Abstract
Amphipols (APols) are a newly designed and milder class of detergent. They have been used primarily in protein structure analysis for membrane protein trapping and stabilization. We have recently demonstrated that APols can be used as an alternative detergent for proteome extraction and digestion, to achieve a “One-stop” single-tube workflow for proteomics. In this workflow, APols are removed by precipitation after protein digestion without depleting the digested peptides. Here, we took further advantage of this precipitation characteristic of APols to concentrate proteins from diluted samples. In contrast with tryptic peptides, a decrease in pH leads to the unbiased co-precipitation of APols with proteins, including globular hydrophilic proteins. We demonstrated that this precipitation is a combined effect of acid precipitation and the APols’ protein interactions. Also, we have been able to demonstrate that APols-aided protein precipitation works well on diluted samples, such as secretome sample, and provides a rapid method for protein concentration.
Electronic supplementary material
The online version of this article (doi:10.1007/s00232-014-9668-6) contains supplementary material, which is available to authorized users.
Keywords: Amphipols, Proteomics, Protein precipitation, Concentrating protein, Mass spectrometry
Introduction
Amphipols (APols) were first introduced by Jean-Luc Popot as a new surfactant to stabilize integral membrane proteins in an aqueous solution (Tribet et al. 1996). APols are a class of amphipathic polymers, with both hydrophobic and hydrophilic moieties, designed to solubilize membrane proteins in detergent-free solutions. The hydrophobic chains anchor to the trans-membrane domains of membrane proteins, allowing membrane proteins to be stable and soluble in water-based solutions. Although APols are milder than other surfactants, they have higher affinity for proteins (Zoonens et al. 2005).
The structure of APols generally consists of a backbone chain connected to 2–3 hydrophobic segments. The most extensively studied APols structure is A8-35 (used in this manuscript). A8-35 is a polyacrylate-based polymer made up of about 35 acrylate residues randomly grafted with octylamine and isopropylamine. It has an average MW of ~4.3 kDa (Giusti et al. 2014). Four A8-35 molecules self-assemble into a ~40 kDa particle (Giusti et al. 2014). APols are unique to other detergents because of their very low critical micelle concentration (CMC). APols are typically used at a concentration of 100–1,000 mg/L, so that the free polymers will self-assemble into particles. At or above a pH of 7, A8-35 is highly soluble in water (240 g/L) (Tehei et al. 2014) because of the many free carboxylate groups present at higher pH. When the pH is lowered, the carboxylate groups get protonated leading to a sharp decrease in the solubility and precipitation of APols. APols have been studied extensively over the past 17 years and have multiple applications, including membrane protein folding, synthesis and immobilization (Charvolin et al. 2009), NMR (Zoonens et al. 2005), and proteomics (Bechara et al. 2012). A review of APols’ structure, properties, and applications can also be found in Amphipols from A to Z (Popot et al. 2011).
We have recently discovered that APols can be used as a mild detergent for general proteome extraction (Ning et al. 2013). Briefly, APols can effectively extract the whole proteome with the help of soniciation. It has been shown that trypsin activity during digestion is not affected by the presence of APols. In addition, APols can be readily removed prior to MS analysis by lowering the pH to form precipitates, without depleting the tryptic peptides. In this work, we further explored the features of APols for proteomic usage. In particular, we have found that APols can be used to co-precipitate intact proteins when the pH of buffer solution is lowered. We further investigated this phenomenon and found that this effect can be used for general protein concentration from a diluted sample. We chose the secretome of HEK cells as a model to test the concentrating ability of APols. Presently, filtration is the most frequently used method for concentrating secretome protein from several mL of sample (Meissner et al. 2013; Boersema et al. 2013; Polacek et al. 2010). Our APols-aided strategy for protein precipitation is rapid and efficient in terms of protein recovery.
Experiment
Chemicals
APol A8-35 was bought from Affymetrix. Urea, dithiothreitol (DTT), iodoacetamide (IAA), ammonium bicarbonate (ABC), formic acid (FA), and sodium dodecyl sulfate (SDS) were obtained from Sigma Aldrich (St. Louis, MO, USA). Water and acetonitrile (ACN) for HPLC were obtained from JT Baker (Phillipsburg NJ, USA). Trypsin was purchased from Worthington Biochemical Corp (USA). DC protein assay kit II (500-0112) was purchased from Bio-Rad. All of the chemicals were of analytical purity grade except ACN and FA, which were of HPLC grade. All the water used in the experiment was prepared using a Milli-Q system (Millipore, Bedford, MA, USA).
Proteome Sample Preparation
Human embryonic kidney (HEK) 293T cells were used throughout the experiment of total lysate and secretome analysis. They were maintained in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10 % (v/v) fetal bovine serum (FBS). All cells were grown in 150 × 25-mm-tissue culture dishes from BD Falcon (San Jose, CA, USA). Cells were washed twice with PBS and scraped off the plate in PBS, aliquoted, centrifuged, and then stored at −80 °C for total lysate analysis. The spent medium used for secretome analysis was taken from HEK 293T cells which were grown to 70 % confluence and were washed twice with PBS, had DMEM media free of FBS added, and incubated for 24 h before collection.
Cell pellets were lysed in 50 mM ABC with varying concentrations of APols for the precipitation test. The solution was vortexed and sonicated to increase the protein recovery. For APols-aided precipitation, 5 % FA was added to the solution until the solution became cloudy. The precipitate was spun down at 16,000×g for 2 min. The pellet was reconstituted in 50 mM ABC of the same volume as the supernatant for protein concentration comparison. Equal volume of original total cell lysate, supernatant, two washes, as well as the reconstituted precipitation were loaded onto NuPAGE 4–12 % bis–tris precast gels (Life Technologies Inc. Burlington, ON, USA) and then stained with Coomassie Blue. The same protocol was applied to tests performed using BSA, lysozyme, myoglobin, and spent medium with adjusted sample concentration and volume.
Acid and APols Precipitation Effectiveness Comparison
Three experiments on serum-free spent cell medium were performed in order to compare the effectiveness of the precipitation. The three scenarios assessed were: (1) the precipitation of protein by acid alone; (2) APols + acid following an acid only precipitation, and (3) APols + acid together for precipitation. In the first scenario, 10 mL of spent medium was acidified to pH 3 with 5 % FA and spun down at 16,000×g for 3 min. The supernatant was kept for scenario 2, and the pellet was washed twice with 0.5 % FA and re-suspended in 50 µL of 50 mM ABC for SDS-PAGE (here by using a large starting volume and small reconstitution volume to increase the protein concentration for the purpose of display). In the second scenario, 200 µL of the supernatant from the first scenario was neutralized to pH 8 with 2 M NaOH, and then APols were added to a final concentration of 1 mg/mL. The medium was then acidified, centrifuged, washed, and then re-suspended in 50 µL of 50 mM ABC. For the third scenario, APols were added to 200 µL of spent medium to a final concentration of 1 mg/mL. The medium was acidified to pH 3 with 5 % FA and spun at 16,000×g for 3 min. The pellet was washed and re-suspended the same way as the other two scenarios.
Protein Digestion and MS Analysis
In-solution digestion protocol was modified from Ning et al. (2013). Briefly, the precipitated protein pellet was re-suspended in 50 mM ABC, then reduced and alkylated by 10 mM DTT and 20 mM IAA in the same solution. Trypsin was added at approximately a 1:20 mass ratio of trypsin to protein. Following digestion, the APols were precipitated. For the large-scale secretome analysis, the tryptic peptides were further subjected to SCX fractionation on StageTip (Rappsilber et al. 2007).
All MS analyses were done by HPLC–ESI–MS/MS. The system consisted of an Agilent 1100 micro-HPLC system (Agilent Technologies, Santa Clara, CA, USA) coupled with an LTQ-Orbitrap mass spectrometer (ThermoFisher Scientific, San Jose, CA, USA) equipped with a nano-electrospray interface operated in positive ion mode. The mobile phases consisted of 0.1 % (v/v) FA in water as buffer A and 0.1 % (v/v) FA in acetonitrile as buffer B. Peptide separation was performed on a 75 μm × 150 mm analytical column packed in-house with reverse phase Magic C18AQ resins (3 μm; 120-Å pore size; Dr. Maisch GmbH, Ammerbuch, Germany). The sample was loaded on the column using 98 % buffer A at a flow rate of 1 µL/min for 20 min. A gradient from 5 to 30 % buffer B was performed in 120 min at a flow rate of ~300 nL/min obtained from splitting a 20 µL/min through a restrictor. The MS method consisted of one full MS scan from 350 to 1,700 m/z followed by data-dependent MS/MS scan of the 5 most intense ions, a dynamic exclusion repeat count of 2, and a repeat duration of 90 s. The full MS scan was performed in the Orbitrap analyzer with R = 60,000 defined at m/z 400, while the MS/MS analysis was performed in the LTQ MS. To improve the mass accuracy, all the measurements in Orbitrap mass analyzer were performed with internal recalibration (“Lock Mass”) (Olsen et al. 2005). On the Orbitrap, the charge state rejection function was enabled, with single and “unassigned” charged ions rejected.
Database Search and Data Analysis
The raw files generated by the LTQ-Orbitrap were processed and analyzed using MaxQuant, version 1.2.2.5 (Cox and Mann 2008) using the Uniprot protein fasta database (2012, July version), with commonly observed protein contaminants. The following parameters were used: cysteine carbamidomethylation as fixed modification; methionine oxidation, protein N-terminal acetylation as variable modification, and enzyme specificity was set to trypsin. Up to two missed cleavages of trypsin were allowed. Precursor ion mass tolerance was 7 ppm, and fragment ion mass tolerance was 0.8 Da. If the identified peptide sequences from one protein were equal to, or covered by, another protein’s peptide set, all these proteins were grouped together and reported as one protein group. The false discovery rate (FDR) for peptide and protein was set at 1 % and a minimum length of six amino acids was used for peptides identification. Data analysis was done in Perseus, which comes with Maxquant. GRAVY value (Kyte and Doolittle 1982) calculations and statistics were done by tools provided in BuildSummary (Sheng et al. 2012). Comparisons between samples or methods were based on the same criteria. The protein abundance index was retrieved from (http://pax-db.org/) (Wang et al. 2012). Figures were plotted in R statistic environment (http://www.r-project.org/).
Result and Discussion
APols: Solubilization and Enrichment of Proteins
In our recently published paper (Ning et al. 2013), we explored a novel application of APols besides their main usage for stabilizing membrane proteins in an aqueous phase. We found that APols could be used as a general surfactant to extract a whole proteome for proteomic sample preparation. Moreover, APols do not need to be removed ahead of digestion because they do not inhibit trypsin activity. Furthermore, following trypsin digestion, APols can be precipitated by lowering the pH of buffer solutions, leaving the peptides in solution. Therefore, APols can be readily removed from digested samples prior to MS analysis. All these advantages were combined to develop a one-stop proteomic sample preparation workflow.
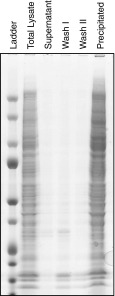
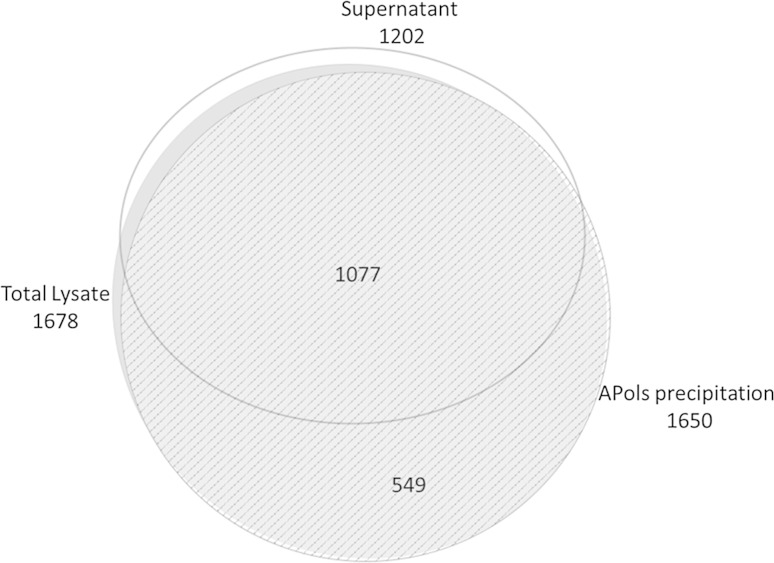
Here, we explored whether APols can be used for the enrichment of proteins by their co-precipitation upon protonation of APols. Our initial focus was the membrane proteome. We first tested whether APols could be used to precipitate membrane proteins from a complex mixture. A final concentration of 1 mg/mL APols was added to 1 mg proteins obtained from a total lysate from HEK 293T cells. The solution was then acidified to pH 3.0 and centrifuged. The supernatant was collected together with two washes of the APols precipitation. The proteins recovered from the supernatant, two washes, and the APols pellets were analyzed by gel electrophoresis (Fig. 1). To our surprise, the vast majority of the proteins appeared to be co-precipitated by APols, with limited protein amounts found in the supernatant and two washes. As well, it appeared that most of the proteins were recovered following reconstitution of the APols precipitation. The same precipitation was readily performed on hydrophilic proteins including BSA, lysozyme, and myoglobin solution, as well as spent cell medium (Figs. S1, S2). Mass spectrometric identification of the proteins co-precipitated by APols, the supernatant, and total HEK 293T cell lysate revealed that APols could precipitate most of the proteins in total lysate (Fig. 2, Fig. S3). The unexpected number of proteins identified from the supernatant fraction is probably from low-abundant proteins and smaller degraded/truncated protein fragments, which cannot be precipitated efficiently by acid. The identification result also showed no significant bias by APols co-precipitation in terms of protein hydrophobicity (GRAVY), molecular weight (MW), and isoelectric point (pI) (Fig. S4). Therefore, it appears that APols can co-precipitate with proteins when the pH is lowered regardless of protein’s hydrophobicity.
Fig. 1.
Comparison of the amount of protein observed at different stages of APols precipitation. The same volumes (adjusted for comparison) of original total cell lysate, supernatant after precipitation, two washes of the pellet with 0.5 % FA, as well as the reconstitution of precipitation pellet were displayed in 1D-SDS-PAGE, and stained by Commassie blue
Fig. 2.
Overlaps between the proteins identified in total lysate, APols precipitate and supernatant. The area of each circle is proportional to the actual number of protein identified. The diagram was plotted using eulerAPE (http://www.eulerdiagrams.org/eulerAPE/)
APols Co-precipitate Proteins
We then tested whether the concentration of APols had an effect on the precipitation of proteins. A dependence on concentration would likely indicate that APols predominantly bind to proteins and then help in their precipitation, whereas, an independence from concentration would indicate that the precipitation of APols predominantly leads to the co-precipitation of proteins. We lowered the APols concentration gradually in the presence of a constant protein concentration, to see whether the concentration of APols (or APols to protein ratio) would have an effect on the quantity of protein precipitated. Our results indicated no significant differences in protein precipitations using 2 mg/mL (14:1 APols:Protein mass ratio) down to 0.05 mg/mL (0.3:1 APols:protein ratio) of APols as shown in Fig. 3. Therefore, the amount of protein precipitated does not appear to be different even when the concentration of APols is approximately three times lower than the proteins. We believe this points to a co-precipitation phenomenon. This also means that less APols can be used for protein precipitation from complex samples, which is important when dealing with large volume and/or dilute protein samples.
Fig. 3.
Effect of APols concentration on protein precipitation efficiency. Aliquot of HEK293T cells was lysed using 200 µL 0.05, 0.2, 1, or 2 mg/mL APols in 50 mM ABC to achieve a concentration of 2.5 mg/mL, then precipitated and reconstituted. The same volumes of the total lysate, supernatant, and reconstituted precipitates were analyzed by SDS-PAGE
APols and Acidification Show Complementary Effects on Protein Precipitation
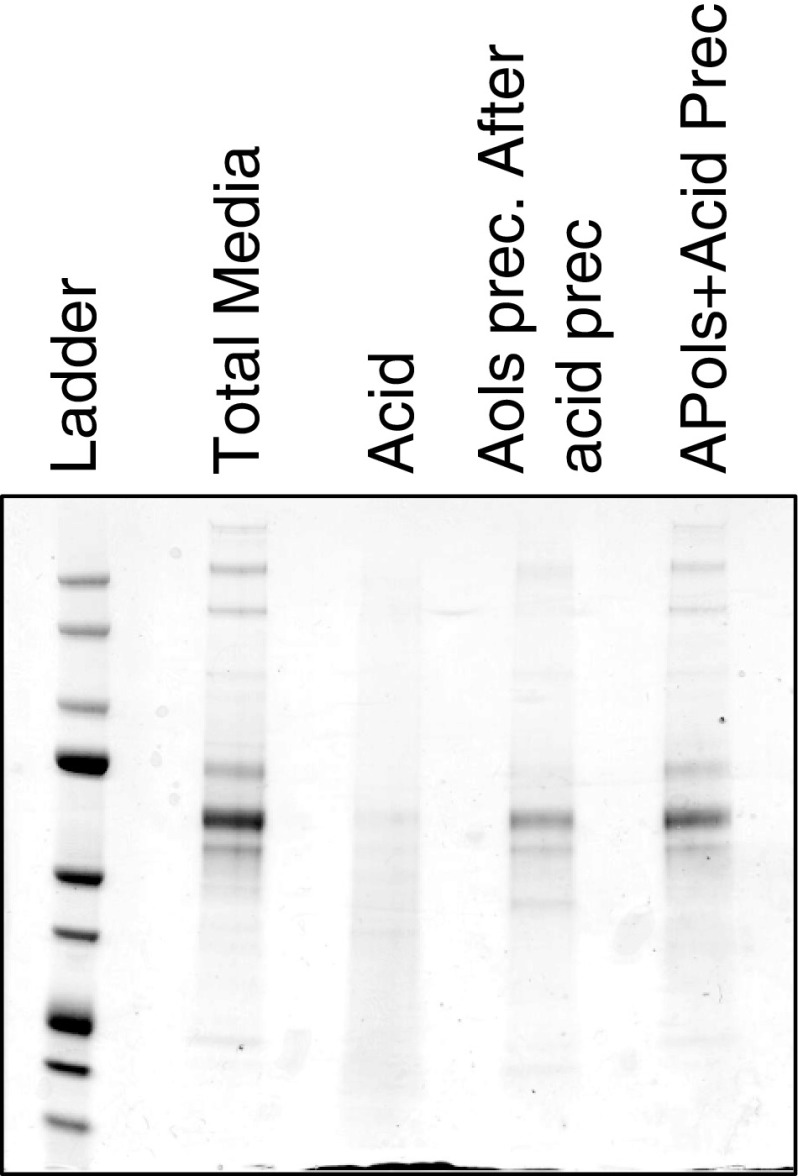
The next step was trying to figure out whether APols and/or the acidification of the samples were the main causes for the precipitation of proteins. Acidification to pH 3.0 is well known to lead to protein precipitation by the formation of insoluble salts between the acid anions, in this case formate, and the positively charged amino groups of the protein molecules. Also, it is known that the hydration layer of proteins is reduced at low pH. Overall, precipitation caused by acidification is particularly efficient for high protein concentrations (Salt et al. 1982; Retz and Steele 1977; Polson et al. 2003). In order to discern the roles that acid and APols play in protein precipitation, a set of protein precipitation experiments was performed with a serum-free spent medium, which only contains less than 0.01 mg/ml of hydrophilic secreted proteins. The spent media were first subjected to acid precipitation by adding 5 % FA to lower the pH of the solution. The resulting pellet was spun down. APols were then added to the supernatant to precipitate the remaining proteins. In the meantime, the “combined” acid plus APols precipitation strategy, which has been used in this work, was also performed for comparison. The amount of acid added and the final pHs were identical for the three experiments. As shown in Fig. 4, the acid only precipitation could hardly produce any protein bands from the spent medium. The starting material was already 50 times more than the other two scenarios, see experimental section for more details. However, after acid precipitation, the addition of APols to the supernatant led to the co-precipitation of proteins. Based on these results, we conclude that both acid and APols are important for the process of protein precipitation. The acid has at least two functions in the precipitation process: firstly, it reduces the solubility of proteins as described above, and secondly, it protonates APols which then become more hydrophobic. We postulate that for sample containing high concentrations of proteins acidification can initialize the nucleation process, and lead to protein aggregation and precipitation. However, for samples with lower concentration of proteins the APols would initialize and accelerate the aggregation process and leads to the co-precipitation of proteins.
Fig. 4.
Effects of acid and APols precipitation on the recovery of proteins from serum-free spent medium. Serum-free cell spent medium was precipitated by 5 % FA only, or followed by APols (1 mg/mL), or by acid + APols
Furthermore, tests were done to determine whether acidification would fully precipitate all the proteins for a sample of high protein concentration. Briefly, a protein extract (2.4 mg/mL) from HEK 293T cells was processed by acid precipitation alone, APols precipitation following acid precipitation, and the combined APols + acid precipitation. The precipitates were then reconstituted, digested with trypsin, and analyzed by mass spectrometry. A total of 4,161 protein groups were identified across the three samples. The acid precipitation alone contributes 3,641 protein groups, which reiterates the important role of acid in the precipitation of higher concentration samples (Fig. S5). However, 2540 proteins were co-precipitated by the addition of APols to the supernatant after the acid precipitation. Therefore, acid precipitation is not sufficient to recover all the proteins even from a concentrated sample. In contrast, APols contributes to the co-precipitation of proteins regardless of the protein concentration, and are more efficient than acid precipitation when dealing with a lower concentration of proteins (Fig. 4). As well, the presence of APols leads to the formation of a visible pellet even when the protein concentration is low, which is beneficial for easy operation. The correlation of the signal intensity between the acid precipitation alone and APols precipitation from the remaining supernatant is not as good as their individual correlation with the combined acid plus APols (Fig. S6), which indicates that the APols and acid precipitation mechanisms are to some extent complementary. Furthermore, we did not find any significant difference in GRAVY, MW or pI, which might be responsible for the complementarity (Fig. S7). Therefore, the APols-aided precipitation, which has the combined effect of acid and APols precipitation, is a preferable protocol for general-purpose protein enrichment.
Case Study: Secretome Analysis by APols Precipitation
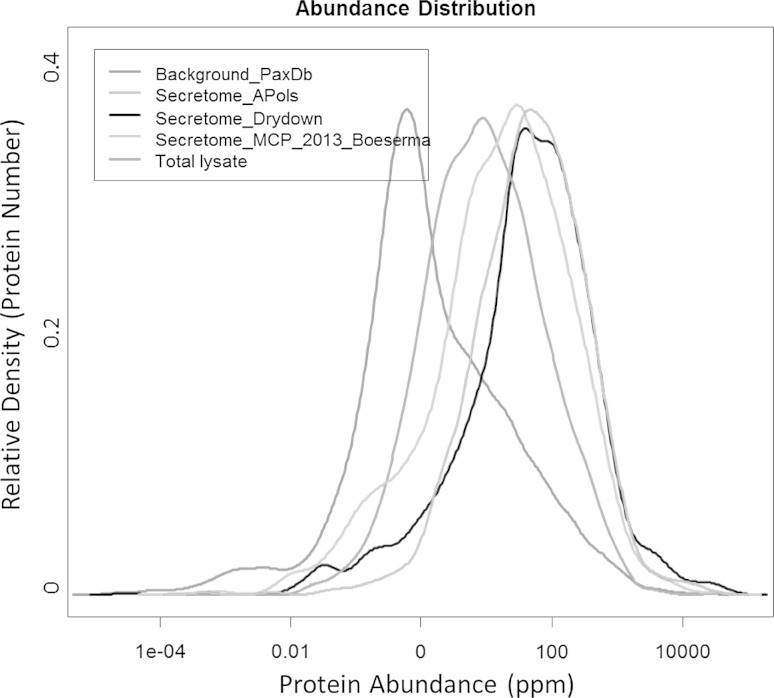
Interestingly, the APols approach for protein co-precipitation is much more efficient than acid precipitation for samples with low concentration of proteins. Therefore this approach appears well suited for diluted biological samples. Typically, the conventional approaches for protein precipitation such as acetone precipitation are less efficient and often need large volume (up to 5 times more volume). An alternative to protein precipitation is ultrafiltration, which is time-consuming when dealing with larger sample volume, and has a low sample recovery rate and high-molecular weight preference, especially when proteins are not denatured. Our approach based on the precipitation of APols is simple and efficient (Fig. 4). To further demonstrate the performance of the APols protein co-precipitation, we used serum-free spent medium, which has an even lower level of secreted proteins. HEK 293T cells were exposed to serum-free medium for 24 h before collection. APols-aided precipitation and protein digestion were performed and the peptides were analyzed by HPLC–ESI–MS/MS. All the manipulations including protein co-precipitation, resolubilization, reduction, alkylation, and digestion were performed in one tube using the APols one-stop approach. 1103 protein groups with 1 % FDR were identified (Table S1). The abundance distribution (Wang et al. 2012) profile of the proteins identified from APols-aided protein precipitation does not show any bias toward protein abundance preference (Fig. 5). It has exactly the same profile as the conventional and presumably non-biased drying down method, by which all secreted proteins were recovered by from the serum-free medium followed by in-solution digestion.
Fig. 5.
Histogram of the protein abundance observed by different protein concentrating techniques. The PaxDb, protein abundance across organisms (Wang et al. 2012), a database of absolute protein abundance was used to assign abundance to each identified protein. (Red) a collection of proteins from the PaxDb illustrating the normal intensity distribution in the whole proteome, (Green) the secretome observed by APols precipitation, (Black) the secretome observed by drying down and classical in-solution digestion, (Purple) (Boersema et al. 2013) a similar size secretome analysis, and (Blue) results from total cell lysate analysis. The X-axis represents the protein abundance in ppm, whereas the Y-axis represents the relative density (Color figure online)
Discussion
APols not only bind to membrane proteins on their hydrophobic regions as reported before but they also appear to co-precipitate proteins when the pH is decreased. The precipitation might be a combined effect of acid precipitation and APols precipitation. The acidification of the protein solution to pH 3.0 causes most proteins to be below their pI (pI 4–6 for most proteins). The protonated amino groups of proteins will form salts (often insoluble) with the acid anion, formate, which decreases the net charge of the proteins. As well, this will be accompanied by a reduction in the protein hydration layer due to the higher concentration of protons. This results in a decrease in repulsive electrostatic forces between proteins, which facilitate their aggregation, and nucleation making further aggregation much easier and faster. APols’ solubility drastically decreases when carboxylate groups are protonated. Our results suggest that APols precipitation accelerates the aggregation of proteins, possibly through co-precipitation.
It is well known that a decrease in the pH can lead to the precipitation of proteins; however, the efficiency is very dependent on the characteristics and concentration of the protein. It is well established that APols at neutral pH interact with hydrophobic regions of proteins; however, little is known about their behavior when the pH decreases. Here, we established that APols-aided precipitation can efficiently precipitate proteins from diluted samples without evident bias on protein size, pI or hydrophobicity. It is worth noting that APols co-precipitation of proteins does not work in the presence of detergents such as SDS, Triton or NP-40 etc. We have not noticed any negative effects of salt from sample buffers. We have applied APols precipitation on secretome analysis and get a decent number of identified peptides compared to work with similar starting material and equipment. We believe that it is promising for membrane proteomic analysis because of the well-established APols’ specificity on membrane protein. In our large-scale data, 452 GO-annotated integral membrane proteins were identified (Table S2). This method can also be a promising alternative strategy for the ultrafiltration, which is time-consuming and has low protein recovery rate.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
D. F. acknowledges a Canada Research Chair in Proteomics and Systems Biology. Funding for this project was provided by NSERC-Canada. Z. N. acknowledges a postdoctoral scholarship from the CIHR Training Program in Neurodegenerative Lipidomics (TGF-96121). Z. N. would also like to acknowledge Dr. Jean-Luc POPOT for the valuable discussion and advices, and Alexandra Therese Star’s help for proofing.
Footnotes
Zhibin Ning and Brett Hawley have contributed equally to this work.
References
- Bechara C, Bolbach G, Bazzaco P, Sharma KS, Durand G, Popot JL, Zito F, Sagan S. MALDI-TOF mass spectrometry analysis of amphipol-trapped membrane proteins. Anal Chem. 2012;84(14):6128–6135. doi: 10.1021/ac301035r. [DOI] [PubMed] [Google Scholar]
- Boersema PJ, Geiger T, Wisniewski JR, Mann M. Quantification of the N-glycosylated secretome by super-SILAC during breast cancer progression and in human blood samples. Mol Cell Proteomics. 2013;12(1):158–171. doi: 10.1074/mcp.M112.023614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charvolin D, Perez JB, Rouviere F, Giusti F, Bazzacco P, Abdine A, Rappaport F, Martinez KL, Popot JL. The use of amphipols as universal molecular adapters to immobilize membrane proteins onto solid supports. Proc Natl Acad Sci USA. 2009;106(2):405–410. doi: 10.1073/pnas.0807132106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26(12):1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- Giusti Fabrice, Rieger Jutta, Catoire Laurent J., Qian Shuo, Calabrese Antonio N., Watkinson Thomas G., Casiraghi Marina, Radford Sheena E., Ashcroft Alison E., Popot Jean-Luc. Synthesis, Characterization and Applications of a Perdeuterated Amphipol. The Journal of Membrane Biology. 2014;247(9-10):909–924. doi: 10.1007/s00232-014-9656-x. [DOI] [PubMed] [Google Scholar]
- Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Meissner F, Scheltema RA, Mollenkopf HJ, Mann M. Direct proteomic quantification of the secretome of activated immune cells. Science. 2013;340(6131):475–478. doi: 10.1126/science.1232578. [DOI] [PubMed] [Google Scholar]
- Ning Z, Seebun D, Hawley B, Chiang CK, Figeys D. From cells to peptides: “one-stop” integrated proteomic processing using amphipols. J Proteome Res. 2013;12(3):1512–1519. doi: 10.1021/pr301064z. [DOI] [PubMed] [Google Scholar]
- Olsen JV, de Godoy LM, Li G, Macek B, Mortensen P, Pesch R, Makarov A, Lange O, Horning S, Mann M. Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol Cell Proteomics. 2005;4(12):2010–2021. doi: 10.1074/mcp.T500030-MCP200. [DOI] [PubMed] [Google Scholar]
- Polacek M, Bruun JA, Johansen O, Martinez I. Differences in the secretome of cartilage explants and cultured chondrocytes unveiled by SILAC technology. J Orthop Res. 2010;28(8):1040–1049. doi: 10.1002/jor.21067. [DOI] [PubMed] [Google Scholar]
- Polson C, Sarkar P, Incledon B, Raguvaran V, Grant R. Optimization of protein precipitation based upon effectiveness of protein removal and ionization effect in liquid chromatography-tandem mass spectrometry. J Chromatogr B Anal Technol Biomed Life Sci. 2003;785(2):263–275. doi: 10.1016/S1570-0232(02)00914-5. [DOI] [PubMed] [Google Scholar]
- Popot JL, Althoff T, Bagnard D, Baneres JL, Bazzacco P, Billon-Denis E, Catoire LJ, Champeil P, Charvolin D, Cocco MJ, Cremel G, Dahmane T, de la Maza LM, Ebel C, Gabel F, Giusti F, Gohon Y, Goormaghtigh E, Guittet E, Kleinschmidt JH, Kuhlbrandt W, Le Bon C, Martinez KL, Picard M, Pucci B, Sachs JN, Tribet C, van Heijenoort C, Wien F, Zito F, Zoonens M. Amphipols from A to Z. Annu Rev Biophys. 2011;40:379–408. doi: 10.1146/annurev-biophys-042910-155219. [DOI] [PubMed] [Google Scholar]
- Rappsilber J, Mann M, Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protoc. 2007;2(8):1896–1906. doi: 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- Retz KC, Steele WJ. Acid precipitation of protein in the presence of Triton X-100 and deoxycholate. Anal Biochem. 1977;79(1–2):457–461. doi: 10.1016/0003-2697(77)90421-3. [DOI] [PubMed] [Google Scholar]
- Salt DJ, Leslie RB, Lillford PJ, Dunnill P. Factors influencing protein structure during acid precipitation: a study of soya proteins. Eur J Appl Microbiol Biotechnol. 1982;14(3):144–148. doi: 10.1007/BF00497890. [DOI] [Google Scholar]
- Sheng Q, Dai J, Wu Y, Tang H, Zeng R. BuildSummary: using a group-based approach to improve the sensitivity of peptide/protein identification in shotgun proteomics. J Proteome Res. 2012;11(3):1494–1502. doi: 10.1021/pr200194p. [DOI] [PubMed] [Google Scholar]
- Tehei M, Giusti, F, Zaccai, G, Popot J-L (2014) Thermal fluctuations in amphipol A8-35 measured by neutron scattering. J Membr Biol (under review) [DOI] [PubMed]
- Tribet C, Audebert R, Popot JL. Amphipols: polymers that keep membrane proteins soluble in aqueous solutions. Proc Natl Acad Sci USA. 1996;93(26):15047–15050. doi: 10.1073/pnas.93.26.15047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Weiss M, Simonovic M, Haertinger G, Schrimpf SP, Hengartner MO, von Mering C. PaxDb, a database of protein abundance averages across all three domains of life. Mol Cell Proteomics. 2012;11(8):492–500. doi: 10.1074/mcp.O111.014704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoonens M, Catoire LJ, Giusti F, Popot JL. NMR study of a membrane protein in detergent-free aqueous solution. Proc Natl Acad Sci USA. 2005;102(25):8893–8898. doi: 10.1073/pnas.0503750102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.