Abstract
Background
Wing discs of B. mori are transformed to pupal wings during the larva-to-pupa metamorphosis with dramatic morphological and structural changes. To understand these changes at a transcriptional level, RNA-seq of the wing discs from 6-day-old fifth instar larvae (L5D6), prepupae (PP) and pupae (P0) was performed.
Results
In total, 12,254 transcripts were obtained from the wing disc, out of which 5,287 were identified to be differentially expressed from L5D6 to PP and from PP to P0. The results of comprehensive analysis of RNA-seq data showed that during larvae-to-pupae metamorphosis, many genes of 20E signaling pathway were up-regulated and those of JH signaling pathway were down-regulated. Seventeen transcription factors were significantly up-regulated. Cuticle protein genes (especially wing cuticle protein genes), were most abundant and significantly up-regulated at P0 stage. Genes responsible for the degradation and de novo synthesis of chitin were significantly up-regulated. There were A and B two types of chitin synthases in B. mori, whereas only chitin synthase A was up-regulated. Both trehalose and D-fructose, which are precursors of chitin synthesis, were detected in the hemolymph of L5D6, PP and P0, suggesting de novo synthesis of chitin. However, most of the genes that are related to early wing disc differentiation were down-regulated.
Conclusions
Extensive transcriptome and DGE profiling data of wing disc during metamorphosis of silkworm have been generated, which provided comprehensive gene expression information at the transcriptional level. These results implied that during the larva-to-pupa metamorphosis, pupal wing development and transition might be mainly controlled by 20E signaling in B. mori. The 17 up-regulated transcription factors might be involved in wing development. Chitin required for pupal wing development might be generated from both degradation of componential chitin and de novo synthesis. Chitin synthase A might be responsible for the chitin synthesis in the pupal wing, while both trehalose and D-fructose might contribute to the de novo synthesis of chitin during the formation of pupal wing.
Electronic supplementary material
The online version of this article (doi:10.1186/1471-2164-15-820) contains supplementary material, which is available to authorized users.
Keywords: B. mori, Wing disc, Metamorphosis, Transcriptome, RNA-Seq
Background
The domestic silkworm, B. mori, is not only an important economic insect, but also a model insect of Lepidoptera. The genome of silkworm has been reported [1, 2], and the genomic resources are available in Silkworm Genome Database: http://silkworm.genomics.org.cn/ and KAIKObase: http://sgp.dna.affrc.go.jp/KAIKObase/ [3, 4].
Wing disc is an imaginal disc of insect wing during larval stage. It consists of undifferentiated and proliferating cells [5]. In B. mori, as larvae develop to pupae, the wing discs undergo dramatic morphological changes and evaginate from the body to form the pupal wing. These changes and processes are regulated by insect hormones and hundreds of genes [6].
20-hydroxyecdysone (20E) and juvenile hormone (JH) are two major hormones in insects and regulate different biological processes, including growth, molting, and reproduction [7, 8]. Both 20E and JH are present at larval stages and they are antagonistic in many aspects. For example, 20E induces periodic ecdysis while JH maintains the "status quo" status of insects at larval stage [9, 10]. Insect metamorphosis is initiated by a surge of 20E at the end of last larval stage when JH titer is low [11]. In the 20E signaling pathway, ecdysone receptor (EcR) binds with ultraspiracle (USP) to form a heterodimer, which binds to the response element of early response genes, such as Broad complex (BR-C), E74 and E75 [12], to initiate the expression. Early-late response genes, including HR3, HR39 and E78, are also involved in the 20E signal transduction pathway [13]. In the JH signal transduction pathway, methoprene-tolerant (Met) was regarded as a potential candidate receptor for JH [14–17]. Knock-out Met led to a down-regulation of JH-response gene Krüppel homolog 1 (Kr-h1), and the insect showed a precocious phenotype in Tribolium castaneum [17]. In B. mori, Met has two isoforms (Met1 and Met2). Met2 combines with steroid receptor coactivator (SRC, also known as "FISC" or "Taiman") to form a heterodimer and induces the expression of Kr-h1 [18]. In B. mori, 20E and JH signaling pathways have been found to be involved in normal wing development [19–21], but no detailed analyses of the two signaling pathways was performed at the transcriptional level.
Genes related to wing disc development at the embryonic stage and early stages were well elucidated in Drosophila. Transcription factors, such as Engrailed (En), Apterous (Ap), Scalloped (Sd) and Vestigial (Vg), secreted proteins, such as Hedgehog (Hh), Decapentaplegic (Dpp), Wingless (Wg) and Fringe (Fng), receptors such as Epidermal growth factor receptor (Egfr) and Notch (N), and their ligands such as Serrate (Ser) and Delta (Dl) were found to be involved in the early differentiation and embryonic development of Drosophila wing discs [5, 22–27]. However their roles in wing transition development from wing disc to pupal wing were not reported.
Cuticle protein and chitin are main structural components of cuticle in insects [28]. Thirteen wing cuticle protein genes (WCP) have been identified in B. mori wing disc, including WCP1a, WCP1b, WCP2, WCP3, WCP4, WCP5, WCP6, WCP71a, WCP7b, WCP8, WCP9, WCP10 and WCP11 [29–31]. Chitin degradation and synthesis in insects were well-studied [28]. Chitinase, β-N-acetylglucosaminidase and chitin deacetylase are three main enzymes that involved in chitin degradation, while four critical enzymes, trehalase, hexokinase, glucose-6-phosphate isomerase and glutamine and fructose-6-phosphate aminotransferase, are needed for the de novo synthesis of chitin (KEGG pathway map: ko00520). There are two kinds of chitin synthases in insects: chitin synthase 1 (class A) is responsible for the chitin synthesis in the epidermis and trachea, while chitin synthase 2 (class B) is critical for the chitin synthesis in the gut peritrophic membrane [28].
For the past decade, a variety of methods such as expressed sequence tags (ESTs) [32, 33], serial analysis of gene expression (SAGE) [34, 35], microarrays [36, 37], and RNA-seq technology [38] have been applied to gene expression analysis in the silkworm, but no information was available on detailed analysis of wing disc development during metamorphosis at the transcriptional level. In this study, RNA-seq method [39, 40] was used to get a comprehensive view on the gene expression of B. mori wing disc during metamorphosis.
Results
Transcriptome sequencing and statistics of gene expression
In total, 4,842,606, 4,741,356 and 4,852,047 clean reads were obtained from the wing disc at 6-day-old fifth instar larvae (L5D6), prepupae (PP) and pupae (P0) stages, respectively. These clean reads were mapped to reference sequences from Silkworm Genome Database using SOAPaligner/soap2 software [41] and their nucleotide sequences were assembled. A total of 12,254 assembled transcripts were obtained and the average length of the transcripts was 1,226 base pairs.
At L5D6, PP and P0 stages 11,316, 11,236, 10,938 assembled transcripts were obtained, respectively (Figure 1). Of the 12,254 assembled transcripts, 9,822 transcripts are commonly shared by the three stages; 607, 373, 312 transcripts are shared by two stages (L5D6 and PP, PP and P0, L5D6 and P0), respectively; 575, 434, 431 transcripts are specifically expressed at L5D6, PP and P0 stage, respectively (Figure 1). These results showed that most transcripts were commonly expressed in each stage, while the stage-specific transcripts might be important for the wing disc development at the specific stage.
Figure 1.
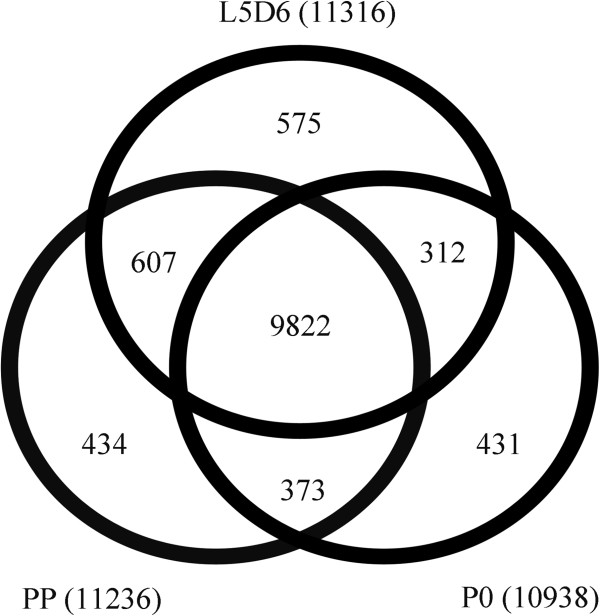
Statistics of transcript numbers and distribution at L5D6, PP and P0.
Differentially expressed transcripts and gene ontology (GO) classification
In total, 5,287 differentially expressed transcripts were identified based on the standard process (see Methods). From L5D6 to PP (L5D6 vs PP), 2,778 transcripts were identified as differentially expressed, among which 926 were up-regulated and 1,852 were down-regulated (Figure 2). From PP to P0 (PP vs P0), 3,864 transcripts were identified as differentially expressed, of which 672 were up-regulated and 3,192 were down-regulated (Figure 2). It is surprising that there were more down-regulated transcripts than up-regulated transcripts during the transition from L5D6 to PP and from PP to P0 (Additional files 1 and 2), implying that with the transitional development of the wing disc during metamorphosis, expression of more and more genes was turned off.
Figure 2.
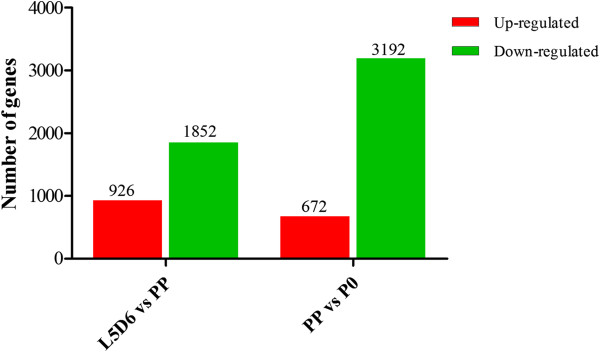
Statistics of differentially expressed transcripts.
It is reasonable to hypothesize that developmental features of the wing disc depend on the activities of differentially expressed transcripts. Therefore, GO classification analysis of differentially expressed transcripts may explain the process of wing disc development. The GO ID numbers of differentially expressed transcripts were obtained from Blast2go software and the GO classification histogram was obtained by uploading GO ID number to BGI WEGO online plot software (see Methods). The results showed that binding and catalytic functions were the most abundant GO function items (>10%), both from L5D6 to PP and PP to P0 developmental stages (Figure 3A and 3B). Specifically, in the cellular component ontology, no dominant GO items of the up-regulated transcripts were found for L5D6 vs PP, while several dominant GO items of the down-regulated transcripts were found, including cell, cell part, macromolecular complex, membrane-enclosed lumen, organelle and organelle part (Figure 3A). The dominant GO items of the significantly up-regulated transcripts over down-regulated transcripts for L5D6 vs PP are catalytic, molecular transducer, structural molecular and transporter in molecular function ontology (Figure 3A). The significantly dominant GO item of the down-regulated transcripts over the up-regulated transcripts for L5D6 vs PP in molecular function ontology was transcription regulator, while the down-regulated transcripts of other functional GO items were not significantly more than the up-regulated transcripts in the same catalogs, including translation regulator, in which no up-regulated transcripts were detected for L5D6 vs PP (Figure 3A). For biological process ontology for L5D6 vs PP, the dominant GO items of the up-regulated transcripts included establishment of localization, localization and post-embryonic development, while the dominant GO items of the down-regulated transcripts included cellular component biogenesis, cellular component organization, cellular process and pigmentation (Figure 3A). Although some GO items in the three ontologies, such as synapse, synapse part, antioxidant, electron carrier and translation regulator, showed obviously distinctive differences in the numbers of transcript expression, they were not statistically significant (p < 0.05) by Pearson test [42] (Figure 3A).
Figure 3.
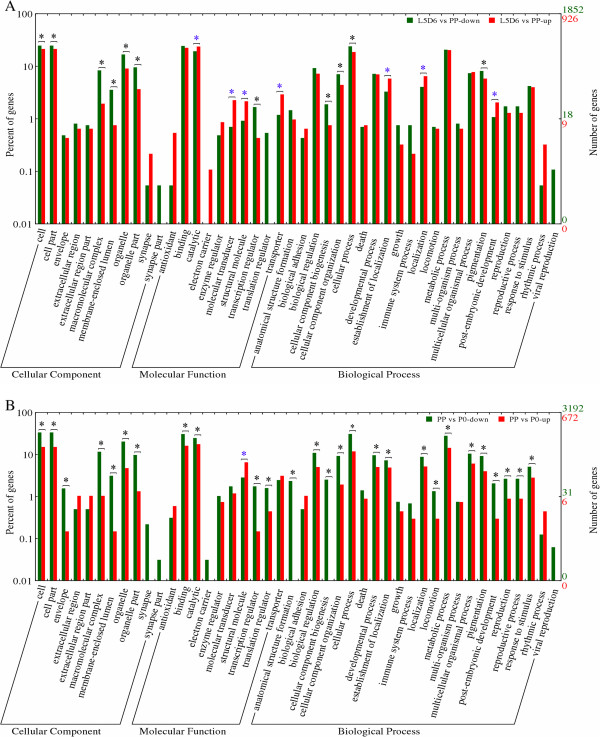
GO classification of the differentially expressed transcripts. The GO classification map was done by uploading the GO ID numbers of genes to BGI WEGO homepage mirror (GO archive: 2009-10-01). Asterisks indicate that the differences between the two stages are significant (p < 0.05). (A) GO classification of the differentially expressed transcripts in L5D6 vs PP. (B) GO classification of the differentially expressed transcripts in PP vs P0.
On the other hand, for PP vs P0, the dominant GO items of significantly down-regulated transcripts in cellular component ontology included most of catalogues except extracellular (part) and synapse (part), while no dominant GO items of significantly up-regulated transcripts were found (Figure 3B). In molecular function ontology, the only dominant GO item of the significantly up-regulated transcripts was structural molecule; the dominant GO items of the significantly down-regulated transcripts included binding, catalytic, transcription regulator and translation regulator, while the other GO items were not significantly different between up- and down-regulated transcripts (Figure 3B). For cellular process, no dominant GO items of the significantly up-regulated transcripts were detected, while most catalogues were significantly dominant for the down-regulated transcripts over the up-regulated transcripts (Figure 3B).
The 20E signal transduction pathway was more active than the JH signal transduction pathway during metamorphosis
To investigate activity of the 20E and JH signal transduction pathways in the wing disc during metamorphosis, the expression patterns of the genes that are involved in the 20E and JH signal transduction pathways were identified and analyzed.
The results revealed that most of the genes that are associated with the 20E signaling pathway, such as E74, E75 and HR3, were significantly up-regulated during metamorphosis, whereas most of the genes that are involved in the JH signaling pathway, such as SRC and Kr-h1, were significantly down-regulated (Tables 1 and 2). The expression levels of the 20E-related genes appeared to be higher than those of the JH-related genes. This implied that 20E signal is actively involved in the wing disc development during the larva-to-pupa metamorphosis. The assembled nucleotide sequences of transcripts (RNA-seq ID) in Tables 1 and 2 were provided in Additional files 3 and 4, respectively.
Table 1.
Changes in expression levels of the genes involved in the 20E signal transduction pathway
| Gene | RNA-Seq ID | L5D6-RPKM | PP-RPKM | P0-RPKM | L5D6 vs PP | PP vs P0 |
|---|---|---|---|---|---|---|
| USP 1 | Bm_nscaf2847_250 | 34.71 | 16.10 | 19.17 | - | + |
| Bm_nscaf2847_251 | 20.51 | 14.60 | 21.78 | - | + | |
| ECR A | Bm_nscaf2855_215 | 0.93 | 4.57 | 0.93 | + | - |
| ECR B2 | Bm_nscaf2855_217 | 21.95 | 2.09 | 8.55 | -3.40* | + |
| ECR B1 | Bm_nscaf2855_219 | 31.24 | 24.62 | 18.58 | - | - |
| E74 1 | Bm_nscaf2888_430 | none | 3.07 | 162.43 | + | 5.73* |
| Bm_nscaf2888_433 | 19.63 | 5.34 | 64.70 | - | 3.60* | |
| E75B | Bm_nscaf2859_001 | 45.67 | 100.29 | 168.69 | -1.13* | + |
| E75C | Bm_nscaf2859_003 | 13.21 | none | 4.83 | - | + |
| HR3 | Bm_nscaf2964_066 | 2.03 | 268.66 | 4.91 | 7.05* | -5.78* |
| BR-C A-NZ1 | Bm_nscaf2970_078 | 178.90 | 235.58 | 67.39 | + | - |
| BR-C Z4 | Bm_nscaf2970_079 | 71.94 | 122.93 | 38.45 | + | - |
| β FTZ-F1 1 | Bm_nscaf1690_227 | 4.32 | 9.94 | none | + | -13.28* |
| Bm_nscaf1690_228 | 8.48 | 7.36 | 1.71 | - | - |
1These genes have more than one RNA-Seq sequences. Gene expression level was calculated by RPKM method. "none" means that the expression of the gene was not detected. Asterisks indicate the gene, whose expression level had more than two folds difference between two stages and False Discovery Rate (FDR) was less than 0.001, was identified as differentially expressed at this developmental stage. "+" represents up-regulation, and "-" represents down-regulation. And the values in the last two columns were calculated by Log2(PP-RPKM/L5D6-RPKM) or Log2(P0-RPKM/PP-RPKM) and only those differentially expressed genes are given.
Table 2.
Changes in expression level of the genes involved in the JH signal transduction pathway
| Gene | RNA-Seq ID | L5D6-RPKM | PP-RPKM | P0-RPKM | L5D6 vs PP | PP vs P0 |
|---|---|---|---|---|---|---|
| Met1 | Bm_nscaf2828_039 | 45.41 | 11.65 | 8.92 | -1.96* | - |
| Met2 1 | Bm_nscaf1690_114 | 1.42 | none | none | - | → |
| Bm_nscaf1690_115 | 1.85 | 1.64 | 0.19 | - | - | |
| SRC 1 | Bm_nscaf3078_04 | none | 3.35 | none | + | - |
| Bm_nscaf3078_05 | 8.99 | 10.38 | 3.22 | + | -1.69 * | |
| Kr-h1 | Bm_nscaf2589_179 | 95.84 | 2.77 | 1.77 | -5.11 * | - |
1These genes have more than one RNA-Seq sequences. Expression level of the genes was calculated by RPKM method. "none" means that the expression of the genes was not detected. "+" represents up-regulation, "-" represents down-regulation, and "→" represents no change. Asterisks indicate the gene, whose expression level had more than two folds difference between two stages and False Discovery Rate (FDR) was less than 0.001, was identified as differentially expressed at this developmental stage. And the values in the last two columns were calculated by Log2(PP-RPKM/L5D6-RPKM) or Log2(P0-RPKM/PP-RPKM) and only those differentially expressed genes are given.
Expression of transcription factors in the wing disc during metamorphosis
Transcription factors play critical roles in regulation of expression of their target genes. To identify transcription factors that may regulate the development of B. mori wing discs during metamorphosis, transcription factors were identified by KEGG analysis. In total, 147 out of 12,254 assembled transcripts were identified as transcription factors, 60 of which were differentially expressed during metamorphosis. Among the 60 differentially expressed transcription factors, 17 were up-regulated at PP and/or P0 stage (Table 3). Some of them were selected for confirmation by qRT-PCR analysis. The results showed that the transcription factors Bm_nscaf2589_261 (hormone receptor HR38), Bm_nscaf1898_501 (D-ETS-4-like isoform X2), Bm_nscaf2876_46 (bHLH protein 15-like), Bm_nscaf2888_249 (nuclear receptor GRF), Bm_nscaf2902_040 (protein delilah-like), Bm_nscaf3090_4 (chorion specific C/EBP) and Bm_scaffold316_2 (protein embryonic gonad-like) were significantly up-regulated at PP stage, which were well consistent with the RNA-Seq results (Table 3 and Figure 4A-C, E-H). The transcription factors Bm_nscaf1898_212 (protein pokkuri-like), Bm_nscaf2770_58 (SOX-4-like), Bm_nscaf2847_110 (Krueppel-like factor 10-like) and Bm_nscaf2930_060 (enhancer of split mbeta) were significantly up-regulated at P0 stage, which were also consistent with its RNA-Seq results (Table 3 and Figure 4I-L). The transcription factor Bm_nscaf2847_349 (protein escargot-like) was highly expressed at W1 stage, which was not consistent with the RNA-Seq result (Table 3 and Figure 4D). The qRT-PCR results showed that the RNA-Seq results were generally consistent. Bm_nscaf2859_001 (E75 isoform B), Bm_nscaf2964_066 (hormone receptor 3C) and Bm_nscaf2888_433 (transcription factor E74) were significantly up-regulated at PP or P0 stage (Table 3), which was consisted with the reported results [43, 44]. ASH2 was essential for scales formation in B. mori wing during pupal stage [45] and we found this transcription factor (Bm_nscaf1898_167) was significantly up-regulated at P0 stage. The assembled nucleotide sequences of transcripts (RNA-seq ID) in Table 3 were provided in Additional file 5, and primers for qRT-PCR were provided in Additional file 6.
Table 3.
Up-regulated transcription factors during the larva-to-pupa metamorphosis
| RNA-Seq ID | log 2(PP/L5D6) | log 2(P0/PP) | Gene description |
|---|---|---|---|
| Bm_nscaf2589_261 | 5.93* | 1.43* | probable nuclear hormone receptor HR38 |
| Bm_nscaf1898_501 | 2.55* | D-ETS-4-like isoform X2 | |
| Bm_nscaf2859_001 | 1.13* | E75 isoform B | |
| Bm_nscaf2876_46 | 15.42* | bHLH protein 15-like | |
| Bm_nscaf2734_12 | 6.05* | -6.63* | hepatic leukemia factor-like isoform X4 |
| Bm_nscaf2847_349 | 1.77* | -2.46* | protein escargot-like |
| Bm_nscaf2888_249 | 8.43* | -1.46* | nuclear receptor GRF |
| Bm_nscaf2902_040 | 4.45* | -4.90* | helix-loop-helix protein delilah-like |
| Bm_nscaf2964_066 | 7.05* | -5.78* | hormone receptor 3C |
| Bm_nscaf3090_4 | 6.65* | -17.02* | chorion specific C/EBP |
| Bm_scaffold316_2 | 2.86* | -2.64* | protein embryonic gonad-like |
| Bm_nscaf1898_167 | 2.41* | achaete-scute-like protein ASH2 | |
| Bm_nscaf1898_212 | 3.88* | ets DNA-binding protein pokkuri-like | |
| Bm_nscaf2770_58 | 2.82* | transcription factor SOX-4-like | |
| Bm_nscaf2847_110 | 3.76* | Krueppel-like factor 10-like | |
| Bm_nscaf2888_433 | 3.60* | transcription factor E74 | |
| Bm_nscaf2930_060 | 2.00* | enhancer of split mbeta |
Asterisks indicate that the gene, whose expression level had more than two folds difference between two stages and False Discovery Rate (FDR) was less than 0.001, was identified as differentially expressed at this developmental stage. The values in the second and third columns were calculated by Log2(PP-RPKM/L5D6-RPKM) or Log2(P0-RPKM/PP-RPKM) and only those differentially expressed genes are given. Genes with black bold ID were selected for qRT-PCR analysis.
Figure 4.
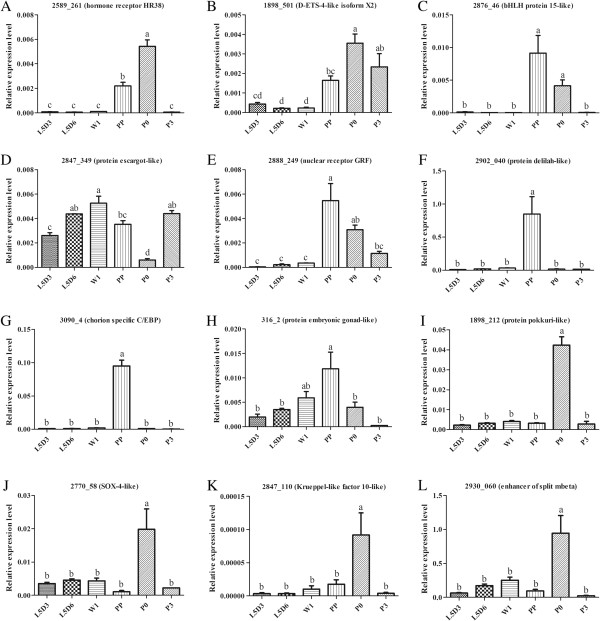
Expression of the selected 12 transcription factors by qRT-PCR. Expression patterns of the selected 12 transcription factors in the wing disc at indicated six developmental stages, which are 3-day-old fifth instar larvae (L5D3), L5D6, 1-day-old wandering (W1), PP, P0 and 3-day-old pupae (P3). (A) Bm_nscaf2589_261 (probable nuclear hormone receptor HR38). (B) Bm_nscaf1898_501 (D-ETS-4-like isoform X2). (C) B m_nscaf2876_46 (bHLH protein 15-like). (D) Bm_nscaf2847_349 (protein escargot-like). (E) Bm_nscaf2888_249 (nuclear receptor GRF). (F) Bm_nscaf2902_040 (helix-loop-helix protein delilah-like). (G) Bm_nscaf3090_4 (chorion specific C/EBP). (H) Bm_scaffold316_2 (protein embryonic gonad-like). (I) Bm_nscaf1898_212 (ets DNA-binding protein pokkuri-like). (J) Bm_nscaf2770_58 (transcription factor SOX-4-like). (K) Bm_nscaf2847_110 (Krueppel-like factor 10-like). (L) Bm_nscaf2930_060 (enhancer of split mbeta). The relative expression levels were normalized to the Bmβ-actin levels. The values are mean ± SEM (n = 3). Different letters indicate statistical significance (p < 0.05).
Changes in expression level of cuticle protein genes in wing disc during metamorphosis
Cuticle proteins are important component of wing disc in the post-larva development. To study the relationship between expression of cuticle proteins and wing development, cuticle protein genes were analyzed. Totally 205 cuticle protein transcripts were identified using Blast search. Ninety-seven transcripts were expressed at all the three stages, while 7, 9 and 58 transcripts were specifically expressed at L5D6, PP and P0 stage respectively (Figure 5A). One hundred forty eight out of 205 transcripts were differentially expressed, with most of them being up-regulated during the larva-to-pupa metamorphosis, particularly from PP to P0 stage (Figure 5B). This result was consistent with the results by microarray analysis [36]. By similarity analysis of sequences, 10 WCPs genes were identified in our RNA-Seq dataset. Expression of WCP1a, WCP1b, WCP3, WCP4, WCP5, WCP8 and WCP9 was slightly up-regulated at the PP stage and then greatly increased at the P0 stage. Expression of WCP10 was slightly up-regulated at the PP stage and then greatly decreased at the P0 stage, whereas the expression of WCP11 was decreased at the PP stage and then greatly increased at the P0 stage (Table 4).
Figure 5.
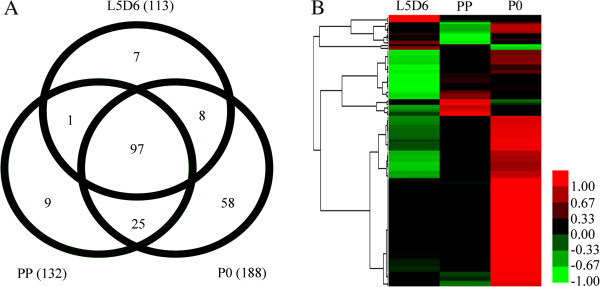
Expression analysis of cuticular or cuticle protein coding genes. The RPKM values of undetected genes are viewed as 0.001. Cluster 3.0 software was used to plot the heat map (Similarity Metric: Euclidean distance, Clustering method: Complete linkage), and Treeview software was used to generate the heat map. The colors in the map display the relative values at the given 3 developmental stages. Green indicates the lowest level of expression, black indicates the intermediate level of expression, and red indicates the highest level of expression. (A) Distribution of cuticular or cuticle protein coding genes at the three stages. (B) Heat map of hierarchical clustering of 148 differentially expressed cuticle protein genes.
Table 4.
Expression level of WCP genes
| WCPs | RNA-Seq ID | L5D6-RPKM | PP-RPKM | P0-RPKM |
|---|---|---|---|---|
| WCP1a | Bm_nscaf1681_169 | 2.72 | 3.76 | 5361.40 |
| WCP1b | Bm_nscaf1681_171 | 1.10 | 72.00 | 4902.09 |
| WCP2 | Bm_nscaf1681_167 | 1.35 | 29.18 | 6441.87 |
| WCP3 | Bm_nscaf1681_163 | 3.68 | 176.15 | 16652.38 |
| WCP4 | Bm_nscaf1681_161 | none | 7.91 | 744.39 |
| WCP5 | Bm_nscaf1681_160 | 21.79 | 88.31 | 48948.29 |
| WCP8 | Bm_nscaf1681_192 | 2.38 | 161.58 | 10340.63 |
| WCP9 | Bm_nscaf1681_048 | 19.65 | 143.49 | 15882.70 |
| WCP10 | Bm_nscaf2136_095 | 437.87 | 699.39 | 17.69 |
| WCP11 | Bm_nscaf1681_049 | 487.93 | 1.05 | 197.21 |
The gene expression level was calculated by RPKM method. "none" means that the expression of the gene was not detected.
Change in expression level of chitin degradation and synthesis related genes
To study the dynamic of chitin in wing disc during the pupal wing transition, the genes that are involved in chitin degradation and synthesis pathways were investigated. Most of the genes that are related to chitin degradation were significantly up-regulated at the PP stage and the genes that are related to chitin synthesis were also up-regulated at the PP or P0 stage (Tables 5 and 6). Two critical chitin degradation related genes [Bm_nscaf2993_299 (chitin deacetylase 4) and Bm_nscaf3031_201 (β-N-acetylglucosaminidase 1 precursor)] were significantly up-regulated at the PP stage, then down-regulated at the P0 stage (Figure 6A, B), which were consistent with the results of RNA-Seq (Table 5). Chitin synthesis related genes including Bm_nscaf3027_063 (hexokinase), Bm_nscaf2887_059 (glutamine:F-6-P aminotransferase), Bm_nscaf2823_132 (glucosamine-6-P-N-acetyltransferase), Bm_nscaf2589_266 (chitin synthase A) were significantly up-regulated at the PP or P0 stage, and Bm_nscaf2589_269 (chitin synthase B) was significantly down-regulated from L5D3 to P3 stage (Figure 7A-E), which were generally consistent with the results of RNA-Seq (Table 6). The assembled nucleotide sequences of transcripts (RNA-seq ID) in Tables 5 and 6 were provided in Additional files 7 and 8, respectively. Primers for qRT-PCR were provided in Additional file 6.
Table 5.
Expression level of the chitin degradation related genes
| RNA-Seq ID | L5D6-RPKM | PP-RPKM | P0-RPKM | Gene description |
|---|---|---|---|---|
| Bm_nscaf2912_25 | 44.80 | 5923.77 | 246.74 | chitinase |
| Bm_nscaf2986_109 | 45.50 | 13836.70 | 477.85 | chitinase precursor |
| Bm_nscaf2829_151 | 82.40 | 230.91 | 246.51 | chitinase 3-like isoform X1 |
| Bm_nscaf2829_153 | 52.70 | 157.91 | 173.42 | chitinase |
| Bm_nscaf2993_229 | 28.70 | 156.08 | 100.65 | chitin deacetylase 4 |
| Bm_nscaf2993_230 | 44.40 | 184.62 | 142.59 | chitin deacetylase 4 |
| Bm_nscaf3031_201 | 3.87 | 186.23 | 14.43 | β-N-acetylglucosaminidase 1 precursor |
| Bm_nscaf463_07 | 1.86 | none | 0.37 | β-N-acetylglucosaminidase 2 precursor |
| Bm_nscaf463_08 | 0.35 | 0.34 | none | β-N-acetylglucosaminidase 3 precursor |
The gene expression level was calculated by RPKM method. "none" means that the expression of the gene was not detected. The genes with black bold ID were selected for qRT-PCR analysis.
Table 6.
Expression levels of the chitin synthesis related genes
| RNA-seq ID | L5D6-RPKM | PP-RPKM | P0-RPKM | Gene description |
|---|---|---|---|---|
| Bm_nscaf2800_23 | 20.24 | 12.65 | 8.76 | trehalase-2 |
| Bm_nscaf2800_24 | 11.56 | 9.11 | 13.95 | trehalase-2 |
| Bm_nscaf2829_133 | 0.21 | none | 1.28 | trehalase precursor |
| Bm_nscaf2829_134 | 0.42 | 1.88 | none | trehalase 1B |
| Bm_nscaf3027_062 | 6.90 | 15.30 | 8.68 | hexokinase type 2-like isoform X3 |
| Bm_nscaf3027_063 | 47.91 | 105.58 | 100.81 | hexokinase type 2-like isoform X3 |
| Bm_nscaf2780_10 | 122.98 | 145.82 | 36.81 | glucose-6-phosphate isomerase |
| Bm_nscaf2887_059 | 53.54 | 51.33 | 675.08 | glutamine:fructose-6-phosphate aminotransferase |
| Bm_nscaf2823_132 | 77.53 | 155.12 | 886.62 | glucosamine-6-phosphate N-acetyltransferase |
| Bm_nscaf3003_158 | 13.61 | 56.53 | 34.73 | phosphoacetylglucosamine mutase |
| Bm_nscaf2176_247 | 17.11 | 14.13 | 436.84 | UDP-N-acetylhexosamine pyrophosphorylase |
| Bm_nscaf2589_266 | 49.30 | 138.56 | 277.03 | chitin synthase A |
| Bm_nscaf2589_269 | 3.55 | 0.27 | none | chitin synthase B |
| Bm_nscaf2589_270 | 5.42 | none | none | chitin synthase B |
The gene expression level was calculated by RPKM method. "none" means that the expression of the gene was not detected. The genes with black bold ID were selected for qRT-PCR analysis.
Figure 6.
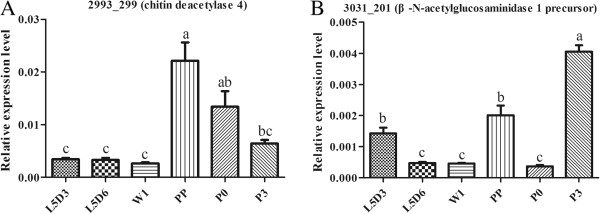
qRT-PCR results of expression of the chitin degradation related genes. The expression patterns of two chitin degradation genes in the wing disc at six developmental stages, which are 3-day-old fifth instar larvae (L5D3), L5D6, 1-day-old wandering (W1), PP, P0 and 3-day-old pupae (P3) The relative expression levels were normalized to the Bmβ-actin levels. (A) Bm_nscaf2993_229 (chitin deacetylase 4). (B) Bm_nscaf3031_201 (β-N-acetylglucosaminidase 1 precursor). The values are the mean ± SEM (n = 3) of three repeat experiments using qRT-PCR. Different letters indicate statistical significance (p < 0.05).
Figure 7.
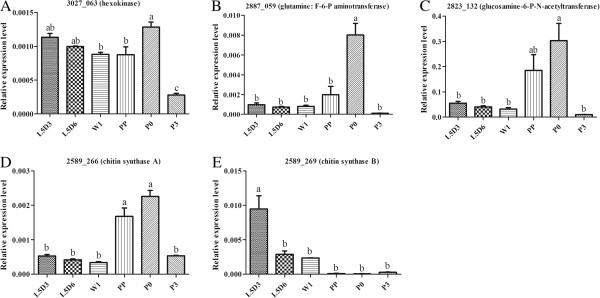
qRT-PCR results of 5 genes relevant to chitin synthesis. The expression patterns of five selected chitin synthesis related genes in the wing disc at six developmental stages. which are 3-day-old fifth instar larvae (L5D3), L5D6, 1-day-old wandering (W1), PP, P0 and 3-day-old pupae (P3). (A) Bm_nscaf3027_063 (hexokinase type 2-like isoform X3). (B) Bm_nscaf2887_059 (glutamine: fructose-6-phosphate aminotransferase). (C) Bm_nscaf2823_132 (glucosamine-6-phosphate N-acetyltransferase). (D) Bm_nscaf2589_266 (chitin synthase A). (E) Bm_nscaf2589_269 (chitin synthase B). The relative expression levels were normalized to the Bmβ-actin levels. The values are the mean ± SEM (n = 3) of three repeat experiments using qRT-PCR. Different letters indicate statistical significance (p < 0.05).
De novo synthesis of chitin in insects usually utilizes trehalose as the starting material. However, base on the pathway map of chitin synthesis (KEGG pathway map: ko00520), D-fructose can also be used for chitin de novo synthesis. In this study, high performance liquid chromatography (HPLC) was used to determine the levels of trehalose, D-glucose and D-fructose in the hemolymph of the silkworm at L5D6, PP and P0 stage. The results showed that trehalose, D-glucose and D-fructose were presented with high concentrations at L5D6 stage, and then their concentrations rapid decreased at the PP stage (Table 7). The concentrations of the three sugars were slightly decreased from the PP to P0 stage (Table 7). These results imply that the rapid use of the three sugars in the hemolymph might be taking place from L5D6 to PP stage.
Table 7.
Concentrations of trehalose, D-glucose and D-fructose in the hemolymph at three stages of B. mori
| Sample | Trehalose (mg/ml) | D-glucose (mg/ml) | D-fructose (mg/ml) |
|---|---|---|---|
| hemolymph of L5D6 | 5.14 | 0.33 | 0.51 |
| hemolymph of PP | 2.71 | 0.27 | 0.40 |
| hemolymph of P0 | 2.56 | 0.23 | 0.39 |
Changes in the expression levels of wing disc development related genes
Expression patterns of the genes homologous to Drosophila wing early development were also analyzed in this study. The results showed that most of the genes, such as En, Ap, Hh, reported to be important for embryonic and early development of Drosophila wing disc were significantly decreased during metamorphosis (Table 8). However, Fng that was necessary for B. mori wing development during larvae to pupae metamorphosis [21], Ash2 that was vital for scales formation of B. mori wing during pupal stage [45], and urbain, an unknown gene that was up-regulated at PP stage in B. mori wing [46], were also significantly up-regulated at the PP or P0 stage in this study. The assembled nucleotide sequences of transcripts (RNA-seq ID) in Table 8 were provided in Additional file 9.
Table 8.
Expression patterns of the genes related to wing disc early development
| Gene | RNA-seq ID | L5D6-RPKM | PP-RPKM | P0-RPKM | L5D6 vs PP | PP vs P0 |
|---|---|---|---|---|---|---|
| engrailed | Bm_nscaf2964_172 | 57.41 | 27.17 | 25.47 | -1.08* | - |
| Hedgehog | Bm_nscaf3048_40 | 13.70 | 5.56 | 3.24 | - | - |
| decapentaplegic | Bm_nscaf2993_221 | 38.81 | 22.75 | 17.10 | - | - |
| apterous AE | Bm_nscaf2210_128 | 117.49 | 57.39 | 27.28 | -1.03* | - |
| apterous AF | Bm_nscaf2210_130 | 31.10 | 27.52 | 8.94 | - | -1.62* |
| apterous Bβ | Bm_nscaf2210_132 | 95.88 | 83.98 | 16.08 | - | -2.38* |
| Fringe 1 | Bm_nscaf2860_94 | 13.82 | 40.86 | 15.17 | 1.56* | -1.43* |
| Bm_nscaf2860_96 | 45.94 | 74.37 | 28.61 | + | -1.37* | |
| delta | Bm_nscaf2986_021 | 12.75 | 6.39 | 3.16 | - | - |
| serrate | Bm_nscaf1705_01 | 83.35 | 35.30 | 18.98 | -1.24* | - |
| Notch 1 | Bm_nscaf2888_347 | 9.04 | 4.05 | 3.31 | - | - |
| Bm_nscaf2888_348 | 5.91 | 4.25 | 9.16 | - | + | |
| Bm_nscaf2888_349 | 17.65 | 14.18 | 7.54 | - | - | |
| Wingless(precursor) | Bm_nscaf2847_167 | 17.20 | 5.51 | 0.86 | -1.64* | - |
| vestigial | Bm_nscaf3056_42 | 17.08 | 7.54 | 7.70 | - | + |
| scalloped | Bm_nscaf1898_116 | 52.42 | 25.22 | 14.10 | -1.06* | - |
| Scalloped isoform 2 | Bm_nscaf1898_117 | 44.05 | 35.26 | 8.31 | - | - |
| achaete-scute 2 | Bm_nscaf1898_167 | 2.04 | 7.54 | 40.06 | + | 2.41* |
| urbain | Bm_nscaf2902_362 | 45.40 | 11497.96 | 161.44 | 7.98* | -6.15* |
1These genes have more than one RNA-Seq sequences. The gene expression level was calculated by RPKM method. "+" represents up-regulation, and "-" represents down-regulation. Asterisks indicate that the gene, whose expression level had more than two folds difference between two stages and False Discovery Rate (FDR) was less than 0.001, was identified as differentially expressed at this developmental stage. And the values in the last two columns were calculated by Log2(PP-RPKM/L5D6-RPKM) or Log2(P0-RPKM/PP-RPKM) and only those differentially expressed genes are given.
Discussion
Insect metamorphosis is mediated by 20E and JH signals. During larval stage, JH exhibits its "status quo" function probably through two transcription factors, E75A and Kr-h1, to prevent the activation of BR-C, a 20E signal transduction gene for larvae-pupae transition [7, 47]. Though a relative high titer of JH was present in hemolymph of B. mori at the end of the final instar larvae [48], the JH does not inhibit the wing disc transformation. Some studies implied that the success of this transition was attributed to the loss of sensitivity of JH signal in B. mori wing disc at the end of the instar larvae [19, 20]. However, the mechanism for the JH insensitiveness in the wing disc is not revealed yet. In this study, during the larva-to-pupa metamorphosis, 20E signal transduction pathway related genes, such as E74, E75 and HR3, were significantly up-regulated in B. mori wing disc. However JH signal transduction pathway related genes, such as Met, SRC and Kr-h1, were significantly down-regulated and their expression levels were relatively low. High expression of BR-C was detected in B. mori wing disc, while the expression of E75A was not detectible before pupation (Tables 1 and 2). These results implied that the loss of sensitivity to JH is probably due to the low expression of JH-related genes during larvae-to-pupae metamorphosis, resulting that the B. mori wing disc can be transformed to pupal wing despite high JH titer in the hemolymph before pupation. However what causes the inactive expression of transmitters of JH signaling during metamorphosis has not been revealed yet.
Out of the 17 up-regulated transcription factors, 10 were significantly up-regulated at the PP stage (Table 3). These genes may be involved in positive regulation of the transition of wing disc to pupal wing. In Drosophila, HR38 was involved in regulating the expression of cuticle protein genes during pupal stage [49]. Cuticle protein genes were actively transcribed in B. mori wing at P0 stage (Figure 5B), so HR38 might be involved in modulating their expression. Previous study indicated that GRF might be a cofactor of βFTZ-F1 and HR39 [50]. βFTZ-F1 was reported to have an essential role in regulating the expression of cuticle protein [51, 52]. In this way, the transcription factor GRF may be positively involved in regulating the expression of cuticle protein, which induces the growth of the wing disc during the larva-to-pupa transformation. Transcription factors HR3 and E75 were involved in 20E signal transduction and they were found to be capable of binding with the response element of many cuticle protein genes [12, 53]. Therefore, they may also be involved in regulating the expression of cuticle proteins in the wing disc during metamorphosis. In Drosophila, gonad, also known as Eagle, is important for the development of nervous system, and egMZ360/egMZ360 (a partial loss-of-function allele) Drosophila showed a held-out wing phenotype and lost the ability of flying [54, 55], implying that Eagle may be involved in the normal development of wing. C/EBP transcription factor was reported to have a role in regulating chorion gene expression in silkmoth, however, its role in wing disc development has not been studied yet [56]. Delilah is a marker gene for epidermal muscle attachment site in Drosophila and may play a role in epidermal muscle attachment site development [57, 58], but its role for B. mori wing development has not been clarified yet.
Out of the 17 transcription factors, except for the PP up-regulated transcription factors, six transcription factors were up-regulated only at the P0 stage (Table 3) which may play a role in pupal wing development in B. mori. For example, ASH2 is required for normal formation of wing scales in B. mori during pupal stage [45]. Transcription factor Pokkuri is also known as Yan and Aop in Drosophila. Yan is regulated by epidermal growth factor (EGF). Ectopic expression of Yan in the A-P boundary of Drosophila wing disc can lead to the mis-expression of Vein, a gene involved in normal development of wing vein and the small wing, and result in the loss of bristles phenotype [59, 60]. This implies that Pokkuri is involved in wing development, but the exact role of Pokkuri in B. mori wing development is still uncovered. SOX-4, a HMG-box transcription factor, is an activator in regulating the expression of neuronal gene and necessary for neuronal maturation [61]. Nervous system can be found in wing vein and the up-regulated SOX-4 at P0 stage may be involved in regulating the neuronal development in B. mori wing vein. Pupal wing of B. mori has reached to the proper size of its adult wing. Krueppel-like factor 10 is an anti-proliferative factor in mammals [62]. It is possible that when B. mori wing disc transforms to pupal wing, cell proliferation is negatively regulated by Krueppel-like factor 10, resulting in the arrest of wing growth when it reaches the proper size. E74 is an early response gene in 20E signaling pathway and is involved in the expression regulation of many cuticle protein genes [12, 52]. Its high level of expression in the wing disc suggests its potential role in regulating the expression of cuticle protein during the wing development. Study in Drosophila suggests that Enhancer of split mbeta [E(spl)mβ] has a function to limit the expression of Rho (Rhomboid), a vein marker gene, in the Drosophila wing vein [59, 63]. In B. mori, wing veins are optimized during pupal stage. Therefore, E(spl)mβ may play similar role as in Drosophila. Functions of the other up-regulated transcription factors remain unknown.
GO classification analyses indicated that the significantly up-regulated transcripts are dominant in structural molecule GO item (Figure 3A and 3B). Actually, cuticle protein genes have high expression levels during the transition of from larval wing disc to pupal wing (Figure 5B and Table 4). Cuticle protein genes were highly expressed at the P0 stage and their RPKM value accounted for about 45.5% of RPKM from all genes (see Additional file 10). These results implied that the large amount of cuticle proteins are synthesized, which is one of the main events related to morphological changes in wing, during larva-to-pupa metamorphosis. In addition, among the 13 WCPs genes identified in B. mori, WCP 1–9 were highly expressed around pupation, while WCP10 and WCP11 have high expression levels before pupation and then significantly down-regulated during pupation [29–31]. In this study, 10 WCPs can be found and their expression patterns are similar to previous studies (Table 4). These results implied that WCP1-9 are the main cuticle proteins for pupal wing, while WCP10 and WCP11 are main cuticle proteins for wing disc in B. mori.
As insects metamorphosize from larvae to pupae, old integument of larvae is degraded and replaced by pupal integument. Wing disc undergoes rapid growth and differentiation during metamorphosis, evaginating to form pupal wing. No cuticle is degraded in B. mori wing disc during the transition from the wing disc to pupal wing. In this study, relevant genes for both chitin degradation and de novo synthesis were significantly up-regulated (Tables 5 and 6 and Figures 6 and 7). This implied that the pupal wing chitin may come from both chitin degradation and de novo synthesis. Chitin deacetylases can catalyze chitin to form chitosan and had a function in organizing tracheal tube length in Drosophila [64, 65]. Chitin deacetylases had higher expression in wing disc at PP stage in B. mori. Whether it involves in extending of tracheal tube in wing is still unknown. In B. mori, there are two kinds of chitin synthase, A and B. While synthase A is responsible for the chitin synthesis in the epidermis, synthase B is responsible for the chitin synthesis in the gut [28]. However it is not clear which synthase is involved in chitin synthesis in insect wing. In this study, synthase A was significantly up-regulated in wing disc during metamorphosis, while the expression of synthase B was almost undetectable (Table 6 and Figure 7D, 7E). This result suggested that the chitin synthesis in pupal wing is majorly performed by synthase A. In Drosophila and Tribolium, Knickkopf (Knk) and Retroactive (Rtv) are essential for chitin synthesis and always along with the expression of chitin synthase A [66, 67]. In Lepidopterans, these two factors had not been studied so far. But Bm_nscaf3003_024 (protein Skeletor, isoforms D/E-like), the homolog of Knk, was found to be significantly up-regulated at PP stage in our data (Additional file 1), whose expression pattern was consistent with that of chitin synthase A (Figure 7D), implying that Protein Skeletor may have similar function as Knk did in Drosophila and Tribolium. The homolog of Rtv was not detected in our data.
In addition, D-fructose can be transformed to D-fructose 6-phosphate by hexokinase, which can be used by chitin synthesis pathway (KEGG pathway map: ko00520). Trehalose, D-glucose and D-fructose were detected in B. mori hemolymph, and their amounts rapidly decreased from L5D6 to PP (Table 7). This suggests that the rapid de novo synthesis of chitin might be taken place in the wing during transition from L5D6 to PP, and both trehalose and D-fructose may be used in de novo synthesis of chitin in the wing of B. mori.
Early gene Fng was also necessary for the B. mori wing development during larva-to-pupa metamorphosis [5, 21, 68]. To investigate other early function genes, expression patterns of these genes were analyzed during metamorphosis in B. mori (Table 8). Results showed that most of them, including Ap and En, which were reported to play important roles in cell fate determination in Drosophila wing disc during early developmental stages, were significantly down-regulated in B. mori wing during metamorphosis, implying that cell fate determination of B. mori wing disc may be decided before pupation.
Conclusion
In this work, total 12,254 assembled transcripts were obtained from the wing disc at L5D6, PP and P0 stages. Totally 5,287 differentially expressed assembled transcripts were identified. From L5D6 to PP (L5D6 vs PP) 2,778 transcripts were differentially expressed and from PP to P0 (PP vs P0) 3,864 transcripts were differentially expressed. More down-regulated transcripts than up-regulated transcripts were identified during the transition from L5D6 to PP and from PP to P0. The transition of wing disc to pupal wing appeared under the control of the 20E signal transduction pathway because high levels of the transcription factors related to 20E signal transduction pathway were found during larva-to-pupa metamorphosis. Seventeen up-regulated transcription factors were screened out, and their functions on B. mori wing development were predicated. High levels of the expression of cuticle proteins and chitin related genes reflect the active chitin metabolism related to the morphological and structural changes during the metamorphosis from the wing disc to pupal wing. Chitin source of pupal wing may come from both degradation of componential chitin and de novo synthesis. Chitin synthase A might be responsible for the chitin synthesis in B. mori pupal wing. De novo synthesis of chitin initiated from D-fructose may exist in B. mori wing.
Methods
Experimental animal and RNA isolation
Silkworm, B. mori, were reared with mulberry leaves at 27°C under a 12 h light/12 h dark cycle. Under this condition, the fifth instar stage lasted about 7 days; the wandering stage (W) lasted about 3 days and pupal stage lasted about 10 days. The stage between the end of larval spinning and the onset of pupation was defined as prepupae (PP), which lasted about 12 hours, equal to the period from 2.5-day-old wandering to 3-day-old wandering. P0 was defined as the stage when B. mori just finished pupation. B. mori wing discs were clearly dissected from larvae or pupae at stage of L5D6, PP and P0. Total RNA was isolated by using Rneasy Kit (QIAGEN) according to the manufacturer’s instructions.
RNA-seq sequencing and de novoassembly of sequences
Five μg of total RNA from L5D6, PP and P0 wing was prepared, respectively, for performing RNA-seq sequencing analysis by using Ilumina HiSeq™ 2000 platform. The clean reads of each gene detected in RNA-seq sequencing analysis was assembled by using Trinity software (de novo assembly) [40] and the assemble results were provided in supplemental files.
Calculation of gene expression level and annotation of predicted proteins
Expression level of genes is calculated by using RPKM (Reads Per kb per Million reads) method [69], and the formula is:
Here C means the number of reads that uniquely aligned to target gene; N means the total number of reads that uniquely aligned to all genes; L means the number of base on target gene.
Genes were annotated by using Blast2go software ( http://www.blast2go.com/b2ghome) according to the translated nucleotide sequences in Blastp.
Isolation and analysis of differentially expressed genes
To compare the differentially expressed genes, the transcripts of PP were compared with those of L5D6 (L5D6 as control), and the transcripts of P0 were compared with those of PP (PP as control). Those genes that had two fold difference between two stages and False Discovery Rate (FDR) was less than 0.001 [70] were identified as differentially expressed genes. The detailed information of their expression patterns can be obtained in the supplemental files (Additional files 1 and 2).
The Cluster 3.0 and Treeview software ( http://www.eisenlab.org/eisen/) were used to analyze the expression pattern of differentially expressed genes. Kyoto Encyclopedia of Genes and Genomes (KEGG: http://www.kegg.jp/) [71] was used to identify transcription factors. BGI Web Gene Ontology Annotation Plotting (WEGO) ( http://wego.genomics.org.cn/cgi-bin/wego/index.pl) [72] were used to classify genes to different GO catalogs and Pearson Chi-Square test was used to justify if the gene numbers of the two input dataset were significantly different.
Quantitative real time -PCR (qRT-PCR)
RNAiso Plus (Takara Dalian, China) was used to isolate total RNA from B. mori wing disc from 3-day-old fifth instar larvae (L5D3), L5D6, 1-day-old wandering (W1), PP, P0 and 3-day-old pupae (P3), respectively. Two μg of total RNAs from each stage was used to synthesize cDNA using reverse transcriptase (Takara) and oligo d (T) primer (Takara). The total volume of real-time quantitative PCR reactions was 20 μl, which contained 10 μl of 2 × SYBR® Premix EX Taq™ (Takara), 0.4 μl of 50 × ROX Reference Dye (Takara) and 0.4 μl of specific primers (10 μM). Detail information of qRT-PCR Primers were provided in Additional file 4. qRT-PCR was performed with an ABI7300 real-time PCR system (Applied Biosystems, Foster City, CA), using the following condition: 95°C for 30 s followed by 40 cycles in 95°C for 5 s, 60°C for 30 s and 72°C for 31 s. The mRNA quantity of each gene was calculated with the 2-△△Ct method and normalized to the abundance of a house-keeping gene, Bmβ-actin (GenBank NO: EU780706.1). The relative mRNA levels of each gene were represented as folds over the expression levels of Bmβ-actin.
Hemolymph collection and sugar detection
The hemolymph of B. mori was collected by cutting its proleg (L5D6 and PP stages) or pupal wing (P0 stage), and quickly transformed to centrifuge tube by pipette. All hemolymph samples were stored at -80°C, until performing the sugar detection. The sugar detection was performed by China National Analytical Center (Guangzhou) by HPLC method.
Availability of supporting data
The assembled unigenes (larger than 200 bp) were deposited in the Transcriptome Shotgun Assembly Sequence Database (TSA) at DDBJ/EMBL/GenBank under the accession GBJR00000000. The version described in this paper is the first version, GBJR01000000.
Electronic supplementary material
Additional file 1: Table S1: Differentially expressed transcripts in L5D6-VS-PP. The criteria applied for significance difference were FDR ≤ 0.001, and estimated absolute |log2Ratio (PP/L5D6)| ≥ 1. L5D6- and PP-RPKM: reads per kb per million reads for each transcript in stages L5D6 and PP, respectively. Log2 Ratio (PP/L5D6): the ratio between the RPKM in PP and the RPKM in L5D6. Up-Down (PP/L5D6) and FDR of each gene are also shown. GO Component, GO Function and Go Process: ontology information of Cellular Components, Molecular Function and Biological Processes of Gene-corresponding GO terms. "-": no hit. Blast nr: identification of homologues in GenBank; E-values are also shown. (XLS 980 KB)
Additional file 2: Table S2: Differentially expressed transcripts in PP-VS-P0. Description of data is similar to Table S1. Log2 Ratio (P0/PP): the ratio between the RPKM in P0 and the RPKM in PP. (XLS 1 MB)
Additional file 3: Assembled nucleotide sequences of transcripts in Table 1. (DOC 36 KB)
Additional file 4: Assembled nucleotide sequences of transcripts in Table 2. (DOC 32 KB)
Additional file 5: Assembled nucleotide sequences of transcripts in Table 3. (DOC 42 KB)
Additional file 6: Table S3: Primers for qRT-PCR. (XLS 21 KB)
Additional file 7: Assembled nucleotide sequences of transcripts in Table 5. (DOC 34 KB)
Additional file 8: Assembled nucleotide sequences of transcripts in Table 6. (DOC 44 KB)
Additional file 9: Assembled nucleotide sequences of transcripts in Table 8. (DOC 44 KB)
Additional file 10: Table S4: All transcripts at P0 stage. All transcripts at stage P0 were listed. Description of data is similar to Table S1. (XLS 3 MB)
Acknowledgements
This research was supported by grants from National Basic Research Program of China (2012CB114602) and National Natural Science Foundation of China (31172265; 31330071).
Abbreviations
- L5D3
3-day-old fifth instar larvae
- W1
1-day-old wandering
- P3
3-day-old pupae
- L5D6
6-day-old fifth instar larvae
- PP
Prepupae
- P0
Pupae
- 20E
20-hydroxyecdysone
- JH
Juvenile hormone
- EcR
Ecdysone receptor
- USP
Ultraspiracle
- BR-C
Broad complex
- Met
Methoprene-tolerant
- Kr-h1
Krüppel homolog 1
- SRC
Steroid receptor coactivator
- En
Engrailed
- Ap
Apterous
- Sd
Scalloped
- Vg
Vestigial
- Hh
Hedgehog
- Dpp
Decapentaplegic
- Wg
Wingless
- Fng
Fringe
- Egfr
Epidermal growth factor receptor
- N
Notch
- Ser
Serrate
- Dl
Delta
- WCP
Wing cuticle protein
- GO
Gene ontology
- RPKM
Reads Per kb per Million reads
- FDR
False Discovery Rate
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- WEGO
BGI Web Gene Ontology Annotation Plotting.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
QLF and LL conceived and designed the study. HMD collected and prepared RNA samples for RNA-seq. JO performed RNA extraction and qRT-PCR, JO, LL and LHH contributed to bioinformatic analysis. JO, QLF, LL and SCZ contributed to writing the manuscript and data analysis. All authors read and approved the final manuscript.
Contributor Information
Jun Ou, Email: oujun198706@126.com.
Hui-Min Deng, Email: denghuiminmin@163.com.
Si-Chun Zheng, Email: sczheng62@126.com.
Li-Hua Huang, Email: huanglh@scnu.edu.cn.
Qi-Li Feng, Email: qlfeng@scnu.edu.cn.
Lin Liu, Email: liul@scnu.edu.cn.
References
- 1.Xia Q, Zhou Z, Lu C, Cheng D, Dai F, Li B, Zhao P, Zha X, Cheng T, Chai C, Pan G, Xu J, Liu C, Lin Y, Qian J, Hou Y, Wu Z, Li G, Pan M, Li C, Shen Y, Lan X, Yuan L, Li T, Xu H, Yang G, Wan Y, Zhu Y, Yu M, Shen W, et al. A draft sequence for the genome of the domesticated silkworm (Bombyx mori) Science. 2004;306(5703):1937–1940. doi: 10.1126/science.1102210. [DOI] [PubMed] [Google Scholar]
- 2.International Silkworm Genome C The genome of a lepidopteran model insect, the silkworm Bombyx mori. Insect Biochem Mol Biol. 2008;38(12):1036–1045. doi: 10.1016/j.ibmb.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 3.Wang J, Xia Q, He X, Dai M, Ruan J, Chen J, Yu G, Yuan H, Hu Y, Li R, Feng T, Ye C, Lu C, Wang J, Li S, Wong GK, Yang H, Wang J, Xiang Z, Zhou Z, Yu J. SilkDB: a knowledgebase for silkworm biology and genomics. Nucleic Acids Res. 2005;33(Database issue):D399–D402. doi: 10.1093/nar/gki116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shimomura M, Minami H, Suetsugu Y, Ohyanagi H, Satoh C, Antonio B, Nagamura Y, Kadono-Okuda K, Kajiwara H, Sezutsu H, Nagaraju J, Goldsmith MR, Xia Q, Yamamoto K, Mita K. KAIKObase: an integrated silkworm genome database and data mining tool. BMC Genomics. 2009;10:486. doi: 10.1186/1471-2164-10-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klein T. Wing disc development in the fly: the early stages. Curr Opinion Genetics Dev. 2001;11(4):470–475. doi: 10.1016/S0959-437X(00)00219-7. [DOI] [PubMed] [Google Scholar]
- 6.Deng H, Zhang J, Li Y, Zheng S, Liu L, Huang L, Xu WH, Palli SR, Feng Q. Homeodomain POU and Abd-A proteins regulate the transcription of pupal genes during metamorphosis of the silkworm, Bombyx mori. Proc Natl Acad Sci U S A. 2012;109(31):12598–12603. doi: 10.1073/pnas.1203149109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dubrovsky EB. Hormonal cross talk in insect development. Trends in Endocrinol Metab TEM. 2005;16(1):6–11. doi: 10.1016/j.tem.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 8.Thummel CS. Molecular mechanisms of developmental timing in C. elegans and Drosophila. Dev Cell. 2001;1(4):453–465. doi: 10.1016/S1534-5807(01)00060-0. [DOI] [PubMed] [Google Scholar]
- 9.Truman JW, Riddiford LM. The origins of insect metamorphosis. Nature. 1999;401(6752):447–452. doi: 10.1038/46737. [DOI] [PubMed] [Google Scholar]
- 10.Truman JW, Riddiford LM. Endocrine insights into the evolution of metamorphosis in insects. Annu Rev Entomol. 2002;47:467–500. doi: 10.1146/annurev.ento.47.091201.145230. [DOI] [PubMed] [Google Scholar]
- 11.Riddiford LM, Hiruma K, Zhou X, Nelson CA. Insights into the molecular basis of the hormonal control of molting and metamorphosis from Manduca sexta and Drosophila melanogaster. Insect Biochem Mol Biol. 2003;33(12):1327–1338. doi: 10.1016/j.ibmb.2003.06.001. [DOI] [PubMed] [Google Scholar]
- 12.Thummel CS. Ecdysone-regulated puff genes 2000. Insect Biochem Mol Biol. 2002;32(2):113–120. doi: 10.1016/S0965-1748(01)00112-6. [DOI] [PubMed] [Google Scholar]
- 13.Huet F, Ruiz C, Richards G. Sequential gene activation by ecdysone in Drosophila melanogaster: the hierarchical equivalence of early and early late genes. Development. 1995;121(4):1195–1204. doi: 10.1242/dev.121.4.1195. [DOI] [PubMed] [Google Scholar]
- 14.Ashok M, Turner C, Wilson TG. Insect juvenile hormone resistance gene homology with the bHLH-PAS family of transcriptional regulators. Proc Natl Acad Sci U S A. 1998;95(6):2761–2766. doi: 10.1073/pnas.95.6.2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Charles JP, Iwema T, Epa VC, Takaki K, Rynes J, Jindra M. Ligand-binding properties of a juvenile hormone receptor, Methoprene-tolerant. Proc Natl Acad Sci U S A. 2011;108(52):21128–21133. doi: 10.1073/pnas.1116123109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Konopova B, Jindra M. Juvenile hormone resistance gene Methoprene-tolerant controls entry into metamorphosis in the beetle Tribolium castaneum. Proc Natl Acad Sci U S A. 2007;104(25):10488–10493. doi: 10.1073/pnas.0703719104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parthasarathy R, Tan A, Palli SR. bHLH-PAS family transcription factor methoprene-tolerant plays a key role in JH action in preventing the premature development of adult structures during larval-pupal metamorphosis. Mech Dev. 2008;125(7):601–616. doi: 10.1016/j.mod.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kayukawa T, Minakuchi C, Namiki T, Togawa T, Yoshiyama M, Kamimura M, Mita K, Imanishi S, Kiuchi M, Ishikawa Y, Shinoda T. Transcriptional regulation of juvenile hormone-mediated induction of Kruppel homolog 1, a repressor of insect metamorphosis. Proc Natl Acad Sci U S A. 2012;109(29):11729–11734. doi: 10.1073/pnas.1204951109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Obara Y, Miyatani M, Ishiguro Y, Hirota K, Koyama T, Izumi S, Iwami M, Sakurai S. Pupal commitment and its hormonal control in wing imaginal discs. J Insect Physiol. 2002;48(10):933–944. doi: 10.1016/S0022-1910(02)00160-9. [DOI] [PubMed] [Google Scholar]
- 20.Koyama T, Obara Y, Iwami M, Sakurai S. Commencement of pupal commitment in late penultimate instar and its hormonal control in wing imaginal discs of the silkworm, Bombyx mori. J Insect Physiol. 2004;50(2–3):123–133. doi: 10.1016/j.jinsphys.2003.09.009. [DOI] [PubMed] [Google Scholar]
- 21.Sato K, Matsunaga TM, Futahashi R, Kojima T, Mita K, Banno Y, Fujiwara H. Positional cloning of a Bombyx wingless locus flugellos (fl) reveals a crucial role for fringe that is specific for wing morphogenesis. Genetics. 2008;179(2):875–885. doi: 10.1534/genetics.107.082784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kubota K, Goto S, Eto K, Hayashi S. EGF receptor attenuates Dpp signaling and helps to distinguish the wing and leg cell fates in Drosophila. Development. 2000;127(17):3769–3776. doi: 10.1242/dev.127.17.3769. [DOI] [PubMed] [Google Scholar]
- 23.Bray S. Drosophila development: Scalloped and Vestigial take wing. Current Biol CB. 1999;9(7):R245–R247. doi: 10.1016/S0960-9822(99)80154-7. [DOI] [PubMed] [Google Scholar]
- 24.Morata G. How Drosophila appendages develop. Nat Rev Mol Cell Biol. 2001;2(2):89–97. doi: 10.1038/35052047. [DOI] [PubMed] [Google Scholar]
- 25.Schwank G, Basler K. Regulation of organ growth by morphogen gradients. Cold Spring Harbor Perspect Biol. 2010;2(1):a001669. doi: 10.1101/cshperspect.a001669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Swarup S, Verheyen EM. Wnt/Wingless signaling in Drosophila. Cold Spring Harbor Perspect Biol. 2012;4:6. doi: 10.1101/cshperspect.a007930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neto-Silva RM, Wells BS, Johnston LA. Mechanisms of growth and homeostasis in the Drosophila wing. Annu Rev Cell Dev Biol. 2009;25:197–220. doi: 10.1146/annurev.cellbio.24.110707.175242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Merzendorfer H, Zimoch L. Chitin metabolism in insects: structure, function and regulation of chitin synthases and chitinases. J Exp Biol. 2003;206(Pt 24):4393–4412. doi: 10.1242/jeb.00709. [DOI] [PubMed] [Google Scholar]
- 29.Takeda M, Mita K, Quan GX, Shimada T, Okano K, Kanke E, Kawasaki H. Mass isolation of cuticle protein cDNAs from wing discs of Bombyx mori and their characterizations. Insect Biochem Mol Biol. 2001;31(10):1019–1028. doi: 10.1016/S0965-1748(01)00050-9. [DOI] [PubMed] [Google Scholar]
- 30.Noji T, Ote M, Takeda M, Mita K, Shimada T, Kawasaki H. Isolation and comparison of different ecdysone-responsive cuticle protein genes in wing discs of Bombyx mori. Insect Biochem Mol Biol. 2003;33(7):671–679. doi: 10.1016/S0965-1748(03)00048-1. [DOI] [PubMed] [Google Scholar]
- 31.Futahashi R, Okamoto S, Kawasaki H, Zhong YS, Iwanaga M, Mita K, Fujiwara H. Genome-wide identification of cuticular protein genes in the silkworm, Bombyx mori. Insect Biochem Mol Biol. 2008;38(12):1138–1146. doi: 10.1016/j.ibmb.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 32.Gan LP, Zhang WY, Niu YS, Xu L, Xi J, Ji MM, Xu SQ. Construction and application of an electronic spatiotemporal expression profile and gene ontology analysis platform based on the EST database of the silkworm, Bombyx mori. J Insect sci. 2010;10:114. doi: 10.1673/031.010.11401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mita K, Morimyo M, Okano K, Koike Y, Nohata J, Kawasaki H, Kadono-Okuda K, Yamamoto K, Suzuki MG, Shimada T, Goldsmith MR, Maeda S. The construction of an EST database for Bombyx mori and its application. Proc Natl Acad Sci U S A. 2003;100(24):14121–14126. doi: 10.1073/pnas.2234984100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Funaguma S, Hashimoto S, Suzuki Y, Omuro N, Sugano S, Mita K, Katsuma S, Shimada T. SAGE analysis of early oogenesis in the silkworm, Bombyx mori. Insect Biochem Mol Biol. 2007;37(2):147–154. doi: 10.1016/j.ibmb.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 35.Zhang Y, Huang J, Jia S, Liu W, Li M, Wang S, Miao X, Xiao H, Huang Y. SAGE tag based cDNA microarray analysis during larval to pupal development and isolation of novel cDNAs in Bombyx mori. Genomics. 2007;90(3):372–379. doi: 10.1016/j.ygeno.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 36.Liang J, Zhang L, Xiang Z, He N. Expression profile of cuticular genes of silkworm, Bombyx mori. BMC Genomics. 2010;11:173. doi: 10.1186/1471-2164-11-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xia Q, Cheng D, Duan J, Wang G, Cheng T, Zha X, Liu C, Zhao P, Dai F, Zhang Z, He N, Zhang L, Xiang Z. Microarray-based gene expression profiles in multiple tissues of the domesticated silkworm, Bombyx mori. Genome Biol. 2007;8(8):R162. doi: 10.1186/gb-2007-8-8-r162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li Y, Wang G, Tian J, Liu H, Yang H, Yi Y, Wang J, Shi X, Jiang F, Yao B, Zhang Z. Transcriptome analysis of the silkworm (Bombyx mori) by high-throughput RNA sequencing. PLoS One. 2012;7(8):e43713. doi: 10.1371/journal.pone.0043713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10(1):57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, Chen Z, Mauceli E, Hacohen N, Gnirke A, Rhind N, di Palma F, Birren BW, Nusbaum C, Lindblad-Toh K, Friedman N, Regev A. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29(7):644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li R, Yu C, Li Y, Lam TW, Yiu SM, Kristiansen K, Wang J. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics. 2009;25(15):1966–1967. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- 42.Curtis K, Youngquist ST. Part 21: categoric analysis: Pearson chi-square test. Air Med J. 2013;32(4):179–180. doi: 10.1016/j.amj.2013.04.007. [DOI] [PubMed] [Google Scholar]
- 43.Matsuoka T, Fujiwara H. Expression of ecdysteroid-regulated genes is reduced specifically in the wing discs of the wing-deficient mutant (fl) of Bombyx mori. Dev Genes Evol. 2000;210(3):120–128. doi: 10.1007/s004270050018. [DOI] [PubMed] [Google Scholar]
- 44.Ote M, Mita K, Kawasaki H, Seki M, Nohata J, Kobayashi M, Shimada T. Microarray analysis of gene expression profiles in wing discs of Bombyx mori during pupal ecdysis. Insect Biochem Mol Biol. 2004;34(8):775–784. doi: 10.1016/j.ibmb.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 45.Zhou Q, Yu L, Shen X, Li Y, Xu W, Yi Y, Zhang Z. Homology of dipteran bristles and lepidopteran scales: requirement for the Bombyx mori achaete-scute homologue ASH2. Genetics. 2009;183(2):619–627. doi: 10.1534/genetics.109.102848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Besson MT, Chareyre P, Deleage G, Demont J, Quennedey B, Courrent A, Fourche J, Bosquet G. Isolation and characterization of Urbain, a 20-hydroxyecdysone-inducible gene expressed during morphogenesis of Bombyx mori wing imaginal discs. Rouxs Arch Dev Biol. 1996;205(7–8):333–343. doi: 10.1007/BF00377213. [DOI] [PubMed] [Google Scholar]
- 47.Abdou M, Peng C, Huang J, Zyaan O, Wang S, Li S, Wang J. Wnt signaling cross-talks with JH signaling by suppressing Met and gce expression. PLoS One. 2011;6(11):e26772. doi: 10.1371/journal.pone.0026772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Muramatsu D, Kinjoh T, Shinoda T, Hiruma K. The role of 20-hydroxyecdysone and juvenile hormone in pupal commitment of the epidermis of the silkworm, Bombyx mori. Mech Dev. 2008;125(5–6):411–420. doi: 10.1016/j.mod.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 49.Kozlova T, Lam G, Thummel CS. Drosophila DHR38 nuclear receptor is required for adult cuticle integrity at eclosion. Dev Dyn Off Publ Am Assoc Anatomists. 2009;238(3):701–707. doi: 10.1002/dvdy.21860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Charles JP, Shinoda T, Chinzei Y. Characterization and DNA-binding properties of GRF, a novel monomeric binding orphan receptor related to GCNF and betaFTZ-F1. Eur J Biochem/FEBS. 1999;266(1):181–190. doi: 10.1046/j.1432-1327.1999.00842.x. [DOI] [PubMed] [Google Scholar]
- 51.Wang HB, Nita M, Iwanaga M, Kawasaki H. betaFTZ-F1 and Broad-Complex positively regulate the transcription of the wing cuticle protein gene, BMWCP5, in wing discs of Bombyx mori. Insect Biochem Mol Biol. 2009;39(9):624–633. doi: 10.1016/j.ibmb.2009.06.007. [DOI] [PubMed] [Google Scholar]
- 52.Deng H, Zheng S, Yang X, Liu L, Feng Q. Transcription factors BmPOUM2 and BmbetaFTZ-F1 are involved in regulation of the expression of the wing cuticle protein gene BmWCP4 in the silkworm, Bombyx mori. Insect Mol Biol. 2011;20(1):45–60. doi: 10.1111/j.1365-2583.2010.01041.x. [DOI] [PubMed] [Google Scholar]
- 53.Ali MS, Iwanaga M, Kawasaki H. Ecdysone-responsive transcriptional regulation determines the temporal expression of cuticular protein genes in wing discs of Bombyx mori. Gene. 2013;512(2):337–347. doi: 10.1016/j.gene.2012.09.126. [DOI] [PubMed] [Google Scholar]
- 54.Lundell MJ, Hirsh J. eagle is required for the specification of serotonin neurons and other neuroblast 7–3 progeny in the Drosophila CNS. Development. 1998;125(3):463–472. doi: 10.1242/dev.125.3.463. [DOI] [PubMed] [Google Scholar]
- 55.Gritzan U, Weiss C, Brennecke J, Bohmann D. Transrepression of AP-1 by nuclear receptors in Drosophila. Mech Dev. 2002;115(1–2):91–100. doi: 10.1016/S0925-4773(02)00116-8. [DOI] [PubMed] [Google Scholar]
- 56.Sourmeli S, Papantonis A, Lecanidou R. A novel role for the Bombyx Slbo homologue, BmC/EBP, in insect choriogenesis. Biochem Biophys Res Commun. 2005;337(2):713–719. doi: 10.1016/j.bbrc.2005.09.103. [DOI] [PubMed] [Google Scholar]
- 57.Armand P, Knapp AC, Hirsch AJ, Wieschaus EF, Cole MD. A novel basic helix-loop-helix protein is expressed in muscle attachment sites of the Drosophila epidermis. Mol Cell Biol. 1994;14(6):4145–4154. doi: 10.1128/mcb.14.6.4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Staudt N, Molitor A, Somogyi K, Mata J, Curado S, Eulenberg K, Meise M, Siegmund T, Hader T, Hilfiker A, Bronner G, Ephrussi A, Rorth P, Cohen SM, Fellert S, Chung HR, Piepenburg O, Schafer U, Jackle H, Vorbruggen G. Gain-of-function screen for genes that affect Drosophila muscle pattern formation. PLoS Genet. 2005;1(4):e55. doi: 10.1371/journal.pgen.0010055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Biehs B, Sturtevant MA, Bier E. Boundaries in the Drosophila wing imaginal disc organize vein-specific genetic programs. Development. 1998;125(21):4245–4257. doi: 10.1242/dev.125.21.4245. [DOI] [PubMed] [Google Scholar]
- 60.Wessells RJ, Grumbling G, Donaldson T, Wang SH, Simcox A. Tissue-specific regulation of vein/EGF receptor signaling in Drosophila. Dev Biol. 1999;216(1):243–259. doi: 10.1006/dbio.1999.9459. [DOI] [PubMed] [Google Scholar]
- 61.Bergsland M, Werme M, Malewicz M, Perlmann T, Muhr J. The establishment of neuronal properties is controlled by Sox4 and Sox11. Genes Dev. 2006;20(24):3475–3486. doi: 10.1101/gad.403406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dang DT, Pevsner J, Yang VW. The biology of the mammalian Kruppel-like family of transcription factors. Int J Biochem Cell Biol. 2000;32(11–12):1103–1121. doi: 10.1016/S1357-2725(00)00059-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.de Celis JF, Bray S, Garcia-Bellido A. Notch signalling regulates veinlet expression and establishes boundaries between veins and interveins in the Drosophila wing. Development. 1997;124(10):1919–1928. doi: 10.1242/dev.124.10.1919. [DOI] [PubMed] [Google Scholar]
- 64.Wang S, Jayaram SA, Hemphala J, Senti KA, Tsarouhas V, Jin H, Samakovlis C. Septate-junction-dependent luminal deposition of chitin deacetylases restricts tube elongation in the Drosophila trachea. Curr Biol CB. 2006;16(2):180–185. doi: 10.1016/j.cub.2005.11.074. [DOI] [PubMed] [Google Scholar]
- 65.Zhao Y, Park RD, Muzzarelli RA. Chitin deacetylases: properties and applications. Marine Drugs. 2010;8(1):24–46. doi: 10.3390/md8010024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Moussian B, Tang E, Tonning A, Helms S, Schwarz H, Nusslein-Volhard C, Uv AE. Drosophila Knickkopf and Retroactive are needed for epithelial tube growth and cuticle differentiation through their specific requirement for chitin filament organization. Development. 2006;133(1):163–171. doi: 10.1242/dev.02177. [DOI] [PubMed] [Google Scholar]
- 67.Chaudhari SS, Arakane Y, Specht CA, Moussian B, Boyle DL, Park Y, Kramer KJ, Beeman RW, Muthukrishnan S. Knickkopf protein protects and organizes chitin in the newly synthesized insect exoskeleton. Proc Natl Acad Sci U S A. 2011;108(41):17028–17033. doi: 10.1073/pnas.1112288108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Milan M, Perez L, Cohen SM. Short-range cell interactions and cell survival in the Drosophila wing. Dev Cell. 2002;2(6):797–805. doi: 10.1016/S1534-5807(02)00169-7. [DOI] [PubMed] [Google Scholar]
- 69.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5(7):621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 70.Benjamini Y, Yekutieli D. The control of the false discovery rate in multiple testing under dependency. Ann Stat. 2001;29:1165–1188. doi: 10.1214/aos/1013699998. [DOI] [Google Scholar]
- 71.Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, Yamanishi Y. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36(Database issue):D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ye J, Fang L, Zheng H, Zhang Y, Chen J, Zhang Z, Wang J, Li S, Li R, Bolund L, Wang J. WEGO: a web tool for plotting GO annotations. Nucleic Acids Res. 2006;34(Web Server issue):W293–W297. doi: 10.1093/nar/gkl031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1: Differentially expressed transcripts in L5D6-VS-PP. The criteria applied for significance difference were FDR ≤ 0.001, and estimated absolute |log2Ratio (PP/L5D6)| ≥ 1. L5D6- and PP-RPKM: reads per kb per million reads for each transcript in stages L5D6 and PP, respectively. Log2 Ratio (PP/L5D6): the ratio between the RPKM in PP and the RPKM in L5D6. Up-Down (PP/L5D6) and FDR of each gene are also shown. GO Component, GO Function and Go Process: ontology information of Cellular Components, Molecular Function and Biological Processes of Gene-corresponding GO terms. "-": no hit. Blast nr: identification of homologues in GenBank; E-values are also shown. (XLS 980 KB)
Additional file 2: Table S2: Differentially expressed transcripts in PP-VS-P0. Description of data is similar to Table S1. Log2 Ratio (P0/PP): the ratio between the RPKM in P0 and the RPKM in PP. (XLS 1 MB)
Additional file 3: Assembled nucleotide sequences of transcripts in Table 1. (DOC 36 KB)
Additional file 4: Assembled nucleotide sequences of transcripts in Table 2. (DOC 32 KB)
Additional file 5: Assembled nucleotide sequences of transcripts in Table 3. (DOC 42 KB)
Additional file 6: Table S3: Primers for qRT-PCR. (XLS 21 KB)
Additional file 7: Assembled nucleotide sequences of transcripts in Table 5. (DOC 34 KB)
Additional file 8: Assembled nucleotide sequences of transcripts in Table 6. (DOC 44 KB)
Additional file 9: Assembled nucleotide sequences of transcripts in Table 8. (DOC 44 KB)
Additional file 10: Table S4: All transcripts at P0 stage. All transcripts at stage P0 were listed. Description of data is similar to Table S1. (XLS 3 MB)


