Abstract
The identification of genes targeted by a specific transcription regulatory factor (TRF) is essential to our understanding of the regulatory mechanism of gene expression. We constructed a system for the comprehensive identification of genes directly regulated by a TRF. It includes a combination of perturbation of gene expression by RNA interference (RNAi) of the TRF, cDNA microarray analysis, computer searches for the putative TRF recognition sequences, and in vivo and in vitro TRF–DNA binding assays. Endogenous hepatocyte nuclear factor-1β (HNF-1β) mRNA was efficiently degraded by transfection of mouse hepatoma cells with short interfering RNAs. Expression profile analysis with 20 K mouse cDNA microarrays detected 243 genes whose expression levels were decreased by >50% upon RNAi of HNF-1β. The upstream regions of the top 26 downregulated genes were searched for the HNF-1β consensus recognition sequences leading to the extraction of 13 candidate genes. Finally, TRF–DNA binding assays identified five novel as well as three known HNF-1β-regulated genes. In combination with quantitative real-time RT–PCR, the present system revealed the existence of a more expanded regulatory network among seven HNF family members, demonstrating its practicability to identify the TRF network as well as genes directly regulated by a specific TRF.
INTRODUCTION
Transcriptional regulation of gene expression is largely due to the actions of specific transcription regulatory factors (TRFs) on their target genes, and thus the identification of genes targeted by a specific TRF is key to understanding the regulatory mechanism of gene expression. A strategy that one can adopt to identify eukaryotic genes controlled by a TRF is to perturb their expression by increasing or reducing the level of the regulator and then analyzing the expression profile with microarrays. Alternatively one may examine the binding of DNA sequences in the nucleus by crosslinking-chromatin immunoprecipitation (X-ChIP) (1–4). For perturbation of gene expression within the cells, tissues or bodies, various techniques can be employed. Overexpression of a specific TRF gene in target cells is a robust way to increase the amount of the transcriptional regulator in the target cells leading to stimulation or repression of its regulated genes. We have already constructed a system for the comprehensive identification of TRF-regulated genes in mammalian cells by using the perturbation of gene expression through TRF overproduction, a large-scale expression analysis with cDNA microarrays, mapping of the candidates to the mouse genome, a computer search for the TRF-recognizable sequences in the upstream regions of the mapped candidate genes, and X-ChIP with the specific antibody against the TRF followed by comparative PCR (1). On the other hand, downregulation of a TRF gene expression also causes stimulation or repression of specific gene sets in the cells. Among the many techniques using gene downregulation, the most efficient ones are gene targeting, and knockout or knockdown of the gene expression through reciprocal recombination, repression of translation of a specific mRNA with antisense RNA or DNA and degradation of a specific mRNA with ribozyme. In addition, the recently developed RNA interference (RNAi) technique is a particularly useful and powerful way to suppress the expression of a specific mammalian gene (5). Using the RNAi system, the target mRNA is specifically cleaved and degraded by the cellular machineries guided by short interfering double-stranded RNA (siRNA). One of the strands is perfectly complementary to a corresponding portion of the target. Increasing evidence shows that RNAi against specific mRNAs in mammalian cells is feasible for the interrogation of the functions of a variety of mammalian genes (6,7).
RNAi against TRFs is becoming a standard approach for examining the downstream genes of a specific regulator in lower animals such as nematodes (8) and flies (9). Recently, RNAi of NF-κB and fra-1 in human cells including HeLa and mesothelioma cells was successfully used to search for genes whose expression levels were affected by the TRFs (10,11). However, RNAi of a TRF affects gene expression in several different ways, i.e. it up- or downregulates the genes directly downstream, and indirectly regulates and incidentally affects some unrelated genes. Therefore, it is very important for understanding the transcriptional regulatory cascades to discriminate the real or directly regulated genes from the genes regulated secondarily or affected consequently to the suppression of the TRF. To perform such a discrimination between the TRF-affected genes, we tried to apply the recently developed system for the identification of the specific TRF-regulated genes (1) in combination with RNAi-induced gene expression perturbation.
To verify the practicality and robustness of this newly devised system, we applied it to find genes regulated by hepatocyte nuclear factor (HNF)-1β (also known as vHNF-1, vAPF and LFB3), which is a homeodomain-containing TRF (12) and encoded by the TCF2 gene (13). HNF-1β is expressed in several tissues such as liver (14), kidney (15) and pancreas (16). More important, mutations of the human HNF-1β gene are associated with maturity onset diabetes of the young (MODY) type 5 (16). Although the important roles of HNF-1β in various physiological events have been established, the mechanism of its action in the regulation of gene expression remains to be elucidated.
MATERIALS AND METHODS
siRNAs
Three 21-nucleotide long siRNAs targeting HNF-1β mRNA (accession no. NM_009330) were designed according to the standard selection criteria (6) and chemically synthesized by Japan BioService Co. Ltd (Saitama, Japan). The synthetic siRNAs with 3′ overhang of 2dTs are as follows: 1β-1, GCCGGUUUUCCAUACUCUCtt (corresponding to 175–195 relative to the first nucleotide of the translation initiation codon of mouse HNF-1β); 1β-2, CAAGAAGAUGCGCC GCAACtt (682–702); and 1β-3, UGGUGGUCACAGA UACCAGtt (1596–1616). Hu 1β-1, a 1β-1 variant, which had a one-nucleotide mismatch at the 7th nucleotide position from the 5′-terminus of the sense strand of the siRNA and was perfectly identical to the human HNF-1β sequence, was also used for RNAi. In addition to these siRNAs, Silencer Negative Control siRNA1 (Ambion, Woodward, TX) having no significant homology to any known gene sequences from mouse, rat or human, was used as a control siRNA. All siRNA sequences designed were checked by BLAST search to preclude sequences with significant similarity to other genes in the mouse genome. Sense and antisense RNA were solved in annealing buffer (100 mM potassium acetate, 30 mM HEPES–KOH pH 7.4, 2 mM magnesium acetate) and incubated for 1 min at 90°C and then for 1 h at 37°C just prior to transfection.
Cell culture and transfection
Mouse hepatoma cell line Hepa 1-6 cells were purchased from RIKEN Bioresource Center (Tsukuba, Japan) and maintained in Dulbecco’s modified Eagle’s medium supplemented with 0.45% glucose and 5% fetal calf serum at 37°C in a 5% CO2 and 95% atmosphere. Hepa 1-6 cell were seeded in 6-well plates at a density of 1.4 × 105 cells/well and cultured for 24 h. Cell were washed with PBS and transfected with 1 ml of Opti-MEM I medium (Invitrogen, Carlsbad, CA) containing 100 nM siRNA using Lipofectamine 2000™ (Invitrogen) in accordance with the manufacturer’s instructions. Transfection was made with cells grown on 10 cm dishes to prepare total RNA used for the cDNA microarray experiments.
Quantitative real-time RT–PCR
Total RNA was extracted from siRNA-transfected cells using TRIzol reagent (Invitrogen) as recommended by the manufacturer and treated with RNase-free DNase I (Takara Shuzo, Kyoto, Japan). The yield and purity of RNA was spectrophotometrically determined. The target mRNAs were quantified by real-time RT–PCR analysis using SYBR Green or TaqMan Probe. The cDNAs prepared with random primers and SuperScript II (Invitrogen) were used as templates for PCR reaction in 25 µl-scale with SYBR Green PCR Core Reagents kit (Applied Biosystems, Foster City, CA) in the presence of 200 nM of the forward and reverse primers. PCR reaction was carried out on the ABI PRISM7700 Sequence Detection System (Applied Biosystems) at 50°C for 2 min and at 95°C for 10 min, followed by running for 40 cycles at 95°C for 15 s and 60°C for 1 min. The measurements of mRNA using TaqMan probes were carried out in 50 µl of reaction mixture containing 200 nM of the target gene-specific forward and reverse primers, 250 nM probe and 1 × TaqMan One-Step RT–PCR Master Mix (Applied Biosystems). The primer set and TaqMan probes used for the real-time PCR analysis are shown in Supplementary Material available at NAR Online. Expression of mRNA was assessed by evaluating threshold cycle (CT) values. The CT values were normalized with the expression level of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and the relative amount of mRNA specific to each of the target genes was calculated using the 2–ΔΔCT method (17).
cDNA microarray analysis
Total RNA was extracted from Hepa 1-6 cells 48 h after the transfection with 1β-1 siRNA or negative control siRNA and labeled by the double step labeling method with aminoallyl-dUTP (Sigma, St Louis, MO). After incubation of 50 µg of the total RNA with oligo (dT) primer (Takara) at 70°C for 10 min, reverse transcription reaction was carried out at 42°C for 1 h in a total volume of 30 µl reaction mixture containing 10 mM DTT (Invitrogen), 0.5 mM each dGTP, dATP and dCTP 0.3 mM dTTP (Amersham Biosciences, Piscataway, NJ, USA), 0.2 mM aminoallyl-dUTP and 400 units of SuperScriptII (Invitrogen) in first strand buffer (Invitrogen). The cDNA was purified using GFX PCR DNA and Band Purification Kit (Amersham Biosciences), dried and then dissolved in 9 µl of 0.1 M sodium bicarbonate buffer (pH 9.0) with either Cy3 or Cy5 monofunctional reactive dye (62 µg each, Amersham Biosciences). The mixture was allowed to stand in the dark at room temperature for 1 h for dye coupling reaction and then the reaction was quenched by addition of 4.5 µl of 4 M hydroxylamine and being kept at room temperature in the dark for 15 min. The resulting cDNA, labeled with either Cy3 or Cy5, was then mixed together and purified using a MicroSpin Column S-200 HR Column (Amersham Biosciences). After concentration of the cDNA solution to 17 µl, blocking solution consisting of 3 µl of 10 µg/µl oligo(dA), 3 µl of 10 µg/µl yeast tRNA, 1 µl of 20 µg/µl Cot1 DNA, 5.1 µl of 20× SSC and 0.9 µl of 10% SDS was added and the resultant solution was used as a probe on the RIKEN mouse full-length cDNA 20 K microarray (18). Hybridization was performed overnight at 65°C in a Hybricassette using aliquots of the probe heated at 95°C for 1 min and then cooled to room temperature. Hybridization with the probe inversely labeled with Cy dyes was also carried out and experiments were done in triplicate. After hybridization, slides were washed in SSC and scanned on a ScanArray 5000 confocal laser scanner (Perkin Elmer, Boston, MA) and then images were analyzed by using Digital Genome (Molecularware, Cambridge, MA). Signal intensity data obtained from each of the spots was processed automatically with a filtering program PRIM (Preprocessing Implementation for Microarray) (19). Student’s t-tests were performed with the Excel program to calculate whether the mean relative intensity for a gene in the siRNA-treated samples was statistically different from that in the negative control siRNA-treated samples.
Analysis of HNF-1β binding consensus sequence
The upstream regions (1 kb) extending from –1000 to +1 relative to the transcription start sites of the primary candidate HNF-1β-regulated genes, which were selected on the basis of the data from cDNA microarray analysis, were extracted from ENSEMBL database. DNA sequences homologous to HNF-1β recognition sequences in the upstream regions were searched with Match program (20) in Transfac 7.3 (Biobase). The cut-off to minimize false positive matches (minFP) was applied for searching the DNA sequences. HNF-1β recognition sequences used for the search were V$HNF1_Q6 WRGTTAATNATTAACNNN and V$HNF1_C DGTTAATKAWTNACCAM (Transfac7.3 accession nos M00790 and M00206, respectively). In addition, the 10-kb upstream regions and the first exons of the primary candidate HNF-1β-regulated genes were searched in the same manner.
Preparation of HNF-1β protein
Poly (A)-RNA was purified from mouse liver total RNA using Oligotex™ –dT30 <Super >mRNA Purification Kit (Takara) and used for the synthesis of full-length HNF-1β cDNA. A set of the specific primers, which contained an attB sequence at their 5′ ends, was as follows: forward 5′-GGGGACAAG TTTGTACAAAAAAGCAGGCTTAATGGTGTCCAAGC TCACGTC-3′; reverse 5′-GGGGACCACTTTGTACAAGA AAGCTGGGTCTCACCAGGCTTGCAGTGG-3′. The amplicon was cloned into the pDEST17 vector (Invitrogen) according to the Gateway protocols provided by the manufacturer. Polyhistidine-tagged HNF-1β protein was produced in Escherichia coli BL21-SI cells and purified using a TALON Ni–NTA Spin Column (Clontech, Palo Alto, CA). The His-tagged HNF-1β protein was eluted with Imidazole Elution Buffer (45 mM NaH2PO4 pH 7.0, 8 M urea, 270 mM NaCl, 150 mM imidazole) and dialyzed against EMSA buffer (10 mM HEPES–KOH pH 7.5, 77 mM KCl, 1 mM DTT, 5 mM MgCl2, 5% glycerol, 1 M urea). HNF-1β protein was quantified using Coomassie Plus Protein Assay Reagent Kit (Pierce, Rockford, IL) and its purity was verified on an SDS–polyacrylamide gel.
Electrophoretic mobility shift assay (EMSA)
The double-stranded 30 bp DNA oligomers, which corresponded to the portions of the upstream regions of the candidate genes and contained a putative or demonstrated HNF-1β recognition sequence, were constructed by annealing chemically synthesized single-stranded DNA pairs and labeled with T4 polynucleotide kinase and [γ-32P]ATP by incubating at 37°C for 30 min. Unincorporated radioactive nucleotides were removed by using QIAquick Nucleotide Removal Kit (Qiagen, Hilden, Germany). HNF-1β protein (800 ng) was incubated with 150 fmol of each of the labeled oligonucleotide probes in a 20 µl reaction mixture containing 10 mM HEPES–KOH (pH 7.5), 77 mM KCl, 1 mM DTT, 5 mM MgCl2, 5% glycerol, 2.5 µg of poly dI-dC and 10 mM spermidine. In DNA competition assays, 50 pmol of each of the competitors (unlabeled oligomers with the same sequences as the probes) or non-competitor (unlabeled oligomer without any HNF-1β recognition sequence) was added to the reaction. The reaction mixture was incubated at 30°C for 1 h, then on ice for 1 h and run on a 6% polyacrylamide gel in 0.5 × Tris–borate–EDTA running buffer at room temperature. The gel was dried and autoradiographed overnight at –80°C.
X-ChIP/comparative PCR
The procedures for X-ChIP were essentially as described (1) with some modifications. The soluble chromatin was incubated with anti-HNF-1β-specific antibody (sc-7411; Santa Cruz Biotechnology, Santa Cruz, CA) for more than 12 h at 4°C. The chromatin–antibody mixture was incubated with Dynabeads Protein G (Dynal Biotech, Oslo, Norway) for 1 h at 4°C and the immunoprecipitates were captured by using magnets. The immunoprecipitates recovered were washed twice with IP wash buffer Low (2 mM EDTA, 20 mM Tris–HCl pH 8.0, 150 mM NaCl, 1% Triton X-100, 0.1% SDS), once with IP wash buffer High (2 mM EDTA, 20 mM Tris–HCl pH 8.0, 500 mM NaCl, 1% Triton X-100, 0.1% SDS), once with IP wash buffer LiCl (1 mM EDTA, 10 mM Tris–HCl pH 8.0, 250 mM LiCl, 0.5% NP-40, 0.5% sodium deoxycholate) and twice with TE buffer (10 mM Tris–HCl pH 8.0, 1 mM EDTA). The washed complexes were treated with 0.05 mg/ml RNase A (Nippon Gene, Tokyo, Japan) in TE buffer at 37°C for 30 min and then SDS was added to a final concentration of 0.25%, followed by being treated with 0.25 mg/ml proteinase K (Nippon Gene) at 37°C for 12 h. The reversal of formaldehyde-induced crosslinks was carried out by heating at 65°C for 6 h and DNA released was extracted with phenol and phenol:chloroform:isoamylalcohol (25:24:1), ethanol-precipitated, and then dissolved in 50 µl of H2O. DNA obtained by ChIP with HNF-1β-specific antibody, from the precipitates ChIP without any antibody or without chromatin (mock) and input DNA (total chromatin DNA) were used as templates for comparative PCR assay. PCR mixture contained 2 µl DNA preparation, 0.2 µM of each of the specific primer set, 1 mM dNTP mixture, 1.5 mM MgCl2, and 1.25 units of Ex Taq DNA polymerase (Takara) in a total volume of 50 µl. The optimum PCR conditions were determined by experiments in which the number of PCR cycles was changed stepwise from 25 to 40 cycles.
RESULTS
Positional effects of siRNAs against HNF-1β mRNA
It has been suggested that accessible siRNA target sites may be rare in some human mRNAs: RNAi with several siRNAs synthesized against different sites on the same target mRNA showed striking differences in silencing efficiency and only a few of the siRNAs resulted in a significant reduction in the mRNA level (21). To assess the positional effect, siRNAs were selected from three different sites on the mouse HNF-1β mRNA on the basis of the conventional criteria for siRNA designing (22).
HNF-1β is coexpressed with HNF-1α, which is highly homologous to HNF-1β in their amino acid sequences, in the normal mouse liver cells as well as murine hepatoma cell line Hepa 1A (23,24). Assuming the nucleotide mismatch tolerance of RNAi (21,25), nucleotide sequence homology between the two TRFs may trigger cross-suppression of HNF-1α by siRNAs that are designed for HNF-1β mRNA destruction. However, no identical sequence stretch expanding over 15 nucleotides long was found in the mouse HNF-1α and 1β mRNA sequences. In addition, there is no significant sequence homology between the three siRNAs and mouse HNF-1α mRNA sequence. Real-time RT–PCR with the primer sets specific for respective genes showed that HNF-1α and 1β genes were expressed roughly at the ratio of 1.0 to 1.9 in Hepa 1-6 cells.
Figure 1 summarizes the results of RNAi of HNF-1β in Hepa 1-6 cells with different siRNAs and an antisense RNA. 1β-1 and 1β-3 siRNAs (100 nM each), which were derived from the exons 1 and 8 of the mouse HNF-1β coding sequence, respectively, were almost equally effective in reducing the HNF-1β mRNA levels. On the other hand, 1β-2 derived from the central region of the HNF-1β coding sequence showed only a marginal inhibitory effect. These data clearly show the positional effects of siRNAs on the mouse HNF-1β mRNA cleavability. When the 1:1 mixture of 1β-1 and 1β-3 (50 nM each) was transfected, no additional reduction in the HNF-1β mRNA level was found. The antisense RNA of 1β-1 reduced appreciably the HNF-1β mRNA level, as shown previously for tissue factor mRNA in mammalian cells treated with the specific antisense RNAs (26,27). We also tested the RNAi effect of Hu 1β-1, a 1β-1 variant which had a one-nucleotide mismatch at the 7th nucleotide position from the 5′-terminus of the sense strand of the siRNA. Transfection of Hepa 1-6 cells with Hu 1β-1 siRNA resulted in only a modest decrease in HNF-1β mRNA level, suggesting a moderate mismatch tolerance of RNAi for HNF-1β mRNA. Since RNAi effect by 1β-1 was found to be most reproducible, we decided to use it for the subsequent RNAi experiments.
Figure 1.
Suppression of HNF-1β gene expression. (A) The locations of the three sites (1β-1, 1β-2 and 1β-3) chosen for preparing synthetic siRNAs are indicated by bars on the mature HNF-1β mRNA divided in the boxes of exons. (B) Reduction of HNF-1β mRNA levels in the siRNA-transfected Hepa 1-6 cells. Hu 1β-1 siRNA is identical to human HNF-1β sequence and different at one nucleotide pair in the central position (7U:A→C:G) from 1β-1 siRNA. AS 1β-1 is the antisense strand of 1β-1 siRNA. Hepa 1-6 cells were harvested 48 h after transfection with 100 nM siRNA and total RNA was extracted. The RNA preparations were analyzed by real-time RT–PCR to determine the level of HNF-1β mRNA relative to that of GAPDH mRNA. HNF-1β expression was normalized to that of GAPDH and normalized expression in the negative control siRNA-transfected cells was set to 1.0. The bars represent the averages ± SD of the data obtained in three independent experiments except the data for Hu 1β-1 and AS 1β-1 which were obtained in duplicated experiments.
Time-course analysis of HNF-1β RNAi
The silencing effect induced by siRNAs is usually transient, suggesting absence of a propagative system for RNAi in mammalian cells (21). Therefore, it is important to determine the time after transfection that will give maximum RNAi effect with a selected siRNA for examining the influence on the expression of the downstream genes regulated by a specific endogenous TRF. Triplicate experiments of the time-course analysis showed that the amount of HNF-1β mRNA fell to the lowest level nearly 48 h after transfection with 1β-1 siRNA followed by being rapidly increased during prolonged culturing (Fig. 2). Suppression of HNF-1β gene expression also resulted in marked reduction of the level of HNF-1β protein 48 h after cotransfection of 1β-1 siRNA and HNF-1β expression plasmids, implying that RNAi of HNF-1β could lead to perturbation of expression of its downstream genes.
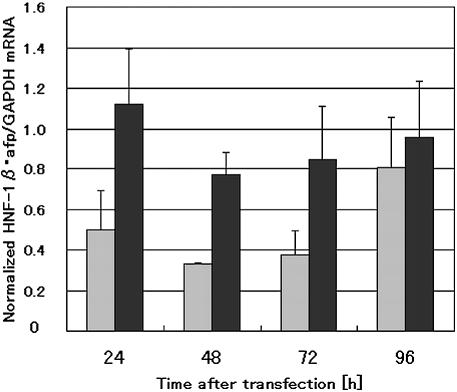
Figure 2.
Time course analysis of HNF-1β and α-fetoprotein mRNA levels after transfection with 1β-1 siRNA. Hepa 1-6 cells were transfected with 100 nM siRNA and harvested for mRNA isolation 24, 48, 72 and 96 h after transfection. The mRNA levels were determined by real-time RT–PCR. Expression levels of HNF-1β and α-fetoprotein were normalized as described in Figure 1. The normalized expression value at the time point of the siRNA addition was then 1.0. The bars represent the averages + SD of the data obtained in three independent experiments Gray bars, HNF-1β; black bars, α-fetoprotein.
To assess whether the RNAi methodology could be used for pursuing genes regulated by a specific TRF, we then evaluated quantitatively the effect of 1β-1 siRNA on the expression of known HNF-1β-regulated genes. Similarly to HNF-1β mRNA, the mRNA specific for α-fetoprotein gene, one of the representative HNF-1β-regulated genes expressed in liver cells, was also reproducibly time-dependently reduced and seemed to approach the lowest level nearly 48 h after 1β-1 siRNA transfection (Fig. 2). In addition, expression levels of β-fibrinogen and vitamin D binding protein genes, other known HNF-1β-regulated genes, were also reduced 48 h after 1β-1 siRNA transfection. On the basis of these data, we used RNA samples prepared from Hepa 1-6 cells 48 h after transfection with 1β-1 siRNA for cDNA microarray and real-time RT–PCR analysis of the candidates for the HNF-1β-regulated genes. In contrast to the rapid regaining of HNF-1β mRNA level, α-fetoprotein mRNA gradually increased during prolonged culturing, suggesting the difference in the mRNA turnover between the TRF and its controlling gene. Dose-dependent increase in the RNAi effect of 1β-1 was found with both HNF-1β and α-fetoprotein mRNAs (data not shown) and the concentration of 1β-1 (100 nM), which gave the maximum suppression of HNF-1β, was applied to perturb expression of endogenous genes in Hepa 1-6 cells.
Comprehensive search for the HNF-1β-regulated gene candidates
RNAi intrinsically does not knockout completely but knockdown gene expression. To assess that the partial impairment of expression of a given TRF may result in down- or upregulation of its controlling genes, we performed comprehensive characterization of the expression perturbation by HNF-1β suppression with the 20 K cDNA microarray representing 19 584 mouse genes. The 20 K cDNA set contains 10 genes whose regulatory sequences have been known to interact with HNF-1α and/or HNF-1β. The sample and control mRNAs were extracted from Hepa 1-6 cells suppressed by 1β-1 48 h after transfection with the siRNA and the negative control siRNA-transfected cells and then used for the calculation of differential expression of the 20 K genes. In these experiments, the levels of HNF-1β–mRNA were reduced by ∼70% on average.
But for one of the three independent cDNA microarray experiments, correlation efficiencies were significantly high (0.760 and 0.721) and then we chose these two data sets for analyzing the perturbation of gene expression. Using these data sets, we detected 243 genes whose mRNA levels were reproducibly decreased to less than half of the control and 58 genes that were upregulated >2-fold. The range of changes in expression levels for all of these appreciably perturbed genes was within 0.25–2.33-fold. Because a threshold of 2.0-fold difference in relative expression levels is usually used for microarray data analysis (28) and downregulation of α-fetoprotein gene expression was ∼50% on average in the cDNA microarray assays, we adopted the 243 genes downregulated >50% as the primary candidates for the HNF-1β-regulated genes. Standard student’s t-tests with the 0.05 significance level were then performed with these 243 genes leading to the removal of genes showing large P-values (>0.05). Because it was expected that a set of genes whose expression levels were greatly influenced by a perturbation treatment would contain the real TRF-regulated genes at a high frequency rate, we selected the top 28 genes that were well-characterized or annotated in order of the extent of downregulation (<50% of the controls) from the primary candidate genes for the subsequent examination (Table 1).
Table 1. Genes markedly perturbed by HNF-1β RNAi.
| Gene name | Fold | GenBank accession number | RIKEN ID |
|---|---|---|---|
| Downregulated genes | |||
| Keratin complex 2, basic, gene 8 | 0.25 | AV074436 | 2210009D21 |
| Ribosomal protein S24 | 0.33 | AV060062 | 1810061E16 |
| Phosphoenolpyruvate carboxykinase 1, cytosolic | 0.38 | AV065203 | 2010013P13 |
| Small proline-rich protein 1A | 0.39 | AV085389 | 2310011K12 |
| Ferritin light chain 1 | 0.39 | AV029790 | 1500006O17 |
| Basigin | 0.40 | AV104097 | 2510002N16 |
| Tubulin, α1 | 0.40 | AV036419 | 1600019E11 |
| Retina and anterior neural fold homeobox | 0.41 | AV311625 | 5730599E04 |
| Stearoyl-coenzyme A desaturase 1 | 0.42 | AV006099 | 1020006A05 |
| Small inducible cytokine A8 | 0.42 | AK007942 | 1810063B20 |
| Homolog to PNAS-110 | 0.43 | AK013934 | 3110001B11 |
| Homolog to leucine-rich α-2-glycoprotein | 0.43 | AK009555 | 2310031E04 |
| Adaptor protein complex AP-1, mu 2 subunit | 0.43 | AV093553 | 2400003B20 |
| Expressed in non-metastatic cells 2, protein (NM23B) | 0.43 | AK012447 | 2700056N14 |
| MAP kinase-interacting serine/threonine kinase 2 | 0.44 | AK008277 | 2010016G11 |
| Kinesin-associated protein 3 | 0.44 | AK013954 | 3110001K12 |
| Small EDRK-rich factor 2 | 0.44 | AK008802 | 2210402L08 |
| Insulin-like 6 | 0.45 | AK005622 | 1700003B04 |
| MYB binding protein (P160) 1a | 0.45 | AV057657 | 1810047C02 |
| Cyclin A2 | 0.45 | AV133568 | 2810001C09 |
| Fatty acid binding protein 5, epidermal | 0.45 | AV129055 | 2700065F13 |
| Lectin, galactose binding, soluble 9 | 0.45 | AV073637 | 2210002H01 |
| Homolog to insulin gene enhancer protein ISL-2 | 0.46 | AK013964 | 3110001N10 |
| Homolog to farnesyl pyrophosphate synthetase | 0.46 | AV087562 | 2310031J03 |
| Neurogenic differentiation factor 4 | 0.46 | AV114025 | 2610027J16 |
| Myeloblastosis oncogene-like 2 | 0.47 | AV093686 | 2400003N16 |
| Cofilin 1, non-muscle | 0.49 | AV134068 | 2810004B05 |
| α-Fetoprotein | 0.49 | AV105248 | 2510015E23 |
| Upregulated genes | |||
| Serine protease inhibitor 14 | 1.19 | AK008931 | 2210414M20 |
| MutS homolog 3 | 1.16 | AV095246 | 2410004O15 |
| Homolog to SCHIP-1 | 1.14 | AV085270 | 2310010P03 |
| Prolactin-like protein C-gamma | 1.13 | AV036496 | 1600019J01 |
| Similar to pregnancy-specific glycoprotein 23 | 1.12 | AV038458 | 1600029O16 |
| Peroxisomal biogenesis factor 3 | 1.11 | AV028672 | 1500001G17 |
| Similar to flavin-containing monooxygenase 2 | 1.09 | AK009224 | 2310008D08 |
| RAB9, member RAS oncogene family (Rab9) | 1.09 | AV036483 | 1600019I03 |
Expression profile analysis of 19 584 genes detected 243 genes downregulated by >50% and 58 genes upregulated >2-fold, and these were regarded as the candidate for HNF-1β-regulated genes. The top 28 and top eight for downregulated and upregulated genes, respectively, were selected from the annotated candidate genes according to the siRNA-induced changes in their mRNA levels. The extents of down- or upregulation were estimated by comparison of the respective mRNA levels in 1β-1-transfected cells with those in the negative control siRNA-transfected ones. Each value (log2-ratio) is an average of the data obtained in two independent experiments.
In the same way, the top eight genes that were upregulated >2-fold and showed significantly low P-values were selected. The P-values of these selected down- and upregulated genes were within a range of 0.00028–0.03029 and 0.01744–0.04658, respectively. Eight out of 10 genes whose upstream sequences were known to interact with HNF-1α and/or HNF-1β were detected as significantly perturbed ones in these cDNA microarray assays. Among the genes that were downregulated by RNAi of HNF-1β were albumin, α-fetoprotein, insulin-like growth factor binding protein 1 and HNF-1 genes. As opposed to these genes, alcohol dehydrogenase 2, α1-antitrypsin, α-fibrinogen and apolipoprotein AII genes were upregulated by 1β-1 siRNA transfection. Expression levels of the remaining two known HNF-1β-regulated genes (C-reactive protein and β-fibrinogen genes) were hardly changed by RNAi with 1β-1 siRNA.
Search for HNF-1β consensus recognition sequences in the 5′-flanking regions of perturbed genes
To exclude the possible non-HNF-1β-regulated genes from the set of genes selected, we then aligned the 28 cDNAs with the mouse genomic sequence followed by retrieving their upstream sequences. As a result, we failed to map two cDNAs (homolog to farnesyl pyrophosphate synthetase and α-1 tubulin genes). Because most of the known HNF-1 binding sequences were found to be located in the proximal promoter regions, the area of the upstream region between –1 and –1000 relative to the transcription start site was retrieved. We used Transfac Match program (20) to search the HNF-1β consensus recognition sequences (V$HNF1_Q6: WRGTTAATNATT AACNNN; V$HNF1_C: DGTTAATKAWTNACCAM) in the upstream regions of genes mapped (26 genes).
In a preliminary test, we searched the upstream –1 to approximately –1000 sequences containing a regulatory region specific for HNF-1β derived from 15 different known HNF-1β-regulated genes and detected 11 sequences (73%). Out of the 26 candidate genes picked up by genomic mapping, 11 genes were found to contain one or more HNF-1β consensus sequences in their proximal upstream regions (Table 2). On the other hand, four out of the randomly selected 20 genes whose expressions were not significantly perturbed by 1β-1 siRNA transfection were found to contain a consensus HNF-1β recognition sequence in their –1 to –1000 regions. The higher frequency of the appearance of genes containing the consensus HNF-1β recognition sequence in the apparently perturbed gene population suggests the validity of this approach to pan the TRF-regulated genes.
Table 2. Possible HNF-1β recognition sites in the proximal upstream regions of HNF-1β-regulated gene candidates.
| Gene name | Possible recognition site | Position | Matrix |
|---|---|---|---|
| Ribosomal protein S24 | attgtaattaaTTTGCt | –516 (–) | C |
| Phosphoenolpyruvate carboxykinase 1, cytosolic | aagttcaatcaTTATCt | –163 (–) | C |
| Ferritin light chain 1 | tagTTAGTagttgttcat | –918 (+) | Q6 |
| Adaptor protein complex AP-1, mu 2 subunit | ttttagaattAATAAccc | –402 (–) | Q6 |
| Kinesin-associated protein 3 | aGATAAtgactacctta | –948 (+) | C |
| aGTTAAtcagtggacga | –544 (+) | C | |
| Insulin-like 6 | aGGCAAtcaattaatag | –866 (+) | C |
| MYB binding protein (P160) 1a | cctTTAATttttaacagg | –483 (+) | Q6 |
| Lectin, galactose binding, soluble 9 | aataaaaattATTAAaaa | –923 (–) | Q6 |
| Neurogenic differentiation factor 4 | aaaTTAATagttacttgt | –935 (+) | Q6 |
| agttcaaaaaATTAActa | –846 (–) | Q6 | |
| Myeloblastosis oncogene-like 2 | aGGTAGtaaatgacaac | –881 (+) | C |
| α-Fetoprotein | tggTTAATgatctacagt | –48 (+) | Q6 |
| cagTTATTggttaaagaa | –34 (+) | Q6 | |
| Homolog to leucine-rich α-2-glycoprotein | aGTTAATgagaggtcct | –773 (+) | |
| Retina and anterior neural fold homeobox | CGTTAA cgctggccca | +1 (+) | |
| Apolipoprotein B | gtggtcattgaTTGGCt | –715 (–) | C |
| HNF3β | ttaactgaaaAGTAAcct | –116 (–) | Q6 |
| HNF4α | gtgattaaccaTTAACt | –100 (–) | C |
| gaggagaatcaTTTACt | –728 (–) | C |
The upstream regions (1 kb) extending from –1000 to –1 relative to the transcription start site (+1) of the genes corresponding to the candidates for the HNF-1β-regulated genes were extracted from ENSEMBL database and used for searching the DNA sequences homologous to HNF-1β recognition sequences (V$HNF1_Q6 WRGTTAATNATTAACNNN and V$HNF1_C DGTTAATKAWTNACCAM) with Match program in Transfac 7.3 (Biobase). The cut-off to minimize false positive matches (minFP) was used for the sequence search. Position numbers refer to the first nucleotides in Possible recognition site on the sense (+) and antisense (–) strands, respectively, of the coding sequences. The core sequences detected are indicated in capital letters. The homolog to leucine-rich α-2-glycoprotein and retina and anterior neural fold homeobox genes were not hit by Match search, but were found to have a core sequence for HNF-1β recognition. No significant expression change was detected for apolipoprotein B gene in the cDNA microarray analysis. Two HNF family members (HNF-3β and 4α) were also found to have an HNF-1β recognition sequence.
Besides these proximal upstream regions, we searched the HNF-1β consensus recognition sequences in the 10-kb upstream regions of the candidate genes and found that most of the candidate genes (26 genes) contained the HNF-1β consensus recognition sequences in their 10-kb upstream regions. Next, the homology of these 10-kb upstream sequences of mouse candidate genes and the corresponding regions of the human orthologs was examined by Harr plot analysis and a most significant sequence homology was found in the upstream regions of mouse (–767 to ∼2700) and human (–1151 to approximately –3050) neurogenic differentiation factor 4 (NeuroD4) genes. In this region, six and four consensus HNF-1β recognition sequences were detected in mouse and human DNA, respectively, and two sets of these sequences were almost perfectly aligned (mouse –1127 to –1144 and human –1530 to –1547; mouse –937 to –951 and human –1335 to –1322). In addition, HNF-1β consensus recognition sequences were detected in the first introns of eight candidate genes. The involvement of these far upstream and intronic sequences in the transcriptional regulation by HNF-1β remains to be demystified.
In addition to searching for HNF-1β recognizable sequences with stringent conditions, we also searched for the core HNF-1β consensus recognition sequence, GTTAAT, in the upstream regions of the 26 genes and found two more genes having the core sequence. A total of 13 genes having one or more HNF-1β-recognizable sequences were selected and designated as the potential HNF-1β-regulated genes (Table 2). Among these genes, two known HNF-1β-regulated genes, α-fetoprotein and phosphoenolpyruvate carboxykinase (PEPCK) genes, were included. In contrast to the genes suppressed by siRNA transfection, only one (a small GTP binding protein gene) out of eight upregulated genes mapped was found to contain a consensus HNF-1β recognition sequence in their upstream 1 kb regions, suggesting that upregulation does not seem to be a reliable indication of the TRF-regulated genes.
To assess the off-target gene regulation by RNAi, we searched the annotated and mapped 26 cDNA sequences for such a sequence identity to 1β-1 siRNA, but no clear significant sequence homology was found, strongly suggested that these could be excluded from off-target gene members.
X-ChIP/PCR analysis and EMSA demonstrated in vivo and in vitro specific HNF-1β binding on the targeted genes
As the next step in the identification of the target genes for HNF-1β, we adopted the X-ChIP/PCR technique. To make an assessment of the validity of our approach, we selected seven out of the 13 potential HNF-1β-regulated genes, which included five putative and two previously identified HNF-1-regulated genes (PEPCK and α-fetoprotein genes), as the representative ones. We also added three HNF-1-related genes, apolipoprotein B, HNF-4α and HNF-3β genes, to the X-ChIP/PCR analysis. The proximal promoter region of mouse apolipoprotein B gene has a consensus HNF-1 binding sequence and is regulated by HNF-1α (29). HNF-1 regulation of the HNF-4α gene has already been reported (30). In addition, our computational search detected a consensus HNF-1 recognition sequence in the proximal promoter region of the mouse HNF-3β gene.
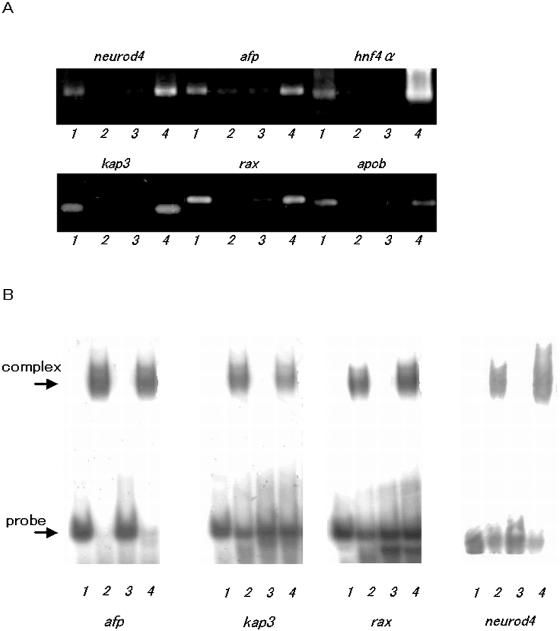
The crosslinked DNA samples were extracted from the nuclei of Hepa 1-6 cells and immunoprecipitated with HNF-1β-specific antibody. By choosing proper PCR amplification cycles as the thresholds, DNA samples recovered from the immunoprecipitates gave a clear band of DNA amplified with each of the specific primer sets for eight genes and presented a sharp contrast to those prepared without antibody, which gave faint or no detectable bands in comparative PCR analysis (Fig. 3A). These results clearly demonstrate that HNF-1β was being complexed in vivo with the upstream regions of eight out of 10 genes examined. The immunoprecipitates and specific primers against the upstream region of insulin-like 6 gene yielded the PCR product almost equivalent to that produced with the DNA sample recovered without any specific antibody, indicating no appreciable enrichment of HNF-1β-specific DNA fragments. On the other hand, no detectable bands were amplified with the primer sets specific for NF-κB gene and albumin mRNA sequence used as negative controls. Although clear DNA bands amplified were detected for two positive control genes, α-fetoprotein and apolipoprotein B genes, any specific amplification was not observed for PEPCK gene, another positive control gene, for some unknown reason. Among the genes that gave positive results in the comparative PCR assay, genes encoding adaptor protein complex AP-1 mu 2 subunit, kinesin-associated protein 3, NeuroD4 and retina and anterior neural fold homeobox (RAX) and HNF-3β genes are previously unrecognized HNF-1β-regulated genes.
Figure 3.
HNF-1β binding to the target sites in the upstream regions of the candidate HNF-1β-regulated genes by X-ChIP/PCR and EMSA analysis. (A) X-ChIP/PCR analysis. DNA recovered from the immunoprecipitates was used as templates for PCR using primer pairs spanning HNF-1β recognition sequences found in the upstream regions of the candidate genes. Lane 1, PCR product amplified with DNA recovered by X-ChIP with the HNF-1β specific antibody; lane 2, PCR product amplified with DNA recovered in the precipitates obtained without any antibody; lane 3, PCR product amplified without template DNA (mock); lane 4, PCR product amplified with aliquots of total chromatin DNA (input). neurod4, neurogenic differentiation factor 4; afp, α-fetoprotein; hnf4α, hepatocyte nuclear factor 4α; kap3, kinesin-associated protein 3; rax, retina and anterior neural fold homeobox; and apob, apolipoprotein B. (B) EMSA analysis: DNA oligomers containing a consensus HNF-1β recognition sequence found in the upstream regions of each of the candidate genes were labeled with [γ-32P]ATP and used as probes. The binding complexes were separated on 5% non-denaturing polyacrylamide gels. Lane 1, probe only; lane 2, probe with HNF-1β; lane 3, probe with HNF-1β and competitor; lane 4, probe with HNF-1β and non-competitor.
The specific recognition of and binding to the upstream regions containing a consensus HNF-1β recognition sequence by HNF-1β was confirmed by EMSA with the polyhistidine-tagged TRF produced in E.coli cells. His6-mouse HNF-1β was produced as insoluble proteins in the form of an inclusion body and solubilized in 8 M urea followed by stepwise dialysis against buffer containing gradually reducing concentrations of the denaturant (Y. Nomura, M. Endo, M. Suzuki and Y. Hayashizaki, manuscript in preparation). The 30-bp oligonucleotide probes for EMSA were designed to contain each of the consensus HNF-1β recognition sequences detected in five previously unrecognized HNF-1β-regulated genes. The authentic and specific binding was observed with all of the probes examined in EMSA (Fig. 3B). The X-ChIP/PCR and EMSA data indicate that the proximal upstream sequences of all of the potential HNF-1β-regulated genes tested can actually bind with HNF-1β in both in vivo and in vitro, demonstrating that these are the real HNF-1β-regulated genes. Insulin-like 6 gene formed only one exception: EMSA detected a clear band shift with the specific probe but X-ChIP/PCR analysis failed to detect significant enrichment of a specific DNA fragment containing the recognition sequence for HNF-1β.
HNF network analysis
Regulation of TRF expression per se is a key factor in the regulation of gene expression. Some of the HNF family members have been suggested to constitute a transcriptional cross-regulatory network among them in embryoid body (31) and pancreatic islet β-cells (32–34). Therefore, we investigated whether the RNAi of HNF-1β would cause any effects on expression of the other HNF family members to get a line on the HNF network in the liver-derived cells. Because only a few HNF family members were on the 20 K cDNA microarray, we examined the effect of RNAi of HNF-1β against the expression of six other HNF family members by quantitative real-time RT–PCR. Discriminating quantification of the HNF-1α and HNF-1β transcripts was carried out with the primer sets specific for respective genes. In addition, HNF-3 family members (HNF-3α, β and γ) having great homology in their amino acid sequences particularly in the DNA binding domains were discriminated from one another by using respective specific primers in real-time RT–PCR.
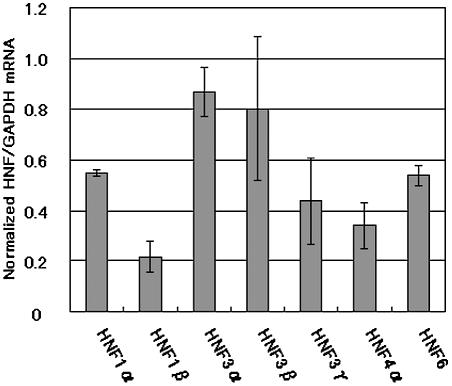
Surprisingly, 1β-1 siRNA-induced HNF-1β suppression resulted in the significant downregulation of all of the HNFs examined (Fig. 4). HNF-4α, which has been suggested to be directly regulated by HNF-1β in human hepatoma cells (35,36), showed the maximum decrease (66%) in the expression level. HNF-3γ, 6 and 1α were also markedly downregulated (45–56%). In contrast to these, HNF-3α showed only a marginal decrease in its expression (13%) and the impairment of HNF-3β expression was found to be modest (20%). Computer search of these mouse HNF genes detected an HNF-1β consensus recognition sequence in the upstream regions of HNF-3β as well as HNF-4α genes. These results strongly suggest that all these HNF family members may be involved and HNF-1β plays an important role in the TRF regulatory network in mouse liver cells.
Figure 4.
Effects of HNF-1β suppression on the mRNA levels of HNF family members. Hepa 1-6 cells were transfected with 1β-1 siRNA and harvested for mRNA isolation 48 h after transfection. The HNF and GAPDH mRNAs were analyzed by real-time RT–PCR using SYBR Green. The mRNA level of each of HNFs was normalized to that of GAPDH and the normalized mRNA level in negative control siRNA-transfected cells was set to 1.0. Data are averages + SD of three independent experiments.
DISCUSSION
The present system for the identification of genes directly regulated by a TRF consists of a combination of perturbation of gene expression through RNAi, comprehensive expression profile analysis, extracting the upstream sequences of the primary candidate genes, searching for the consensus TRF-recognition sequences, demonstration of in vivo and in vitro TRF–target DNA binding by X-ChIP/PCR and EMSA. To minimize the possible contamination by HNF-1β-unrelated genes into the primary candidates, we selected the candidate genes that exhibited reproducibly high reduction in their expression levels. Nevertheless, the results of searching for the HNF-1β consensus recognition sequences in the upstream regions suggested that genes whose expressions may be perturbed indirectly, secondarily and/or non-specifically may be included besides the bona fide TRF-regulated genes in this primary candidate gene set. By combinatorial use of the three types of TRF consensus recognition sequences, we detected 13 out of 26 annotated candidate genes as potential HNF-1β-regulated genes. The subsequent examination of in vivo binding by X-ChIP/comparative PCR of a part of these successfully led to the finding that at least five out of the top 26 genes selected on the basis of their perturbations are real HNF-1β-regulated genes. The overall X-ChIP/PCR analysis detected the in vivo binding of HNF-1β to the upstream sequences of eight genes including five novel and three known HNF-1β-regulated genes. This fact strongly supports the practicability of the present step-by-step system for the identification of genes regulated by a TRF.
Perturbation of gene expression is a critical step of the identification of TRF-regulated genes. To do this efficiently with RNAi, selection of highly potent siRNA is the most important step. A positional effect of siRNAs has already been reported for other target mRNAs (21,22,37,38). Two out of the three siRNAs designed according to conventional rules have proven to potently suppress HNF-1β expression as compared to a negative control siRNA. In our experiments, it has been clearly shown that RNAi with 1β-1 siRNA results in extensive perturbation of expressions of a number of genes and enabled us to select the primary candidates for the TRF-regulated genes. RNAi perturbation seems to lead preferentially to downregulation of genes whose expressions are influenced. This makes a clear contrast to our previous finding that overexpression of the HNF-3α gene preferentially caused upregulation of a huge number of genes over a reasonable threshold of 2-fold increase in expression levels (1).
Until now there have been only a few investigations on the effects of RNAi on genome-wide expression profiles. One report has pointed out off-target gene regulation using RNAi targeting of a growth factor receptor and a protein kinase in human cells (39). In this case, siRNA-specific rather than target-specific silencing seemed to be caused by partial sequence identity to the siRNA used. However, our search of the selected 26 cDNA sequences for such a sequence identity to 1β-1 siRNA did not offer any corroborative evidence for off-target gene regulation. Although the reason for this discrepancy is unclear at present, it is possible that our TRF-targeted suppression might have caused direct and emphasized effects on transcriptional regulation of the downstream genes.
It is interesting to note that HNF-1β can regulate two genes encoding putative neuronal transcriptional regulators, Rax and NeuroD4 containing a homeobox and basic helix–loop–helix domain, respectively. Microarray analysis showed marked downregulation of these genes by 1β-1 siRNA transfection. Moreover, in vitro and in vivo binding of HNF-1β to their upstream sequences containing a consensus HNF-1β recognition element was also clearly demonstrated. These findings strongly suggest that HNF-1β could also play some physiological roles via its transcription regulatory cascades in the neuronal system. It has been known that HNF-3α and HNF-3β function in early mammalian neurogenesis (40). Our X-ChIP/PCR data revealed the direct interaction of HNF-1β with the regulatory sequence of the HNF-3β gene, suggesting that HNF-1β could also regulate expressions of these neuron-specific genes through the intermediary of HNF-3β gene regulation.
HNF family members seem to constitute the transcription regulatory network. Indeed, very recently, tissue-specific transcription regulatory circuits containing HNF-4α, 1α and 6 have been identified in human liver and pancreatic islets (41). In the present study, RNAi suppression of HNF-1β mRNA led to the downregulation of all other HNF family members examined. Concomitant preferential downregulation of HNF-1α, 4α and 3γ by suppression of HNF-1β in the hepatoma cells is highly analogous to the phenomenon observed in mouse HNF-1β (TCF2) null embryoid bodies, in which there is no activation of these HNFs (42). HNF-4α is a known HNF-1β-regulated gene (36) and positively regulates expression of HNF-1α (43); in turn HNF-1α is required for maintenance of the HNF-3γ gene expression (44). Therefore, the concomitant downregulation of HNF-4α, 1α and 3γ is thought to be performed according to the cascade in which HNF-1β is located at the top of the hierarchy. HNF family members are involved in regulation of liver-specific genes that function in various important physiological events such as sugar and lipid metabolism. Moreover, some of them are responsible for human diseases, for example: HNF-4α, 1α and 1β are identified as causal genes of MODY 1, 3 and 5, respectively, and HNF-1β mutations are associated with familial hypoplastic glomerulocystic kidney disease (45). Diabetes mellitus is a typical multifactorial disorder and one may expect that the mechanism of this devastating disease can be deciphered at least partially through understanding the details of the HNF network.
In contrast to the clear binding of HNF-1β to the upstream region of the mouse HNF-3β gene in the Hepa 1-6 cells, only modest downregulation of HNF-3β by 1β-1 siRNA transfection was observed. This discrepancy may reflect that the in vivo binding of a TRF to its target sequence does not always lead to an appreciable perturbation of the target gene expression. It should be noted that the silencing of a TRF gene will influence differentially the expression patterns of its controlling genes and its influence may depend on the extent of contribution of the TRF to the transcriptional control of each gene regulated by the regulatory factor. Indeed, several known HNF-1β-regulated genes did not show any appreciable changes in their expression levels. Nevertheless, we were able to find several novel target genes for HNF-1β by using our integrated system, leading to the conclusion that this is a useful tool for the identification of genes regulated by a specific TRF and the characterization of the TRF network.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We thank N. Tominaga-Goto and Y. Tsujimura for their help in microarray analysis; M. Endoh for preparation HNF-1β protein; S. Kondo for help in consensus sequence search and Y. Ueda, W. Hashizume and M. Hashimoto for technical support. We also thank Julian Gough for critical reading of our manuscript. This work was supported by the Research Grant for the RIKEN Genome Exploration Research Project from the Ministry of Education, Culture, Sports, Science and Technology of the Japanese Government to Y.H.
REFERENCES
- 1.Tomaru Y., Kondo,S., Suzuki,M. and Hayashizaki,Y. (2003) A comprehensive search for HNF-3α-regulated genes in mouse hepatoma cells by 60K cDNA microarray and chromatin immunoprecipitation/PCR analysis. Biochem. Biophys. Res. Commun., 310, 667–674. [DOI] [PubMed] [Google Scholar]
- 2.Young R.A. (2000) Biomedical discovery with DNA arrays. Cell, 102, 9–15. [DOI] [PubMed] [Google Scholar]
- 3.Devaux F., Marc,P. and Jacq,C. (2001) Transcriptomes, transcription activators and microarrays. FEBS Lett., 498, 140–144. [DOI] [PubMed] [Google Scholar]
- 4.Futcher B. (2002) Transcriptional regulatory networks and the yeast cell cycle. Curr. Opin. Cell Biol., 14, 676–683. [DOI] [PubMed] [Google Scholar]
- 5.Elbashir S.M., Harborth,J., Lendeckel,W., Yalcin,A., Weber,K. and Tuschl,T. (2001) Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature, 411, 494–498. [DOI] [PubMed] [Google Scholar]
- 6.Elbashir S.M., Harborth,J., Weber,K. and Tuschl,T. (2002) Analysis of gene function in somatic mammalian cells using small interfering RNAs. Methods, 26, 199–213. [DOI] [PubMed] [Google Scholar]
- 7.Harborth J., Elbashir,S.M., Bechert,K., Tuschl,T. and Weber,K. (2001) Identification of essential genes in cultured mammalian cells using small interfering RNAs. J. Cell Sci., 114, 4557–4565. [DOI] [PubMed] [Google Scholar]
- 8.Murphy C.T., McCarroll,S.A., Bargmann,C.I., Fraser,A., Kamath,R.S., Ahringer,J., Li,H. and Kenyon,C. (2003) Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature, 424, 277–283. [DOI] [PubMed] [Google Scholar]
- 9.Dimova D.K., Stevaux,O., Frolov,M.V. and Dyson,N.J. (2003) Cell cycle-dependent and cell cycle-independent control of transcription by the Drosophila E2F/RB pathway. Genes Dev., 17, 2308–2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou A., Scoggin,S., Gaynor,R.B. and Williams,N.S. (2003) Identification of NF-κB-regulated genes induced by TNFα utilizing expression profiling and RNA interference. Oncogene, 22, 2054–2064. [DOI] [PubMed] [Google Scholar]
- 11.Ramos-Nino M.E., Scapoli,L., Martinelli,M., Land,S. and Mossman,B.T. (2003) Microarray analysis and RNA silencing link fra-1 to cd44 and c-met expression in mesothelioma. Cancer Res., 63, 3539–3545. [PubMed] [Google Scholar]
- 12.De Simone V., De Magistris,L., Lazzaro,D., Gerstner,J., Monaci,P., Nicosia,A. and Cortese,R. (1991) LFB3, a heterodimer-forming homeoprotein of the LFB1 family, is expressed in specialized epithelia. EMBO J., 10, 1435–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abbott C., Piaggio,G., Ammendola,R., Solomon,E., Povey,S., Gounari,F., De Simone,V. and Cortese,R. (1990) Mapping of the gene TCF2 coding for the transcription factor LFB3 to human chromosome 17 by polymerase chain reaction. Genomics, 8, 165–167. [DOI] [PubMed] [Google Scholar]
- 14.Bach I., Mattei,M.G., Cereghini,S. and Yaniv,M. (1991) Two members of an HNF1 homeoprotein family are expressed in human liver. Nucleic Acids Res., 19, 3553–3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lazzaro D., De Simone,V., De Magistris,L., Lehtonen,E. and Cortese,R. (1992) LFB1 and LFB3 homeoproteins are sequentially expressed during kidney development. Development, 114, 469–479. [DOI] [PubMed] [Google Scholar]
- 16.Horikawa Y., Iwasaki,N., Hara,M., Furuta,H., Hinokio,Y., Cockburn,B.N., Lindner,T., Yamagata,K., Ogata,M., Tomonaga,O. et al. (1997) Mutation in hepatocyte nuclear factor-1 beta gene (TCF2) associated with MODY. Nature Genet., 17, 384–385. [DOI] [PubMed] [Google Scholar]
- 17.Livak K.J. and Schmittgen,T.D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- 18.Miki R., Kadota,K., Bono,H., Mizuno,Y., Tomaru,Y., Carninci,P., Itoh,M., Shibata,K., Kawai,J., Konno,H. et al. (2001) Delineating developmental and metabolic pathways in vivo by expression profiling using the RIKEN set of 18 816 full-length enriched mouse cDNA arrays. Proc. Natl Acad. Sci. USA, 98, 2199–2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kadota K., Miki,R., Bono,H., Shimizu,K., Okazaki,Y. and Hayashizaki,Y. (2001) Preprocessing implementation for microarray (PRIM): an efficient method for processing cDNA microarray data. Physiol. Genomics, 4, 183–188. [DOI] [PubMed] [Google Scholar]
- 20.Matys V., Fricke,E., Geffers,R., Gossling,E., Haubrock,M., Hehl,R., Hornischer,K., Karas,D., Kel,A.E., Kel-Margoulis,O.V. et al. (2003) TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res., 31, 374–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holen T., Amarzguioui,M., Wiiger,M.T., Babaie,E. and Prydz,H. (2002) Positional effects of short interfering RNAs targeting the human coagulation trigger Tissue Factor. Nucleic Acids Res., 30, 1757–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elbashir S.M., Martinez,J., Patkaniowska,A., Lendeckel,W. and Tuschl,T. (2001) Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J., 20, 6877–6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kalkuhl A., Kaestner,K., Buchmann,A. and Schwarz,M. (1996) Expression of hepatocyte-enriched nuclear transcription factors in mouse liver tumours. Carcinogenesis, 17, 609–612. [DOI] [PubMed] [Google Scholar]
- 24.Mendel D.B., Hansen,L.P., Graves,M.K., Conley,P.B. and Crabtree,G.R. (1991) HNF-1 alpha and HNF-1 beta (vHNF-1) share dimerization and homeo domains, but not activation domains and form heterodimers in vitro. Genes Dev., 5, 1042–1056. [DOI] [PubMed] [Google Scholar]
- 25.Boutla A., Delidakis,C., Livadaras,I., Tsagris,M. and Tabler,M. (2001) Short 5′-phosphorylated double-stranded RNAs induce RNA interference in Drosophila. Curr. Biol., 11, 1776–1780. [DOI] [PubMed] [Google Scholar]
- 26.Martinez J., Patkaniowska,A., Urlaub,H., Luhrmann,R. and Tuschl,T. (2002) Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell, 110, 563–574. [DOI] [PubMed] [Google Scholar]
- 27.Holen T., Amarzguioui,M., Babaie,E. and Prydz,H. (2003) Similar behaviour of single-strand and double-strand siRNAs suggests they act through a common RNAi pathway. Nucleic Acids Res., 31, 2401–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ichikawa J.K., Norris,A., Bangera,M.G., Geiss,G.K., van ‘t Wout,A.B., Bumgarner,R.E. and Lory,S. (2000) Interaction of Pseudomonas aeruginosa with epithelial cells: identification of differentially regulated genes by expression microarray analysis of human cDNAs. Proc. Natl Acad. Sci. USA, 97, 9659–9664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brooks A.R. and Levy-Wilson,B. (1992) Hepatocyte nuclear factor 1 and C/EBP are essential for the activity of the human apolipoprotein B gene second-intron enhancer. Mol. Cell. Biol., 12, 1134–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thomas H., Jaschkowitz,K., Bulman,M., Frayling,T.M., Mitchell,S.M., Roosen,S., Lingott-Frieg,A., Tack,C.J., Ellard,S., Ryffel,G.U. et al. (2001) A distant upstream promoter of the HNF-4α gene connects the transcription factors involved in maturity-onset diabetes of the young. Hum. Mol. Genet., 10, 2089–2097. [DOI] [PubMed] [Google Scholar]
- 31.Duncan S.A., Navas,M.A., Dufort,D., Rossant,J. and Stoffel,M. (1998) Regulation of a transcription factor network required for differentiation and metabolism. Science, 281, 692–695. [DOI] [PubMed] [Google Scholar]
- 32.Mitchell S.M. and Frayling,T.M. (2002) The role of transcription factors in maturity-onset diabetes of the young. Mol. Genet. Metab., 77, 35–43. [DOI] [PubMed] [Google Scholar]
- 33.Shih D.Q. and Stoffel,M. (2001) Dissecting the transcriptional network of pancreatic islets during development and differentiation. Proc. Natl Acad. Sci. USA, 98, 14189–14191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamagata K. (2003) Regulation of pancreatic beta-cell function by the HNF transcription network: lessons from maturity-onset diabetes of the young (MODY). Endocr. J., 50, 491–499. [DOI] [PubMed] [Google Scholar]
- 35.Bois-Joyeux B., Thomassin,H., Richard,F., Ikonomova,R., Denissenko,M. and Danan,J.L. (1995) Several transcription factors participate in the functioning of the α-fetoprotein gene promoter. Bull. Cancer, 82, 541–550. [PubMed] [Google Scholar]
- 36.Hatzis P. and Talianidis,I. (2001) Regulatory mechanisms controlling human hepatocyte nuclear factor 4α gene expression. Mol. Cell. Biol., 21, 7320–7330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miyagishi M. and Taira,K. (2002) U6 promoter-driven siRNAs with four uridine 3′ overhangs efficiently suppress targeted gene expression in mammalian cells. Nat. Biotechnol., 20, 497–500. [DOI] [PubMed] [Google Scholar]
- 38.Hohjoh H. (2002) RNA interference (RNAi) induction with various types of synthetic oligonucleotide duplexes in cultured human cells. FEBS Lett., 521, 195–199. [DOI] [PubMed] [Google Scholar]
- 39.Jackson A.L., Bartz,S.R., Schelter,J., Kobayashi,S.V., Burchard,J., Mao,M., Li,B., Cavet,G. and Linsley,P.S. (2003) Expression profiling reveals off-target gene regulation by RNAi. Nat. Biotechnol., 21, 635–637. [DOI] [PubMed] [Google Scholar]
- 40.Jacob A., Budhiraja,S. and Reichel,R.R. (1997) Differential induction of HNF-3 transcription factors during neuronal differentiation. Exp. Cell Res., 234, 277–284. [DOI] [PubMed] [Google Scholar]
- 41.Odom D.T., Zizlsperger,N., Gordon,D.B., Bell,G.W., Rinaldi,N.J., Murray,H.L., Volkert,T.L., Schreiber,J., Rolfe,P.A., Gifford,D.K. et al. (2004) Control of pancreas and liver gene expression by HNF transcription factors. Science, 303, 1378–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barbacci E., Reber,M., Ott,M.O., Breillat,C., Huetz,F. and Cereghini,S. (1999) Variant hepatocyte nuclear factor 1 is required for visceral endoderm specification. Development, 126, 4795–4805. [DOI] [PubMed] [Google Scholar]
- 43.Tian J.M. and Schibler,U. (1991) Tissue-specific expression of the gene encoding hepatocyte nuclear factor 1 may involve hepatocyte nuclear factor 4. Genes Dev., 5, 2225–2234. [DOI] [PubMed] [Google Scholar]
- 44.Boj S.F., Parrizas,M., Maestro,M.A. and Ferrer,J. (2001) A transcription factor regulatory circuit in differentiated pancreatic cells. Proc. Natl Acad. Sci. USA, 98, 14481–14486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bingham C., Bulman,M.P., Ellard,S., Allen,L.I., Lipkin,G.W., Hoff,W.G., Woolf,A.S., Rizzoni,G., Novelli,G., Nicholls,A.J. et al. (2001) Mutations in the hepatocyte nuclear factor-1β gene are associated with familial hypoplastic glomerulocystic kidney disease. Am. J. Hum. Genet., 68, 219–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.