Abstract
Somatic mosaicism of repeat length is prominent in repeat expansion disorders such as Huntington disease and myotonic dystrophy. Somatic mosaicism is age-dependent, tissue-specific and expansion-biased, and likely contributes toward the tissue-specificity and progressive nature of the symptoms. We propose that therapies targeted at somatic repeat expansion may have general utility in these disorders. Specifically, suppression of somatic expansion would be expected to be therapeutic, whilst reversion of the expanded mutant repeat to within the normal range would be predicted to be curative. However, the effects of genotoxic agents on the mutational properties of specific nuclear genes are notoriously difficult to define. Nonetheless, we have determined that chronic exposure over a three month period to a number of genotoxic agents can alter the rate of triplet repeat expansion in whole populations of mammalian cells. Interestingly, high doses of caffeine increased the rate of expansion by ∼60%. More importantly, cytosine arabinoside, ethidium bromide, 5-azacytidine and aspirin all significantly reduced the rate of expansion by from 35 to 75%. These data establish that drug induced suppression of somatic expansion is possible. These data also suggest that highly unstable expanded simple sequence repeats may act as sensitive reporters of genotoxic assault in the soma.
INTRODUCTION
A growing number of inherited human disorders, including Huntington disease (HD), myotonic dystrophy (DM), fragile X syndrome, Friedreich ataxia and several spinocerebellar ataxias have been associated with the expansion of simple sequence DNA repeats (1,2). The number of repeats in the array of the associated loci is polymorphic in the general population, but usually less than about 30 repeats. Disease-associated alleles have expanded beyond this range, typically 40 to 60 repeats, although into the hundreds or thousands of repeats for some disorders. In each case, longer arrays are typically associated with a more severe phenotype and an earlier age of onset of symptoms. Disease-associated alleles are genetically highly unstable and frequently change length upon transmission from one generation to the next with mutation rates approaching 100% (3). Such germline mutations are not random, but are highly biased toward expansion at most loci, thus accounting for the genetic anticipation observed in many of these disorders (1).
In addition to germline instability, somatic mosaicism of disease length alleles is often prominent and tends to be age-dependent, expansion-biased and highly tissue-specific (see for example 1,4–8). In particular, DM1 patients always have much larger expansions in their muscle cells, the primary affected tissue, than in circulating leukocytes (4,9–11). Similarly, strikingly large expansions have recently been observed to accumulate specifically in the commonly affected brain regions of HD patients (12). It is likely that somatic mosaicism contributes, at least in part, to the tissue-specificity and progressive nature of many of the disorders associated with unstable DNA expansions. Indeed, consistent with the late age of onset in most of these disorders, we believe somatic expansion may actually be required to precipitate late onset symptoms in many affected individuals who inherit relatively small expansions.
Despite the similarity in their underlying genetic aetiology, pathology in these disorders is mediated via a variety of routes. These include: gain of function effects of polyglutamine tracts (13) and expanded CUG/CCUG RNA molecules (2,14); loss of function of associated genes through either RNA mis-localisation (15), transcriptional interference of ‘sticky’ DNA (16), and chromatin structure (17) and DNA methylation-associated loss of promoter activity (18); and other as yet uncharacterised pathogenic mechanisms (19,20). Thus, therapies targeted at the downstream effects of the expansion will most likely be disease-specific. We propose that therapies directed at modifying repeat dynamics in the soma may have general utility in these disorders. Specifically, in disorders where expansion-biased somatic mosaicism is prominent, strategies that result in suppression of somatic repeat expansion would be expected to be of therapeutic benefit. Whilst for all the disorders, strategies that result in reversion of the expanded mutant repeat in somatic tissues to within the length range observed in the general population would be predicted to be curative (at least if applied prior to cell loss and/or fixation of developmental abnormalities). In support of this idea, disease progression in a mouse model of HD has been slowed genetically by introduction of an Msh2 null allele that suppresses somatic repeat expansion (21).
The molecular mechanisms that give rise to somatic mosaicism in patients are not yet fully understood. Age-dependent, expansion-biased, tissue-specific somatic mosaicism has been fully replicated in transgenic mouse (5,22–24) and cell culture models of unstable DNA (25). Data indicate that somatic mosaicism accumulates through multiple small mutations (4) that require components of the mismatch repair machinery (21,26–28) and may be independent of cell division (5,23–25).
Although numerous agents are known to affect genomic integrity and modify DNA mutation rates, it has nevertheless proven particularly difficult to detect environmentally induced mutations at specific unique sequence nuclear loci in mammalian cells. Indeed, even after exposure to a highly genotoxic agent, the likelihood of finding an induced mutation at a specific unique sequence nuclear locus is very low. Consequently, detection of rare mutants requires either a selection system and/or the necessity to screen hundreds or thousands of cells/individuals. For instance, even using doses of mutagens so high that >75% of cells are killed, induced mutations in the HPRT/hprt gene are detected in <1 in 104 cells (29,30). Similarly low induced germline mutation rates (∼1 × 10–3 per locus) were reported in male mice treated with multiple doses of ENU (N-ethyl-N-nitrosourea), the most potent germline mutagen described (31). These very low mutation rates reflect the general stability of DNA sequences in mammalian cells and the very active DNA repair systems that operate to maintain genomic integrity. However, if an environmentally induced effect on somatic DNA mutational dynamics is to be of utility in treating the human disorders associated with repeat expansions, it will most likely be required to influence repeat length dynamics in most, if not all, cells. Nonetheless, it is possible that expanded simple sequence repeats that have high basal mutation rates in normal cells, may be more sensitive to the effects of genotoxic agents. Support for such an idea is provided by the detection of radiation and chemical-induced increases in germline length change mutation frequencies of hypermutable minisatellite sequences (32–34). In this study we have sought to test the hypothesis that expanded trinucleotide repeat dynamics in somatic cells can be modified by environmental effects. To this end, we have examined the effects on somatic expansion of chronic exposure to a number of genotoxic agents over a 3-month period in a mammalian cell model that replicates the unstable DNA phenotype.
MATERIALS AND METHODS
Dmt-D mouse kidney cell cultures and drug treatment
The D2763Kc2 cell line is a clonal transgenic mouse cell line [clone 2 from Gomes-Pereira et al. (25)]. It was cloned by limiting dilution from the D2763K kidney cell line (25). The D2763K line was a spontaneously transformed kidney cell line derived from a 6-month old Dmt-D transgenic mouse (24,35). Cells were maintained and passaged as previously described (25). For drug treatment experiments the progenitor culture was split into multiple aliquots: six replicate no-drug controls and six replicate cultures for each test-compound. All cultures were maintained in parallel throughout the course of the experiment. Control cultures were supplied with fresh medium every 2–3 days and cells were passaged when confluent at a 1:40 dilution approximately weekly. For treated cultures, the drugs were supplied to the cultures dissolved in the medium at the concentrations indicated (Table 1). Treated cultures were supplied with fresh drug-supplemented medium every 2–3 days and the cells were passaged the same as for the no-drug controls. Control and treated cultures were maintained for the time periods indicated (Table 1).
Table 1. Chemical treatment and expanded CAG·CTG repeat dynamics.
| Conc. of drug in cell culture | Time [days] | PDa | Increase in PDTb [%] | Comparisons relative to time controlc | Comparisons relative to PD controlc | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Control | Significance, P | Median rate of expansion [repeats per day] (relative to control [%]) | Control | Significance, P | Median rate of expansion [repeats per PD] (relative to control [%]) | |||||
| Control: C70/82 | – | 82 | 70 | – | – | – | 0.759 | – | – | 0.889 |
| Control: C82/97 | – | 97 | 82 | – | – | – | 0.887 | – | – | 1.050 |
| Control: C106/122 | – | 122 | 106 | – | – | – | 0.615 | – | – | 0.708 |
| Novobiocin | 60 µM | 99 | 80 | 5 | C82/97 | 0.3785 | 0.792 (89) | C82/97 | 0.5752 | 0.980 (93) |
| Caffeine | 2 mM | 130 | 80 | 41 | C106/122 | 0.0202 | 1.005 (163) | C82/97 | 0.0453 | 1.634 (156) |
| Ethidium bromide | 250 nM | 128 | 66 | 69 | C106/122 | 0.0051 | 0.310 (50) | C70/82 | 0.0202 | 0.602 (68) |
| H2O2 | 100 µM | 126 | 80 | 37 | C106/122 | 0.0202 | 0.180 (29) | C82/97 | 0.0131 | 0.283 (27) |
| Rhodamine 6G | 50 nM | 116 | 80 | 26 | C106/122 | 0.0051 | 0.298 (48) | C82/97 | 0.0082 | 0.432 (41) |
| Aspirin | 5.6 µM | 99 | 80 | 5 | C82/97 | 0.0453 | 0.565 (64) | C82/97 | 0.0453 | 0.699 (67) |
| AraC | 500 nM | 95 | 71 | 13 | C82/97 | 0.0082 | 0.219 (25) | C70/82 | 0.0082 | 0.293 (33) |
| 5-azacytidine | 10 µM | 99 | 80 | 5 | C82/97 | 0.0202 | 0.475 (54) | C82/97 | 0.0386 | 0.587 (56) |
aPD, population doublings.
bPDT, population doubling time.
cSee Materials and Methods for details of the calculations and statistical comparisons.
Assessment of trinucleotide repeat size variability and statistical analysis
At the end of the experimental period DNA samples were extracted from the cultured treated cells using standard procedures. DNA was also extracted from the control cultures at each passage. The degree of repeat length variation that had accumulated in each sample was then assessed using sensitive small pool PCR (SP-PCR) analysis, performed as previously described (25). Single molecule SP-PCR analysis of 20–80 molecules per culture was used to determine the median repeat length in individual cultures. The median rates of expansion for each individual culture were calculated relative to the measured median repeat length in the progenitor culture at time zero. Median expansion rates for each culture were calculated in terms of both rate of change per day, and rate of change per population doubling (Supplementary Material, Table 2). To determine if a given treatment had an effect on mutational dynamics, we ranked the 12 median rates of expansion from the six control cultures and the six treated cultures. Under the null hypothesis that a drug treatment would have no effect on median expansion rates, the measured median expansion rates in the six treated and six control cell cultures should be randomly distributed within the rank order. However, if a drug treatment resulted in an increased rate of expansion, then we would predict that the measured expansion rates of the treated cells would, on average, be higher in the rank order than the non-treated control cells. Conversely, if a drug treatment resulted in a decreased rate of expansion, then we would predict that the measured expansion rates of the treated cells would, on average, be lower in the rank order than the non-treated control cells. The statistical significance of deviations from the expected random distribution of measured expansion rates within the rank order in treated and control cells was determined using the two-tailed Mann-Whitney U test. This conservative non-parametric test compares two independent groups of sampled data and uses the ranks of the medians rather than their raw values to calculate the statistic. For each treatment we performed a two tailed Mann-Whitney U test comparing the measured median expansion rates in terms of both repeats gained per day, and separately, in terms of repeats gained per population doubling. For each comparison, we used the data from the replicate control cultures at the time point closest to the treated culture, both in terms of days in culture, and separately, in terms of population doubling numbers. Statistical analyses were performed using Microsoft Excel and MINITAB (v10.51, Minitab Inc.). For each treatment we also calculated a single median rate of expansion from the medians of the replicates and compared this to the closest control to derive a relative expansion rate for a given treatment.
RESULTS
Previously, we generated transgenic mice containing a large expanded CAG·CTG array (∼160 repeats) randomly integrated into the mouse genome (24,35) in which age-dependent, tissue-specific, expansion-biased somatic repeat instability is reproduced (24,35). Particularly frequent and large expansions were observed in the kidney. We then used these animals to create cell lines in which the in vivo expansion-biased, tissue-specific mutational dynamics were also faithfully reproduced (25). Of particular utility, a subclone (D2763Kc2) of one of the kidney cell lines we created was shown to display very high levels of instability. Repeat length change mutations in the D2763Kc2 cell line are highly biased toward expansion, with the CAG·CTG repeat tract gaining on average ∼0.8 repeats per day when maintained in culture under standard conditions. The D2763Kc2 cell line therefore represented an ideal resource for testing whether environmental agents could modify these dynamics.
Before we commenced the study a number of experimental design points were considered, specifically, the time and level of exposure to the compound, the number and nature of the controls, and the nature of the reagents to test. In experimental terms, an expansion rate of 0.8 repeats per day requires a considerable period of time to result in clearly measurable shifts in the average repeat length within a population of cells. Therefore, rather than treat cells with a single acute dose of a genotoxic agent, we reasoned that chronic exposure to a genotoxic agent would more likely induce measurable changes in the rate of expansion. Clearly, the choice of a time period of exposure during which untreated control cells accumulated repeats at a measurable rate would allow the identification of reagents that either increased or decreased the rate of expansion. The precise dose of reagents used was also carefully considered. In an attempt to avoid cell selection effects that might be facilitated by overt toxicity, we determined concentrations of drugs that did not have detectable effects on cell viability after a 12-h exposure. Nonetheless, many of the treatments did result in detectable shifts in the population doubling time (PDT) (Table 1 and Supplementary Material, Table 3). Although previous studies have indicated that cell proliferation rates are not the most critical factor underlying differences in expansion rates between cell lines (25), we nevertheless compared treated cells with controls grown for the same number of population doublings, in addition to control cells cultured for the same period of time. Although preliminary studies strongly suggested some agents could modulate repeat dynamics, we also noted some stochastic differences within and between control and treated cultures. Such stochastic variations in cell line mutational dynamics have been noted before (25,36). Therefore, to ensure that any differences observed were mediated by the test chemical and were not due to stochastic variation between cultures or sampling error, we monitored repeat dynamics in multiple parallel control and treated cultures.
Thus, we chose to treat D2763Kc2 cells by chronic exposure to a range of genotoxic chemicals for up to 80 population doublings or 130 days. A single progenitor culture was split to generate all subsequent control and treated cell cultures at time point zero. For each treatment six cultures were established and maintained in parallel with six untreated controls. We tested a variety of genotoxic reagents that have effects on DNA repair and/or cell cycle control: caffeine, which inhibits ATM kinase activity and thus uncouples DNA repair and replication from the progression of the cell cycle (37); ethidium bromide, a DNA intercalating agent with known mutagenic properties (38); hydrogen peroxide, a free radical producer that can damage DNA directly and has been shown to increase microsatellite mutation rates (39); rhodamine 6G, which inhibits oxidative phosphorylation and also leads to an increase in the levels of oxidative stress (40); aspirin, a non-steroidal anti-inflammatory that reduces the proliferative capacity of cells resulting in an accumulation of cells at the G0/G1 boundary, as well as suppressing the mutator phenotype associated with hereditary non-polyposis colon cancer (HNPCC) (41); novobiocin, a potent inhibitor of DNA topoisomerase II (42); and two deoxynucleotide analogues, 5-azacytidine and cytosine arabinoside (araC). 5-Azacytidine is a potent inhibitor of CpG methylation and results in global hypomethylation (43). AraC is incorporated into DNA and is a robust inhibitor of chain elongation by DNA polymerases α, δ and ε and thus suppresses DNA synthesis, particularly on the leading strand (44).
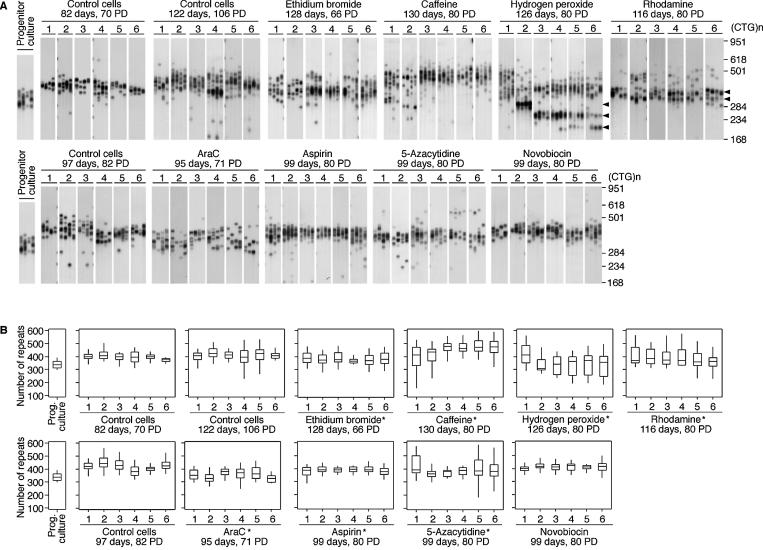
All of the treatments resulted in an increase in the PDT, up to ∼70% for ethidium bromide (Table 1). However, variation in PDT between parallel cultures with the same treatment was very low and the observed PDTs changed only very slightly through the course of the experiment (Table 1 and Supplementary Material, Table 3). Repeat length variation in the control cells grown in parallel with the treated cultures was determined at three time points to provide two controls for each treatment, i.e. one control grown for the same amount of time and one control grown for the same number of population doublings (Fig. 1). Median repeat lengths in each culture were measured and compared to the progenitor culture to calculate the median length change and rate of expansion (Table 1 and Supplementary Material, Table 2). As expected, the transgenic repeat length continued to expand rapidly in the control cells. The small differences in expansion rates observed between the control cells sampled at different times were not statistically significant. At the end of the treatment period we also quantified the degree of repeat length variation, and calculated the rate of expansion, in each of the individual treated cultures (Fig. 1, Table 1 and Supplementary Material, Table 2). As expected, we observed variation in the measured rate of expansion between independent controls (Supplementary Material, Table 4), and variation between control and treated cultures. In order to determine if the apparent differences between control and treated cultures were statistically significant, we performed a Mann-Whitney U test on the rank order of median rates of expansion for control and treated cultures (see Materials and Methods for a more detailed explanation of the statistical analyses). Although the ∼10% suppression observed in the novobiocin treated cells was not statistically significant, all the other treatments resulted in statistically significant differences (P < 0.05) in the rate of expansion when compared with both time and population doubling controls (Fig. 1B, Table 1). Interestingly, high doses of caffeine resulted in an ∼63% increase in the rate of expansion. In contrast, hydrogen peroxide, rhodamine 6G, ethidium bromide, aspirin, 5-azacytidine and araC treatments all resulted in highly significant decreases in the rate of expansion, ranging from ∼25 to 75% (Table 1). Interestingly, the frequency of cells containing large deletions was dramatically increased with hydrogen peroxide. However, some of the hydrogen peroxide and rhodamine treated cell cultures showed evidence of reduced variability in the length of the CAG·CTG repeat tract (Fig. 1A), suggesting that some of this effect may have been mediated by clonal growth of cells selected for enhanced viability under conditions of oxidative stress. No evidence for clonal expansion was observed in the treatments with ethidium bromide, araC, aspirin or 5-azacytidine, confirming that some agents affecting DNA metabolism can indeed suppress the rate of expansion of an unstable CAG·CTG repeat sequence by a mechanism that influences the mutational dynamics of the expanded repeat in the whole population of cells.
Figure 1.
Chemical treatment and expanded CAG·CTG repeat dynamics. (A) The autoradiographs shown are representative SP-PCR analyses of DNA samples extracted from replicate D2763Kc2 cells cultured for up to 130 days with the drugs indicated (see Materials and Methods and Table 1 for details) and the progenitor culture from which all cells were derived at day zero. Evidence for clonal growth in the hydrogen peroxide and rhodamine 6G treated cultures is indicated with arrows. (B) The box plots show the degree of repeat length variation observed in treated and control cells at the time points indicated. The top and bottom of the boxes correspond to the third (Q3) and first quartiles (Q1), respectively and the line across the box displays the median. The lines extending from the top and the bottom of the boxes, include values that fall inside the lower and upper limits: Q1-1.5(Q3-Q1) and Q3+1.5(Q3-Q1), respectively. Statistically significant differences (*, P < 0.05) in the median rates of expansion were observed for some treatments relative to both time and PD control cultures.
DISCUSSION
The main aim of these experiments was to determine if environmental factors could modify the somatic mutational dynamics of expanded CAG·CTG repeat sequences. Our findings reveal that chronic exposure to agents affecting DNA metabolism is capable of inducing statistically significant changes in the expansion rate of this simple sequence repeat in a mammalian cell culture model. We have identified one agent, caffeine, that increased the rate of expansion, and, uniquely, several including araC, ethidium bromide and 5-azacytidine that decreased the rate of expansion. Importantly, these effects were observed at a single nuclear locus in the absence of a selective system for detecting mutants. Indeed the extremely high basal mutation rates of this type of locus provide a new approach for detecting environmental modifiers. Usually, basal mutation rates are so low that the effects of environmental modifiers are detected as changes in the absolute mutation rate. For a classic unique sequence nuclear mutation target, such as the HPRT gene, even the most genotoxic agents induce mutation rates that are so low as to preclude direct detection and necessitate a selective system for recovering the very rare mutants. In contrast, the basal mutation rates of the expanded simple sequence repeats utilised in this study are so high that the mutation frequency in the control cells is essentially 100% per cell. In fact, most cells will have undergone multiple small mutations to yield the observed final length changes. Thus, the effects of environmental modifiers can be measured as differences in the absolute rate of repeat length change in the whole population of cells. This enables both enhancers and suppressors of the expansion rate to be identified. The measured differences directly reflect effects on the mutational dynamics of a single locus within individual cells, as opposed to measuring the mutation rate in a population of cells, where the vast majority of cells will have acquired no mutations at the locus of interest. In practical terms, this means that the status of the expanded repeat sequence will need to be assessed in a relatively small number of cells (20–80) to detect an effect, compared to the many thousands of cells that must be screened with a traditional unique sequence single copy nuclear marker. Thus, expanded simple sequence repeats with high basal mutation rates, such as those utilised in this study, are excellent reporters of genotoxic assault. These types of loci may enable the measurement of environmentally induced somatic mutations in cell cultures and/or individuals in a manner that is similar to the way that hypervariable minisatellites are currently being used to measure germline DNA damage (33). These data also highlight the possibility that variation in expansion rates observed in patients (45), and variation in disease severity not accounted for by allele length (46), might already be being modified by as yet unknown naturally occurring environmental modifiers.
Theoretically, treatment of a culture with a drug could lead to the preferential expansion by a selective process of a subset of cells with altered repeat length profiles from the average, thus compromising our ability to detect true drug induced changes in repeat metabolism. The most obvious route through which such an effect could be mediated is if a given cell were to acquire a mutation elsewhere in the genome that gave that cell a growth advantage under the culture conditions used independent of the repeat length that cell carried. Selective clonal expansion of that cell would then result in a loss of variability in repeat length from the cell population thus compromising measures of repeat length change. In the experiments described here we have obtained evidence for such an effect in the cultures treated with hydrogen peroxide and rhodamine 6G which both show sub-populations of cells with dramatically reduced variation. The other treatments do not have such clonal sub-populations and do not appear to have been compromised by such effects. An alternative scenario that could be imagined is that the expansion bias in repeat length observed over time within a culture is driven not by a bias in the mutation process, but rather the selective growth of cells with longer alleles. Indeed, support for such ‘mitotic drive’ has been previously provided in EBV transformed lymphoblastoid cell lines from DM patients in which cells carrying longer alleles do have a growth advantage over cells with shorter arrays (47). This effect is correlated with shorter population doubling times in cells with longer arrays and a dysregulation of the cell cycle control gene p21 (47). Under such a scenario, a drug could theoretically mediate an apparent effect on the rate of repeat expansion by modifying such a selective process, rather than affecting repeat dynamics directly. However, we consider this possibility unlikely and a number of lines of evidence suggest that this is not the case in the experiments we have described here. First, expansion biased somatic mutations accumulate in our transgenic model in vivo in post-mitotic tissues such as brain (35), indicating that the basal mechanism of mutation is indeed expansion biased, obviating the need to invoke a selective component to the process. Secondly, the transgene used in our model does not contain any coding DNA, promoter elements or a transcriptional start site that could result in the production of a transcript to mediate any selective effect (35). Although very low amounts of RNA transcripts containing the transgene can be detected by RT–PCR and Southern blot hybridisation, the predominant hnRNA transcripts derived from the locus contain the CAG strand rather than the CUG repeat strand. No polyadenylated cytoplasmic mRNA transcripts containing the transgene have been detected (Fortune and D.G.Monckton, unpublished observations). Moreover, we have been unable to detect any pathogenic effect in mice hemi- or homozygous for the transgene which segregates in the expected 1:1 ratio from hemizygous parents (35). Thirdly, the p21 gene through which the selective advantage of DM1 patient cell lines is mediated (47), is not expressed in the spontaneously immortalised cell lines we have used (Supplementary Material, Fig. 2). Fourthly, measurement of population doubling times of new subclones of clonal cell lines differing in repeat length by up to 200 repeats do not have detectable differences in their population doubling times (M.Gomes-Pereira and D.G.Monckton, unpublished observations). And, fifthly, selective sweeps previously observed in non-treated cells were bi-directional (i.e. cells carrying smaller alleles than the original major population sometimes overgrew the culture) (25) arguing against a bias in favour of cells carrying larger alleles. Thus, with the exception of the effects observed for hydrogen peroxide and rhodamine 6G, we are confident that the effects on repeat expansion rates that we have measured here reflect genuine drug induced differences in the mutational properties of the cells. Nonetheless, selection remains an important issue in this type of experiment and one that we might hope to eliminate completely in future studies by using non-cycling cell populations.
There are still major gaps in our understanding of the molecular mechanisms that operate to generate somatic expansion at CAG·CTG repeat loci and, whilst it was not the main aim of these experiments, the system we have developed offers the potential to investigate the molecular pathways mediating somatic expansions by interfering with specific aspects of DNA metabolism. Such studies are inevitably limited by the complex cellular response to genotoxic assault and the consequent difficulty in assigning an observed effect to a specific mechanism. Nonetheless, it is possible to make some tentative suggestions as to how certain drugs may mediate the effects we have observed. The caffeine induced ∼63% increase in the rate of expansion might be mediated by forced progression through the G2/M DNA-damage checkpoint and the accumulation of non-repaired DNA replication slippage errors during mitosis. Alternatively, it might be mediated via caffeine’s inhibitory effect on DNA replication and slowing of S-phase (48). However, araC also inhibits DNA replication and had the opposite effect on repeat dynamics. Most intriguingly, araC also inhibits DNA polymerase δ, the main polymerase involved in DNA mismatch repair (49). Given the requirement for certain mismatch repair proteins to mediate somatic expansion in mouse studies (21,26–28), it is possible that araC might suppress repeat expansion by inhibiting the DNA synthesis stage of mismatch repair. Ethidium bromide, which reduces the rate of repeat expansion, intercalates into nuclear DNA and can inhibit enzymes that bind to DNA, including topoisomerase II (50). Direct inhibition of topoisomerase II using novobiocin had a mild, statistically non-significant effect on the rate of expansion in this experiment, suggesting this is not the main route of action of ethidium bromide. It has previously been reported that the physical properties of expanded CGG·CCG repeats were specifically modified by the presence of ethidium bromide as evidenced by shifts in the gel mobility of repeat containing DNA fragments (51). Thus, it is possible that ethidium bromide exerts its effect on repeat length dynamics via a direct interaction with the unusual DNA structures (52) adopted by expanded repeats. However, ethidium bromide also affects mitochondrial metabolism and we determined that mRNA levels for the mitochondrial enzyme cytochrome oxidase II were reduced by ∼66% in our treated cells (Supplementary Material, Fig. 3), consistent with ethidium bromide-induced oxidative stress (53). Thus, three different treatments that increase levels of oxidative stress in cells, hydrogen peroxide, rhodamine 6G and ethidium bromide, surprisingly resulted in a reduction in the rate of expansion. This effect might be mediated by a suppression of mismatch repair activity by oxidative stress (54) and alternative processing of mutation intermediates by components of the base excision repair pathway which is altered in cells exposed to reactive oxygen species (55). The chromosomal context of a repeat tract is critical in mediating its stability (35) and there is a striking correlation between the GC content of the flanking DNA and the stability of the repeat (56). Such an effect might be mediated by the methylation status and/or local chromatin structure, both of which are disrupted by 5-azacytidine treatment (43). Aspirin’s effect on microsatellite instability in HNPCC cell lines is thought to be mediated by apoptotic induction (41). However, our experiments revealed no evidence for clonal selection in aspirin-treated cells. Aspirin also results in an increase in the proportion of cells in G0/G1 (57) and the effect we have observed here might be mediated by such an alteration in cell cycle dynamics.
Recently, two groups have reported similar experiments using human myotonic dystrophy cell lines (58,59). In one study an apparent increase in the rate of expansion was observed with chronic exposure to mitomycin C (59) and in the other study multiple acute applications of the replication inhibiting drugs emetine and aphidicolin also resulted in increases in the rate of expansion (58). Unfortunately however, in neither study was the effect of the increased amount of time that exposed cells were grown for accounted for. As discussed above, it is also possible that the mitotic drive effect previously reported in human DM1 cell lines (47) could also have compromised these analyses. Even more importantly, the mitomycin C exposure was not replicated. We and others have shown previously that repeat expansion in cell lines is not always uniformly synchronous and is liable instead to stochastic variation (25,36,47,58,60). Indeed, we have shown here that even identical duplicate cultures maintained in parallel do not expand synchronously (Fig. 1) and that such stochastic variation can result in repeat length distributions that are highly statistically significantly different from each other (Supplementary Material, Table 4). Thus, it is insufficient to compare a single control and experimental culture even if the measured repeat length distributions in the two cultures are highly statistically different from each other. Rather, as was performed in the replication inhibitor study (58) and we have performed here, it is instead necessary to show that any putative difference between control and treated cultures are systematically reproduced in multiple experiments and/or multiple replicates. Regardless of the potential limitations of these studies, they both only identified agents that appeared to lead to increased rates of expansion and none that lead to decreased rates of expansion.
It is our central hypothesis that tissue-specific, age-dependent, expansion-biased somatic mosaicism contributes toward the tissue-specificity and progressive nature of the symptoms in the repeat expansion disorders. Thus, management of repeat length in the soma presents as a potential therapeutic target and it is our long-term aim to develop treatments for these disorders using such an approach. Treatment of disease by chemical gene therapy will be achieved by modifying the nature of endogenous DNA sequences by the application of exogenous chemicals. En route to this goal we have shown that treatment of cell lines with exogenous chemical agents can modify expanded repeat dynamics. Importantly, we have demonstrated that agents that modify the dynamics of a single locus in the whole population of cells can be identified. Crucially, for the first time, we have identified agents that result in lower rates of expansion, an effect that if reproduced in vivo would be predicted to be therapeutically beneficial. In this regard, it is of note that the concentrations of reagents we have used are not dissimilar to those achieved in vivo in other unrelated studies (with the exception of caffeine which was used at a dose approximately 50 times typical in vivo doses—equivalent to a human ingesting approximately 200 cups of coffee a day) (Supplementary Material, Table 5). This important proof of principle study provides a rational basis for more extended screens. Promising agents identified in vitro, could be further assessed for their utility in vivo in the many transgenic mice that model the unstable DNA phenotype (5,6,24). Given the complex downstream pathology and dominant nature of most of these disorders, it is not yet clear how standard gene therapy approaches might be used for beneficial effect in these disorders. Limiting somatic expansions at the site of the primary mutation, or more ideally, decreasing the size of the mutation by chemical gene therapy is thus very attractive, particularly since it might be achievable with small molecule drugs for which delivery methods would be less problematic than those associated with current gene therapy protocols.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank Sanam Mustafa for technical assistance with preliminary studies. We are also grateful to Peggy F. Shelbourne, Marshall Stark and the Dynamic Mutation Group at the University of Glasgow and Christopher E. Pearson at the University of Toronto for helpful discussions during the course of this work. M.G.P. was funded for part of this work by a studentship from the Fundação para a Ciência e Tecnologia and Fundação Calouste Gulbenkian (Portugal). Awards to D.G.M. from the Lister Institute, Medical Research Council (UK), Hereditary Disease Foundation (USA), Association Française Contre Les Myopathies (France) and Wellcome Trust (UK) also supported this study.
REFERENCES
- 1.Richards R.I. (2001) Dynamic mutations: a decade of unstable expanded repeats in human genetic disease. Hum. Mol. Genet., 10, 2187–2194. [DOI] [PubMed] [Google Scholar]
- 2.Liquori C.L., Ricker,K., Moseley,M.L., Jacobsen,J.F., Kress,W., Naylor,S.L., Day,J.W. and Ranum,L.P. (2001) Myotonic dystrophy type 2 caused by a CCTG expansion in intron 1 of ZNF9. Science, 293, 864–867. [DOI] [PubMed] [Google Scholar]
- 3.Richards R.I. and Sutherland,G.R. (1992) Dynamic mutations: a new class of mutations causing human disease. Cell, 70, 709–713. [DOI] [PubMed] [Google Scholar]
- 4.Monckton D.G., Wong,L.-J.C., Ashizawa,T. and Caskey,C.T. (1995) Somatic mosaicism, germline expansions, germline reversions and intergenerational reductions in myotonic dystrophy males: small pool PCR analyses. Hum. Mol. Genet., 4, 1–8. [DOI] [PubMed] [Google Scholar]
- 5.Kennedy L. and Shelbourne,P.F. (2000) Dramatic mutation instability in HD mouse striatum: does polyglutamine load contribute to cell-specific vulnerability in Huntington’s disease? Hum. Mol. Genet., 9, 2539–2544. [DOI] [PubMed] [Google Scholar]
- 6.Sato T., Oyake,M., Nakamura,K., Nakao,K., Fukusima,Y., Onodera,O., Igarashi,S., Takano,H., Kikugawa,K., Ishida,Y. et al. (1999) Transgenic mice harboring a full-length human mutant DRPLA gene exhibit age-dependent intergenerational and somatic instabilities of CAG repeats comparable with those in DRPLA patients. Hum. Mol. Genet., 8, 99–106. [DOI] [PubMed] [Google Scholar]
- 7.Maciel P., Lopes-Cendes,I., Kish,S., Sequeiros,J. and Rouleau,G.A. (1997) Mosaicism of the CAG repeat in CNS tissue in relation to age at death in spinocerebellar ataxia type 1 and Machado-Joseph disease patients. Am. J. Hum. Genet., 60, 993–996. [PMC free article] [PubMed] [Google Scholar]
- 8.Monckton D.G., Cayuela,M.L., Gould,F.K., Brock,G.J.R., de Silva,R. and Ashizawa,T. (1999) Very large (CAG)n DNA repeat expansions in the sperm of two spinocerebellar ataxia type 7 males. Hum. Mol. Genet., 8, 2473–2478. [DOI] [PubMed] [Google Scholar]
- 9.Anvret M., Ahlberg,G., Grandell,U., Hedberg,B., Johnson,K. and Edstrom,L. (1993) Larger expansions of the CTG repeat in muscle compared to lymphocytes from patients with myotonic dystrophy. Hum. Mol. Genet., 2, 1397–1400. [DOI] [PubMed] [Google Scholar]
- 10.Ashizawa T., Dubel,J.R. and Harati,Y. (1993) Somatic instability of CTG repeat in myotonic dystrophy. Neurology, 43, 2674–2678. [DOI] [PubMed] [Google Scholar]
- 11.Thornton C.A., Johnson,K.J. and Moxley,R.T. (1994) Myotonic dystrophy patients have larger CTG expansions in skeletal muscle than in leukocytes. Ann. Neurol., 35, 104–107. [DOI] [PubMed] [Google Scholar]
- 12.Kennedy L., Evans,E., Chen,C.M., Craven,L., Detloff,P.J., Ennis,M. and Shelbourne,P.F. (2003) Dramatic tissue-specific mutation length increases are an early molecular event in Huntington disease pathogenesis. Hum. Mol. Genet., 12, 3359–3367. [DOI] [PubMed] [Google Scholar]
- 13.Gusella J.F. and MacDonald,M.E. (2000) Molecular genetics: unmasking polyglutamine triggers in neurodegenerative disease. Nature Rev. Neurosci., 1, 109–115. [DOI] [PubMed] [Google Scholar]
- 14.Mankodi A., Logigian,E., Callahan,L., McClain,C., White,R., Henderson,D., Krym,M. and Thornton,C.A. (2000) Myotonic dystrophy in transgenic mice expressing an expanded CUG repeat. Science, 289, 1769–1773. [DOI] [PubMed] [Google Scholar]
- 15.Davis B.M., McCurrach,M.E., Taneja,K.L., Singer,R.H. and Housman,D.E. (1997) Expansion of a CUG trinucleotide repeat in the 3′ untranslated region of myotonic dystrophy protein kinase transcripts results in nuclear retention of transcripts. Proc. Natl Acad. Sci. USA, 94, 7388–7393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sakamoto N., Chastain,P.D., Parniewski,P., Ohshima,K., Pandolfo,M., Griffith,J.D. and Wells,R.D. (1999) Sticky DNA: self-association properties of long GAA·TTC repeats in R·R·Y triplex structures from Friedreich’s ataxia. Mol. Cell, 3, 465–475. [DOI] [PubMed] [Google Scholar]
- 17.Klesert T.R., Otten,A.D., Bird,T.D. and Tapscott,S.J. (1997) Trinucleotide repeat expansion at the myotonic dystrophy locus reduces expression of DMAHP. Nature Genet., 16, 402–406. [DOI] [PubMed] [Google Scholar]
- 18.Jin P. and Warren,S.T. (2000) Understanding the molecular basis of fragile X syndrome. Hum. Mol. Genet., 9, 901–908. [DOI] [PubMed] [Google Scholar]
- 19.Koob M.D., Moseley,M.L., Schut,L.J., Benzow,K.A., Bird,T.D., Day,J.W. and Ranum,L.P. (1999) An untranslated CTG expansion causes a novel form of spinocerebellar ataxia (SCA8). Nature Genet., 21, 379–384. [DOI] [PubMed] [Google Scholar]
- 20.Matsuura T., Yamagata,T., Burgess,D.L., Rasmussen,A., Grewal,R.P., Watase,K., Khajavi,M., McCall,A.E., Davis,C.F., Zu,L. et al. (2000) Large expansion of the ATTCT pentanucleotide repeat in spinocerebellar ataxia type 10. Nature Genet., 26, 191–194. [DOI] [PubMed] [Google Scholar]
- 21.Wheeler V.C., Lebel,L.A., Vrbanac,V., Teed,A., Te Riele,H. and MacDonald,M.E. (2003) Mismatch repair gene Msh2 modifies the timing of early disease in Hdh(Q111) striatum. Hum. Mol. Genet., 12, 273–281. [DOI] [PubMed] [Google Scholar]
- 22.Mangiarini L., Sathasivam,K., Mahal,A., Mott,R., Seller,M. and Bates,G.P. (1997) Instability of highly expanded CAG repeats in mice transgenic for the Huntington’s disease mutation. Nature Genet., 15, 197–200. [DOI] [PubMed] [Google Scholar]
- 23.Lia A.S., Seznec,H., Hofmann Radvanyi,H., Radvanyi,F., Duros,C., Saquet,C., Blanche,M., Junien,C. and Gourdon,G. (1998) Somatic instability of the CTG repeat in mice transgenic for the myotonic dystrophy region is age dependent but not correlated to the relative intertissue transcription levels and proliferative capacities. Hum. Mol. Genet., 7, 1285–1291. [DOI] [PubMed] [Google Scholar]
- 24.Fortune M.T., Vassilopoulos,C., Coolbaugh,M.I., Siciliano,M.J. and Monckton,D.G. (2000) Dramatic, expansion-biased, age-dependent, tissue-specific somatic mosaicism in a transgenic mouse model of triplet repeat instability. Hum. Mol. Genet., 9, 439–445. [DOI] [PubMed] [Google Scholar]
- 25.Gomes-Pereira M., Fortune,M.T. and Monckton,D.G. (2001) Mouse tissue culture models of unstable triplet repeats: in vitro selection for larger alleles, mutational expansion bias and tissue specificity, but no association with cell division rates. Hum. Mol. Genet., 10, 845–854. [DOI] [PubMed] [Google Scholar]
- 26.Manley K., Shirley,T.L., Flaherty,L. and Messer,A. (1999) Msh2 deficiency prevents in vivo somatic instability of the CAG repeat in Huntington disease transgenic mice. Nature Genet., 23, 471–473. [DOI] [PubMed] [Google Scholar]
- 27.vanDenBroek W.J., Nelen,M.R., Wansink,D.G., Coerwinkel,M.M., te Riele,H., Groenen,P.J. and Wieringa,B. (2002) Somatic expansion behaviour of the (CTG)(n) repeat in myotonic dystrophy knock-in mice is differentially affected by Msh3 and Msh6 mismatch-repair proteins. Hum. Mol. Genet., 11, 191–198. [DOI] [PubMed] [Google Scholar]
- 28.Savouret C., Brisson,E., Essers,J., Kanaar,R., Pastink,A., te Riele,H., Junien,C. and Gourdon,G. (2003) CTG repeat instability and size variation timing in DNA repair-deficient mice. EMBO J., 22, 2264–2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Diaz-Llera S., Podlutsky,A., Osterholm,A.M., Hou,S.M. and Lambert,B. (2000) Hydrogen peroxide induced mutations at the HPRT locus in primary human T-lymphocytes. Mutat. Res., 469, 51–61. [DOI] [PubMed] [Google Scholar]
- 30.Greber B., Lehrach,H. and Himmelbauer,H. (2003) Characterization of trimethylpsoralen as a mutagen for mouse embryonic stem cells. Mutat. Res., 525, 67–76. [DOI] [PubMed] [Google Scholar]
- 31.Barnett L.B., Tyl,R.W., Shane,B.S., Shelby,M.D. and Lewis,S.E. (2002) Transmission of mutations in the lacI transgene to the offspring of ENU-treated Big Blue male mice. Environ. Mol. Mutagen., 40, 251–257. [DOI] [PubMed] [Google Scholar]
- 32.Sadamoto S., Suzuki,S., Kamiya,K., Kominami,R., Dohi,K. and Niwa,O. (1994) Radiation induction of germline mutation at a hypervariable mouse minisatellite locus. Int. J. Radiat. Biol., 65, 549–557. [DOI] [PubMed] [Google Scholar]
- 33.Dubrova Y.E., Nesterov,V.N., Krouchinsky,N.G., Ostapenko,V.A., Neumann,R., Neil,D.L. and Jeffreys,A.J. (1996) Human minisatellite mutation-rate after the Chernobyl accident. Nature, 380, 683–686. [DOI] [PubMed] [Google Scholar]
- 34.Vilarino-Guell C., Smith,A.G. and Dubrova,Y.E. (2003) Germline mutation induction at mouse repeat DNA loci by chemical mutagens. Mutat. Res., 526, 63–73. [DOI] [PubMed] [Google Scholar]
- 35.Monckton D.G., Coolbaugh,M.I., Ashizawa,K., Siciliano,M.J. and Caskey,C.T. (1997) Hypermutable myotonic dystrophy CTG repeats in transgenic mice. Nature Genet., 15, 193–196. [DOI] [PubMed] [Google Scholar]
- 36.Ashizawa T., Monckton,D.G., Vaishnav,S., Patel,B.J., Voskova,A. and Caskey,C.T. (1996) Instability of the expanded (CTG)n repeats in the myotonin protein kinase gene in cultured lymphoblastoid cell lines from patients with myotonic dystrophy. Genomics, 36, 47–53. [DOI] [PubMed] [Google Scholar]
- 37.Blasina A., Price,B.D., Turenne,G.A. and McGowan,C.H. (1999) Caffeine inhibits the checkpoint kinase ATM. Curr. Biol., 9, 1135–1138. [DOI] [PubMed] [Google Scholar]
- 38.McCann J., Choi,E., Yamasaki,E. and Ames,B.N. (1975) Detection of carcinogens as mutagens in the Salmonella/microsome test: assay of 300 chemicals. Proc. Natl Acad. Sci. USA, 72, 5135–5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jackson A.L. and Loeb,L.A. (2000) Microsatellite instability induced by hydrogen peroxide in Escherichia coli. Mutat. Res., 447, 187–198. [DOI] [PubMed] [Google Scholar]
- 40.Gear A.R. (1974) Rhodamine 6G. A potent inhibitor of mitochondrial oxidative phosphorylation. J. Biol. Chem., 249, 3628–3637. [PubMed] [Google Scholar]
- 41.Ruschoff J., Wallinger,S., Dietmaier,W., Bocker,T., Brockhoff,G., Hofstadter,F. and Fishel,R. (1998) Aspirin suppresses the mutator phenotype associated with hereditary nonpolyposis colorectal cancer by genetic selection. Proc. Natl Acad. Sci. USA, 95, 11301–11306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fortune J.M. and Osheroff,N. (2000) Topoisomerase II as a target for anticancer drugs: when enzymes stop being nice. Prog. Nucleic Acid Res. Mol. Biol., 64, 221–253. [DOI] [PubMed] [Google Scholar]
- 43.Juttermann R., Li,E. and Jaenisch,R. (1994) Toxicity of 5-aza-2′-deoxycytidine to mammalian cells is mediated primarily by covalent trapping of DNA methyltransferase rather than DNA demethylation. Proc. Natl Acad. Sci. USA, 91, 11797–11801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carbone G.M., Catapano,C.V. and Fernandes,D.J. (2001) Imbalanced DNA synthesis induced by cytosine arabinoside and fludarabine in human leukemia cells. Biochem. Pharmacol., 62, 101–110. [DOI] [PubMed] [Google Scholar]
- 45.Martorell L., Monckton,D.G., Gamez,J., Johnson,K.J., Gich,I., Lopez de Munain,A. and Baiget,M. (1998) Progression of somatic CTG repeat length heterogeneity in the blood cells of myotonic dystrophy patients. Hum. Mol. Genet., 7, 307–312. [DOI] [PubMed] [Google Scholar]
- 46.Hunter A., Tsilfidis,C., Mettler,G., Jacob,P., Mahadevan,M., Surh,L. and Korneluk,R. (1992) The correlation of age of onset with CTG trinucleotide repeat amplification in myotonic dystrophy. J. Med. Genet., 29, 774–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Khajavi M., Tari,A.M., Patel,N.B., Tsuji,K., Siwak,D.R., Meistrich,M.L., Terry,N.H. and Ashizawa,T. (2001) Mitotic drive of expanded CTG repeats in myotonic dystrophy type 1 (DM1). Hum. Mol. Genet., 10, 855–863. [DOI] [PubMed] [Google Scholar]
- 48.Deplanque G., Ceraline,J., Mah-Becherel,M.C., Cazenave,J.P., Bergerat,J.P. and Klein-Soyer,C. (2001) Caffeine and the G2/M block override: a concept resulting from a misleading cell kinetic delay, independent of functional p53. Int. J. Cancer, 94, 363–369. [DOI] [PubMed] [Google Scholar]
- 49.Longley M.J., Pierce,A.J. and Modrich,P. (1997) DNA polymerase delta is required for human mismatch repair in vitro. J. Biol. Chem., 272, 10917–10921. [DOI] [PubMed] [Google Scholar]
- 50.Thielmann H.W., Popanda,O., Gersbach,H. and Gilberg,F. (1993) Various inhibitors of DNA topoisomerases diminish repair-specific DNA incision in UV-irradiated human fibroblasts. Carcinogenesis, 14, 2341–2351. [DOI] [PubMed] [Google Scholar]
- 51.Cummins J.H. (1997) The unique alteration of electrophoretic mobility of fragile-X-expanded fragments in the presence of ethidium bromide. Tech. Tips Online, S016895259601054. [Google Scholar]
- 52.Pearson C.E. and Sinden,R.R. (1996) Alternative structures in duplex DNA formed within the trinucleotide repeats of the myotonic dystrophy and fragile X loci. Biochemistry, 35, 5041–5053. [DOI] [PubMed] [Google Scholar]
- 53.Miko M. and Chance,B. (1975) Ethidium bromide as an uncoupler of oxidative phosphorylation. FEBS Lett., 54, 347–352. [DOI] [PubMed] [Google Scholar]
- 54.Chang C.L., Marra,G., Chauhan,D.P., Ha,H.T., Chang,D.K., Ricciardiello,L., Randolph,A., Carethers,J.M. and Boland,C.R. (2002) Oxidative stress inactivates the human DNA mismatch repair system. Am. J. Physiol. Cell Physiol., 283, C148–C154. [DOI] [PubMed] [Google Scholar]
- 55.Chen K.H., Yakes,F.M., Srivastava,D.K., Singhal,R.K., Sobol,R.W., Horton,J.K., Van Houten,B. and Wilson,S.H. (1998) Up-regulation of base excision repair correlates with enhanced protection against a DNA damaging agent in mouse cell lines. Nucleic Acids Res., 26, 2001–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brock G.J.R., Anderson,N.H. and Monckton,D.G. (1999) Cis-acting modifiers of expanded CAG/CTG triplet repeat expandability: associations with flanking GC content and proximity to CpG islands. Hum. Mol. Genet., 8, 1061–1067. [DOI] [PubMed] [Google Scholar]
- 57.Shiff S.J., Koutsos,M.I., Qiao,L. and Rigas,B. (1996) Nonsteroidal antiinflammatory drugs inhibit the proliferation of colon adenocarcinoma cells: effects on cell cycle and apoptosis. Exp. Cell Res., 222, 179–188. [DOI] [PubMed] [Google Scholar]
- 58.Yang Z., Lau,R., Marcadier,J.L., Chitayat,D. and Pearson,C.E. (2003) Replication inhibitors modulate instability of an expanded trinucleotide repeat at the myotonic dystrophy type 1 disease locus in human cells. Am. J. Hum. Genet., 73, 1092–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pineiro E., Fernandez-Lopez,L., Gamez,J., Marcos,R., Surralles,J. and Velazquez,A. (2003) Mutagenic stress modulates the dynamics of CTG repeat instability associated with myotonic dystrophy type 1. Nucleic Acids Res., 31, 6733–6740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sharma R., Bhatti,S., Gomez,M., Clark,R.M., Murray,C., Ashizawa,T. and Bidichandani,S.I. (2002) The GAA triplet-repeat sequence in Friedreich ataxia shows a high level of somatic instability in vivo, with a significant predilection for large contractions. Hum. Mol. Genet., 11, 2175–2187. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.