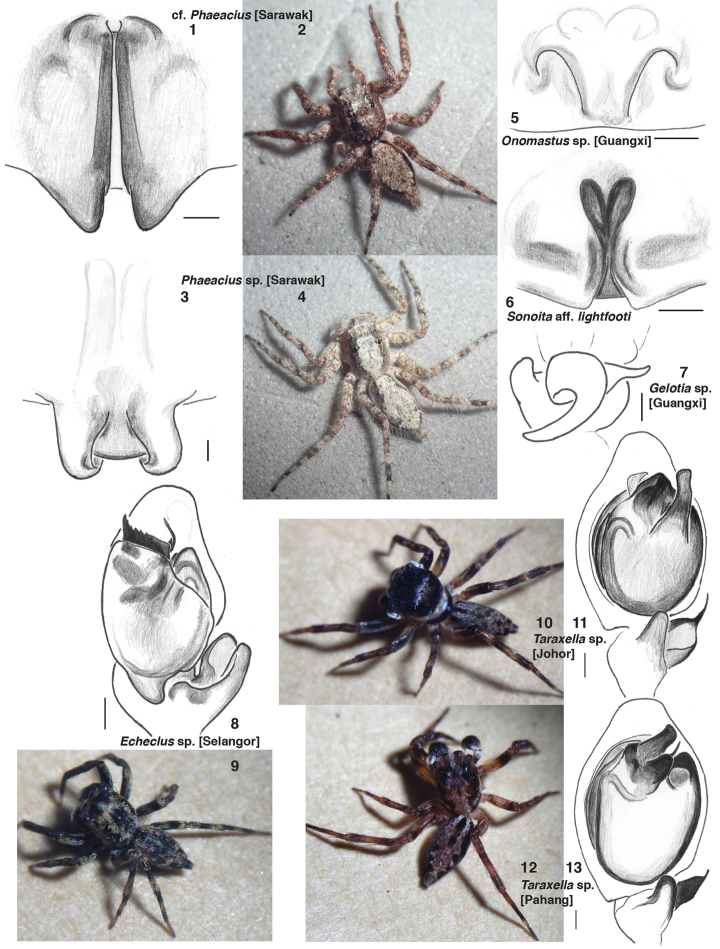
Some of the species studied appear to be undescribed, or are doubtfully the same as described species. Following the usual convention, the names of some of our specimens includes “cf.” to indicate that they may be the same as the mentioned species, “aff.” to indicate that they are close to, but distinctly different from, the mentioned species. Figures 1–13 give illustrations of some of the undescribed species, in order to facilitate future association of our data with a species name. The species we refer to as “cf. Phaeacius [Sarawak]” (Figs 1, 2) is known from a single female and juvenile from Lambir Hills, Sarawak. It resembles Phaeacius but the legs are shorter, and the epigynum is distinctively different. Phaeacius sp. [Sarawak] (Figs 3, 4) is a fairly typical Phaeacius whose epigynum resembles that of Phaeacius leytensis Wijesinghe, 1991, but with the atria elongated posteriorly. Onomastus sp. [Guangxi] is shown in Fig. 5. Sonoita aff. lightfooti (Fig. 6) has longer grooves for the openings of the epigynum than Sonoita lightfooti, and is distinctive in gene sequences as well. Gelotia sp. [Guangxi] (Fig. 7) has a palp resembling Gelotia syringopalpis, but the tibial apophyses are much shorter. Echeclus sp. [Selangor] (Figs 8, 9) was identified as an Echeclus by the distinctive form of the palp tibia, and the embolus hidden behind a ledge of the tegulum, through which several dark sclerites can be seen (Prószyński 1987). It might equally well have been identified, by the same features, as a Curubis species (Zabka 1988). Indeed, the two genera are likely synonyms. “Echeclus” is used as that is the older name. Taraxella sp. [Johor] (Figs 10, 11) and Taraxella sp. [Pahang] (Figs 12, 13) are typical species of Taraxella. The specimen MRB024 identified as Cotinusa sp. is the same as that named “unidentified thiodinine” by Bodner and Maddison (2012). The Hypaeus specimen (d130) was formerly identified as Acragas sp. (Bodner and Maddison 2012). The specimen d105 labeled as “Nannenus lyriger” by Maddison et al. (2008) is not Nannenus lyriger, but another apparently undescribed species of Nannenus. The data for Cheliceroides longipalpis comes from two specimens, d222 which is clearly Cheliceroides longipalpis, and d415 which may be a different but very closely related species. Notes on the undescribed hisponines are given by Maddison and Piascik (2014), whose data we use.
Figures 1–13.
Specimens of undescribed species. 1, 3, 5, 6 are of epigyna, ventral view; 8, 11, 13 of left palps, ventral view; 7 of the right palp tibia, retrolateral view. Scale bar 0.1 mm. 1–2 Female cf. Phaeacius [Sarawak], voucher SWK12–3728 3–4 Female Phaeacius sp. [Sarawak], voucher SWK12–4541 5 female Onomastus sp. [Guangxi], voucher MRB085 6 Female Sonoita aff. lightfooti, voucher MRB200. 7 male Gelotia sp. [Guangxi], voucher MRB199. The drawing is reversed so as to appear to be the left palpus 8–9 Male Echeclus sp. [Selangor], voucher MRB089 10–11 Male Taraxella sp.[Johor], voucher d246 for the palpus. The photo of the living male may or may not be of the same specimen 12–13 Male Taraxella sp. [Pahang], voucher d248. The photo of the living male may or may not be of the same specimen. Figures 1–13 are copyright ©2014 W.P. Maddison, released under a Creative Commons Attribution (CC-BY) 3.0 license.
Specimens whose voucher ID’s (Table 1, Suppl. material 1) are of the form S###, d###, MRB###, or JXZ###, SWK12-####, or ECU11-####, where # is a digit, are deposited in the Spencer Entomological Collection of the Beaty Biodiversity Museum, University of British Columbia. The remaining vouchers are in the Lee Kong Chian Natural History Museum (formerly Raffles Museum for Biodiversity Research or RMBR), National University of Singapore.
In addition to analyses done on all 176 sampled taxa (“Complete”), subsets of taxa were analyzed alone. A first subset (“Salticoida”) of 78 taxa highlighted the Salticoida, with just 7 non-salticoid outgroup taxa (4 hisponines, 1 spartaeine, 1 cocalodine, 1 lapsiine), in order to obtain an alignment that was less perturbed by highly divergent non-salticoids. A second subset highlighted the non-salticoids (“Non-salticoid”, 120 taxa), to obtain an alignment primarily for non-salticoid salticids, and also to be able to explore their relationships in more detail.
Gene choice and sequencing
Eight genes were used for this analysis. Two are nuclear ribosomal genes, 28s and 18s (Maddison and Hedin 2003, Maddison et al. 2008). Four are nuclear protein coding genes: actin 5C (Vink et al. 2008, Bodner and Maddison 2012), wingless (Blackledge et al. 2009), myosin heavy chain (“myosin HC”, Blackledge and Hayashi, unpublished), and histone 3 (Su et al. 2007). Two mitochondrial regions were also used, CO1 and another region including 16s and NADH1 ("16sND1", Hedin and Maddison 2001, Maddison and Hedin 2003). Following Bodner and Maddison (2012), the intron region of actin 5C was deleted from the analyses as it is highly variable and difficult to align.
The sequencing protocols for wingless and myosin HC are described below. For other genes, sequences marked “S” in Table 1 and Suppl. material 1 were obtained by the protocols of Su et al. (2007), all others by the protocols of Bodner and Maddison (2012) and Zhang and Maddison (2013).
For most wingless sequences, the forward and reverse primers used were respectively Spwgf1 and Spwgr1 (Blackledge et al. 2009). PCR amplification included a 2 min 94 °C denaturation and 35 cycles of 30 s at 94 °C, a 30 s annealing step at 52–57 °C, 30 s at 72 °C and one 3 min extension step at 72 °C. For some specimens this did not succeed in amplifying wingless, and for those we used a nested protocol starting with outer primers wg550F and wgABRz (Wild and Maddison 2008). The resulting product was then amplified using two internal primers, forward Wnt8MBf1 5’-TGTGCTACTCARACKTGYTGG-3’ and reverse Wnt8MBr3 5’-ACAAWGTTCTGCA ACTCATRCG-3’. For both the external and internal reactions amplification was done with 2 min 94 °C denaturation and 37 cycles of 20 s at 94 °C, a 20 s annealing step at 52 °C (wg550F/wgABRz) or 56 °C (wnt8MBf1/wnt8MBr3), and 2 min at 72 °C, and no final extension. The nested protocol obtained sequences for Bavia aff. aericeps (voucher d389), Hasarius adansoni (d295), Philodromus sp. (GR011), Simaetha sp. (d027), and Yllenus arenarius (JXZ173). In other specimens, the nested protocol often resulted in amplification of a different member of the wingless family (e.g. WNT-8), but these were readily detected (and excluded) by BLASTing them to other genes in the NCBI database (http://www.ncbi.nlm.nih.gov).
The region of myosin HC sequenced corresponds mostly to an intron. Primers used are (forward) Myhc1f 5'-ACAACAATTCTTCAACCATCAC-3' and (reverse) Myhc5r 5'-CTTCCTCAAGGATGGACA-3' (Blackledge and Hayashi, unpublished). PCR amplification included a 2 min 95 °C denaturation and 35 cycles of 20–45 s at 95 °C, a 45 s annealing step at 52 °C, 1 min at 72 °C and one 10 min extension step at 72 °C. The boundary between the exon and intron was determined by aligning the salticid implicit amino acid translations against the known transcript for myosin HC in Cyrtophora citricola (Genbank accession AAM97635.1; Ruiz-Trillo et al. 2002).
Two small single-nucleotide errors in the sequences were corrected after the analyses but before submission to Genbank. These are near the ends of CO1 of MRB199 (Gelotia sp. [Guangxi]) and MRB231 (Eburneana sp. [Gabon]). Given that CO1 had little resolution, these are unlikely to have affected the results.
Sequence alignment
Automatic multiple sequence alignment was performed by MAFFT (Katoh et al. 2002, 2005), run via the align package of Mesquite (prerelease of version 3, Maddison and Maddison 2014), aided by Mesquite for manual corrections and for alignment by amino acid. Coding regions were easily aligned by hand according to amino acid translations. This was done starting with an initial automated nucleotide alignment, followed by hand correction in Mesquite using the Color Nucleotide By Amino Acid function to reveal amino acid translation. Non-coding regions (28s, 18s, noncoding region of 16sND1, myosin HC intron) were aligned by MAFFT using the L-INS-i option (--localpair --maxiterate 1000). Mesquite was used to color the matrix via the option ‘‘Highlight Apparently Slightly Misaligned Regions’’ so as to identify regions that needed correction. These were almost always near the ends of sequences.
Alignment was done separately on the Complete, Non-salticoid and Salticoida datasets. Following the MAFFT alignment, the Salticoida dataset required 5 small realignments by hand in 18s. The first 60 positions in the initial alignment of 16s were also realigned locally, and in addition 8 minor shifts by one or two positions were made by hand. The Non-salticoid dataset required three simple hand fixes in 28s. The first 24 positions of 16s in the initial alignment were realigned by MAFFT in isolation because of several obvious misalignments. The Complete dataset appeared poorly aligned in 28s from sites 375 to 489 in the initial alignment, which were therefore realigned by MAFFT in isolation. The first 60 positions in the initial alignment of 16s were also realigned locally, and in addition 8 minor shifts by one or two positions were made by hand. Five small shifts were performed by hand for 18s. Many analyses were done with different variants of the alignments as this study was progressing, and the phylogenetic trees remained substantially consistent.
Phylogenetic analysis
Phylogenetic analyses using maximum likelihood were run using RAxML version 7.2.8alpha (Stamatakis 2006a, 2006b). The protein coding genes and 16sND1 were each divided into partitions. Protein coding regions were divided into one partition for 1st and 2nd codon positions, and another partition for third codon positions. Introns and non-coding regions were treated as separate partitions. For the fused 8 gene analyses, there were 7 partitions total: (1) 1st + 2nd codon positions in nuclear genes, (2) 3rd codon position nuclear, (3) nuclear intron, (4) nuclear ribosomal, (5) 1st + 2nd codon positions mitochondrial, (6) 3rd codon position mitochondrial, (7) noncoding mitochondrial. Each partition was permitted to have its own model parameters.
Analyses were done for each gene region separately with the Complete taxon set. In addition, analyses fusing all 8 genes were done for the Non-salticoid and Salticoida taxon sets. For all of these, RAxML runs assuming the GTRCAT model were used with 100 search replicates, to seek maximum likelihood trees. In addition, likelihood bootstrap analysis was performed with 500-1500 bootstrap replicates (as indicated in the figures), each involving a single search replicate. Phylogenetic analyses using GARLI version 1.0.699 (Zwickl 2006) under the model GTR+gamma+I were also done but are not reported; they resulted in substantially similar trees.
Data resources
The data underpinning the analyses reported in this paper are deposited in the Dryad Data Repository at doi: 10.5061/dryad.v53h1.
Results
Sequences obtained and used in analyses are indicated in Table 1 and Suppl. material 1, along with those sequences taken from the literature.
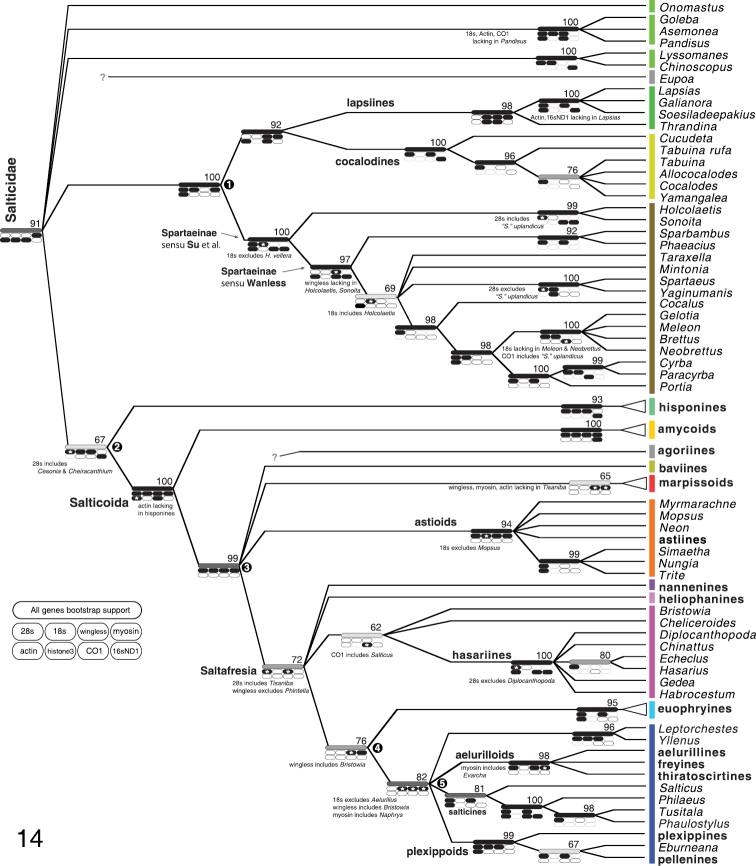
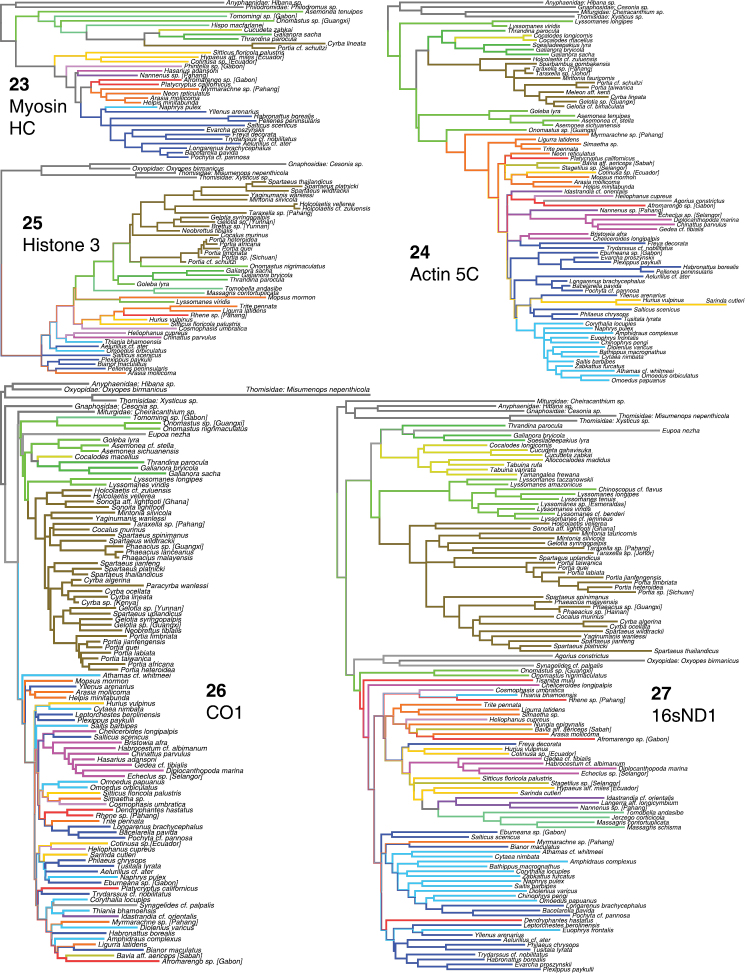
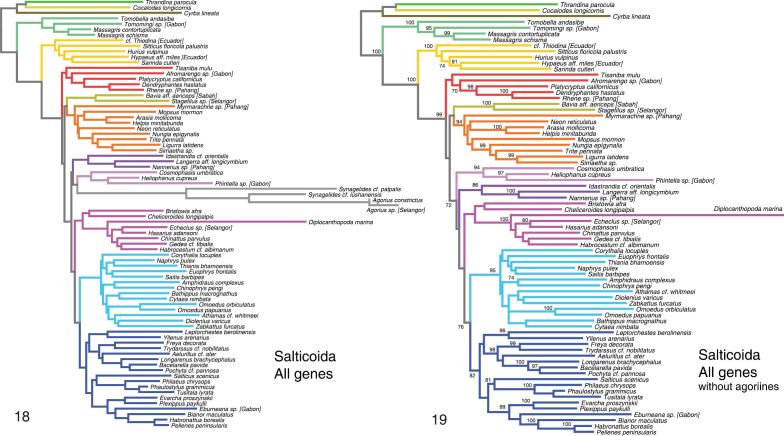
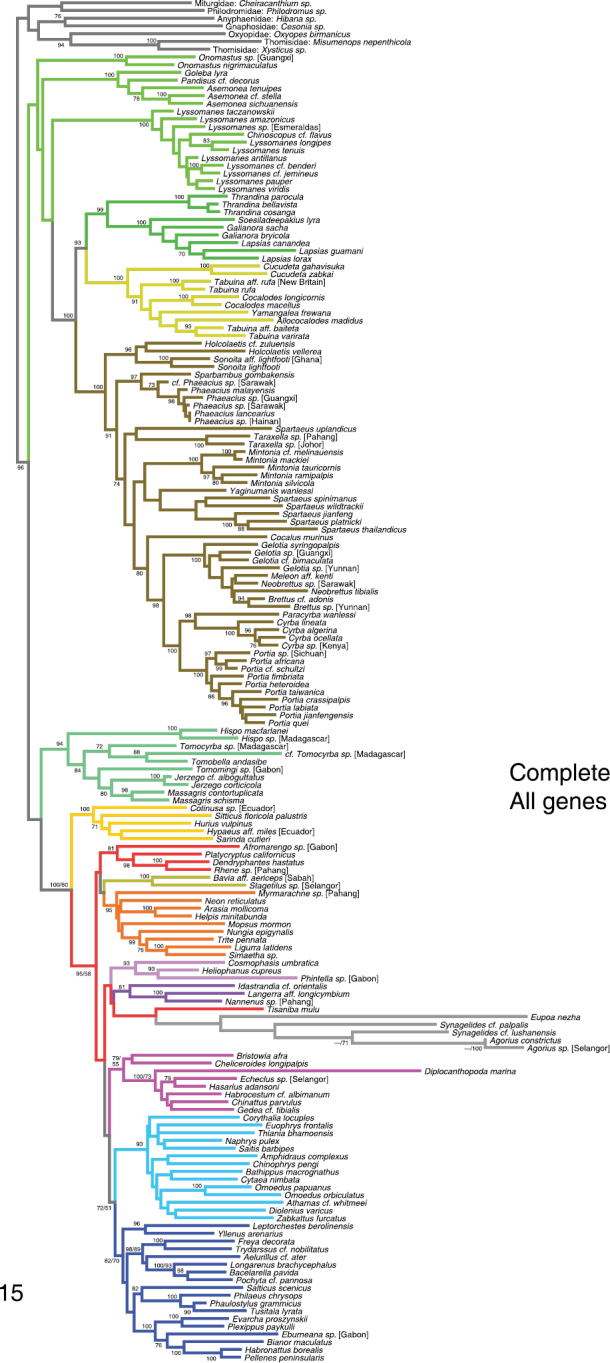
Figure 14 summarizes the results of the phylogenetic analyses, which are given in more detail in Figures 15–27. Colors assigned to clades in Figure 14 are shown in the remaining figures. Figures 15–19 show the All Genes results for the Complete, Non-salticoid and Salticoida datasets. Figures 20–27 show the results for individual genes analyzed separately.
Figure 14.
Summary of phylogenetic results. Number above branch shows percentage of maximum likelihood bootstrap replicates with clade. For clades outside the Salticoida, these percentages come from the Non-salticoid dataset with 1500 replicates; within the Salticoida, these come from the Salticoida dataset with 1000 replicates; the Salticoida percentage comes from the Complete dataset with 1000 replicates. Long bar on branch shows same percentage graphically: black 91–100%; dark gray 81–90%; gray 71–80%; light gray 51–70%. Oval spots show presence of clade in maximum likelihood tree for individual genes, with exceptions noted by * and adjacent notes. The notes about wingless on the Spartaeinae node and actin on the Salticoida node are ambiguous in placement; they could equally well have been placed one node deeper because of missing data. Pale gray outline indicates no conclusion because of inadequate taxon sampling. All indications of support are from analyses excluding Eupoa, agoriines, Spartaeus spinimanus and “Spartaeus” uplandicus. Bars show colors used to highlight taxa in Figs 15–27.
Figure 15.
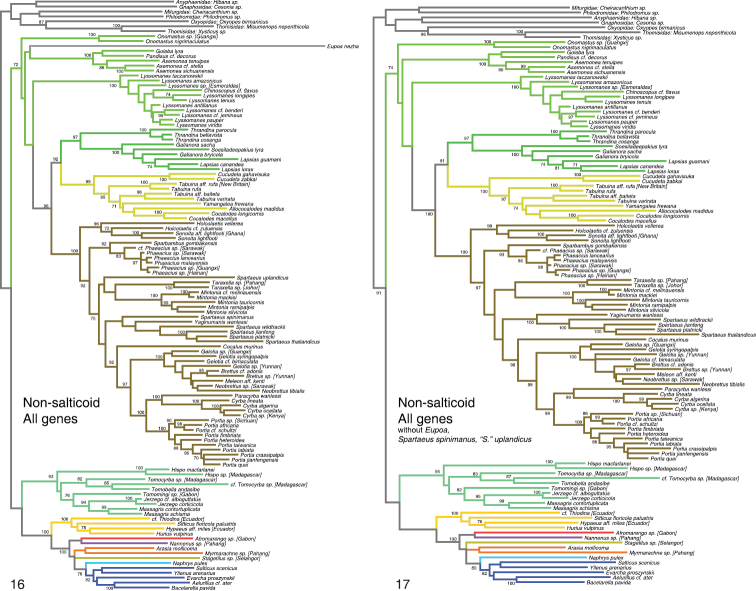
Phylogeny from complete taxon sample, All Genes analysis. Numbers beside branches show percentage of 1000 RAxML likelihood bootstrap replicates with clade in analysis with Eupoa and agoriines excluded. In analyses with these taxa included (500 bootstrap replicates), bootstrap percentages are within 5 of those shown, except for branches with two values (e.g. “100/60”), in which case the first value is from an analysis with Eupoa and agoriines excluded, the second value with them included. Colors of branches are the same as those highlighting taxa in Fig. 14.
Figures 23–27.
Phylogeny from gene regions analyzed alone, complete taxon sample. 23 myosin HC 24 actin 5C 25 Histone 3 26 CO1 27 16sND1. Colors of branches are the same as those highlighting taxa in Fig. 14.
Figures 18–19.
Phylogeny from Salticoida dataset, All Genes analysis. 18 Salticoida analysis with all taxa included 19 Salticoida analysis with Agorius and Synagelides excluded. Numbers beside branches show percentage of 1000 RAxML likelihood bootstrap replicates with clade. Colors of branches are the same as those highlighting taxa in Fig. 14.
Figures 20–22.
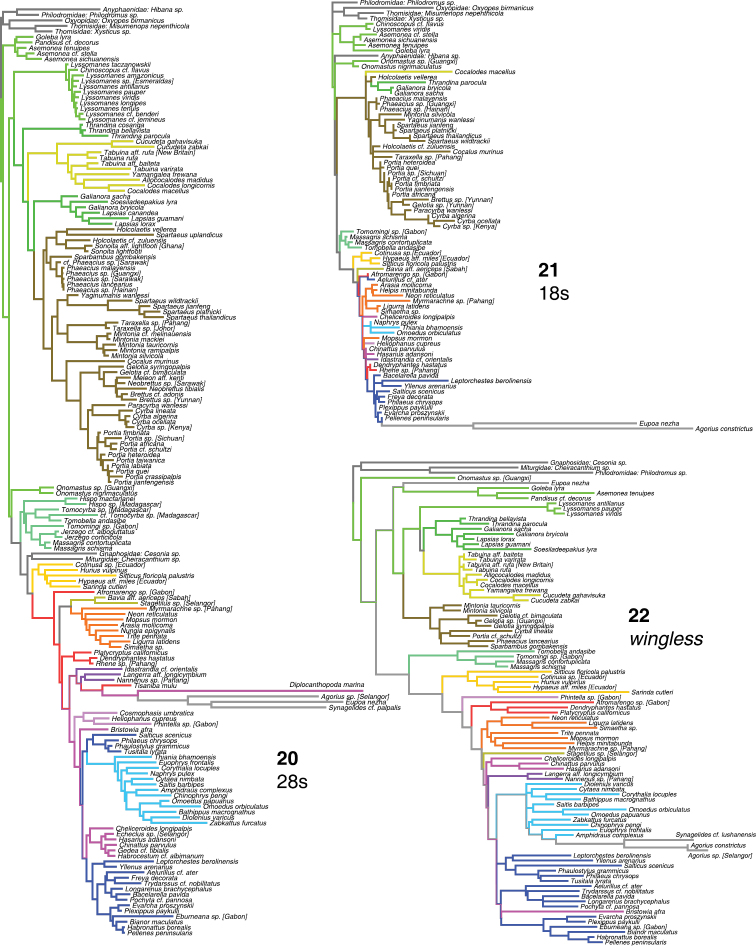
Phylogeny from gene regions analyzed alone, complete taxon sample. 20 28s 21 18s 22 wingless. Colors of branches are the same as those highlighting taxa in Fig. 14.
Several taxa stood out as being problematical, especially for nuclear ribosomal genes. Eupoa was not only difficult to sequence (Maddison et al. 2007) but its 28s and 18s genes stand as outliers in alignments, remarkably different from other salticids. The same holds for the agoriines Agorius and Synagelides and, in 28s, for the hasariine Diplocanthopoda. These sequences do not appear to be contaminants, as they BLAST in the NCBI database to salticids. In analyses with just 28s or 18s, these taxa tend to appear on long branches, wandering to different parts of the salticid phylogeny in different analyses, attaching themselves together and to clearly inappropriate relatives (e.g. within the pellenines, Fig. 21). This instability and unexpected placement are likely artifacts due to long branch attraction (Felsenstein 1978), possibly related to compositional bias (Hasegawa and Hashimoto 1993). Eupoa and the agoriines have the highest GC bias of the sample (0.72–0.78, compared to 0.60–0.69 for all other species) in 28s, and are similar outliers in 18s. With wingless, Eupoa appears on a normal-length branch (Fig. 22). However, the agoriines with wingless are on a long branch in an unlikely place, within the euophryines (Fig. 22). Their placement is unstable: in slight variants of the analyses they come out in other places. There is nothing obviously unusual about the wingless sequences in agoriines, but whatever has shifted the GC bias in the nuclear ribosomal genes might also be affecting the rest of the genome. When Eupoa and the agoriines are excluded from analyses, bootstrap percentages rise through much of the tree, suggesting their instability is adding noise to the other relationships in the tree. For this reason, the reported bootstrap percentages and other indications of support are generally those for analysis with Eupoa and the agoriines excluded. Diplocanthopoda was left in the bootstrap analyses, because CO1, actin 5C and 16sND1 all agree on a clear placement in the hasariines.
Discussion
Many of the salticid clades now recognized by molecular data had been previously recognized by morphological data. For instance, Wanless (1980, 1984, 1985) recognized the three distinct lyssomanine groups and the Spartaeinae. The Salticoida was strongly supported by many morphological characters (Maddison 1988, 1996, Maddison and Hedin 2003), except that the status of the hisponines was unclear. Wanless (1981) implicitly included the hisponines within the salticoids, while Maddison (1996) did not consider the hisponines in his listing of salticoid synapomorphies. Other groups whose previous formulation by morphology mostly or entirely matches their current boundaries by molecular data are the marpissines (Barnes 1958), euophryines (Prószyński 1976), amycines (Galiano 1968), heliophanines (Maddison 1987), dendryphantines (Maddison 1996), and plexippines (Maddison 1988). At the finer scale, morphological systematics gave us concepts for many genera that are concordant with more recent data.
However, the first molecular data for salticid phylogeny as a whole (Maddison and Hedin 2003) uncovered several unanticipated groups, including the Amycoida, Plexippoida, and Marpissoida. Further data revealed the Astioida and Aelurilloida (Maddison et al. 2008), and later the Saltafresia (Bodner and Maddison 2012). These are major groups within the Salticoida, each uniting several subfamilies.
Deepest relationships
Our results help resolve or add strength to relationships at the deepest level of salticid phylogeny. Wanless (1980) recognized three major subdivisions of lyssomanines: (1) the New World genera Lyssomanes and Chinoscopus, (2) the Asian Onomastus, and (3) the remaining Old World genera including Asemonea. He suggested these three groups are so distinct that they may not belong together. The molecular data agree: the three groups’ divisions are so deep that their relationships have not yet been recovered, and it is possible, even likely, that they do not form a monophyletic group. Different analyses give different results of the relationships of these three, with some showing the New World genera as sister to the spartaeine-lapsiine-cocalodine clade (as recovered by Su et al. 2007), other results showing Onomastus in that role, and others showing the three lyssomanine groups together.
Spartaeines, lapsiines and cocalodines form a clade (node 1, Fig. 14). Although Rodrigo and Jackson (1992) concluded that spartaeines, Holcolaetis and the Cocalodes group form a clade (they were unaware of lapsiines), our analysis provides the first support for such an arrangement – their analysis included only a single taxon outside the group, and therefore it could not speak to the monophyly of the group. Our new result is intuitively appealing, as it groups together all of the extant medium-sized generalized non-salticoids/non-hisponines that are typically brown or gray. However, these presumably are or could be plesiomorphic traits; there had been no obvious reason to expect the spartaeines, lapsiines and cocalodines should have fallen together. There is no known morphological synapomorphy of this clade.
Within this spartaeine-lapsiine-cocalodine clade, the subclade historically best known by morphology is Wanless’s (1984) narrow version of the Spartaeinae, delimited by the presence of a tegular furrow (Wanless 1984). The Spartaeinae sensu stricto is primarily Afro-Eurasian, with a few Australasian species. Outside of this clade, there are no clear morphological synapomorphies defining subclades, and yet there is a striking geographical pattern: all of the Neotropical species belong to a clade, thus forming the lapsiines, while all of the Australasian species belong to a clade, thus forming the cocalodines. It is unsatisfying that we lack morphological synapomorphies for the lapsiines or cocalodines. The data suggest that the lapsiines and cocalodines are sister groups, with spartaeines more distant (Fig. 14).
Our results continue to support the relationship of hisponines with the Salticoida (node 2, Fig. 14; Figs 15–17; Maddison and Needham 2006, Bodner and Maddison 2012).
Figures 16–17.
Phylogeny from Non-salticoid dataset, All Genes analysis. Numbers beside branches show percentage of RAxML likelihood bootstrap replicates with clade. 16 Non-salticoid analysis with all taxa included (1500 bootstrap replicates used) 17 Non-salticoid analysis with Eupoa, Spartaeus spinimanus, and “Spartaeus” uplandicus excluded (500 bootstrap replicates used). Colors of branches are the same as those highlighting taxa in Fig. 14.
The placement of Eupoa remains unclear. As noted under Results, the 28s and 18s genes of Eupoa may be unreliable phylogenetically, although Maddison et al. 2007 found those genes to place Eupoa among non-salticoid salticids. In our results Eupoa likewise has no clear placement, except for being outside the clade of Salticoida + Hisponinae. This result appears in the Non-salticoid and Complete datasets, and with the separate analyses of wingless, CO1, and 16sND1.
Spartaeinae
Our results strongly support the monophyly of the Spartaeinae sensu Su et al. (2007), placing Holcolaetis and Sonoita together with the Spartaeinae in the narrow sense. This is concordant with Wanless’s (1985) hypothesis that Holcolaetis and Sonoita formed a clade with the spartaeines to the exclusion of Cocalodes. The analyses of Su et al. (2007) did not sample Sparbambus, Taraxella, Brettus or Meleon, but otherwise their results were largely concordant with ours, which are: (1) Phaeacius (with Sparbambus) diverge deep, (2) Yaginumanus is sister to Spartaeus, (3) Gelotia, Neobrettus, Brettus and Meleon are monophyletic, (4) Paracyrba and Cyrba are sisters, (5) Portia is sister to Cyrba and Paracyrba. There is strong support for Gelotia through Cyrba as a monophyletic group, and for their relationship with Cocalus. By our data the exact placements of Taraxella and Mintonia are unclear.
A few spartaeine taxa in our analyses were problematical in appearing unstable, having different placements by different analyses. One of these is Spartaeus spinimanus, for which we have only 16sND1 and CO1 data, both gene regions that appear to evolve too quickly for reliable phylogenetic placement at this level (Bodner and Maddison 2012, Zhang and Maddison 2013). The other is “Spartaeus” uplandicus, whose 28s sequence appears strongly divergent from others. This sequence is from Maddison and Hedin (2003, as “unidentified spartaeine”, vouchers 185 and 186), and it groups “Spartaeus” uplandicus with one species of Holcolaetis, against the placements by morphology, CO1 and 16sND1. There is a chance that this gene was mis-sequenced in “Spartaeus” uplandicus. Because of the instability generated, we excluded Spartaeus spinimanus and “Spartaeus” uplandicus from our analyses giving bootstrap results.
Because of the concordance of our phylogenetic results with those of Su et al. (2007), our phylogeny continues to support their conclusions on the stepwise evolution of a complex predatory strategy in spartaeines.
Deep Salticoid relationships
The Salticoida’s basal divergence places the primarily-Neotropical Amycoida as sister group to an unnamed clade (node 3, Fig. 14) that contains most of salticid diversity. This surprising result, first discovered by Maddison and Hedin (2003), had very strong support in the analyses of Bodner and Maddison (2012). We here add support from two new genes, wingless and myosin HC, both of which independently resolve both the Amycoida and its sister group as monophyletic.
There have been hints of a clade uniting the Marpissoida, Astioida and baviines (Bodner and Maddison 2012). In our analyses the clade does not receive bootstrap support above 50% in the Complete or Salticoida analyses. The maximum likelihood trees either show the three as monophyletic or not, depending on taxon inclusion and details of the analysis (e.g., Figs 15 and 18). At present we must conclude the relationship between these three and the Saltafresia is unresolved.
Astioida
The astioids as delimited by Maddison et al. (2008) continue to be resolved as a clade, with new support from myosin HC and wingless (Figs 18, 22, 23). Although the body form of Nungia resembles that of baviines and the marpissoid Metacyrba, our data clearly place it as an astioid.
Saltafresia
Bodner and Maddison (2012) proposed a clade, the Saltafresia, containing salticoids other than amycoids, astioids, baviines and marpissoids. They found this clade reasonably well supported – 0.78 likelihood bootstrap and 1.0 posterior probability – but no single gene supported it on its own. Our data here continue to support it when all genes are combined. Two genes support it separately, with the exception of single taxa: 28s (but Tisaniba is included) and wingless (but Phintella is excluded).
Hasarieae
Previous work had established Habrocestum and Chinattus as close relatives of Hasarius (Maddison et al. 2008). We here add several more genera to the group, all Asian. These are Gedea, Echeclus and Diplocanthopoda. The relationships among these genera are not clearly resolved except for a well-supported relationship between Hasarius and Echeclus (Figs 14, 19).
Salticinae
The relationship between Salticus and the Philaeus group proposed by Maddison et al. (2008) receives additional support from wingless, along with previously-demonstrated support from 28s and actin. With high posterior probabilities (Bodner and Maddison 2008) and reasonable likelihood bootstrap values (Figs 15, 19), and supported by different genes independently (Figs 20, 22, 24), this relationship can now be considered sufficiently secure that we here formally place the genera of the Philaeus group into a subfamily — the Salticinae. In addition to genera previously analyzed (Salticus, Philaeus, Carrhotus, Tusitala, Mogrus, and Pignus) the subfamily also includes Phaulostylus, which is related to Tusitala (Fig. 14).
Plexippoida + Aelurilloida + Leptorchesteae + Salticinae (Node 5)
A set of four major groups (plexippoids, aelurilloids, leptorchestines and the Salticinae) form a clade in our analyses (node 5, Fig. 14). This group is resolved in the All Genes analyses with high bootstrap values, and it appears, almost, in the independent analyses of each of three genes (18s, wingless, myosin HC). We say “almost” because three of the genes have one or two taxa missing from or added to the group (Fig. 14). While we believe the evidence is good that these form a clade, there is a possibility that the Euophryinae might also fall nested within it. For instance, in the analyses of Bodner and Maddison (2012) the euophryines were placed as sister to the plexippoids. In our analyses the Euophryinae is placed as sister to the Plexippoida + Aelurilloida + Leptorchesteae + Salticinae.
This major clade is almost entirely Afro-Eurasian, with the plexippoid Habronattus being the only exception with more than a handful of species (others are Pellenes, Sibianor, Evarcha, Phlegra, Paramarpissa and Salticus, each with fewer than 15 described New World species).
Euophryinae
The 14 euophryine taxa in the analyses are resolved strongly as a monophyletic group. This is a stronger test of monophyly than that of Zhang and Maddison (2013), because it includes additional genes and more non-euophryine taxa. The All Genes analyses, along with wingless and myosin HC individually, suggest that the euophryines are the sister group to node 5 (Fig. 14).
Agoriines
Morphologically, the antlike agoriines Agorius and Synagelides are puzzling, with strangely contorted legs and unusual genitalia (Szüts 2003, Logunov and Hereward 2006, Prószyński 2009). While they appear to be salticoids, morphology has given little guidance as to their placement. As noted already, their 28s and 18s genes appear anomalous, and give no clear indication as to their relationships. In the All Genes analysis their placement is ambiguous, though they appear to be salticoids. In an attempt to determine their placement, an additional analysis was done, using a dataset that included Agorius constrictus and a chimera of Synagelides cf. lushanensis and Synagelides cf. palpalis (to have a single Synagelides taxon with three genes). The aberrant nuclear ribosomal genes of agoriines were excluded from the analysis. The other taxa included were the 70 taxa having at least 4 genes other than CO1 and histone 3. A RAxML likelihood analyses placed Agorius and Synagelides within the sister group of the Amycoida (node 3, Figure 14) with high support (bootstrap percentage 88), but exactly where was highly unstable. Among the 100 likelihood non-bootstrap search replicates were 7 different placements: sister to leptorchestines, sister to baviines, sister to node 5 in Figure 14, sister to the Saltafresia, sister to astioids + marpissoids + baviines, sister to node 3, or sister to node 3 without the baviines. While a relationship with the leptorchestines is appealing, as it would allow their antlike body forms to be homologous, the best we can say at present is that agoriines likely belong within the sister group of amycoids (node 3).
Generic limits
Most of the genera for which we have multiple species – e.g., Asemonea, Portia, Mintonia, Phaeacius, Cyrba – are inferred to be monophyletic in our analyses, corroborating existing concepts based on morphology. The clearest exception is Tabuina, in which Tabuina rufa and the similar Tabuina aff. rufa fall apart from the type species Tabuina varirata, which had been anticipated as a possibility by Maddison (2009). Lyssomanes, Galianora, and Gelotia are reconstructed as paraphyletic, but in each case the bootstrap values are low.
The placement of cf. Phaeacius [Sarawak] as sister to Phaeacius, with strong molecular divergence from the other species, would justify establishing a new genus for it.
Behaviour of individual genes
Previous work (Maddison and Hedin 2003, Bodner and Maddison 2012) has suggested that 28s and actin 5C are phylogenetically informative to a reasonable degree for deeper salticid phylogeny, insofar as their results are concordant with summed genes analyses, morphological resemblances, and biogeographical patterns. 16sND1 is useful at the shallower levels (Hedin and Maddison 2001) but has difficulties recovering deeper relationships, while CO1 struggles through both shallow and deep levels (Maddison and Hedin 2003, Bodner and Maddison 2012).
One surprise in our analyses was the informative behaviour of CO1 in deeper relationships among the non-salticoid salticids. Although CO1 is almost nonsensical in its inferred relationships within the Salticoida, it succeeds in recovering the Spartaeinae, the Spartaeineae sensu Wanless, the lapsiines, and the Salticoida as monophyletic.
Two new genes added, wingless, myosin HC, both show clear concordance with the 28s and previous all genes analyses. Wingless supports many of the previously recognized clades, including the Salticoida, Amycoida, the sister clade to Amycoida, Plexippoida, Marpissoida (in part), Astioida (in part), Spartaeinae sensu Wanless, and lapsiines. We find it encouraging that a haphazardly chosen protein-coding gene, independent from 28s, supports previous molecular results in Salticidae. There are still, however, many aspects of salticid relationships yet to be resolved, such as the deepest relationships in the family, including the relationships among the three subgroups of lyssomanines, the placement of Eupoa and the agoriines, and the relationships among astioids, marpissoids, baviines and the Saltafresia. With the coming era of genomic data, we expect large quantities of new data will be available for exploring these relationships.
Acknowledgments
We thank David Maddison for obtaining the sequences of wingless and 28s in Lapsias bellavista and Lapsias guamani. Geneviéve Leduc-Robert and Teresa Maddison assisted with molecular laboratory work. We thank Todd Blackledge and Cheryl Hayashi for their unpublished protocols for myosin HC. Gustavo Ruiz, Martín Ramírez, and G.B. Edwards gave helpful comments on the manuscript. For assistance on the 2008 expedition to Papua New Guinea, we thank Stephen Richards, Bruce Beehler, William Thomas, Luc Fimo Tuki, Aislan Tama Wanakipa Indiaf, Pingisa Saiké, Yainé Ribson, Agustus Kore, Muse Opiango, Banak Gamui, Robert Sine, Conservation International, Porgera Joint Venture, and the PNG Department of Environment and Conservation (for more details, see Maddison 2009). For assistance on the 2010 expedition to Ecuador, we thank David Maddison, Mauricio Vega, Edyta Piascik. Marco Reyes, the Ecuadorian Ministry of the Environment and the Museum of Zoology of the Pontificia Universidad Católica de Ecuador. Dmitri Logunov assisted with identifications of the Lyssomanes, Gustavo Ruiz with the Hypaeus. We also thank the staff of the Centre for Behavioural Ecology and Evolution (CBEE, Hubei University) and of the Behavioural Ecology and Sociobiology Lab (DBS, NUS) for all their help and support throughout this study, in particularly Jian Chen, Zhanqi Chen, Xiaoguo Jiao, Jie Liu, Yu Peng, Xiaoyan Wang, and Zengtao Zhang. This work was supported by an NSERC Discovery grant to WPM, and a NSFC grant (31272324) as well as a Singapore Ministry of Education (MOE) AcRF grant (R-154-000-591-112) to DL.
Citation
Maddison WP, Li D, Bodner M, Zhang J, Xu X, Liu Q, Liu F (2014) The deep phylogeny of jumping spiders (Araneae, Salticidae). ZooKeys 440: 57–87. doi: 10.3897/zookeys.440.7891
Supplementary material
Specimens used in phylogenetic analyses, with localities and GenBank numbers of sequences indicated.
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Wayne Maddison, Daiqin Li, Melissa Bodner, Junxia Zhang, Xu Xin, Qinqing Liu, Fengxiang Liu
Data type: Occurence; geographic locality; sex.
References
- Barnes RD. (1958) North American jumping spiders of the subfamily Marpissinae (Araneae, Salticidae). American Museum Novitates 1867: 1–50 [Google Scholar]
- Blackledge TA, Scharff N, Coddington JA, Szüts T, Wenzel JW, Hayashi CY, Agnarsson I. (2009) Reconstructing web evolution and spider diversification in the molecular era. Proceedings of the National Academy of Sciences (USA) 106: 5229–5234. doi: 10.1073/pnas.0901377106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blest AD, O'Carroll DC, Carter M. (1990) Comparative ultrastructure of layer-I receptor mosaics in principal eyes of jumping spiders – The evolution of regular arrays of light guides. Cell and Tissue Research 262: 445–460. doi: 10.1007/BF00305241 [Google Scholar]
- Bodner MR, Maddison WP. (2012) The biogeography and age of salticid spider radiations (Araneae: Salticidae). Molecular Phylogenetics and Evolution 65: 213–240. doi: 10.1016/j.ympev.2012.06.005 [DOI] [PubMed] [Google Scholar]
- Felsenstein J. (1978) Cases in which parsimony and compatibility methods will be positively misleading. Systematic Zoology 27: 401–410. doi: 10.2307/2412923 [Google Scholar]
- Galiano ME. (1968) Revisión de los generos Acragas, Amycus, Encolpius, Hypaeus, Mago y Noegus (Salticidae, Araneae). Revista del Museo Argentino de Ciencias Naturales, Entomología 2: 2267–360 [Google Scholar]
- Hasegawa M, Hashimoto T. (1993) Ribosomal RNA trees misleading? Nature 361: 23. doi: 10.1038/361023b0 [DOI] [PubMed] [Google Scholar]
- Hedin MC, Maddison WP. (2001) A combined molecular approach to phylogeny of the jumping spider subfamily Dendryphantinae (Araneae, Salticidae). Molecular Phylogenetics and Evolution 18: 386–403. doi: 10.1006/mpev.2000.0883 [DOI] [PubMed] [Google Scholar]
- Katoh K, Kuma K, Toh H, Miyata T. (2005) MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Research 33: 511–518. doi: 10.1093/nar/gki198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T. (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Research 30: 3059–3066. doi: 10.1093/nar/gkf436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Land MF. (1969) Structure of the retinae of the principal eyes of jumping spiders (Salticidae: Dendryphantinae) in relation to visual optics. Journal of Experimental Biology 51: 443–470 [DOI] [PubMed] [Google Scholar]
- Logunov DV, Hereward J. (2006) New species and synonymies in the genus Synagelides Strand in Bösenberg & Strand, 1906 (Araneae: Salticidae). Bulletin of the British Arachnological Society 13: 281–292 [Google Scholar]
- Maddison WP. (1987) Marchena and other jumping spiders with an apparent leg-carapace stridulatory mechanism (Araneae: Salticidae: Heliophaninae and Thiodininae). Bulletin of the British Arachnological Society 7: 101–106 [Google Scholar]
- Maddison WP. (1988) A revision of jumping spider species groups formerly placed in the genus Metaphidippus, with a discussion of salticid phylogeny (Araneae). PhD thesis, Harvard University [Google Scholar]
- Maddison WP. (1996) Pelegrina Franganillo and other jumping spiders formely placed in the genus Metaphidippus (Araneae: Salticidae). Bulletin of the Museum of Comparative Zoology 154: 215–368 [Google Scholar]
- Maddison WP. (2009) New cocalodine jumping spiders from Papua New Guinea (Araneae: Salticidae: Cocalodinae). Zootaxa 2021: 1–22 [DOI] [PubMed] [Google Scholar]
- Maddison WP, Bodner MR, Needham K. (2008) Salticid spider phylogeny revisited, with the discovery of a large Australasian clade (Araneae: Salticidae). Zootaxa 1893: 49–64 [Google Scholar]
- Maddison WP, Hedin MC. (2003) Jumping spider phylogeny (Araneae: Salticidae). Invertebrate Systematics 17: 529–549. doi: 10.1071/IS02044 [Google Scholar]
- Maddison WP, Maddison DR. (2014) Mesquite: A modular system for evolutionary analysis. version 2.75+. http://mesquiteproject.org
- Maddison WP, Needham K. (2006) Lapsiines and hisponines as phylogenetically basal salticid spiders (Araneae: Salticidae). Zootaxa 1255: 37–55 [Google Scholar]
- Maddison WP, Piascik EK. (2014) Jerzego, a new hisponine jumping spider from Borneo (Araneae: Salticidae). Zootaxa 3852: 569–578 [DOI] [PubMed] [Google Scholar]
- Maddison WP, Zhang JX, Bodner MR. (2007) A basal phylogenetic placement for the salticid spider Eupoa, with descriptions of two new species (Araneae: Salticidae). Zootaxa 1432: 22–33 [Google Scholar]
- Platnick NI. (2014) The World Spider Catalog, Version 14.5. American Museum of Natural History; http://research.amnh.org/iz/spiders/catalog [Google Scholar]
- Prószyński J. (1976) Studium systematyczno-zoogeograficzne nad rodzina Salticidae (Aranei) Regionow Palearktycznego i Nearktycznego. Wyzsza Szkola Pedagogiczna w Siedlcach Rozprawy 6: 1–260 [Google Scholar]
- Prószyński J. (1987) Atlas rysunkow diagnostycznych mniej znanych Salticidae 2. Zeszyty Naukowe WSRP, Siedlce, 172 pp [Google Scholar]
- Prószyński J. (2009) Comments on the Oriental genera Agorius and Synagelides (Araneae: Salticidae). In: Makarov SE, Dimitrijevic RN. (Eds) Advances in Arachnology and Developmental Biology. Inst. Zool., bulg. Acad. Sci. Monographs 12: 311–325 [Google Scholar]
- Prószyński J. (2013) Monograph of Salticidae (Araneae) of the World. http://www.peckhamia.com/salticidae/
- Rodrigo AG, Jackson RR. (1992) Four jumping spider genera of the Cocalodes-group are monophyletic with genera of the Spartaeinae (Araneae: Salticidae). New Zealand Natural Sciences 19: 61–67 [Google Scholar]
- Ruiz G, Maddison WP. (2012) DNA sequences corroborate Soesiladeepakius as a non-salticoid genus of jumping spiders: placement with lapsiines, phylogeny and description of six new species (Araneae, Salticidae). Zoological Journal of the Linnean Society 165: 274–295. doi: 10.1111/j.1096-3642.2012.00815.x [Google Scholar]
- Ruiz-Trillo I, Paps J, Loukota M, Ribera C, Jondelius U, Baguna J, Riutort M. (2002) A phylogenetic analysis of myosin heavy chain type II sequences corroborates that Acoela and Nemertodermatida are basal bilaterians. Proceedings of the National Academy of Sciences (USA) 99: 11246–11251. doi: 10.1073/pnas.172390199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. (2006a) RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690. doi: 10.1093/bioinformatics/btl446 [DOI] [PubMed] [Google Scholar]
- Stamatakis A. (2006b) Phylogenetic models of rate heterogeneity: A high performance computing perspective. In: Proceedings of 20th IEEE/ACM International Parallel and Distributed Processing Symposium (IPDPS2006), High Performance Computational Biology Workshop, Rhodos, Greece [Google Scholar]
- Su KF, Meier R, Jackson RR, Harland DP, Li D. (2007) Convergent evolution of eye ultrastructure and divergent evolution of vision-mediated predatory behaviour in jumping spiders. European Society for Evolutionary Biology 20: 1478–1489. doi: 10.1111/j.1420-9101.2007.01335.x [DOI] [PubMed] [Google Scholar]
- Szüts T. (2003) New species of Agorius Thorell, 1877 (Araneae: Salticidae) from New Guinea. Acta Zoologica Academiae Scientiarum Hungaricae 49: 61–69 [Google Scholar]
- Vink CJ, Hedin M, Bodner MR, Maddison WP, Hayashi CY, Garb JE. (2008) Actin 5C, a promising nuclear gene for spider phylogenetics. Molecular Phylogenetics and Evolution 48: 377–382. doi: 10.1016/j.ympev.2008.03.003 [DOI] [PubMed] [Google Scholar]
- Wanless FR. (1980) A revision of the spider genera Asemonea and Pandisus (Araneae: Salticidae). Bulletin of the British Museum of Natural History (Zoology) 39: 213–257 [Google Scholar]
- Wanless FR. (1981) A revision of the spider genus Hispo (Araneae, Salticidae). Bulletin of the British Museum of Natural History (Zoology) 41: 179–198 [Google Scholar]
- Wanless FR. (1982) A revision of the spider genus Cocalodes with a description of a new related genus (Araneae: Salticidae). Bulletin of the British Museum of Natural History (Zoology) 42: 263–298 [Google Scholar]
- Wanless FR. (1984) A review of the spider subfamily Spartaeinae nom. n. (Araneae: Salticidae) with descriptions of six new genera. Bulletin of the British Museum of Natural History (Zoology) 46: 135–205 [Google Scholar]
- Wanless FR. (1985) A revision of the spider genera Holcolaetis and Sonoita (Araneae: Salticidae). Bulletin of the British Museum of Natural History 48: 249–278 [Google Scholar]
- Wild AL, Maddison DR. (2008) Evaluating nuclear protein-coding genes for phylogenetic utility in beetles. Molecular Phylogenetics and Evolution 48: 877–891. doi: 10.1016/j.ympev.2008.05.023 [DOI] [PubMed] [Google Scholar]
- Zabka M. (1988) Salticidae (Araneae) of Oriental, Australian and Pacific regions, III. Annales zoologici, Warszawa 41: 421–479 [Google Scholar]
- Zhang JX, Maddison WP. (2013) Molecular phylogeny, divergence times and biogeography of spiders of the subfamily Euophryinae (Araneae: Salticidae). Molecular Phylogenetics and Evolution 68: 81–92. doi: 10.1016/j.ympev.2013.03.017 [DOI] [PubMed] [Google Scholar]
- Zhang JX, Maddison WP. (2014) Tisaniba, a new genus of marpissoid jumping spiders from Borneo (Araneae: Salticidae). Zootaxa 3852: 252–272 [DOI] [PubMed] [Google Scholar]
- Zwickl DJ. (2006) Genetic Algorithm Approaches for the Phylogenetic Analysis of Large Biological Sequence Datasets under the Maximum Likelihood Criterion. PhD Dissertation, The University of Texas at Austin [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Specimens used in phylogenetic analyses, with localities and GenBank numbers of sequences indicated.
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Wayne Maddison, Daiqin Li, Melissa Bodner, Junxia Zhang, Xu Xin, Qinqing Liu, Fengxiang Liu
Data type: Occurence; geographic locality; sex.