Abstract
CsdA, a DEAD-box protein from Escherichia coli, has been proposed to participate in a variety of processes, such as translation initiation, gene regulation after cold-shock, mRNA decay and biogenesis of the small ribosomal subunit. Whether the protein really plays a direct role in these multiple processes is however, not clear. Here, we show that CsdA is involved in the biogenesis of the large rather than the small ribosomal subunit. Deletion of the csdA gene leads to a deficit in free 50S subunits at low temperatures and to the accumulation of a new particle sedimenting around 40S. Analysis of the RNA and protein contents of this particle indicates that it corresponds to a mis-assembled large subunit. Sucrose gradient fractionation shows that in wild-type cells CsdA associates mainly with a pre50S particle. Presumably the RNA helicase activity of CsdA permits a structural rearrangement during 50S biogenesis at low temperature. We showed previously that SrmB, another DEAD-box RNA helicase, is also involved in 50S assembly in E.coli. Our results suggest that CsdA is required at a later step than SrmB. However, over-expression of CsdA corrects the ribosome defect of the srmB-deleted strain, indicating that some functional overlap exists between the two proteins.
INTRODUCTION
DEAD-box proteins form a large family of putative RNA helicases conserved from bacteria to humans (1,2). Together with the related DExD/H-box proteins, they play important roles in many cellular processes involving RNA, such as RNA processing, transport, degradation, translation or ribosome biogenesis (2,3). In vitro studies have shown that these proteins possess an RNA-dependent ATPase activity, and that some of them can catalyse the ATP-dependent separation of RNA duplexes into single strands. Based on these biochemical characteristics, it is generally believed that they use the energy of ATP hydrolysis to unwind local RNA secondary structures, i.e. that they behave as ‘RNA helicases’. However, it is becoming apparent that, in many cases, they may have a broader role in remodelling RNA structures, i.e. they may assist correct RNA folding [‘RNA chaperones’ (4)] or help in dissociating RNA–protein interactions (5,6).
During ribosome biogenesis in Escherichia coli, a single rRNA precursor is concomitantly synthesized, processed into 23S, 16S and 5S rRNAs and assembled with the 54 ribosomal proteins (r-proteins) in an ordered pathway to form mature 30S and 50S ribosomal subunits (7,8). Whereas this process takes only minutes at 37°C in vivo, reconstitution of ribosomal subunits from their isolated components in vitro requires much more drastic conditions (i.e. long incubation times, high temperature and high ionic concentrations). This difference suggests that non-ribosomal factors such as RNA helicases or chaperones may assist assembly in vivo (see 8,9). Interest ingly, mutations in the DnaK chaperone affect ribosome biogenesis in vivo (10) and the purified protein has been shown to facilitate 30S assembly in vitro (11), a finding that, however, has been controversially discussed (12,13). In contrast, until recently, no DEAD-box helicases had been implicated in ribosome biogenesis in E.coli, although in yeast at least 15 of them are required for this process (14). Last year, we showed that SrmB, one of the five DEAD-box proteins present in E.coli (15), is in fact involved in the assembly of the 50S ribosomal subunit (16). The deletion of the srmB gene leads to a deficit in free 50S subunits and to the accumulation of a new 40S particle corresponding to an incompletely assembled 50S subunit. The protein composition of this particle suggests that SrmB is involved during an early step of 50S assembly (16).
Another candidate for a role in ribosome biogenesis is the E.coli DEAD-box protein DeaD, renamed CsdA (cold-shock DEAD-box protein A) following the discovery that its expression is cold-shock inducible (17). The deaD/csdA gene was initially identified as a multi-copy suppressor of a mutation in the rpsB gene encoding r-protein S2 (18). The manner in which it was discovered suggested that it may play a role in the biogenesis of the small ribosomal subunit, and indeed it was subsequently reported that over-expression of CsdA in an S2 mutant restores the incorporation of r-proteins S2 and S1 into the ribosome (19). Another role proposed for CsdA is to assist translation: in vitro studies suggest that it promotes translation initiation of structured mRNAs (20) and one of its roles during cold-shock might be to de-repress the synthesis of heat-shock proteins by facilitating their translation (17). In addition to ribosome biogenesis and translation, CsdA has also been implicated in the stabilization and degradation of mRNAs, particularly when the protein is over-expressed (21–23). Altogether, these data suggest that CsdA plays overlapping roles in several important processes in vivo. However, it is often unclear whether these multiple roles reflect direct or indirect effects of the protein, and whether they can be observed under normal physiological conditions, i.e. when CsdA is expressed in wild-type (wt) cells and from its single-copy chromosomal gene.
Here, we show that under these conditions, CsdA is involved in the biogenesis of the 50S rather than the 30S ribosomal subunits. The deletion of the corresponding gene leads to a deficit in free 50S subunits and accumulation of a 40S-like particle, as for SrmB. However, the composition of this particle is different from the ΔsrmB 40S particle and suggests that the defect in biogenesis occurs at a later step. Our results also indicate an association of CsdA with 50S precursors at low temperature and provide evidence for a functional link between SrmB and CsdA.
MATERIALS AND METHODS
Bacterial strains and plasmids
A derivative of the E.coli K12 strain WJW45 [W3110 lacU169 (24)] deleted for csdA was constructed according to (25). Approximately 815 bp of flanking DNA 5′ and 3′ of the csdA gene were amplified in two separate PCR reactions (Pwo DNA polymerase from Boehringer Mannheim), using the following pairs of primers: csdA-No, 5′ CGCGGATTCAATCCGACCGGATATGCCTG 3′ and csdA-Ni, 5′ CACGCAATAACCTTCACACTCCAAATTTATAACGGTTTCGAAT TCAGCCATGTAG 3′; csdA-Co, 5′ CGCGGATTCCCGTGATAACCAAACGATGCC 3′ and csdA-Ci, 5′ GTTATAAATTTGGAGTGTGAAGGTTATTGCGTGGATTCTACCGGTCGTCGTCG 3′. The two fragments were then annealed via their overlapping region (underlined in the primer sequences) and re-amplified using the outer primers (Co and No). The fusion product resulted in an in-frame fusion between the first 6 and the last 13 codons of the csdA gene, i.e. removing 1833 bp or 97% of the coding region. The product was then cloned into the SmaI site of pKO3 (25) and sequenced. The resulting plasmid (pKO3-ΔcsdA) was transformed into the WJW45 strain. Co-integration, resolution and elimination of the plasmid were performed as described (25). Colonies were tested for the csdA deletion by PCR, using primers flanking the gene.
The rna-30 mutation (RNase I–) was P1-transduced from strain DK533 (rna-30::kan, Ecoli Genetic Stock Center) into both WJW45 and WJW45ΔcsdA strains. KanR transductants were tested by PCR, using primers specific for rna.
The WJW45ΔsrmB strain has been described in (16).
Strains BL21-FLAG-csdA and WJW45-FLAG-csdA were constructed by replacing the chromosomal csdA gene of BL21(DE3) or WJW45 by an N-terminal FLAG-tagged csdA allele, using the gene replacement technique (25). Approximately 800 bp of DNA 5′ and 3′ of the csdA ATG were amplified in two separate PCR reactions using the following pairs of primers: csdA-5′ext: 5′ CGCGGATCCAATCCGACCGGATATGCCTG 3′ and FLAG-5′: 5′ TTTATCGTCATCGTCTTTGTAGTCCATGTAGTACGTGTGCC TCA 3′; csdA-3′ ext: 5′ CGCGGATCCTACGCTCAAGAGCTTCAGCC 3′ and FLAG-3′: 5′ GACTACAAAGACGATGACGATAAAGCTGAATTCGAAACCACTTTTGC 3′ (the BamHI sites are underlined and the FLAG sequences are in italics). The two fragments were annealed and re-amplified by fusion PCR using the csdA external primers, resulting in an in-frame insertion of the eight FLAG codons after the csdA start codon. The product was then cloned into the BamHI site of pKO3 and transferred onto the chromosome of BL21(DE3) and WJW45 as above. The fusion CsdA protein, which could be detected by western analysis using anti-FLAG antibodies (anti-FLAG M2 monoclonal antibody from Sigma), was over-expressed after a shift at 16°C, like the genuine CsdA; moreover, strains expressing the fusion protein were not cold sensitive (data not shown). These results indicate that the FLAG-CsdA protein was functional.
To construct the WJW45-FLAG-csdA ΔsrmB strain, the srmB deletion of WJW45ΔsrmB strain was tagged with Tn10 using the CAG18480 strain (nadB51::Tn10) and then P1-transduced into the WJW45-FLAG-csdA strain. TetR transductants were tested for the presence of the srmB deletion by PCR.
Plasmid pBD1 is a pBR322 derivative containing the csdA gene under the control of its own promoter (gift of P. Lesage and M. Springer, IBPC, Paris). Cells containing pBR322 or pBD1 were grown in the presence of 10 µg/ml tetracycline.
Polysome profile analysis
Cells were grown at 20°C in LB medium to an absorbance at 600 nm (A600nm) of 0.2–0.7. Extracts were prepared as in (16). About 7 A260nm units of lysate were layered onto 10–40% (w/v) sucrose gradients in buffer A (10 mM Tris–HCl pH 7.5, 50 mM NH4Cl, 10 mM MgCl2, 1 mM DTT), and centrifuged at 35 000 r.p.m. for 2 h 45 min to 3 h at 4°C in a Beckman SW41 or Kontron TST41 rotor. Gradients were analysed with an ISCO UA-6 detector with continuous monitoring at 254 nm.
Purification of ribosomal particles
Between 80 and 100 A260nm units of lysates were layered onto 5–20% (w/w) sucrose gradients in buffer A, centrifuged at 28 000 r.p.m. for 5 h 30 min to 6 h at 4°C in a Beckman SW 28 rotor and analysed as above. Fractions containing ribosomal particles were collected, combined and pelleted overnight by centrifugation in a Kontron TFT 50-38 rotor at 35 000 r.p.m. at 4°C. Ribosomal pellets were re-suspended in buffer A and fractionated again on 5–20% sucrose gradients. Fractions corresponding to clearly separated species were collected.
RNA and r-protein analysis
For RNA analysis, purified ribosomal particles were precipitated overnight with 0.7 vol of ethanol at –20°C. RNA was extracted and analysed by northern blotting and primer extension as in (16).
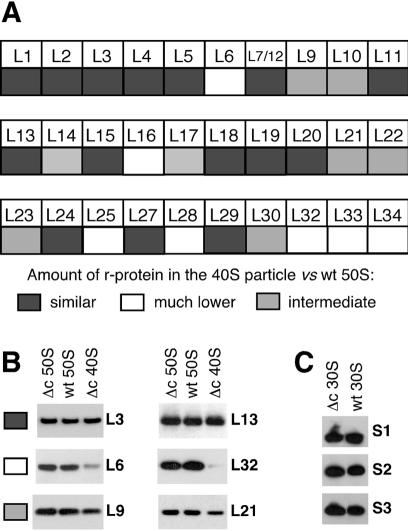
r-Proteins from purified 30S, 40S and 50S particles were precipitated by TCA as described in (16). Based on the A260nm of each ribosomal species, pellets were re-suspended in different volumes of loading buffer so that the protein concentrations were nearly identical for each species (equivalent protein concentrations were also verified on Coomassie-stained polyacrylamide gels). Identical amounts of proteins were then separated on 10% Bis–Tris NuPAGE polyacrylamide gels in MES buffer (Invitrogen) and transferred either to nitrocellulose or PVDF membranes. Membranes were then incubated with polyclonal sheep antibodies against individual r-proteins (a generous gift from R. Brimacombe, Max-Planck Institute, Berlin). However, for r-protein S1, a rabbit polyclonal antibody was used (gift of I. Boni, Academy of Sciences from Russia), and antibodies against L31, L35 and L36 were not available. Blots were decorated with horseradish peroxidase (HRP)-conjugated secondary antibodies (Sigma) and signals were revealed by ECL-Plus kit (Amersham). The reaction yielded a chemiluminescent signal that was revealed by autoradiography and a fluorescent product that was quantified using the FLA-3000 system (Fuji). Different amounts of wt 30S or 50S subunits were loaded on each polyacrylamide gel to validate the correlation between signal and amount of r-protein (data not shown). Quantification of each r-protein was performed at least twice, using different membranes. A protein was regarded as present in either normal or greatly reduced amount in the 40S particle, when its abundance was >65 or <35%, respectively, of its abundance in the 50S subunit (from wt or ΔcsdA strain). Other proteins were considered to be present at intermediate levels. We noticed that for the less abundant proteins, such as L32 (see Fig. 4), the chemiluminescent signal was barely detectable although quantification of the fluorescent signal still indicated a relative abundance of ∼25%. This approach gave results consistent with the mass spectrometry technique used previously to analyse the r-protein content of the ΔsrmB 40S particle (16). For instance, L13, which was not found in the ΔsrmB particle using mass spectrometry, was not found either by western blot analysis (data not shown). In contrast, L1 was found in normal amount by both techniques.
Figure 4.
r-Protein analysis of the 40S and 30S particles from ΔcsdA strain. The ΔcsdA and wt strains were grown at 20°C. r-Proteins were extracted from purified particles and identified and quantified by western blot as described in Materials and Methods. (A) Relative abundance of individual r-proteins in the 40S particle versus the 50S subunit. Grey squares, faint grey squares and white squares correspond to proteins present in similar, lower or much lower amount in the 40S particle, respectively. (B) Western blots illustrating the three classes of proteins defined in (A). Equivalent amounts of 40S particles or 50S subunits (see Materials and Methods) were analysed by western blotting using antibodies against the indicated r-proteins. Δc = ΔcsdA. (C) Western blots showing the presence of r-proteins S1–S3 in the 30S subunits from wt and ΔcsdA strains.
Immunolocalization of CsdA on ribosome profiles
Between 20 and 30 A260nm of lysates were fractionated on sucrose gradients as described above. Proteins from individual fractions (1 ml) were TCA-precipitated as in (16) and re-suspended in 20 µl of gel loading buffer. Aliquots of 2 or 5 µl were loaded on SDS–polyacyrylamide gels and analysed by western blot. Anti-S3 and anti-L20 antibodies were gifts from Drs R. Brimacombe and C. Chiaruttini, respectively. Anti-FLAG M2 monoclonal antibody was from Sigma. Blots were decorated with HRP-conjugated secondary antibodies and signals revealed by ECL (Amersham).
RESULTS
Deletion of csdA leads to a cold-sensitive phenotype
To study the function of csdA, we deleted this gene from the E.coli chromosome using a gene replacement procedure (25). This method creates an in-frame fusion between the first and last few codons of the open reading frame without using a resistance marker, thereby avoiding polar effects on the expression of distal genes. A derivative of the E.coli K12 strain W3110, carrying a deletion of the rna gene encoding the non-specific periplasmic endoribonuclease RNase I, was used as the parental strain for most experiments presented here (see Materials and Methods). This strain and its csdA-deleted derivative will be referred to as wt and ΔcsdA strains respectively.
The growth of the wt and ΔcsdA strains was compared at different temperatures. On LB plates, no difference in growth was observed at 37°C, but at lower temperature the ΔcsdA strain displayed a slow-growth phenotype, which was exacerbated as the temperature decreased (Fig. 1 and data not shown). Plasmid pBD1, a derivative of pBR322 carrying the csdA gene under control of its own promoter, could correct the growth defect of ΔcsdA (Fig. 1), confirming that the cold-sensitive phenotype results from the absence of the csdA gene. The doubling time of the ΔcsdA cells in liquid LB medium was 6 h 45 min at 20°C versus 2 h 45 min for the wt, consistent with the growth phenotype on plates. These results indicate that, in agreement with earlier findings (17), CsdA plays a critical role in cells grown at low temperature.
Figure 1.
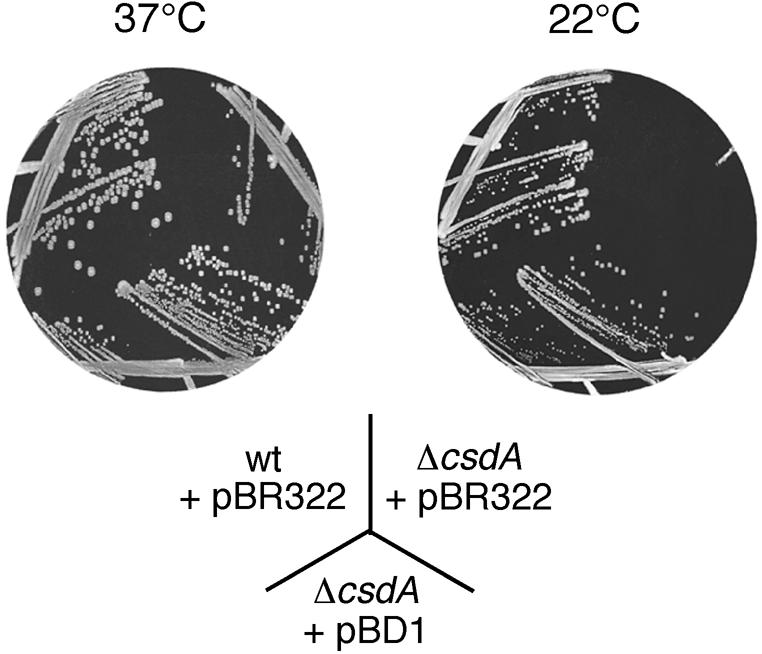
Deletion of csdA leads to a reduced growth at low temperature. wt and ΔcsdA strains transformed with either pBR322 or a derivative bearing the csdA gene (pBD1) were grown on tetracycline-LB plates at 37°C for 15 h or at 22°C for 2 days (at 22°C, the ΔcsdA/pBR322 colonies only become visible after longer incubation times).
At low temperature, deletion of csdA affects the biogenesis of the 50S ribosomal subunit
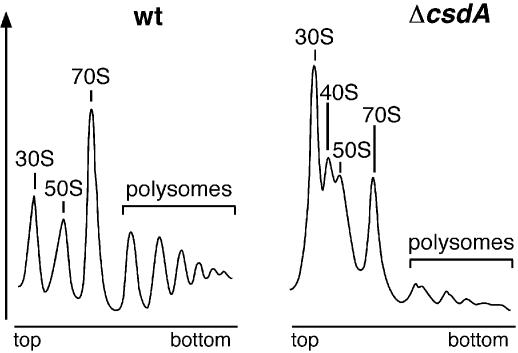
From experiments in which CsdA was over-expressed in an rpsB mutant background, a link between CsdA and ribosome biogenesis has been proposed (see Introduction), but evidence that this link exists when CsdA is expressed at physiological levels and in wt cells was still lacking. To address this question, we compared the polysome profiles of the ΔcsdA and wt strains. After growth in LB medium at 20°C, cells were lysed and ribosomal particles were separated by sucrose gradient sedimentation under conditions favouring ribosome association (10 mM Mg2+). The ribosomal particles of the wt strain were found mainly in polysomes (mRNA harbouring multiple bound ribosomes) and 70S ribosomes (mixture of monosomes and free ribosomes) (Fig. 2, left). In contrast, the ΔcsdA strain showed a considerable increase in the proportion of free ribosomal subunits, with a dramatic reduction in polysomes (Fig. 2, right). Moreover, free 30S subunits were much more abundant than free 50S subunits, indicating a deficiency in 50S subunit formation. Finally, a new particle with a sedimentation coefficient around 40S appeared between the 50S and 30S peaks. This profile resembles that obtained with a strain lacking another DEAD-box protein, SrmB (16). In particular, the ΔsrmB strain also exhibited a deficit in 50S subunit and an accumulation of a particle sedimenting around 40S. We conclude that CsdA, like SrmB, plays a role in the biogenesis of 50S ribosomal subunits.
Figure 2.
Deletion of csdA leads to a deficiency in free 50S ribosomal subunits and to the accumulation of a new particle (40S). wt and ΔcsdA strains were grown in LB medium at 20°C. Polysome profiles were analysed as described in Materials and Methods. Ordinates refer to A254nm. Peaks corresponding to free 30S and 50S subunits, 70S ribosomes (free couples and monosomes), polysomes and 40S particles are indicated.
When the cells were grown at 37°C, no difference between the ribosome profiles of the ΔcsdA and wt strains was observed (data not shown), indicating that the role of CsdA in ribosome biogenesis is critical at low temperature only.
A precursor of 23S rRNA accumulates in the ΔcsdA 40S particle
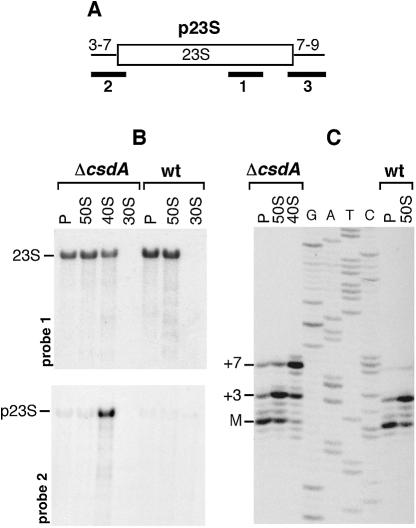
To further characterize the ribosome defect in the ΔcsdA strain, we analysed the rRNA species present in the different ribosomal particles by northern blot and primer extension. Analysis of 23S rRNA is shown in Figure 3. Hybridization with oligonucleotide 1, which is complementary to the mature sequence of 23S (Fig. 3A), revealed a signal in the 50S and polysome fractions of both wt and mutant strains, as well as in the 40S fraction of the mutant (Fig. 3B, top). To test whether this signal corresponds to the mature 23S rRNA or its precursor p23S (see Fig. 3A), the same membrane was hybridized with two oligonucleotides that are specific for p23S. Oligonucleotides 2 and 3, which are complementary to the 5′ and 3′ ends of p23S, respectively, revealed a signal in the 40S particle of the ΔcsdA strain only (Fig. 3B, bottom; hybridization with probe 3 is not shown). Thus, p23S accumulates in the 40S particle.
Figure 3.
The ΔcsdA 40S particle contains a 23S rRNA precursor. Ribosomal particles from wt and ΔcsdA strains grown at 20°C were purified on two sucrose gradients. RNA extracted from these isolated particles was subjected to northern blot (B) and primer extension (C) analysis. (A) The p23S precursor results from RNase III cleavage of the initial 30S rrn transcript. Compared with mature 23S rRNA (open box), it caries 3 or 7 and 7–9 extra-nucleotides at its 5′ or 3′ ends, respectively [thin lines (26)]. The probes used for northern analysis are indicated by solid bars. (B) Equal amounts of RNA from 30S, 40S, 50S and polysome (P) fractions were separated on a 1% agarose gel, transferred to a nylon membrane and probed with the 5′-end-labeled oligonucleotides shown in (A). Upper panel, probe 1; lower panel, probe 2. Data obtained with probe 3 are not shown. The positions of precursor and mature 23S rRNA are indicated. (C) RNA from 40S, 50S and polysome (P) fractions was analysed by primer extension using a 33P-end-labelled primer. A sequencing ladder obtained with the same primer is shown (GATC). The 5′ end of mature 23S rRNA (M) and that of the p23S precursor (+3 and +7) are indicated.
To test whether the rRNA present in the 40S particle consisted of pure p23S or a mixture of both 23S and p23S, we performed primer extension experiments. As shown in Figure 3C, 50S subunits of both strains contained mainly two products, a minor one corresponding to the mature 5′ end of 23S rRNA (‘M’) and the major one with three extra nucleotides (‘+3’). In contrast, the 40S particle isolated from the ΔcsdA mutant contained mostly a +7 species with minor amounts of the +3 species. The latter may be due to a contamination by the 50S subunits during purification on sucrose gradients. These results clearly show that the 40S particle contains only precursor 23S rRNA that is not matured at its 5′ end. The different precursors (i.e. the +3 and +7 species) have been described previously (26,27), and their distribution within the different ribosomal particles is similar to that we observed with the ΔsrmB strain (16). The fact that polysomes of both wt and ΔcsdA strains contained predominantly mature 5′ ends is in agreement with previous data showing that the final maturation of 23S occurs within polysomes (28).
5S rRNA was present in normal quantities and correctly processed in the 40S particle (data not shown). Together, these results indicate that the 40S particle of the ΔcsdA strain is a 50S-related particle in which the 23S rRNA has not been completely matured.
Protein composition of the 40S particle
To further characterize the ΔcsdA 40S particle, we compared its protein content with that of the 50S subunit from the wt strain. r-Proteins from the purified particles were separated by gel electrophoresis, and individual proteins were detected and quantified by western blot analysis as described in Materials and Methods. Membranes were stripped and re-probed several times with antibodies against different r-proteins. This approach gave results consistent with the mass spectrometry technique that we used previously (16; see Material and Methods). Proteins L31, L35 and L36 could not be analysed because the corresponding antibodies were not available. As shown in Figure 4A, all other r-proteins of the large subunit were detected in the 40S particle, but in different amounts. From this analysis, they were classified into three groups according to their abundance, i.e. normal, reduced and very low (see Materials and Methods). An example of two proteins from each group is shown in Figure 4B. The fact that many r-proteins are present in substoichiometric amounts indicates that the 40S particles are either genuinely heterogeneous or that they are loosely assembled, leading to the loss of proteins during particle purification. Nevertheless, proteins with very low abundance may in fact be completely absent from the 40S particle, since contamination by trace amounts of 50S subunits cannot be completely avoided despite two successive purification steps.
It is noteworthy that, according to this analysis, the 40S particle of the ΔcsdA strain is different from the 40S particle isolated from the ΔsrmB strain. For example, L13, a protein essential for an early step of 50S assembly in vitro (29), is absent from the ΔsrmB 40S particle but present at normal levels in the ΔcsdA particle. In fact, most of the proteins that were less abundant in the ΔcsdA particle are incorporated late in the assembly process in vitro (8). We therefore propose that SrmB and CsdA are required at different steps during 50S assembly, with SrmB acting earlier in the process than CsdA.
The csdA deletion does not affect the biogenesis of the 30S ribosomal subunit
CsdA has been suggested to play a role in the biogenesis of the 30S subunit by facilitating the incorporation of r-proteins S1 and S2 (19). To test this hypothesis further, we characterized the 30S particle from the ΔcsdA strain at low temperature. Its sedimentation behaviour was indistinguishable from that of the wt 30S subunit (Fig. 2), and northern blot and primer extension experiments on the 16S rRNA did not reveal any processing defect (data not shown). Likewise, analysis of the r-proteins by two-dimensional gel electrophoresis did not reveal any difference between the ΔcsdA and wt 30S subunits (J.Charollais and K.H.Nierhaus, unpublished results). Further analysis by western blot confirmed that r-proteins S1 and S2 are present in similar amounts in both subunits (Fig. 4C). Together, these results indicate that the biogenesis of the 30S ribosomal subunit is not affected by the csdA deletion.
CsdA is associated with 50S precursors at low temperature
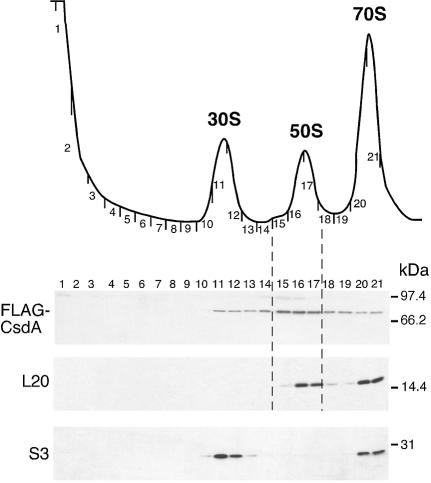
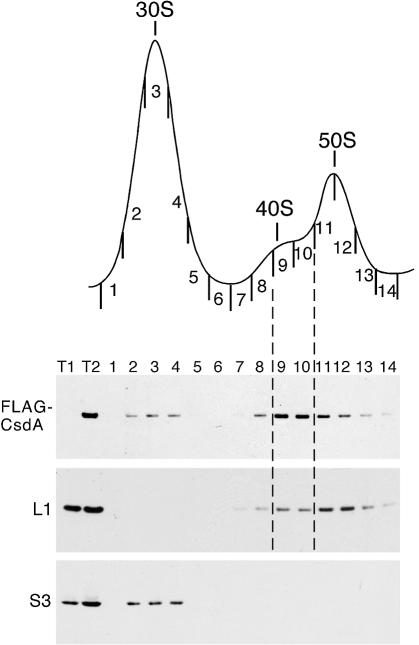
As CsdA appears to be involved in the biogenesis of 50S subunits, we tested whether it might co-purify with ribosomal particles in wt cells. For this purpose, we used a strain harbouring a FLAG-tagged chromosomal copy of the csdA gene (the corresponding fusion protein was shown to be functional, see Materials and Methods). An extract from cells grown at 16°C was fractionated by sucrose gradient centrifugation and proteins from individual fractions were subjected to western blot analysis. The membrane was successively probed with anti-FLAG antibodies and with polyclonal antibodies against L20 and S3, which were used as markers for the 50S and 30S subunits, respectively. As shown in Figure 5, the FLAG-CsdA fusion protein was detected in fractions 11–21, with the strongest signal in fractions 15–17 encompassing the 50S subunit. However, the comparison with the signal corresponding to L20 indicates that fraction 15, corresponding to the lagging edge of the 50S peak, is comparatively strongly enriched in FLAG-CsdA. This distribution is consistent with CsdA co-purifying with a 50S precursor. Interestingly, the SrmB RNA helicase also sedimented predominantly at the same position of the gradient (16). However, SrmB was more restricted to the 50S region compared with CsdA, with no signal detected in the 30S or monosome fractions. We do not know whether the presence of CsdA in the 30S peak reflects an association with the 30S subunit, with an early 50S precursor, or with a non-ribosomal complex co-sedimenting with the 30S subunit. No FLAG signal was detected at the top or the bottom of the gradient (data not shown), indicating that there are no detectable free or polysome-bound CsdA pools. When the same experiment was repeated with cells grown at 37°C, no FLAG-CsdA was detected anywhere in the gradient (data not shown) consistent with the fact that CsdA is over-expressed at low temperature (17).
Figure 5.
CsdA associates with ribosomes in wt cells at low temperature. (Top) Sedimentation profile showing the free subunits and 70S ribosomes from wt cells [BL21(DE3)-FLAG-csdA strain] grown at 16°C. (Bottom) Equal volumes of each fraction were subjected to western blotting using anti-FLAG, anti-L20 and anti-S3 antibodies.
During the mass spectrometry analysis of the 40S particle of the ΔsrmB strain (16), we noticed that CsdA was present in the 40S fractions (unpublished results). To confirm this result, we analysed the localization of FLAG-CsdA on a ΔsrmB ribosomal profile. A strain carrying both a chromosomal FLAG-csdA gene and the srmB deletion was constructed (see Materials and Methods), and an extract of the corresponding cells grown at 20°C was fractionated on a sucrose gradient. FLAG-CsdA was again localized by western blotting, with L1 and S3 being used as markers for the 50S and 30S subunits, respectively (L1 is also present in the ΔsrmB 40S particles; 16). As expected, a prominent 40S particle was detected in this strain (Fig. 6). Interestingly, the FLAG-CsdA protein now peaks in fractions 9 and 10 encompassing this particle, with the signals corresponding to the 50S fractions being comparatively fainter. Thus, in the ΔsrmB strain, CsdA co-purifies with the incompletely assembled, 50S-related 40S particle, much as it does with a near-50S precursor in the wt strain (Fig. 5). The fact that the localization of CsdA on the gradient varies with the presence/abundance of 50S precursors or 50S-related particles, suggests that CsdA interacts with these species, rather than merely sedimenting in the same region of the gradient. Such an interaction, in turn, supports the idea that CsdA plays a direct role in 50S biogenesis.
Figure 6.
CsdA associates with the 40S ribosomal particle from the ΔsrmB extract. (Top) Sedimentation profile showing the free subunits from ΔsrmB cells carrying a FLAG-tagged csdA gene (WJW45ΔsrmB FLAG-csdA). Cells were grown at 20°C. (Bottom) Equal volumes of each fraction were subjected to western blotting using anti-FLAG, anti-L1 and anti-S3 antibodies. Total proteins isolated from WJW45ΔsrmB (T1) and WJW45-FLAG-csdA (T2) strains were used as negative and positive controls for FLAG detection, respectively.
Over-expression of CsdA corrects the ribosome defect in ΔsrmB strains
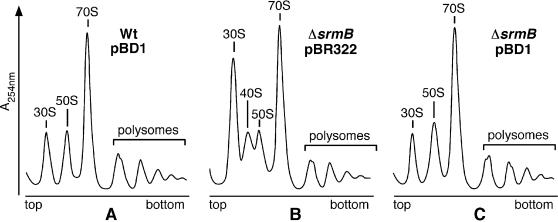
The association of CsdA with the ΔsrmB 40S particle (Fig. 6) led us to test whether CsdA over-expression might correct the ribosome defect in the ΔsrmB strain. To test this idea, we analysed the polysome profile of the ΔsrmB strain transformed with the csdA-bearing plasmid pBD1. Cells were grown at 20°C, and the wt strain transformed with pBD1 and the ΔsrmB strain transformed with pBR322 were used as controls. In the presence of pBD1, the ΔsrmB ribosome defects—excess of 30S over 50S subunit and accumulation of the 40S particle—were corrected (compare Fig. 7B and C), so that the profiles of ΔsrmB and wt strains now became similar (Fig. 7A and C). In contrast, the slow-growth phenotype of the ΔsrmB strain, whether on plates or in liquid medium, was not corrected by pBD1. This result may be due to the fact that CsdA over-expression is toxic on its own. Indeed, the growth of wt cells was slowed by the presence of pBD1, the more so as the temperature was decreased (data not shown).
Figure 7.
Over-expression of csdA suppresses the ΔsrmB ribosome defect. wt and ΔsrmB strains transformed with pBD1 (pBR322-csdA) or pBR322 plasmids were grown in tetracycline-LB medium at 20°C. Polysome profiles show the free ribosomal subunits, 70S ribosomes, polysomes and 40S particle.
Whereas the above result suggests that CsdA can replace SrmB in 50S biogenesis when over-expressed, it gives no clue as to whether these two proteins play similar roles when expressed at their physiological level. To address this question, we asked whether the ΔsrmB and ΔcsdA phenotypes were additive. The growth defect is stronger for the ΔcsdA than for ΔsrmB mutant (see Discussion), and we observed that it is not exacerbated further by combining the two deletions, regardless of the temperature; moreover, the defect of the ribosome profile was not exacerbated either (data not shown). This result fits the view that, when expressed at their normal levels, CsdA and SrmB are involved at different stages of the 50S subunit biogenesis, rather than playing redundant roles.
DISCUSSION
A role for CsdA in 50S biogenesis at low temperature
We have observed that the deletion of csdA results in a deficiency in free 50S subunits at low temperature (20°C) and in the accumulation of a 50S-related particle sedimenting around 40S (Fig. 2). Sucrose gradient fractionation indicates that in wt cells, CsdA co-sediments with a pre-50S particle (Fig. 5). Moreover, in another mutant (ΔsrmB) that also accumulates a 50S-related particle of about 40S, CsdA co-sediments with this particle (Fig. 6). Together, our results show that CsdA participates in the biogenesis of the 50S ribosomal subunit at low temperature, presumably by binding to and altering the RNA structure of a 50S precursor.
The 40S particle that accumulates in the ΔcsdA strain contains a 23S rRNA precursor with 7 extra-nucleotides at its 5′ end (Fig. 3). Moreover, although it contains all r-proteins of the large subunit, several of them are present in reduced or very low amounts (Fig. 4), suggesting that in the absence of CsdA these proteins bind loosely or slowly to the particle. In vivo the assembly of the large ribosomal subunit proceeds via three intermediates sedimenting at 34S, 43S and near-50S (8,30). These species contain precursors of 23S rRNA and a subset of r-proteins, except for the near-50S particle that contains all of them. It seems that the 40S particle differs from the genuine 43S intermediate, since protein L2, which is absent from 43S (31) is present in normal quantities in 40S particles. Thus, the 40S particle probably corresponds to a mis-assembled 50S subunit rather than to a genuine precursor.
The particular composition of the ΔcsdA 40S particle does not necessarily mean that CsdA is implicated in both assembly of r-proteins and 23S rRNA processing. On its own, a defect in assembly is likely to hamper processing, because processing is only completed in polysomes (7,28). The reverse, however, is not true: even the gross processing defects observed in RNase III– mutants do not hinder assembly and formation of functional 50S subunits [see (16) for discussion]. A plausible scenario, then, is that the defect in assembly observed with the 40S particle is a direct consequence of the absence of CsdA, whereas the processing defect stems secondarily from the assembly defect.
Whatever its precise role in 50S assembly, CsdA is only required below 30°C. This feature is consistent with an RNA chaperone or helicase activity, since the rRNA rearrangements that accompany ribosome biogenesis are probably more dependent upon such activities at low temperature, where RNA structures are more stable.
CsdA and SrmB play different roles in 50S biogenesis
The phenotype associated with the csdA deletion resembles that described previously for the deletion of srmB, which encodes another DEAD-box protein involved in the biogenesis of the 50S subunit (16). Beside cold-sensitive growth, both deletion strains show a deficit in 50S subunits and the appearance of a 40S particle. Although both 40S particles contain the same 23S precursor, their protein compositions differ. Most remarkably, L13, one of the five earliest proteins incorporated during ribosome assembly in vitro (8) is absent from the ΔsrmB particle. In contrast, most of the proteins that are present in reduced amount in the ΔcsdA particle are incorporated late during ribosomal assembly in vitro (8). It is therefore likely that SrmB affects 50S assembly at an earlier stage than CsdA. However, we cannot exclude a more complicated scenario, i.e. that in the absence of CsdA, some early proteins may not be assembled correctly leading to a conformational defect impeding the binding of late proteins. Incidentally, when extracts of wt cells are analysed on sucrose gradients, CsdA and SrmB co-migrate with a sedimentation coefficient close to 50S [Fig. 5 and (16)]. We hypothesize that SrmB and CsdA associate with different precursor intermediates but remain bound to the pre-50S until a late stage, explaining their co-sedimentation.
Consistent with the view that the two proteins play non-equivalent roles in 50S biogenesis, the phenotypes associated with the ΔsrmB and ΔcsdA deletions are not additive. The over-expression of CsdA corrects the ΔsrmB ribosome defect, however (Fig. 7). This result suggests that over-expression of CsdA relaxes its specificity so that it can now functionally replace SrmB. We have found that purified CsdA possesses ATPase and unwinding activities towards a variety of RNA substrates in vitro (32), consistent with a non-specific activity of CsdA when present in excess over its genuine partners (see below). As discussed below, the suppression of S2 mutants by over-expression of CsdA may be another example of this relaxed specificity.
Alternate roles for CsdA in E.coli
In addition to the biogenesis of the 50S subunit, CsdA has been reported to play a role in several other important physiological processes (see Introduction), but whether these effects reflect a genuine role of the protein is not always clear. The csdA gene was originally identified as a multi-copy suppressor of an rpsB mutation affecting the r-protein S2 (18). Recently, using another rpsB mutation [rpsB1ts (33)], Moll et al. (19) reported that over-expressing csdA restores the binding of r-proteins S2 and S1 to the 30S subunit at the non-permissive temperature. While these findings suggested that CsdA is involved in 30S subunit biogenesis, we have found that the composition of the 30S subunit, and in particular the amounts of proteins S1 and S2 (see Fig. 4C), was not affected by the deletion of the chromosomal csdA gene. These results indicate that CsdA plays no role in the biogenesis of the 30S subunit when expressed at a physiological level in a wt strain.
Previous experiments have suggested that, in addition to ribosome biogenesis, CsdA plays other genuine roles in the cell, particularly at low temperature. We (21) and others (22) have reported that CsdA can affect mRNA stability when over-expressed. Subsequently, CsdA was shown to interact in vitro with poly(A) polymerase I (34) and with the essential endonuclease RNase E (V.Khemici, I.Toesca, L.Poljak, N.F.Vanzo and A.J.Carpousis, submitted for publication). RNase E is known to associate with PNPase and the DEAD-box protein RhlB to form an ‘RNA degradosome’ (35,36). When expressed at its physiological level CsdA can substitute for RhlB in this complex (37; A.Prud’homme-Généreux, R.K.Beran,II, S.Ramey, G.A.Mackie and R.W.Simons, submitted for publication), consistent with a genuine role of the protein in mRNA degradation. Finally, although evidence is so far less extensive, it seems that CsdA also participates in translation initiation (20). That a single protein can participate in different biological process seems common amongst DExD/H-box proteins, particularly those of eucaryotic origin (38). In addition to their catalytic cores, these proteins possess unique N- or C-terminal extensions that are thought to interact with specific partners, either proteins (38) or RNAs (39). These interactions, in turn, could direct the RNA helicases to specific targets. According to this view, it is the association of CsdA with RNase E within a ‘cold-adapted degradosome’ that would target it to mRNA degradation. Likewise, its interaction with specific ribosomal components, r-proteins or rRNA regions, would direct it to 50S biogenesis. However, the nature of these components remains undefined.
Can the deficit in 50S subunits explain the growth defect of the ΔcsdA strain? Intriguingly, this growth defect is more severe than that of the ΔsrmB strain (the difference in growth rate is over 2-fold at 20°C), whereas the reverse is true for the alteration of ribosome profile. For ΔsrmB, 50S ribosome deficiency is already marked at 30°C whereas for ΔcsdA it only becomes visible at lower temperature (e.g. 20°C). It could thus be argued that the cold-sensitive phenotype of the ΔcsdA strain does not reflect the impaired 50S biogenesis but rather another defect, e.g. in mRNA degradation or translation initiation. This is not necessarily the case, however. By careful examination of the ribosome profiles, we noticed that the 50S subunits from the ΔcsdA cells sedimented slightly slower than their counterparts from either wt or ΔsrmB strains (see Fig. 2). Although tiny, this difference in sedimentation was reproducible. Since the ΔcsdA 50S subunit has a full r-protein complement, its slower sedimentation may reflect a slightly altered shape. Thus, in the ΔcsdA strain, even fully assembled 50S subunits may not be completely genuine, which may in part explain the observed growth defect. Additional work is needed to characterize these subunits further.
Concluding remarks
In the yeast Saccharomyces cerevisiae, as many as 15 DEAD-box helicases are involved in ribosome biogenesis, i.e. six in 40S subunit biogenesis and nine in 60S subunit biogenesis (14). In contrast, in E.coli, the involvement of DEAD-box proteins appears far more modest: only two of them, CsdA and SrmB, are known to be required for ribosome biogenesis, and this requirement holds at lower temperature only. This difference between E.coli and yeast may reflect the reduced complexity of ribosome biogenesis in the case of E.coli and/or the higher optimal growth temperature. Interestingly, E.coli possesses another DEAD-box protein, DbpA, which may also be involved in the biogenesis of the 50S subunit since its ATPase and helicase activities are stimulated by a specific region of the 23S rRNA (40,41). DbpA is evolutionarily conserved (39) but non-essential and the exact conditions under which it may be required for 50S biogenesis remain to be elucidated.
Acknowledgments
ACKNOWLEDGEMENTS
We are much indebted to Dr R. Brimacombe for the kind gift of antibodies against 50S ribosomal proteins. We thank G. Church for the pKO3 plasmid, P. Lesage and M. Springer for the pBD1 plasmid and I. Boni and C. Chiaruttini for the gift of S1 and L20 antibodies. We thank Drs C. Condon and G. A. Mackie for their critical reading of the manuscript, and Dr K. H. Nierhaus for his constant interest and support. J.C. is a recipient of MENRT and ARC fellowships. This work was supported by CNRS, ENS and by grants from ARC (No. 4633) and MENRT (programme PRFMMIP and Action Concertée ‘Dynamique et Réactivité des Assemblages Biologiques’) to M.D..
REFERENCES
- 1.Linder P., Lasko,P.F., Ashburner,M., Leroy,P., Nielsen,P.J., Nishi,K., Schnier,J. and Slonimski,P.P. (1989) Birth of the DEAD box. Nature, 337, 121–122. [DOI] [PubMed] [Google Scholar]
- 2.Tanner N.K. and Linder,P. (2001) DExD/H box RNA helicases: from generic motors to specific dissociation functions. Mol. Cell., 8, 251–262. [DOI] [PubMed] [Google Scholar]
- 3.de la Cruz J., Kressler,D. and Linder,P. (1999) Unwinding RNA in Saccharomyces cerevisiae: DEAD-box proteins and related families. Trends Biochem. Sci., 24, 192–198. [DOI] [PubMed] [Google Scholar]
- 4.Lorsch J.R. (2002) RNA chaperones exist and DEAD box proteins get a life. Cell, 109, 797–800. [DOI] [PubMed] [Google Scholar]
- 5.Linder P., Tanner,N.K. and Banroques,J. (2001) From RNA helicases to RNPases. Trends Biochem. Sci., 26, 339–341. [DOI] [PubMed] [Google Scholar]
- 6.Schwer B. (2001) A new twist on RNA helicases: DExH/D box proteins as RNPases. Nature Struct. Biol., 8, 113–116. [DOI] [PubMed] [Google Scholar]
- 7.Srivastava A.K. and Schlessinger,D. (1990) Mechanism and regulation of bacterial ribosomal RNA processing. Annu. Rev. Microbiol., 44, 105–129. [DOI] [PubMed] [Google Scholar]
- 8.Nierhaus K.H. (1991) The assembly of prokaryotic ribosomes. Biochimie, 73, 739–755. [DOI] [PubMed] [Google Scholar]
- 9.Williamson J.R. (2003) After the ribosome structures: how are the subunits assembled? RNA, 9, 165–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.El Hage A., Sbai,M. and Alix,J.H. (2001) The chaperonin GroEL and other heat-shock proteins, besides DnaK, participate in ribosome biogenesis in Escherichia coli. Mol. Gen. Genet., 264, 796–808. [DOI] [PubMed] [Google Scholar]
- 11.Maki J.A., Schnobrich,D.J. and Culver,G.M. (2002) The DnaK chaperone system facilitates 30S ribosomal subunit assembly. Mol. Cell., 10, 129–138. [DOI] [PubMed] [Google Scholar]
- 12.Alix J.H. and Nierhaus,K.H. (2003) DnaK-facilitated ribosome assembly in Escherichia coli revisited. RNA, 9, 787–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maki J.A., Southworth,D.R. and Culver,G.M. (2003) Demonstration of the role of the DnaK chaperone system in assembly of 30S ribosomal subunits using a purified in vitro system. RNA, 9, 1418–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kressler D., Linder,P. and de la Cruz,J. (1999) Protein trans-acting factors involved in ribosome biogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol., 19, 7897–7912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kalman M., Murphy,H. and Cashel,M. (1991) rhlB, a new Escherichia coli K-12 gene with an RNA helicase-like protein sequence motif, one of at least five such possible genes in a prokaryote. New Biologist, 3, 886–895. [PubMed] [Google Scholar]
- 16.Charollais J., Pflieger,D., Vinh,J., Dreyfus,M. and Iost,I. (2003) The DEAD-box RNA helicase SrmB is involved in the assembly of 50S ribosomal subunits in Escherichia coli. Mol. Microbiol., 48, 1253–1265. [DOI] [PubMed] [Google Scholar]
- 17.Jones P.G., Mitta,M., Kim,Y., Jiang,W. and Inouye,M. (1996) Cold shock induces a major ribosomal-associated protein that unwinds double-stranded RNA in Escherichia coli. Proc. Natl Acad. Sci. USA, 93, 76–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Toone W.M., Rudd,K.E. and Friesen,J.D. (1991) deaD, a new Escherichia coli gene encoding a presumed ATP-dependent RNA helicase, can suppress a mutation in rpsB, the gene encoding ribosomal protein S2. J. Bact., 173, 3291–3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moll I., Grill,S., Gründling,A. and Bläsi,U. (2002) Effects of ribosomal proteins S1, S2 and the DeaD/CsdA DEAD-box helicase on translation of leaderless and canonical mRNAs in Escherichia coli. Mol. Microbiol., 44, 1387–1396. [DOI] [PubMed] [Google Scholar]
- 20.Lu J., Aoki,H. and Ganoza,M.C. (1999) Molecular characterization of a prokaryotic translation factor homologous to the eukaryotic initiation factor eIF4A. Int. J. Biochem. Cell Biol., 31, 215–229. [DOI] [PubMed] [Google Scholar]
- 21.Iost I. and Dreyfus,M. (1994) mRNAs can be stabilized by DEAD-box proteins. Nature, 372, 193–196. [DOI] [PubMed] [Google Scholar]
- 22.Brandi A., Spurio,R., Gualerzi,C.O. and Pon,C.L. (1999) Massive presence of the Escherichia coli ‘major cold-shock protein’ CspA under non-stress conditions. EMBO J., 18, 1653–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamanaka K. and Inouye,M. (2001) Selective mRNA degradation by polynucleotide phosphorylase in cold shock adaptation in Escherichia coli. J. Bact., 183, 2808–2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Britton R.A., Powell,B.S., Dasgupta,S., Sun,Q., Margolin,W., Lupski,J.R. and Court,D.L. (1998) Cell cycle arrest in Era GTPase mutants: a potential growth rate-regulated checkpoint in Escherichia coli. Mol. Microbiol., 27, 739–750. [DOI] [PubMed] [Google Scholar]
- 25.Link A.J., Phillips,D. and Church,G.M. (1997) Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: application to open reading frame characterization. J. Bact., 179, 6228–6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sirdeshmukh R. and Schlessinger,D. (1985) Ordered processing of Escherichia coli 23S rRNA in vitro. Nucleic Acids Res., 13, 5041–5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Allas U., Liiv,A. and Remme,J. (2003) Functional interaction between RNase III and the Escherichia coli ribosome. BMC Mol. Biol., 4, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Srivastava A.K. and Schlessinger,D. (1988) Coregulation of processing and translation: mature 5′ termini of Escherichia coli 23S ribosomal RNA form in polysomes. Proc. Natl Acad. Sci. USA, 85, 7144–7148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Herold M. and Nierhaus,K.H. (1987) Incorporation of six additional proteins to complete the assembly map of the 50 S subunit from Escherichia coli ribosomes. J. Biol. Chem., 262, 8826–8833. [PubMed] [Google Scholar]
- 30.Lindahl L. (1975) Intermediates and time kinetics of the in vivo assembly of Escherichia coli ribosomes. J. Mol. Biol., 92, 15–37. [DOI] [PubMed] [Google Scholar]
- 31.Nierhaus K.H., Bordasch,K. and Homann,H.E. (1973) Ribosomal proteins. XLIII. In vivo assembly of Escherichia coli ribosomal proteins. J. Mol. Biol., 74, 587–597. [DOI] [PubMed] [Google Scholar]
- 32.Bizebard T., Ferlenghi,I., Iost,I. and Dreyfus,M. (2004) Studies on three E. coli DEAD-box helicases point to an unwinding mechanism different from that of model DNA helicases. Biochemistry, in press. [DOI] [PubMed] [Google Scholar]
- 33.Bollen A., Lathe,R., Herzog,A., Denicourt,D., Lecocq,J.P., Desmarez,L. and Lavallé,R. (1979) A conditionally lethal mutation of Escherichia coli affecting the gene coding for ribosomal protein S2 (rpsB). J. Mol. Biol., 132, 219–233. [DOI] [PubMed] [Google Scholar]
- 34.Raynal L.C. and Carpousis,A.J. (1999) Poly(A) polymerase I of Escherichia coli: characterization of the catalytic domain, an RNA binding site and regions for the interaction with proteins involved in mRNA degradation. Mol. Microbiol., 32, 765–775. [DOI] [PubMed] [Google Scholar]
- 35.Py B., Higgins,C.F., Krisch,H.M. and Carpousis,A.J. (1996) A DEAD-box RNA helicase in the Escherichia coli RNA degradosome. Nature, 381, 169–172. [DOI] [PubMed] [Google Scholar]
- 36.Miczak A., Kaberdin,V.R., Wei,C.L. and Lin-Chao,S. (1996) Proteins associated with RNase E in a multicomponent ribonucleolytic complex. Proc. Natl Acad. Sci. USA, 93, 3865–3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beran R.K. and Simons,R.W. (2001) Cold-temperature induction of Escherichia coli polynucleotide phosphorylase occurs by reversal of its autoregulation. Mol. Microbiol., 39, 112–125. [DOI] [PubMed] [Google Scholar]
- 38.Silverman E., Edwalds-Gilbert,G. and Lin,R.J. (2003) DExD/H-box proteins and their partners: helping RNA helicases unwind. Gene, 312, 1–16. [DOI] [PubMed] [Google Scholar]
- 39.Kossen K., Karginov,F.V. and Uhlenbeck,O.C. (2002) The carboxy-terminal domain of the DExDH protein YxiN is sufficient to confer specificity for 23S rRNA. J. Mol. Biol., 324, 625–636. [DOI] [PubMed] [Google Scholar]
- 40.Nicol S.M. and Fuller-Pace,F.V. (1995) The ‘DEAD box’ protein DbpA interacts specifically with the peptidyltransferase center in 23S rRNA. Proc. Natl Acad. Sci. USA, 92, 11681–11685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Diges C.M. and Uhlenbeck,O.C. (2001) Escherichia coli DbpA is an RNA helicase that requires hairpin 92 of 23S rRNA. EMBO J., 20, 5503–5512. [DOI] [PMC free article] [PubMed] [Google Scholar]