Abstract
We investigated expression patterns of DNA repair genes such as the CPD photolyase, UV-DDB1, CSB, PCNA, RPA32 and FEN-1 genes by northern hybridization analysis and in situ hybridization using a higher plant, rice (Oryza sativa L. cv. Nipponbare). We found that all the genes tested were expressed in tissues rich in proliferating cells, but only CPD photolyase was expressed in non-proliferating tissue such as the mature leaves and elongation zone of root. The removal of DNA damage, cyclobutane pyrimidine dimers and (6–4) photoproducts, in both mature leaves and the root apical meristem (RAM) was observed after UV irradiation under light. In the dark, DNA damage in mature leaves was not repaired efficiently, but that in the RAM was removed rapidly. Using a rice 22K custom oligo DNA microarray, we compared global gene expression patterns in the shoot apical meristem (SAM) and mature leaves. Most of the excision repair genes were more strongly expressed in SAM. These results suggested that photoreactivation is the major DNA repair pathway for the major UV-induced damage in non-proliferating cells, while both photoreactivation and excision repair are active in proliferating cells.
INTRODUCTION
All living organisms have to protect the integrity of their genomes from a wide range of genotoxic stresses. Numerous environmental mutagenic agents such as UV light, chemical mutagens, fungal and bacterial toxins, and ionizing radiation can cause damage to DNA in the cells. Recently, ozone depletion in the stratosphere has resulted in increased UV-B radiation at the earth’s surface, which induces various DNA lesions. The major lesions are cyclobutane pyrimidine dimers (CPDs) and pyrimidine (6–4) pyrimidinone dimers [(6–4) photoproducts], while the minor includes oxidized or hydrated bases, single-strand breaks and others (1–3). The repair of DNA damage caused by UV-B radiation is essential for the survival of organisms (4). If repair does not take place, genomic integrity will not be maintained. The maintenance of the genome requires coordination between DNA replication and repair. At present, little is known about the underlying mechanisms, although the factors in each individual process have been well studied.
Plants are interesting systems for such studies because cell proliferation occurs only in meristematic tissues, and not in non-meristematic tissues such as mature leaves. Plants use sunlight for photosynthesis and subsequently must remain exposed to solar UV-B radiation. DNA repair and replication have been examined in rice, Arabidopsis, wheat and maize (5–7), although not as extensively as in animals and yeast.
Through evolution, plants have acquired two main protective strategies to avoid the adverse effects of UV, the shielding by flavonoids and phenolic compounds (8–10) and the DNA repair such as photoreactivation and dark repair (5–7,11). Photoreactivation, which is mediated by photolyase, is thought to be the major DNA repair pathway for CPDs and (6–4) photoproducts in higher plants (12). Photolyases bind specifically to these DNA lesions and remove them directly by absorbing light in the 300–600 nm range (13,14). Photoreactivation and photolyases have been reported in several plant species (3,12–20). The dark repair includes nucleotide excision repair (NER), base excison repair (BER), mismatch repair (MMR) and other DNA repair pathways. Dark repair has been observed in several plant species, and some of the genes required for dark repair have been identified (5–7,21–29). The wide class of helix-distorting lesions such as CPDs and (6–4) photoproducts are repaired by NER (6,7). The NER sequentially involves recognition of DNA damage, incision on damaged strand, excision of damage containing oligonucleotides, DNA synthesis and ligation. There are two subpathways of NER, designated global genomic repair (GGR) and transcription-coupled repair (TCR). While GGR repairs the DNA damage over the entire genome, TCR is selective for the transcribed DNA strand in expressed genes. Oxidized or hydrated bases and single-strand breaks are repaired by BER. DNA glycosylases initiate this process by releasing the damaged base, with cleavage of the sugar-phosphate chain, excision of the abasic residue or of the abasic residue containing oligonucleotides, and DNA synthesis and ligation (6,7). There are two subpathways for BER. The short-patch BER is DNA polymerase beta-dependent, while the long-patch BER is DNA polymerase delta/epsilon-dependent. Excision repair (NER and BER) is very important for maintaining genome stability and essential for survival for organisms.
We have recently investigated several plant genes related to DNA repair and replication (22,30–40). Interestingly, the expression levels of excision repair genes were found to be high in meristematic tissues where DNA replication actively occurs (22,35). In this study, to ascertain whether this is indeed the case for dark repair, expression patterns of plant genes were analyzed by northern hybridization, in situ hybridization and oligo DNA microarray analysis. We also investigated the DNA repair activity in different tissues (proliferating and non-proliferating) using an in vivo DNA repair assay. The results indicate that excision repair is correlated with cell proliferation while photoreactivation is not.
MATERIALS AND METHODS
Plant materials
Rice plants (Oryza sativa L. cv. Nipponbare) were grown in a growth cabinet under a 16 h day/8 h night cycle at 28°C. Suspended cells of rice (O. sativa L. cv. Nipponbare) were cultured as described previously (41).
Rice DNA repair genes used in this study
We have reported the isolation of cDNAs of UV-damaged DNA binding protein large subunit (UV-DDB1) (22), proliferating cell nuclear antigen (OsPCNA) (35), replication protein A 32 kDa subunit (OsRPA32) (36), flap endonuclease-1a (OsFEN-1a) (30,35,37), origin recognition complex 1 (OsORC1) (38) and Cockayne syndrome WD-repeat protein (OsCSB) from rice, O.sativa, previously. Rice cDNA for the CPD photolyase was referenced from the Knowledge-based Oryza Molecular Biological Encyclopedia (KOME) (http://cdna01.dna.affrc.go.jp/cDNA/) (42) provided by the Rice Genome Resource Center (RGRC; http://www.rgrc.dna.affrc.go.jp/index.html.en). The DDBJ/EMBL/GenBank accession numbers of these sequences are shown in Table 1.
Table 1. Plant DNA repair genes used in this study.
| Gene | Function/remarks | Accession No. | Publication | |
|---|---|---|---|---|
| CPD photolyase | Photolyase | Photolyactivation; removal of CPD | AK111418 | |
| OsUV-DDB1 | UV-DDB large subunit | Dark repair; NER (GGR); damage recognition | AB037144 | (22) |
| OsCSB | Cockayne syndrome WD-repeat protein | Dark repair; NER (TCR); damage recognition | AB111944 | |
| OsPCNA | Proliferating cell nuclear antigen | Dark repair; NER; BER; accessory protein of DNA polymerases | X54046 | (35) |
| OsRPA32 | Replication protein A 32 kDa subunit | Dark repair; NER; ssDNA binding protein; damage recognition | AB037145 | (36) |
| OsFEN-1a | Flap endonuclease-1a | Dark repair; BER; structure-specific endonuclease | AB021666 | (30) |
| OsORC1 | Origin recognition complex 1 | DNA replication; recognition of DNA replication origin | AB037135 | (38) |
CPD, cyclobutane pyrimidine dimer; NER, nucleotide excision repair; GGR, global genome NER; TCR, transcription-coupled NER; BER, base excision repair.
Northern hybridization
Aliquots of 20 µg of total RNA isolated from plant tissues were resolved on 1.2% formaldehyde agarose gels and transferred onto nylon membranes (Hybond-N, Amersham). After prehybridization, the filters were probed with 32P-labeled cDNA at 42°C for 16 h then washed twice with 2× SSC + 0.1% SDS at room temperature for 15 min, and three times with 0.1× SSC + 0.1% SDS at 65°C for 20 min.
In situ hybridization
Riboprobes for in situ hybridization were labeled with digoxigenin-11-rUTP using a DIG RNA Labeling Kit (Boehringer-Mannheim) according to the manufacturer’s protocol. Anti-sense and sense probes were subjected to mild alkaline hydrolysis by heating at 60°C in carbonate buffer and used at a concentration of 2 µg/ml. Plant tissues from 10-day-old rice seedlings were fixed for 16 h at 4°C with a mixture of 4% (w/v) paraformaldehyde and 0.25% (v/v) glutaraldehyde in 50 mM sodium phosphate buffer (pH 7.2). The fixed tissues were dehydrated through a series of xylene and ethanol and embedded in paraffin (HISTPREP 568, Wako). Embedded tissues were sectioned at a thickness of 10 µm and placed on microscope slides precoated with 3-aminopropyltriethoxysilane (APS). Sections were deparaffinized with xylene and rehydrated through a graded ethanol series. They were subsequently pretreated with proteinase K in 100 mM Tris–HCl pH 7.5 and 50 mM EDTA at 37°C for 30 min, dehydrated, and dried under vacuum for 2 h. The hybridization and detection of hybridized riboprobes were performed as described by Sato et al. (43).
In vivo DNA repair assay
To analyze the DNA repair activity in vivo, the amount of DNA damage was determined by ELISA using TDM-2 and 64M-2 monoclonal antibodies (44,45), recognizing CPDs and (6–4) photoproducts, respectively. The rice plant was irradiated by UV-B using a UV-B lamp (TL 20 W/01RS UV-B Medical; Philips). The UV-B radiation intensity was 4 W/m2, as measured using a spectroradiometer (model DRC100X; Spectronics Corporation). The rice tissues were harvested after UV irradiation and ground to a fine powder in liquid nitrogen. DNA was isolated using a DNeasy Plant Kit (Qiagen) and the yield was determined with reference to spectrophotometric absorbance at 260 nm. In the ELISA procedure, heat-denatured DNA was plated onto microtiter well plates and allowed to dry at 37°C. The plates were washed with PT buffer (PBS containing 0.05% Tween) to remove any non-bound DNA, and blocked with a 2% solution of fetal bovine serum for 30 min. The blocking solution was removed by washing in PT. Then, the first antibody (TDM-2 or 64M-2) was added as a dilute solution in PBS and the plates incubated in the dark for 30 min. After washing, the second antibody (biotinylated goat anti-mouse antibody diluted in PBS) was added and incubation continued for 30 min in the dark. Following removal of the second antibody by washing, the final antibody (streptavidin-linked horseradish peroxidase in PBS) was added with incubation for 30 min and the plates were given a final wash with PT, then a single wash in citrate-phosphate buffer (pH 5.0). For the final color development, o-phenylenediamine (1,2-benzenediamine) was used and the color formation stopped by addition of sulfuric acid. The plates were read at 492 nm to give a quantitative evaluation of DNA damage levels.
Oligo DNA microarray analysis
A rice 22K custom oligo DNA microarray kit was used which contains 21 938 oligonucleotides synthesized based on the sequence data of the rice full-length cDNA project (42). Total RNA was purified from shoot apical meristem (SAM) or mature leaves using an RNeasy Plant Kit according to the manufacturer’s instructions (Qiagen) and the yield and RNA purity determined spectrophotometrically. Integrity was checked using an Agilent 2100 Bioanalyzer (Agilent Technology). Total RNA (200 ng) was labeled with Cy-3 or Cy-5 using an Agilent Low RNA Input Fluorescent Linear Amplification Kit (Agilent Technology). Fluorescently labeled targets were hybridized to Agilent rice 22K custom oligo DNA microarrays. Hybridization and wash processes were performed according to the manufacturer’s instructions and hybridized microarrays were scanned using an Agilent Microarray Scanner (Agilent Technology). Feature Extraction software (Agilent Technology) was employed for the image analysis and data extraction processes.
RESULTS
Rice DNA repair genes examined in this study
We have earlier investigated the functions of DNA repair genes such as the genes for UV-DDB1 (22), OsCSB (Y. Tahira, T. Ishibashi, S. Kimura and K. Sakaguchi, unpublished results), OsPCNA (35), OsRPA32 (36) and OsFEN-1a from rice, O.sativa (30,35,37), all of which are thought to be directly related to excision repair. We also described the OsORC1 (38) which is related to replication. We also succeeded in isolating a cDNA of CPD photolyase contributing to photoreactivation. Therefore, cDNAs of all these genes are available in our laboratory and were used here to analyze the expression pattern of DNA repair genes. Table 1 lists the rice repair-related genes investigated. CPD photolyase and OsORC1a were used as representative markers of photoreactivation and DNA replication, respectively. OsPCNA, OsRPA32 and OsFEN-1a are thought to be related to both DNA repair and replication. UV-DDB1, OsCSB, OsPCNA and OsRPA32 are known to be factors for NER, and OsPCNA and OsFEN-1a are factors for BER (5–7,46).
Expression of DNA repair genes in different organs of rice
Total RNA samples were isolated from various organs of 50-day-old rice plants such as shoot apexes (containing SAM), young leaves, mature leaves, flag leaves, panicles and roots, and were blotted and probed with 32P-labeled cDNAs of CPD photolyase, OsUV-DDB1, OsCSB, OsPCNA, OsRPA32, OsFEN-1a and OsORC1 (Fig. 1). Only CPD photolyase was expressed in the mature leaves, and transcripts for factors involved in NER and BER were not detected in mature leaves. The transcripts of these excision repair genes were not detected in the mature leaves even after severe exposure to UV (data not shown). CPDs in DNA from plants grown in natural solar radiation were measured as described by Stapleton et al. (3). According to that study, green leaves had detectable DNA damage, and mature leaves might have efficient repair mechanisms for daily UV-induced DNA damage. On the other hand, all the genes used were expressed in the SAM, the young leaves, the panicles, flag leaves and the roots (Fig. 1). Thus, they all might be expressed in the meristem where cell proliferation is very active.
Figure 1.
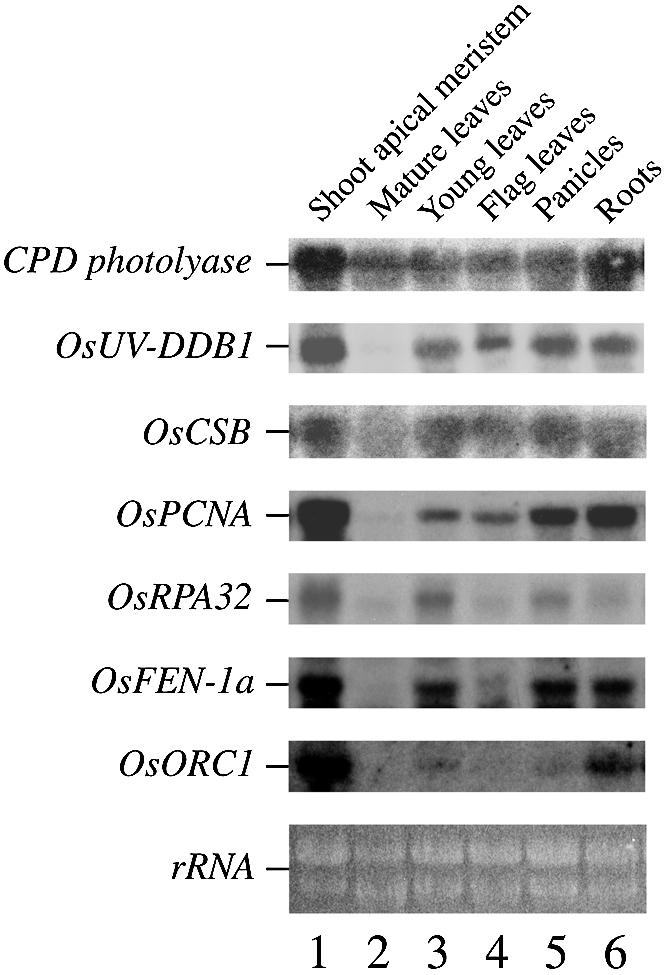
Expression of DNA repair genes in various plant tissues. Each lane was loaded with 20 µg of total RNA isolated from SAM (lane 1), mature leaves (lane 2), young leaves (lane 3), flag leaves (lane 4), panicles (lane 5) or roots (lane 6). The blot was probed with 32P-labeled cDNA. Similar amounts of RNA were loaded in each lane as confirmed by ethidium bromide staining (lowest panel).
Spatial expression patterns of DNA repair genes
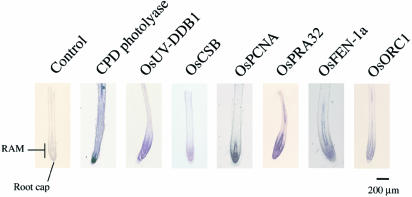
To analyze the expression patterns of the DNA repair genes more precisely, the spatial expression patterns in the regions of the shoot apex (Fig. 2) and root tip (Fig. 3) were investigated by in situ hybridization analysis using digoxigenin-labeled antisense RNA as a probe (Figs 2 and 3). In situ hybridization was performed on paraffin sections of plant tissues from 10-day-old rice seedlings, which were fixed for 16 h at 4°C as described in the Materials and Methods. No hybridization signals were detected when digoxigenin-labeled sense probes of any genes were used (controls in Figs 2 and 3, and data not shown).
Figure 2.
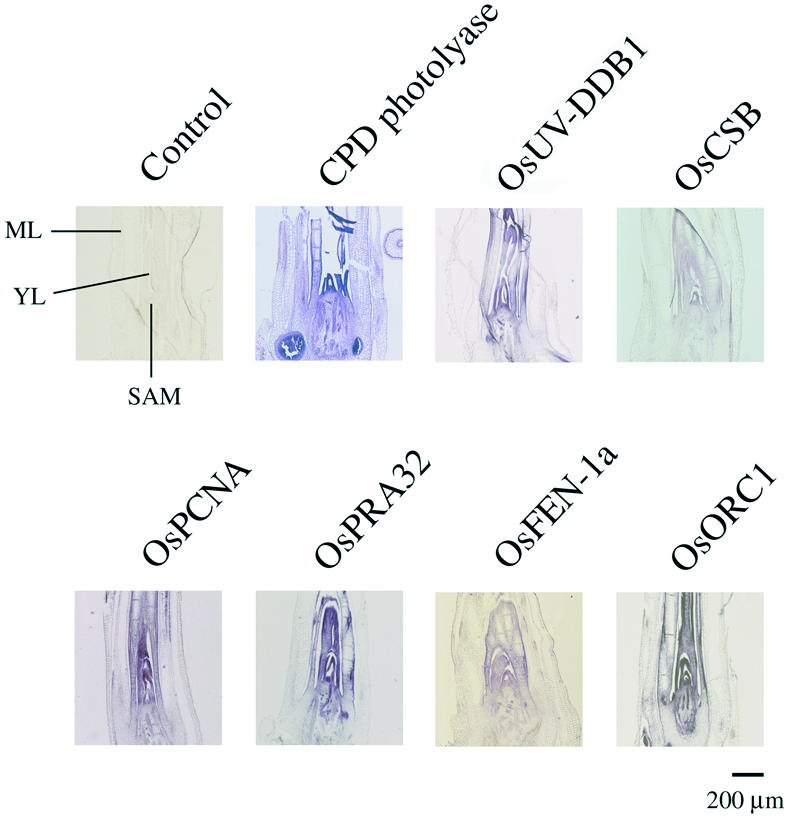
Spatial expression pattern of DNA repair genes in the shoot apex region. In situ hybridization analysis was performed using longitudinal sections from the shoot apex region of 10-day-old rice seedlings. The plant tissues were probed with antisense riboprobe labeled with digoxigenin-UTP. The result using the sense probe for CPD photolyase is shown as a control. No hybridization signals were detected when sense probes for the other genes were used (data not shown).
Figure 3.
Spatial expression pattern of DNA repair genes in the root tip region. In situ hybridization analysis was performed using longtitudinal sections from the root tip region of 10-day-old rice seedlings. The plant tissues were probed with antisense riboprobe labeled with digoxigenin-UTP. The result using the sense probe for CPD photolyase is shown as a control. No hybridization signals were detected when sense probes for the other genes were used (data not shown).
Figures 2 and 3 show the in situ hybridization patterns for CPD photolyase, OsUV-DDB1, OsCSB, OsPCNA, OsRPA32, OsFEN-1a and OsORC1. The antisense probes for each of the genes showed strong hybridization signals in the SAM and root apical meristem (RAM) (Figs 2 and 3). OsUV-DDB1, OsCSB, OsPCNA, OsRPA32, OsFEN-1a and OsORC1 were expressed in SAM but not in the mature leaves where cell proliferation does not occur (Fig. 2). OsUV-DDB1, OsCSB, OsPCNA, OsRPA32, OsFEN-1a and OsORC1 were expressed abundantly in the RAM but not in the root cap (Fig. 3). Only the CPD photolyase signal appeared in non-proliferating tissues such as the mature leaves and elongation zone of roots (Figs 2 and 3). These spatial expression patterns well coincided with the results of the northern hybridization analysis in Figure 1. These results confirmed that all the genes, except CPD photolyase, were mainly expressed mostly in actively proliferating tissues.
In vivo DNA repair assay
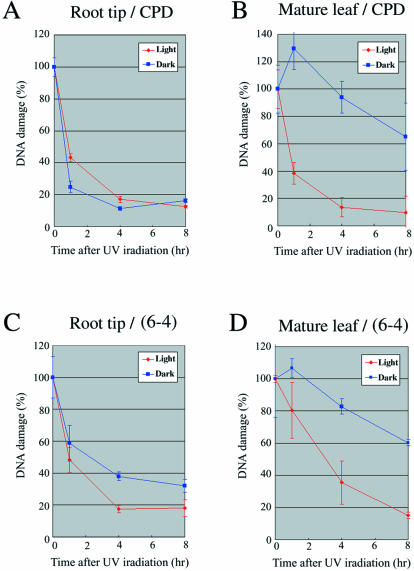
DNA repair activities in proliferating (root tip) and non-proliferating tissues (mature leaves) were measured using an in vivo DNA repair assay. After the irradiation of UV-B at 2000 J/m2, the rice plants were left to stand for 1–8 h under light or in darkness, and then DNA was isolated from the root tip or a mature leaf for the in vivo DNA repair assay. Under light, both CPDs (Fig. 4A and B) and (6–4) photoproducts (Fig. 4C and D) were significantly and time-dependently repaired in root tips (Fig. 4A and C) and mature leaves (Fig. 4B and D). On the other hand, the DNA damage could not be repaired in the mature leaves under darkness (Fig. 4B and D). As shown in Figure 4A and C, however, the repair in the roots did not require light. These results suggested that photoreactivation is the main pathway for removal of DNA damage in mature leaves (non-proliferating cells) and dark repair is coordinated with cell proliferation.
Figure 4.
DNA repair activity of meristematic (root tips) and non-meristematic (mature leaves) tissues. The removal of DNA damage induced by UV-B irradiation was measured under light and in total darkness. (A) Removal of CPDs in the root tips. (B) Removal of CPDs in the mature leaves. (C) Removal of (6–4) photoproducts in the root tips. (D) Removal of (6–4) photoproducts in the mature leaves. Points represent an average of at least three independent experimental values (error bars are 1 SD), expressed as the percentage of the original amount of damage.
Confirmation of gene expression patterns by oligo DNA microarray analysis
To confirm the results of the expression analyses, analysis using the Agilent 22K custom oligo DNA microarray was performed to identify global gene expression patterns (Figs 5–7). This microarray contains 21 938 oligonucle otides synthesized based on the sequence data of the rice full-length cDNA project (42).
Figure 5.
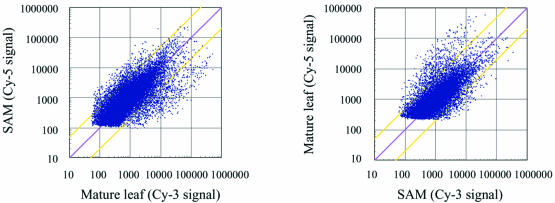
Scatter plots comparing gene expression levels in SAM and mature leaves. An Agilent rice 22K custom oligo DNA microarray was used. Each spot on the array is represented by a dot in the scatter plot. The results of color-swap experiments (left, mature leaf-Cy 3/SAM-Cy 5; right, SAM-Cy 3/mature leaf-Cy 5) are shown. The red line represents the diagonal (equal expression in the two tissues). The yellow line above represents a Cy-3/Cy-5 ratio of 5:1, while that below represents a Cy-3/Cy-5 ratio of 1:5.
Figure 7.
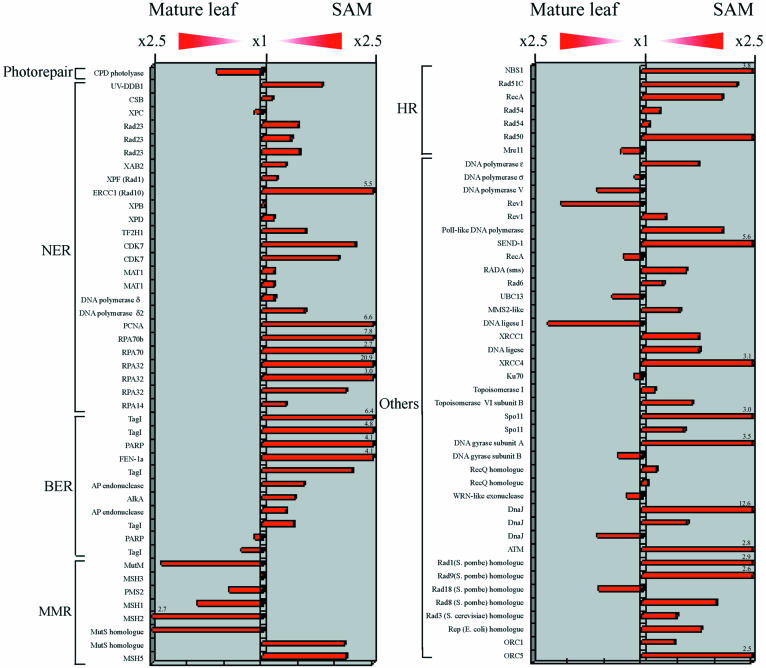
Comparison of expression levels of DNA repair genes between SAM and mature leaves. The expression levels of 90 DNA repair genes are shown in a bar chart. Figures on the bars represent the fold expression.
Total RNA from mature leaves or SAM was hybridized to the oligo DNA microarray to examine the expression of all genes. Figure 5 shows scatter plots comparing total gene expression levels in mature leaf and SAM. Unexpectedly, greater numbers of genes were more strongly expressed in the mature leaves than in the SAM.
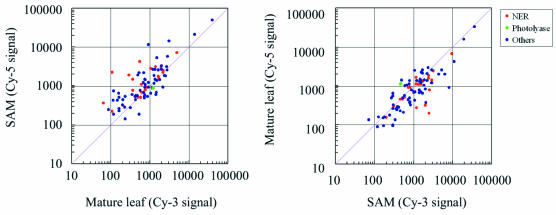
Figure 6 shows scatter plots drawn based on expression data for 90 DNA repair genes spotted on the rice 22K custom oligo DNA microarray slide. The DNA repair genes are involved in photoreactivation, NER, BER, MMR, homologous recombination (HR) and the other repair systems. As shown in Figure 6, CPD photolyase (green dot) was strongly expressed in the mature leaves, but the NER-related genes (red dots) were actively transcribed in SAM. Most of the other DNA repair genes (blue dots) were also actively transcribed in SAM.
Figure 6.
Expression data for rice DNA repair genes. Scatter plots were drawn based on expression data for 90 DNA repair genes spotted on the rice 22K custom oligo DNA microarray slide. Red and green dots represent spots for NER genes and the CPD photolyase gene, respectively. Blue dots represent spots for other dark repair genes (BER, MMR, HR, etc.). The red line represents the diagonal line.
The expression levels of the 90 DNA repair genes are shown in a bar chart in Figure 7. Most of the excision repair genes were strongly expressed in SAM. The expression of dark repair genes in mature leaves was not induced by UV irradiation (data not shown). These results demonstrated that dark repair systems are active in proliferating cells. On the other hand, many of the MMR genes were markedly more expressed in the mature leaves (Figs 6 and 7). Similarly, some of the other DNA repair genes were also transcribed in the mature leaves (Figs 6 and 7). These genes might be involved in DNA repair in mature leaves.
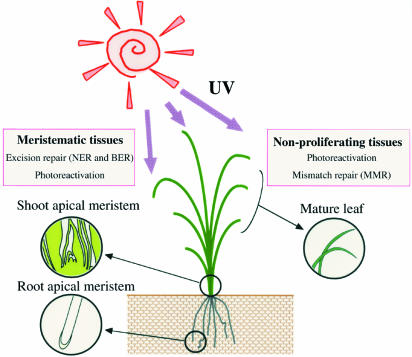
The oligo DNA microarray data strongly support our conclusion that excision repair is active in proliferating cells and UV-induced DNA damage in mature leaves is repaired mostly by photoreactivation (Fig. 8).
Figure 8.
Overview of DNA repair in plants.
DISCUSSION
The present study provided clear evidence that excision repair is active in proliferating cells. This might be critical for maintaining genome stability. In higher plants, the majority of UV-B radiation is absorbed by the epidermis, resulting in heterogeneity in exposure within organs, a feature that may have generated heterogeneity in the biological responses. The plant meristems on the ground must proliferate under UV bombardment because plants utilize sunlight for photosynthesis. Unlike mammals in which the germ cells are set aside early and are completely shielded from UV radiation, plant cells enter meiosis only after significant vegetative growth. Mutations occurring in the SAM may be passed to the gametophytes. Moreover, in higher plants, cells proliferate only in the meristematic tissues.
The present results are summarized in Figure 8. They showed that transcripts for excision repair genes accumulated mainly in the meristem of the shoots and the roots, which actively proliferate. Transcripts of UV-DDB1, OsCSB, OsPCNA, OsRPA32, OsFEN-1a and OsORC1 were not detected in mature leaves despite continued exposure to high levels of UV. Only the CPD photolyase transcript was present in both mature leaves and proliferating tissues. Using the in vivo DNA repair assay, we found photoreactivation to be the main pathway for removal of DNA damage in mature leaves (non-proliferating cells) and dark repair to be coordinated with cell proliferation. These observations indicate that the dark repair gene might have roles in DNA repair during DNA replication. This conclusion was strongly supported by the oligo DNA microarray analysis, revealing that many of the approximately 90 dark repair genes were expressed more in the SAM than in mature leaves. Interestingly, most of the MMR genes were expressed more in mature leaves than the SAM. MMR is necessary for the maintenance of genomic stability and targets mispaired bases that arise through replication errors, during HR, and as a result of DNA damage (47,48). These results suggested that MMR has important roles in DNA repair in mature leaves. Photoreactivation appears to be the main pathway for removal of DNA damage in mature leaves. In contrast, the meristem may need both photoreactivation and excision repair to remove CPDs and (6–4) photoproducts. If one can extrapolate the results obtained from plants to animal systems, then excision repair must work with cell proliferation. Further study of the biological function of DNA repair genes should provide clues to understanding the DNA repair mechanism of higher plants.
Acknowledgments
ACKNOWLEDGEMENTS
We thank the Rice Genome Resource Center for providing the full-length cDNA clone AK111418 and for technical advice on the oligo DNA microarray analysis. We also express our appreciation to Drs Y. Nagamura and H. Motoyama of the National Institute of Agrobiological Sciences and T. Yamamoto, T. Furukawa, Y. Uchiyama, A. Saotome, M. Isogai, N. Sakaguchi, A. Koga, and Y. Sakakibara of Tokyo University of Science for technical advice and helpful discussions. This work was supported by a grant from the Ministry of Agriculture, Forestry and Fisheries of Japan (Rice Genome Project IP-5006) and by a grant from the Ministry of Education, Science, Sports and Culture of Japan [Grant-in-Aid for Young Scientists (B), 15770031]. This work was also supported by a grant from Futaba Electronics Memorial Foundation (Japan) and from the Asahi Glass Foundation (Japan).
REFERENCES
- 1.Ballare C.L., Rousseau,M.C., Searles,P.S., Zaller,J.G., Giordano,C.V., Robson,T.M., Caldwell,M.M., Sala,O.E. and Scopel,A.L. (2001) Impacts of solar ultraviolet-B radiation on terrestrial ecosystems of Tierra del Fuego (southern Argentina). An overview of recent progress. J. Photochem. Photobiol. B, 62, 67–77. [DOI] [PubMed] [Google Scholar]
- 2.Rousseaux M.C., Ballare,C.L., Giordano,C.V., Scopel,A.L., Zima,A.M., Szwarcberg-Bracchitta,M., Searles,P.S., Caldwell,M.M. and Diaz,S.B. (1999) Ozone depletion and UVB radiation: impact on plant DNA damage in southern South America. Proc. Natl Acad. Sci. USA, 96, 15310–15315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stapleton A.E., Thornber,C.S. and Walbot,V. (1997) UV-B component of sunlight causes measurable damage in field-grown maize (Zea mays L.): developmental and cellular heterogeneity of damage and repair. Plant Cell Environ., 20, 279–290. [Google Scholar]
- 4.Ries G., Heller,W., Puchta,H., Sandermann,H., Seidlitz,H.K. and Hohn,B. (2000) Elevated UV-B radiation reduces genome stability in plants. Nature, 406, 98–101. [DOI] [PubMed] [Google Scholar]
- 5.Hays J.B. (2002) Arabidopsis thaliana, a versatile model system for study of eukaryotic genome-maintenance functions. DNA Repair, 1, 579–600. [DOI] [PubMed] [Google Scholar]
- 6.Tuteja N., Singh,M.B., Misra,M.K., Bhalla,P.L. and Tuteja,R. (2001) Molecular mechanisms of DNA damage and repair: progress in plants. Crit. Rev. Biochem. Mol. Biol., 36, 337–397. [DOI] [PubMed] [Google Scholar]
- 7.Britt A.B. (1999) Molecular genetics of DNA repair in higher plants. Trends Plant Sci., 4, 20–25. [DOI] [PubMed] [Google Scholar]
- 8.Schmitz-Hoerner R. and Weissenbock,G. (2003) Contribution of phenolic compounds to the UV-B screening capacity of developing barley primary leaves in relation to DNA damage and repair under elevated UV-B levels. Phytochemistry, 64, 243–255. [DOI] [PubMed] [Google Scholar]
- 9.Rozema J., Bjorn,L.O., Bornman,J.F., Gaberscik,A., Hader,D.P., Trost,T., Germ,M., Klisch,M., Groniger,A., Sinha,R.P., Lebert,M., He,Y.Y., Buffoni-Hall,R., de Bakker,N.V., van de Staaij,J. and Meijkamp,B.B. (2002) The role of UV-B radiation in aquatic and terrestrial ecosystems—an experimental and functional analysis of the evolution of UV-absorbing compounds. J. Photochem. Photobiol. B, 66, 2–12. [DOI] [PubMed] [Google Scholar]
- 10.Stapleton A.E. and Walbot,V. (1994) Flavonoids protect maize DNA from the induction of ultraviolet radiation damage. Plant Physiol., 105, 881–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Landry L.G., Stapleton,A.E., Jim,J., Hoffman,P., Hays,J.B., Walbot,V. and Last,R.L. (1997) An Arabidopsis photolyase mutant is hypersensitive to ultraviolet-B radiation. Proc. Natl Acad. Sci. USA, 94, 328–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dany A.L., Douki,T., Triantaphylides,C. and Cadet,J. (2001) Repair of the main UV-induced thymine dimeric lesions within Arabidopsis thaliana DNA: evidence for the major involvement of photoreactivation pathways. J. Photochem. Photobiol. B, 31, 127–135. [DOI] [PubMed] [Google Scholar]
- 13.Takeuchi Y., Murakami,M., Nakajima,N., Kondo,N. and Nikaido,O. (1996) The photorepair of photoisomerization of DNA lesions in etiolated cucumber cotyledons after irradiation by UV-B depends on wavelength. Plant Cell Physiol., 39, 745–750. [Google Scholar]
- 14.Pang Q. and Hays,J.B. (1991) UV-B inducible and temperature sensitive photoreactivation of cyclobutane pyrimidine dimers in Arabidopsis thaliana. Plant Physiol., 95, 536–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hidema J. and Kumagai,T. (1998) UVB-induced cyclobutyl pyrimidine dimer and photorepair with progress of growth and leaf age in rice. J. Photochem. Photobiol. B, 43, 121–127. [Google Scholar]
- 16.Takahashi S., Nakajima,N., Saji,H. and Kondo,N. (2002) Diurnal change of cucumber CPD photolyase gene (CsPHR) expression and its physiological role in growth under UV-B irradiation. Plant Cell Physiol., 43, 342–349. [DOI] [PubMed] [Google Scholar]
- 17.Tanaka A., Sakamoto,A., IshigakiY., Nikaido,O., Sun,G., Hase,Y., Shikazono,N., Tano,S. and Watanabe,H. (2002) An ultraviolet-B-resistant mutant with enhanced DNA repair in Arabidopsis. Plant Physiol., 129, 64–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Britt A.B., Chen,J.J., Wykoff,D. and Mitchell,D. (1993) A UV-sensitive mutant of Arabidopsis defective in the repair of pyrimidine-pyrimidinone (6–4) dimers. Science, 261, 1571–1574. [DOI] [PubMed] [Google Scholar]
- 19.Hirouchi T., Nakajima,S., Najrana,T., Tanaka,M., Matsunaga,T., Hidema,J., Teranishi,M., Fujino,T., Kumagai,T. and Yamamoto,K. (2003) A gene for a Class II DNA photolyase from Oryza sativa: cloning of the cDNA by dilution-amplification. Mol. Genet. Genomics, 269, 508–516. [DOI] [PubMed] [Google Scholar]
- 20.Hidema J., Kumagai,T. and Southerland,B.M. (2000) UV radiation-sensitive Norin 1 rice contains defective cyclobutane pyrimidine dimer photolyase. Plant Cell, 12, 1569–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hefner E., Preuss,S.B. and Britt,A.B. (2003) Arabidopsis mutants sensitive to gamma radiation include the homologue of the human repair gene ERCC1. J. Exp. Bot., 54, 669–680. [DOI] [PubMed] [Google Scholar]
- 22.Ishibashi T., Kimura,S., Yamamoto,T., Furukawa,T., Takata,K., Uchiyama,Y., Hashimoto,J. and Sakaguchi,K. (2003) Rice UV-DDB homologues are most abundant in proliferating tissues. Gene, 308, 79–87. [DOI] [PubMed] [Google Scholar]
- 23.Liu Z., Hong,S.W., Escobar,M., Vierling,E., Mitchell,D.L., Mount,D.W. and Hall,J.D. (2003) Arabidopsis UVH6, a homolog of human XPD and yeast RAD3 DNA repair genes, functions in DNA repair and is essential for plant growth. Plant Physiol., 132, 1405–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dubest S., Gallego,M.E. and White,C.I. (2002) Role of the AtRad1p endonuclease in homologous recombination in plants. EMBO Rep., 3, 1049–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li A., Schuermann,D., Gallego,F., Kovalchuk,I. and Tinland,B. (2002) Repair of damaged DNA by Arabidopsis cell extract. Plant Cell, 14, 263–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gallego F., Fleck,O., Li,A., Wyrzykowska,J. and Tinland,B. (2000) AtRAD1, a plant homologue of human and yeast nucleotide excision repair endonuclease, is involved in dark repair of UV damages and recombination. Plant J., 21, 507–518. [DOI] [PubMed] [Google Scholar]
- 27.Liu Z., Hossain,G.S., Islas-Osuna,M.A., Mitchell,D.L. and Mount,D.W. (2000) Repair of UV damage in plants by nucleotide excision repair: Arabidopsis UVH1 DNA repair gene is a homolog of Saccharomyces cerevisiae Rad1. Plant J., 21, 519–528. [DOI] [PubMed] [Google Scholar]
- 28.Xu. H., Swoboda,I., Bhalla,P.L., Sijbers,A.M., Zhao,C., Ong,E.K., Hoeijmakers,J.H. and Singh,M.B. (1998) Plant homologue of human excision repair gene ERCC1 points to conservation of DNA repair mechanisms. Plant J., 13, 823–829. [DOI] [PubMed] [Google Scholar]
- 29.Sturm A. and Lienhard,S. (1998) Two isoforms of plant RAD23 complement a UV-sensitive rad23 mutant in yeast. Plant J., 13, 815–821. [DOI] [PubMed] [Google Scholar]
- 30.Kimura S., Furukawa,T., Kasai,N., Mori,M., Kitamoto,H.K., Sugawara,F., Hashimoto,J. and Sakaguchi,K. (2003) Functional characterization of two flap endonuclease-1 (FEN-1) homologues in rice. Gene, 314, 63–71. [DOI] [PubMed] [Google Scholar]
- 31.Yamamoto T., Kimura,S., Mori,Y., Uchiyama,Y., Ishibashi,T., Hashimoto,J. and Sakaguchi,K. (2003) Interaction between proliferating cell nuclear antigen and a JUN-activated-domain-binding proteins 1 in the meristem of rice, Oryza sativa L. Planta, 217, 175–183. [DOI] [PubMed] [Google Scholar]
- 32.Furukawa T., Kimura,S., Ishibashi,T., Hashimoto,J. and Sakaguchi,K. (2003) OsSEND-1: a new Rad2 nuclease family member in higher plant. Plant Mol. Biol., 51, 59–70. [DOI] [PubMed] [Google Scholar]
- 33.Uchiyama Y., Hatanaka,M., Kimura,S., Ishibashi,T., Furukawa,T., Ueda,T., Sakakibara,Y., Matsumoto,T., Hashimoto,J. and Sakaguchi,K. (2002) Characterization of DNA polymerase δ from a higher plant, rice (Oryza sativa L.). Gene, 295, 19–26. [DOI] [PubMed] [Google Scholar]
- 34.Kimura S., Uchiyama,Y., Kasai,N., Saotome,A., Ueda,T., Ando,T., Ishibashi,T., Namekawa,S., Oshige,M., Furukawa,T., Yamamoto,T., Hashimoto,J. and Sakaguchi,K. (2002) A novel DNA polymerase homologous to E. coli DNA polymerase I from a higher plant, rice (Oryza sativa L.). Nucleic Acids Res., 30, 1585–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kimura S., Suzuki,T., Yanagawa,Y., Yamamoto,T., Nakagawa,H., Tanaka,I., Hashimoto,J. and Sakaguchi,K. (2001) Characterization of plant proliferating cell nuclear antigen (PCNA) and flap endonuclease-1 (FEN-1) and their distribution in mitotic and meiotic cell cycles. Plant J., 28, 643–654. [DOI] [PubMed] [Google Scholar]
- 36.Ishibashi T., Kimura,S., Furukawa,T., Hatanaka,M., Hashimoto,J. and Sakaguchi,K. (2001) Two types of replication protein A 70 kDa subunit in rice, Oryza sativa: molecular cloning, characterization and cellular and tissue distribution. Gene, 272, 335–343. [DOI] [PubMed] [Google Scholar]
- 37.Kimura S., Ueda,T., Hatanaka,M., Hashimoto,J. and Sakaguchi,K. (2000) Plant homologue of flap endonuclease-1: molecular cloning, characterization and evidence for expression in meristematic tissues. Plant Mol. Biol., 42, 415–427. [DOI] [PubMed] [Google Scholar]
- 38.Kimura S., Ishibashi,T., Hatanaka,M., Sakakibara,Y., Hashimoto,J. and Sakaguchi,K. (2000) Molecular cloning and characterization of a plant homologue of the origin recognition complex 1 (ORC1). Plant Sci., 158, 33–39. [DOI] [PubMed] [Google Scholar]
- 39.Furukawa T., Ishibashi,T., Kimura,S., Tanaka,H., Hashimoto,J. and Sakaguchi,K. (2003) Characterization of all the subunits of replication factor C from a higher plant, rice (Oryza sativa L.) and their relation to the development. Plant Mol. Biol., 53, 15–25. [DOI] [PubMed] [Google Scholar]
- 40.Yamamoto T., Mori,Y., Ishibashi,T., Uchiyama,Y., Sakaguchi,N., Furukawa,T., Hashimoto,J., Kimura,S. and Sakaguchi,K. (2004) Characterization of Rad6 from a higher plant, rice (Oryza sativa L.) and its interaction with Sgt1, a subunit of the SCF ubiquitin ligase complex. Biochem. Biophys. Res. Commun., 314, 434–439. [DOI] [PubMed] [Google Scholar]
- 41.Baba A., Hasegawa,S. and Syono,K. (1986) Cultivation of rice protoplasts and their transformation mediated by Agrobacterium spheroplasts. Plant Cell Physiol., 27, 463–471. [Google Scholar]
- 42.Kikuchi S. et al. (2003) Collection, mapping and annotation of over 28,000 cDNA clones from japonica rice. Science, 301, 376–379. [DOI] [PubMed] [Google Scholar]
- 43.Sato Y., Hong,S.-K., Tagiri,A., Kitano,H., Yamamoto,N., Nagato,Y. and Matsuoka,M. (1996) A rice homeobox gene, OSH1, is expressed before organ differentiation in a specific region during early embryogenesis. Proc. Natl Acad. Sci. USA, 93, 8117–8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mori T., Nakane,M., Hattori,T., Matsunaga,T., Ihara,M. and Nikaido,O. (1991) Simultaneous establishment of monoclonal antibodies specific for either cyclobutane pyrimidine dimer or (6–4) photoproduct from the same mouse immunized with ultraviolet-irradiated DNA. Photochem. Photobiol., 54, 225–232. [DOI] [PubMed] [Google Scholar]
- 45.Roza L., Van Der Wulp,K.J.M., MacFarlane,S.J., Lohman,P.H.M. and Baan,R.A. (1988) Detection of cyclobutane thymine dimers in DNA of human cells with monoclonal antibody raised against a thymine dimer containing tetranucleotide. Photochem. Photobiol., 48, 627–633. [DOI] [PubMed] [Google Scholar]
- 46.Wood R.D., Mitchell,M., Sgouros,J. and Lindahl,T. (2001) Human DNA repair genes. Science, 291, 1284–1289. [DOI] [PubMed] [Google Scholar]
- 47.Yang W., Junop,M.S., Ban,C., Obmolova,G. and Hsieh,P. (2000) DNA mismatch repair: from structure to mechanism. Cold Spring Harbor Symp. Quant. Biol., 65, 225–232. [DOI] [PubMed] [Google Scholar]
- 48.Culligan K.M. and Hays,J.B. (1997) DNA mismatch repair in plants. Plant Physiol., 115, 833–839. [DOI] [PMC free article] [PubMed] [Google Scholar]