Abstract
Sinorhizobium meliloti RmInt1 is an efficient mobile group II intron that uses an unknown reverse transcriptase priming mechanism as the intron ribonucleoprotein complex can reverse splice into DNA target substrates but cannot carry out site-specific second strand cleavage due to the lack of a C- terminal DNA endonuclease domain. We show here that, like other mobile group II introns, RmInt1 moves around by an efficient RNA-based retrohoming mechanism. We found evidence of two distinct RmInt1 retrohoming pathways for mobility depending on the orientation of the target site relative to the direction of DNA replication. The preferred retrohoming pathway is consistent with reverse splicing of the intron RNA into single-stranded DNA at a replication fork, using a nascent lagging DNA strand as the primer for reverse transcription. This strand bias is the opposite of that reported for mobility of the lactococcal Ll.ltrB intron in the absence of second strand cleavage. The mobility mechanism found here for RmInt1 may be used for dissemination by many bacterial group II introns encoding proteins lacking the DNA endonuclease domain.
INTRODUCTION
Group II introns are a unique class of retroelements initially identified in organelles of lower eukaryotes and plants, but later found in bacteria (1–6) and in some archaebacterial species (7,8). These genetic elements are large catalytic RNAs, which splice via a lariat intermediate, in a mechanism similar to that of spliceosomal introns (9). Some group II introns are mobile elements that insert at intron-specific locations via a mechanism known as retrohoming (10,11). They can also transpose at low frequency to ectopic sites that resemble the normal homing site, primarily by reverse splicing directly into a DNA target site similar to retrohoming (12–17). This process facilitates intron dispersal in nature and may shed light on the origin of nuclear pre-mRNA introns (9) and non-LTR retrotransposons, both of which are thought to be descendents of mobile group II introns (18,19). Three main phylogenetic classes (IIA, IIB and IIC) of group II introns have been described based on intron-encoded protein (IEP) and conserved intron RNA structures (1,5,8,20–22).
The mechanism for group II intron retrohoming has been clearly established for the Lactococcus lactis Ll.ltrB intron and for the yeast mitochondrial aI1 and aI2 introns (10,11), all of which belong to the IIA subclass. Mobility of these group II introns occurs by means of a target DNA-primed reverse transcription mechanism involving a ribonucleoprotein (RNP) complex containing both the intron RNA and the IEP. The yeast and lactococcal group II intron IEPs have four conserved domains, including an N-terminal reverse transcriptase (RT) domain, domain X, a putative RNA-binding domain associated with RNA splicing or maturase activity, and C-terminal DNA-binding and DNA endonuclease domains (Fig. 1A). The conserved DNA endonuclease domain contains two pairs of cysteine residues, as in the zinc finger motif, interspersed with amino acid sequences characteristic of the H-N-H family of endonucleases (23,24). RNPs bind DNA non-specifically and then scan for target sites (25). The IEP first recognizes a small number of fixed positions in duplex DNA in the distal 5′-exon region, triggering local DNA unwinding, which enables the intron RNA exon-binding sites (EBS1 and EBS2) and δ sequence to base pair to the complementary DNA target sequences IBS1 and IBS2 in the 5′-exon and δ′ at the start of the 3′-exon. The intron RNA cleaves one strand by a partial or complete reverse splicing reaction at the intron insertion site. There is then a short time lag, followed by additional interactions between the IEP and the 3′-exon and then the IEP (DNA endonuclease domain) cleaves the opposite strand at a precise position within the 3′-exon (+9 in the case of the Ll.ltrB intron and +10 for yeast introns). The 3′-end of the cleaved site is then used as the primer for reverse transcription of the inserted intron RNA. The resulting cDNA copy of the intron is integrated into the host DNA by repair mechanisms, which differ according to the organism considered (26–28). Retrohoming of the Ll.ltrB intron occurs via complete reverse splicing of the intron RNA into DNA, independent of homologous recombination (27).
Figure 1.
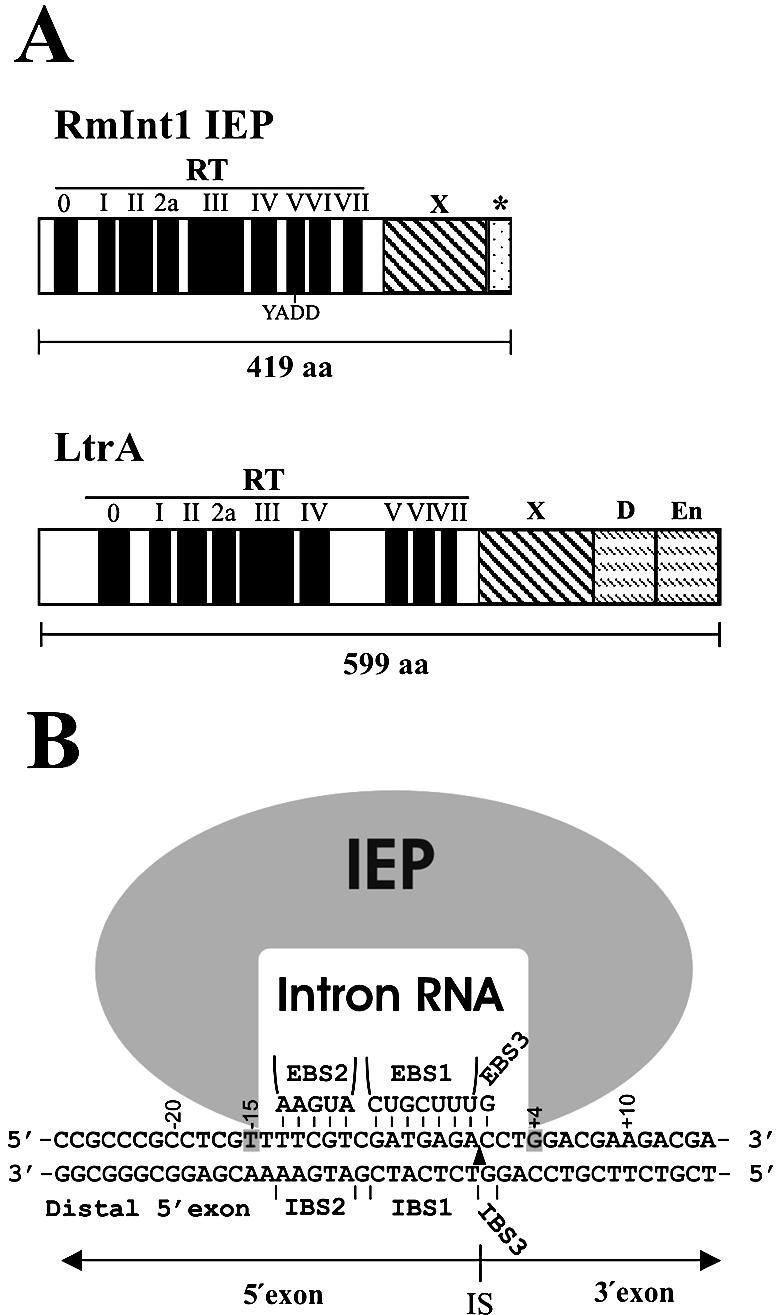
Intron-encoded proteins and DNA target site for RmInt1. (A) Comparison between L.lactis LltrB (LtrA) and S meliloti RmInt1 IEPs. Schematics (drawn to scale) show the N-terminal RT domain with conserved RT motifs 0–VII, the X domain associated with maturase activity and C-terminal DNA-binding (D) and conserved DNA endonuclease (En) domains (30,34). The asterisk (*) denotes an uncharacterized C-terminal extension of 20 amino acid residues in the RmInt1 IEP. (B) DNA target site recognized by the RmInt1 RNP complex. The DNA target site in the ISRm2011-2 insertion sequence is recognized by an RNP complex containing the IEP and the excised intron lariat RNA. Outside the IBS–EBS interaction, only position –15T of the DNA target is strictly required for intron homing, with position +4G having a less important effect (highlighted with a gray background) (33). The arrowhead pointing to the top strand indicates the intron insertion site (IS).
Many bacterial group II introns encode a protein that lacks the C-terminal endonuclease domain, which is responsible for second-strand cleavage in yeast aI1 and aI2 introns and the L.lactis Ll.ltrB intron (10,11). The best studied intron of this type is the Sinorhizobium meliloti RmInt1 intron (Fig. 1), which belongs to the IIB3 subclass (8) [also classified by others as belonging to class D (5)]. In addition, the intron-encoded protein of RmInt1, like some other bacterial group II intron IEPs, lacks a cognate C-terminal DNA-binding domain (Fig. 1A) (5,6,8,29,30), which is thought to interact with the distal 5′-exon region to promote DNA unwinding (31). Nevertheless, RmInt1 is an efficient mobile element with a homing frequency (fraction of cells containing at least one homing event) approaching 100%, similar to that of the fungal mtDNA introns and the lactococcal Ll.ltrB intron, with a homing efficiency (fraction of recipient targets invaded by the intron within a single cell) ranging from 20 to 45% (29,32). The homing pathway of the mobile RmInt1 intron has not yet been fully determined but, in contrast to those of yeast and the lactococcal introns, it is known to require an as yet undetermined reverse transcriptase priming mechanism, as the intron RNP can reverse splice into double- or single-stranded DNA substrates but cannot carry out site-specific second strand cleavage, as expected given the absence of the DNA endonuclease domain (33). The homing of RmInt1, like that of Ll.ltrB, requires modifiable base pairing interactions, involving 13 nt, between the intron RNA and the DNA target. However, instead of the δ–δ′ interaction typical of subgroup IIA introns, RmInt1 recognizes the first nucleotide within the 3′-exon of the target site by means of the EBS3–IBS3 interaction (32) typical of subgroup IIB self-splicing introns (Fig. 1B).
Data recently obtained for a mutant of Ll.ltrB (YRT) affected in the C-terminal region with no detectable DNA endonuclease activity suggest that Ll.ltrB can retrohome at low frequency without second strand cleavage, predominantly by reverse splicing of the intron RNA into double-stranded DNA, followed by use of a nascent leading DNA strand generated during DNA replication as a primer for reverse transcription of the inserted intron RNA (34). The YRT mutant also shows a lower level of mobility with target sites in the opposite orientation, which likely occurs by reverse splicing into double-stranded DNA followed by the use of a nascent lagging strand to prime reverse transcription (34). Ichiyanagi et al. (16,17) found that Ll.ltrB retrotransposition was not dependent on endonuclease activity and preferentially used the template for lagging strand DNA synthesis, consistent with reverse splicing into single-stranded DNA at a replication fork, followed by use of a nascent lagging DNA strand as a primer. It has therefore been suggested, but not demonstrated, that group II introns encoding proteins lacking a DNA endonuclease domain may use a similar mobility mechanism, using a nascent strand (leading or lagging) at a DNA replication fork as the primer for reverse transcription (34).
We show here that RmInt1 mobility occurs via an RNA intermediate and that this retrohoming process is predominantly biased towards the template for lagging strand DNA synthesis, consistent with reverse splicing of the intron RNA into single-stranded DNA at a replication fork, followed by the use of a nascent lagging DNA strand as a primer for reverse transcription This replication orientation bias is opposite of that reported for mobility of the Ll.ltrB intron in the absence of second strand cleavage and is similar to the ectopic transposition of group II introns.
MATERIALS AND METHODS
Bacterial strains, media and growth conditions
The rhizobial strains used in this work were Sinorhizobium meliloti GR4 (this laboratory), RMO17 (35) and the genome sequenced strain 1021 (J.Dénarié). Escherichia coli (DH5α) was routinely cultured at 37°C in Luria–Bertani medium and rhizobial strains were cultured at 28°C on TY or defined minimal medium (36). Antibiotics were used as required, at the following concentrations: tetracycline, 10 µg ml–1; kanamycin, 180 µg ml–1 for S.meliloti and 50 µg ml–1 for E.coli.
DNA manipulation and plasmid constructs
General molecular biology techniques were used, as described in Sambrook et al. (37). The construction of the RmInt1 donor plasmid pKG2.5 and recipient plasmid pJB0.6s (+), containing a 640 bp fragment encompassing the intron insertion site of ISRm2011-2, and pJBΔ129, carrying an internal deletion of the target site (used as a negative control for retrohoming) has been described elsewhere (29). pJB0.6as is a pJBtc19 derivative (38) in which the DNA target was cloned as a SphI–SacI DNA fragment, but in the opposite orientation to that found in pJB0.6s.
pJB9s and pJB10as were generated by insertion into the SphI site of pJBTc19, the amplified product obtained with primers S/up (5′-gggggcatgccgtagcgccga) and SS/down (5′-gggcatgcgtcgacattcgagcctcggtacca), using pTD0.6 as the template. pTD0.6 is a pIC20H (39) derivative that carries the intron insertion site (–149 to +425 position) flanked by E.coli rrnb transcription terminators functional in S.meliloti, upstream by one T1 and downstream by three T2 terminators. T1 was obtained by amplification of pKK232-8 (40) with primers T1F (5′-GCATGCCGTAGCGCCGATGGTAGTGTG) and T1R (5′-CTGCAGCCGTTGCTTCGCAACGTTCA). The product was inserted into pGEM-T (Promega), resulting in the pGE-T1 construct. A 200 bp SphI–PstI fragment from pGE-T1 containing T1 was inserted into pIC20H to generate pIC-T1. T2 was obtained in a similar fashion to T1, using primers T2F (5′-GCATCCGGAGGGTGGCGGGCAGGACGC) and T2R (5′-GGTACCAGATCTTGGGGGGATGGCTTGTAGATATGA). The resulting fragment was inserted into pGEM-T to create pGE-T2, which was subjected to BamHI and BglII digestion and ligation in pGE-T2 to generate pGE-T2X3. BamHI and KpnI digestion of this plasmid generated a 420 bp fragment containing three T2 units in tandem that was inserted into pIC-T1 to produce pICT12. A 575 bp target-containing fragment obtained from pGEM0.6 (29) by PCR amplification with primers SalT600 (5′-ACATGTCCACCTTCGTGCACGAAGA) and XbaT600 (5′-ACTCTAGAATGTGGCGCAGTTCGTCA) was inserted into pICT12 to generate pTD0.6. Constructs pJB9s and pJB10as generated a 1.05 kb fragment on digestion with SalI, which hybridizes with the exon probe.
To generate the twintron containing the group I intron td interrupting the RmInt1 sequence, we constructed pKGBx by generating a XhoI site downstream from the IEP coding sequence. We carried out inverse PCR (41) using the divergent primers X-1 (5′-gccgctcgagtcaggtaaacgtgttcgttccgaac) and X-2 (5′-GCCGCTCGAGCCTGATGGGAGCGGTGTGAATCGAG) with Pfu DNA polymerase (Promega) and pKG2 (a pKG2.5 derivative in which the intron is flanked by shorter exons –50/+146) as the template. The resulting 7.1 kb PCR product was XhoI digested, self-ligated and used to isolate pKGBx, which was used as a donor plasmid in homing assays to check that the modified intron was functional. A 429 bp DNA fragment containing the tdΔ1-3 sequence (27), along with 12 bp of each of the td flanking exons, was generated by PCR using primers td up (5′-GGTCGACGTTTTCTTGGGTTAATTGAG) and td down (5′-GCTCGAGATTAAACGGTAGCATTATGTTC) and pTZtdIΔ1-3 as the template (42). The twintron construct pKGAD1 (td+) was then generated by inserting this td-containing DNA fragment into the XhoI site of pKGBx in the sense orientation with respect to the direction of RmInt1 transcription.
Retrohoming analysis
RmInt1 retrohoming analysis was performed in S.meliloti host cells. We investigated the transfer of intron copies from the genome of the GR4 strain (S.meliloti) to the recipient plasmids or between compatible donor pKG2.5, pKGBx or pKGAD1 and recipient plasmids in the RmInt1-less strain RMO17 (29). Plasmids were transferred from E.coli (DH5α) to S.meliloti (GR4 or RMO17) by triparental mating (43). Briefly, a donor culture (E.coli cells containing the plasmid to be transferred) and another culture containing the helper plasmid pRK2013 were mixed in exponential growth phase with a recipient culture (S.meliloti cells) in late exponential phase in a 1:1:1 ratio. The mixture was plated on TY agar and incubated for 24 h at 28°C. Appropriate dilutions were then plated on minimal medium supplemented with the corresponding antibiotic. The plates were incubated for 2–3 days at 28°C (depending on whether one or two antibiotics were used). We then picked 5–10 single colonies, which were cultured in 5 ml of TY medium until stationary phase (∼30–40 generations). Clones selected for studies of the effect of the number of generations on homing efficiency were cultured in exponential phase for 60 additional generations. Plasmids extracted from harvested cells (1.5 ml) were analyzed for group II intron retrohoming by agarose gel electrophoresis and hybridization (29). Southern blots were carried out using probes specific for the intron (RmInt1) and the insertion site (ISRm2011-2) reported elsewhere (3,29). The td intron probe was generated by PCR using primers tdIa (5′-TAATTGAGGCCTGAGTATAA) and tdIb (5′-GAGCAGACTATATCTCCAAC).
Densitometry of the bands was carried out with Image Quantity One (Bio-Rad system software package). Retrohoming efficiency (percentage homing) was measured as the percentage of recipient plasmids invaded by RmInt1 after a given number of generations (29), and this dependent variable was calculated as (H forms/H + R forms) × 100.
RESULTS
RmInt1 mobility occurs via an RNA-based retrohoming mechanism
In vivo and in vitro data obtained with lactococcal and yeast introns have indicated that the mobility mechanisms of these introns predominantly involve an RNA intermediate (retrohoming). Although, it has been assumed that RmInt1 also moves by a retrohoming pathway, this has yet to be unequivocally demonstrated. We investigated whether RmInt1 mobility events corresponded to RNA-based retrohoming by constructing a twintron donor based on the twintron engineered with Ll.ltrB (27). We did this by creating a XhoI site downstream from the intron open reading frame (ORF) and adding 5 nt to preserve the stem of ribozyme domain IV (dIV) (pKGBx, Fig. 2A). This modification did not affect intron homing efficiency (data not shown). A group I intron, the td intron, lacking its own mobility apparatus, was amplified by PCR, along with its exon sequences (12 nt each side) and the product was inserted in the sense (pKGAD1) orientation into the newly created XhoI site (Fig. 2A). In RNA-based mobility events, the group I intron, when cloned in the sense orientation, should be spliced out of a fraction of the twintron RNAs, resulting in group I intron loss in the homing product (pJB2.5* in Fig. 2A). Mobility assays performed with the intron donor plasmid pKGAD1, which contains the engineered twintron construct, and the target recipient plasmid pJB0.6as (Fig. 2A) revealed a homing product when pKGAD1 was used (Fig. 2B, lane 2). This product was similar in size to that generated by the wild-type construct pKGBx (Fig. 2B, lane 1), indicating that the group I intron was missing. This was confirmed by hybridization with specific probes for RmInt1 and td (data not shown) and by analysis of the sequences of invaded recipient plasmids, which showed the presence of 24 nt corresponding to the expected td ligated exons (see pJB2.5* in Fig. 2A). As shown in Figure 2B, the homing efficiency was only slightly reduced for the construct pKGAD1. Thus, even though the possibility of other mobility pathways may exist, retrohoming appears to be the predominant pathway for RmInt1 mobility.
Figure 2.
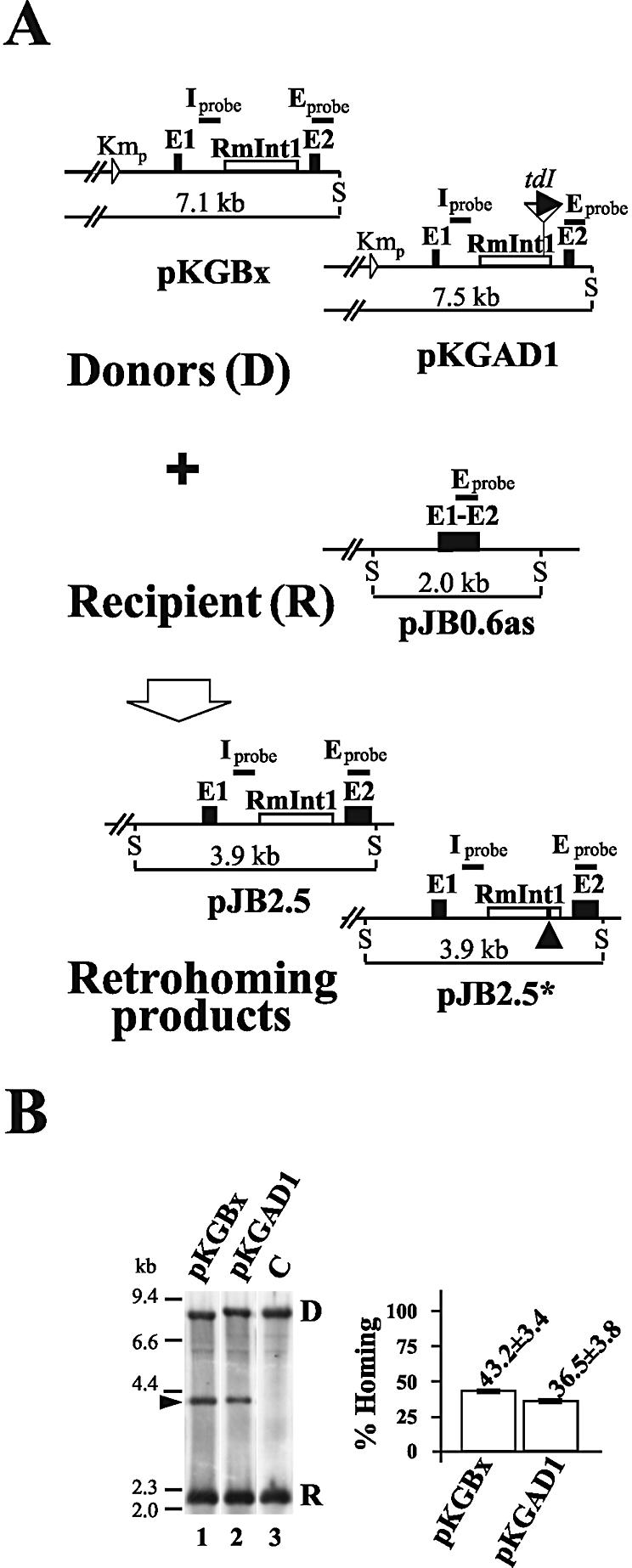
RmInt1 retrohoming. (A) Schematic diagram of mobility assays. The intron donors (D), with pKGBx as the wild-type and pKGAD1 as the twintron donor, recipient (pJB0.6as) and detected retrohoming products (pJB2.5 and pJB2.5*) are illustrated. The arrowhead on pJB2.5* indicates the exon junctions of group I intron td. Relevant SalI restriction sites (S) and the size of the corresponding DNA fragments are indicated. Exon- (E) and RmInt1-specific (I) probes are indicated by thick lines. (B) Plasmid pool analysis by Southern blot hybridization with the DNA target probe E indicated above. RMO17 cells containing intron donor plasmids pKGBx (lane 1) and pKGAD1 (lane 2) together with the target recipient plasmid pJB0.6as were harvested and their plasmid content was further analyzed. Plasmids were digested with SalI, resulting in several DNA fragments hybridizing to the indicated exon-specific probe: a 7.1 or 7.5 kb band corresponding to the donors (D), a 2.0 kb band corresponding to the recipient plasmid (R) and a 3.9 kb band corresponding to the intron-invaded plasmid (arrowhead). A negative control (C) for the mobility assays using a splicing-defective intron as a donor plasmid is shown (lane 3). DNA molecular size markers are indicated at the left of the panel. A bar graph showing the percentage homing achieved with the wild-type (pKGBx) and the twintron (pKGAD1) constructs is shown on the right. Data are the means of four determinations for plasmid pools in two independent assays, with the SD indicated.
Retrohoming of RmInt1 displays a replication orientation bias
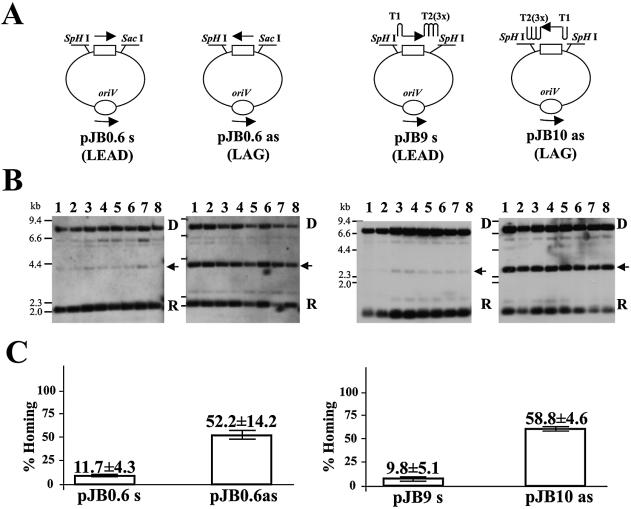
Recent studies performed with the Ll.ltrB intron have reported a replication orientation bias with a mutant affected in the C-terminal region (YRT) lacking endonuclease activity or with DNA target sites that do not support second strand cleavage (34). Biochemical studies have shown that the RmInt1 IEP has RT activity and that the intron RNP can reverse splice into double- or single-stranded DNA substrates, but that this intron cannot perform second strand cleavage (33). We investigated whether RmInt1 mobility displayed strand orientation bias by carrying out mobility assays in S.meliloti strain RMO17 (lacking genomic RmInt1 intron copies) harboring the intron donor plasmid pKG2.5 with recipient plasmids in which the target site was cloned in both orientations with respect to the direction of DNA replication. The target site (–175 to +456 of the intron insertion site) was cloned such that reverse transcription of the inserted intron RNA could make use of either a nascent leading (LEAD) or a nascent lagging (LAG) DNA strand as the primer, giving plasmids pJB0.6s and pJB0.6as, respectively (Fig. 3A). In other mobility assays, the DNA target was flanked by upstream T1 and downstream T2 (three) transcription terminators, giving the pJB9s (LEAD) and pJB10as (LAG) constructs (Fig. 3A), to rule out any effect of target transcription in retrohoming events. Homing efficiency was similar for the pJB0.6s and pJB9s (LEAD) constructs, and also for the pJB0.6as and pJB10as (LAG) constructs (Fig. 3B andC). However, homing efficiency was five times higher in LAG (pJB0.6as and pJB10as) than in LEAD (pJB0.6s or pJB9s) constructs. Similar results were obtained in a RecA– host background (data not shown). The homing efficiency (although efficiency was lower overall) was also higher in LAG than in LEAD constructs when recipient plasmids were introduced into strain GR4, in which retrohoming from genomic copies of introns occurs (data not shown). The RT mutant YAHH and an intron with a mutation affecting the RNA domain V D5-CGA, which inhibits RNA splicing (33), displayed no detectable mobility in either recipient plasmid construct. We also confirmed that homing efficiency did not differ according to whether the intron donor plasmid was introduced into the host cell before or after the recipient plasmid (data not shown). The lower homing efficiency in LEAD constructs was additionally confirmed with the intron donor plasmid pKGAD1. Moreover, the td intron was absent in the corresponding homing product and thus retrohoming appears to be the predominant mobility pathway for both target site orientations (data not shown).
Figure 3.
RmInt1 intron retrohoming with DNA target sites cloned in opposite orientations in RK2-derived plasmids. (A) Schematic representation of target recipient plasmids pJB0.6s, pJB0.6as, pJB9s and pJB10as. pJB0.6s and pJB0.6as contain the RmInt1 DNA target site cloned in opposite orientations in pJBTc19, between the SphI and SacI cloning sites. Similarly, pJB9s and pJB10as contain the RmInt1 DNA target site flanked with transcription terminators T1 and T2 cloned in opposite orientations (see Materials and Methods). The target was cloned such that reverse transcription of the inserted intron RNA could potentially use either a nascent leading (LEAD) or lagging (LAG) DNA strand as a primer at the replication fork. The location and direction of oriV are indicated. (B) Plasmid pool analysis by Southern blot hybridization performed with an exon-specific probe. Sinorhizobium meliloti RMO17 cells containing pKG2.5 were transformed with the compatible recipient plasmids described above and the plasmid pool obtained from eight single colonies for each cross were digested with SalI, resulting in several hybridizing fragments: 7.6 kb for intron donor (D) pKG2.5; non-invaded target (R) of 2.0 or 1.2 kb for pJB0.6s/pJB0.6as or pJB9/pJB10. Intron invasion of the recipient plasmid (indicated with an arrow) results in hybridizing DNA fragment of 3.73 and 3.0 kb for pJB0.6s/pJB0.6as and pJB9/pJB10, respectively. Other hybridizing bands are derived from the donor plasmids. (C) The amounts of recipient target DNA and intron-invaded DNA in each lane were estimated and the mean ± SD is shown.
In S.meliloti strain 1021, previous experiments (44) showed that RmInt1 displayed no detectable retrohoming into the recipient plasmid pJB0.6+(s) (target site in the leading strand orientation). To assesses the possibility that in this host retrohoming occurs in a target when cloned in the lagging strand orientation, mobility assays were carried out with the LAG (pJB0.6as) and LEAD (pJB0.6s) constructs. As shown in Figure 4, in this host background we detected retrohoming in the LAG (pJB0.6as) but not in the LEAD construct (pJB0.6s). The retrohoming efficiency in 1021 was half that in RMO17, consistent with a general lower level of mobility in this host, revealing that this strain supports intron homing events. These results additionally confirmed that RmInt1 mobility shows a replication orientation bias. Together, these findings indicate that RmInt1 mobility depends on intron RNA splicing and RT activity and has a pronounced replication orientation bias, opposite to that found for Ll.ltrB in the absence of second strand cleavage and consistent with the preferred use of a nascent lagging DNA strand as a primer for reverse transcription.
Figure 4.
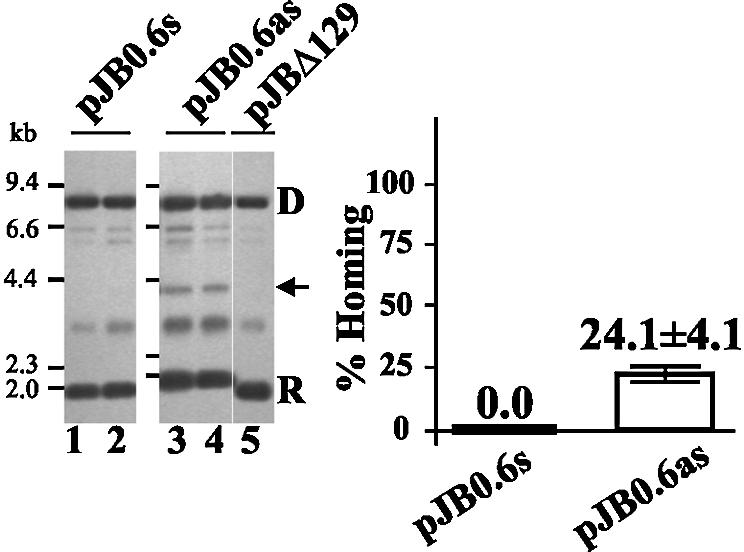
RmInt1 intron retrohoming in S.meliloti strain 1021. Plasmid pool analysis by Southern blot hybridization performed with an exon-specific probe is shown. Sinorhizobium meliloti 1021 cells containing pKG2.5 were transformed with the compatible recipient plasmid pJB0.6s (lanes 1 and 2), pJB0.6as (lanes 3 and 4) or pJBΔ129 (as a negative control; lane 5) and the plasmid pool was analyzed as described in Figure 3. The amounts of recipient target DNA and intron-invaded DNA in each lane were estimated and the mean ± SD is shown.
Evidence for two different RmInt1 retrohoming pathways
Preference for the lagging strand orientation over the leading strand orientation may be due to differences in the mechanisms of RmInt1 retrohoming to the two strands. A replication-dependent process would generate a copy of wild-type recipient plasmid in addition to the homing product (34). However, continuous retrohoming in the host cell would lead to the accumulation of invaded recipient plasmids in the bacterial population. We tested this hypothesis by culturing a clone of RMO17 containing the intron donor plasmid pKG2.5 and carrying ∼20% copies of recipient plasmid pJB0.6s (LEAD) invaded by the intron for 60 generations (see Materials and Methods) and estimating the percentage of homing occurring in the plasmid population. A clone containing donor pKG2.5 and carrying ∼50% copies of recipient plasmid pJB0.6as (LAG) invaded by the intron was cultured in parallel (Fig. 5A). Similar behavior was observed for distinct ratios of invaded and non-invaded recipient plasmids (data not shown). The percentage of invaded recipient plasmids did not increase on culturing the bacteria containing the clone with the DNA target in the leading strand orientation (pJB0.6s), as was previously observed for recipient pBB-derived plasmids (29). However, the percentage of homing for the target site cloned in the lagging strand orientation (pJB0.6as) increased linearly with the number of generations, approaching 100% after 60 generations (Fig. 5B). Using the equation shown in Figure 5B (LAG), the rate of retrohoming into the lagging strand template can be estimated at 0.745% per generation, implying that 134 generations would be required for full recipient plasmid invasion. It should be noted that the intron RNPs displayed RT activity in cells from both clones after 60 generations and that we have not observed any selection for intron-containing plasmids (data not shown). These findings provide additional evidence of two different RmInt1 retrohoming pathways for mobility depending on the orientation of the target site relative to the direction of DNA replication.
Figure 5.
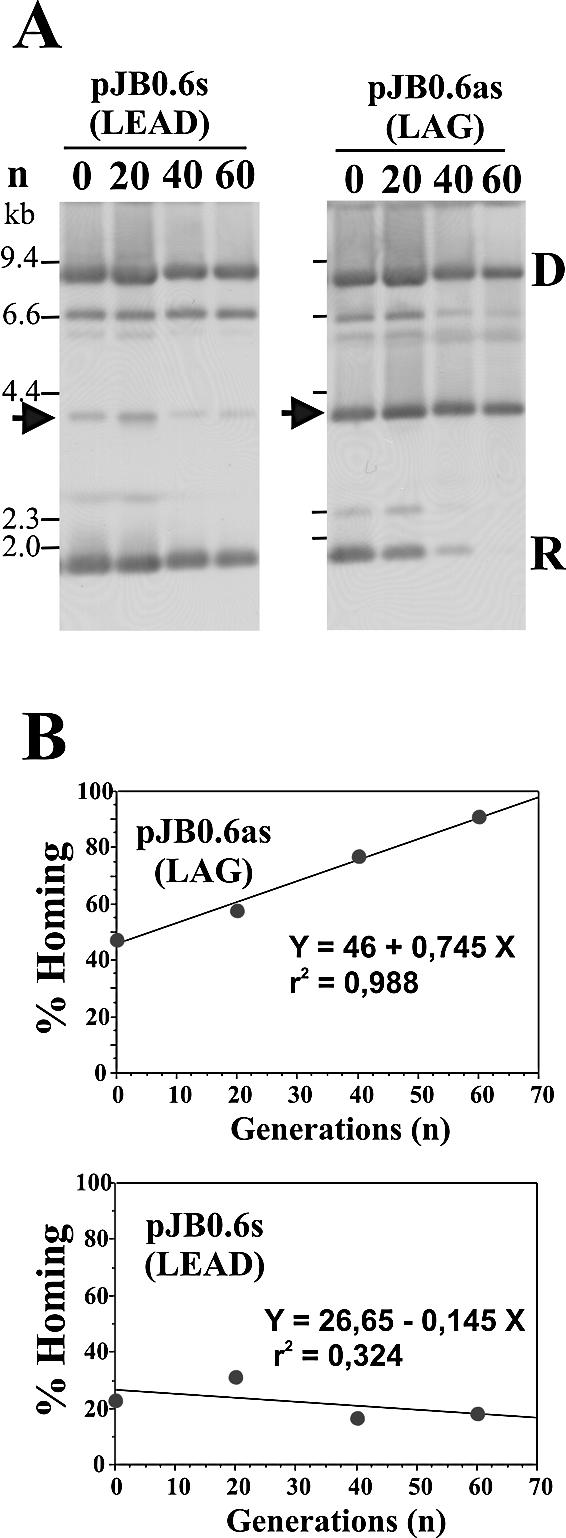
Retrohoming of RmInt1 intron biased to the template for lagging strand DNA synthesis is linked to plasmid replication. (A) Two different RMO17 clones containing intron donor pKG2.5 and target recipient plasmid pJB0.6s or pJB0.6as were grown in exponential growth phase for 60 additional generations (see Materials and Methods). The plasmid pool at 0, 20, 40 and 60 generations was analyzed by Southern blotting, as described in Figure 3. (B) Graphic representation of homing efficiency [y-axis, (H/H + R) × 100] versus the number of generations (x-axis, n) of the cultures and the corresponding regression lines. The rectilinear equations are also indicated, as well as the coefficient of determination r2.
DISCUSSION
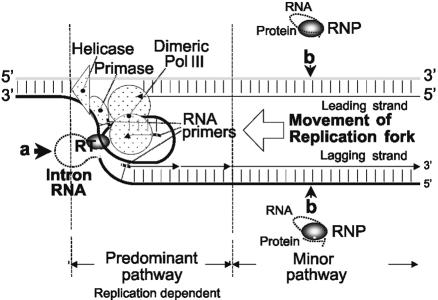
The S.meliloti RmInt1 group II intron is an efficient mobile retroelement, with an IEP lacking the DNA endonuclease domain and a cognate DNA-binding domain (29,30). Our results indicate that RmInt1 mobility occurs by retrohoming through an RNA intermediate, as previously demonstrated for lactococcal and yeast introns, but with a predominant bias for the lagging strand as template for DNA synthesis, in contrast to the situation for the Ll.ltrB intron in the absence of second strand cleavage (34) and similar to the ectopic transposition of group II introns (16,17). This mobility pathway may be coupled to DNA replication. This bias is consistent with reverse splicing into single-stranded DNA and the use of a nascent lagging strand as a primer for reverse transcription (Fig. 6). RmInt1 retrohoming also occurred, albeit less efficiently, into the leading strand template, but this other mobility pathway, which may also occur with the lagging strand template, does not increase with cell division (Fig. 6). Further, we found that RT and intron RNA splicing activities were required for both retrohoming pathways.
Figure 6.
A model for RmInt1 mobility. The major mobility pathway (a, thick arrow) may be linked to DNA replication involving invasion of the DNA strand that serves as a template for lagging strand synthesis (black strand), which is then used to prime reverse transcription of the inserted intron RNA. This priming process could involve either the RNA primer synthesized by the primase or partial polymerization of the Okazaki fragment by DNA polymerase III. A second possible minor retrohoming pathway (b, thin arrows) may occur independently of the strand into which the target is inserted. Alternative mechanisms for priming may include random non-specific opposite strand nicks, a nascent leading strand (34) or de novo initiation priming (48).
We showed, using group I–group II twintron constructs, that RmInt1 mobility occurs by an RNA-mediated retrohoming pathway, as previously demonstrated for the lactococcal Ll.ltrB intron (27). Since the homing efficiency of the twintron construct pKGAD1 is only slightly reduced with respect to the wild-type intron donor plasmid (pKGBx), it would appear that RmInt1 mobility occurs predominantly by an RNA intermediate. Retrohoming of RmInt1 is extremely efficient because all recipient cells undergo homing events (100% homing frequency) and up to 50% of the recipient target plasmids are invaded. We found here that the retrohoming of RmInt1 displayed a replication orientation bias (∼5 times higher for one orientation than for the other), consistent with preferential use of the lagging DNA strand as a primer for reverse transcription. This bias was particularly evident in host backgrounds such as that of strain 1021, in which invasion was observed only with a target cloned in the lagging strand orientation. In addition, the three copies of RmInt1 present in the genome of 1021 are all located in DNA targets oriented such that the lagging strand could have been used as a primer for reverse transcription of the inserted intron RNA (44). Similarly, Ichiyanagi et al. (16) noted that a number of group II introns that lack a C-terminal DNA endonuclease domain are found on the lagging strand template. Retrohoming into the leading or lagging strand template requires both intron RNA and IEP RT activity (33; this work). Thus, the IEP probably copies the inserted intron RNA in target sites cloned in both orientations.
Interestingly, we observed a continuous increase in invaded recipient plasmids containing the target cloned in the lagging strand orientation with the number of generations. This was not the case for the target cloned into the leading strand orientation, which provides further evidence of two different RmInt1 retrohoming pathways for mobility. In addition to the lack of second strand cleavage, previous observations have suggested that the RmInt1 RNPs may have some difficulty in unwinding DNA (33). In addition, recognition of the DNA target by the RmInt1 IEP appears to be more limited than in lactococcal and yeast aI2 introns, with just two key nucleotide (–15T and +4G) (32) residues flanking the region base pairing with the intron RNA (Fig. 1B). Moreover, in contrast to what has been observed for the yeast aI2 and lactococcal Ll.ltrB introns, we recently found that recognition of the critical key nucleotide in the distal 5′-exon (–15T) is not involved in DNA unwinding (Molina-Sánchez and Toro, unpublished results). The preference found here for lagging strand DNA suggests a retrohoming model for group II introns lacking the DNA endonuclease domain different from that of Ll.ltrB intron in the absence of second strand cleavage (34) and similar to the model proposed for the ectopic transposition of group II introns (16,17). This preference implies the insertion of intron RNA into single-stranded DNA once the replication fork has passed the DNA target and before use of the nascent lagging DNA strand as a primer (Fig. 6). Further evidence for this RmInt1 mobility mechanism involving a single-stranded DNA intermediate is provided by the lower likelihood of mobility events over the DNA target in the leading strand template, where the target site is less likely to be found in single-stranded than in the lagging strand template. The mobility of RmInt1 in the lagging DNA strand may also be stimulated by the host replication machinery, increasing the efficiency of the retrohoming process, either by improving the targeting of single-stranded regions or by stimulating IEP RT activity, which could in turn facilitate intron insertion (25).
The target recipient plasmid used here, pJB, is a broad host range RK2 derivative of the IncP-1 incompatibility group, which displays theta (θ) unidirectional replication using host replication proteins (DnaA, DnaB, DnaC, Dna gyrase and DnaG) (for a review see 45), making it a useful model for examining the involvement of host DNA replication in the retrohoming of RmInt1. Interestingly, the replication orientation bias for the lagging DNA strand found with recipient pJB derivatives contrasts with previous data obtained with the DNA target site cloned into pBBR1MCS-2 (46) derivatives (pBB0.6+/–) (29), for which no strand bias was observed. pBBR1 is a broad host range plasmid isolated from Bordetella bronchiseptica (47), for which the replication model and host proteins involved are unknown. Efficient homing in pBB recipient plasmids has been shown to be limited to initial generations (29), for both orientations of the cloned target site (Martinez-Abarca and Toro, unpublished results). This behavior resembles that reported here for pJB0.6s (LEAD). The lack of the predominant retrohoming pathway observed here in recipient pBB derivatives may be due to specific features of the former plasmid replication machinery yet to be identified. This minor retrohoming pathway (Fig. 6) may involve reverse splicing into either double-stranded DNA or transiently single-stranded DNA target sites. Mechanisms for priming may include random non-specific opposite strand nicks, a nascent leading strand or de novo initiation priming (48). These events may be facilitated during DNA uptake because they do not appear to increase with further cell divisions.
The efficient retrohoming pathway preferentially targeting the template for lagging strand synthesis at a DNA replication fork shown here for RmInt1 is important because it may be used by other bacterial group II introns encoding proteins lacking the DNA endonuclease domain (60% of the bacterial group II intron IEPs annotated to date) (5,6) for propagation. The reason for the different strand biases observed for RmInt1 and Ll.ltrB intron mobility remains to be elucidated. The mechanisms of retrohoming and retrotransposition of group II introns, the IEPs of which lack the DNA endonuclease domain, may be identical, with the frequency of each process determined purely by the efficiency of RNPs to recognize the site-specific target or ectopic sequences.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank Dr Marlene Belfort for kindly providing us with pTZtdIΔ1-3 for the construction of twintron donors. We also thank Miss Dolores Molina-Sánchez for her help in mobility assays and Dr Jose Ignacio Jimenez-Zurdo for critical reading of the manuscript and Miss Ascensión Martos Tejera for her technical assistance. This work was funded by research project BIO2002-02579 from the Ministerio de Ciencia y Tecnología.
REFERENCES
- 1.Michel F., Umesoko,K. and Ozeki,H. (1989) Comparative and functional anatomy of group II catalytic introns: a review. Gene, 82, 5–30. [DOI] [PubMed] [Google Scholar]
- 2.Ferat J.L. and Michel,F. (1993) Group II self-splicing introns in bacteria. Nature, 364, 358–361. [DOI] [PubMed] [Google Scholar]
- 3.Martínez-Abarca F., Zekri,S. and Toro,N. (1998) Characterization and splicing in vivo of a Sinorhizobium meliloti group II intron associated with particular insertion sequences of the IS630-Tc1/IS3 retroposon superfamily. Mol. Microbiol., 28, 1295–1306. [DOI] [PubMed] [Google Scholar]
- 4.Martínez-Abarca F. and Toro,N. (2000) Group II introns in the bacterial world. Mol. Microbiol., 38, 917–926. [DOI] [PubMed] [Google Scholar]
- 5.Zimmerly S., Hausner,G. and Wu,X.C. (2001) Phylogenetic relationships among group II intron ORFs. Nucleic Acids Res., 29, 1238–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dai L. and Zimmerly,S. (2002) Compilation and analysis of group II intron insertions in bacterial genomes: evidence for retroelement behaviour. Nucleic Acids Res., 30, 1091–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dai L. and Zimmerly,S. (2003) ORF-less and reverse-transcriptase-encoding group II introns in archaebacteria, with a pattern of homing into related group II intron ORFs. RNA, 9, 14–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Toro N. (2003) Bacteria and Archaea Group II introns: additional mobile genetic elements in the environment. Environ. Microbiol., 5, 143–151. [DOI] [PubMed] [Google Scholar]
- 9.Michel F. and Ferat,J.L. (1995) Structure and activities of group II introns. Annu. Rev. Biochem., 64, 435–461. [DOI] [PubMed] [Google Scholar]
- 10.Lambowitz A.M., Caprara,M.G., Zimmerly,S. and Perlman,P.S. (1999) Group I and group II ribozymes as RNPs: clues to the past and guides to the future. In Gesteland,R.F., Cech,T.R. and Atkins,J.F. (eds), The RNA World, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 451–485. [Google Scholar]
- 11.Belfort M., Derbyshire,V., Parker,M.M, Cousineau,B. and Lambowitz,A.M. (2002). Mobile introns: pathways and proteins. In Craig,N.L., Craigie,R., Gellert,M. and Lambowitz,A.M. (eds), Mobile DNA, 2nd Edn. ASM Press, Washington, DC, pp. 761–783. [Google Scholar]
- 12.Yang J., Mohr,G., Perlman,P.S. and Lambowitz,A.M. (1998) Group II intron mobility in yeast mitochondria: target DNA primed reverse transcription activity of aI1 and reverse splicing into DNA transposition sites in vitro. J. Mol. Biol., 282, 505–523. [DOI] [PubMed] [Google Scholar]
- 13.Martínez-Abarca F. and Toro,N. (2000) RecA-independent ectopic transposition in vivo of a bacterial group II intron. Nucleic Acids Res., 28, 4397–4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dickson L., Huang,H.R., Liu,L., Matsuura,M., Lambowitz,A.M. and Perlman,P.S. (2001) Retrotransposition of a yeast group II intron occurs by reverse splicing directly into ectopic DNA sites. Proc. Natl Acad. Sci. USA, 98, 13207–13212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muñoz E., Villadas,P.J. and Toro,N. (2001) Ectopic transposition of a group II intron in natural bacterial populations. Mol. Microbiol., 41, 645–652. [DOI] [PubMed] [Google Scholar]
- 16.Ichiyanagi K., Beauregard,A., Lawrence,S., Smith,D., Cousineau,B. and Belfort,M. (2002) Retrotransposition of the Ll.ltrB group II intron proceeds predominantly via reverse splicing into DNA targets. Mol. Microbiol., 46, 1259–1271. [DOI] [PubMed] [Google Scholar]
- 17.Ichiyanagi K., Beauregard,A. and Belfort,M. (2003) A bacterial group II intron favors retrotransposition into plasmid targets. Proc. Natl Acad. Sci. USA, 100, 15742–15747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zimmerly S., Guo,H., Eskes,R., Yang,J., Perlman,P.S. and Lambowitz,A.M. (1995) A group II intron RNA is a catalytic component of a DNA endonuclease involved in intron mobility. Cell, 83, 529–538. [DOI] [PubMed] [Google Scholar]
- 19.Eickbush T. (1999) Mobile introns: retrohoming by complete reverse splicing. Curr. Biol., 9, 11–14. [DOI] [PubMed] [Google Scholar]
- 20.Toor N., Hausner,G. and Zimmerly,S. (2001) Coevolution of group II intron RNA structures with their intron-encoded reverse transcriptases. RNA, 7, 1142–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Toro N., Molina-Sánchez,M.D. and Fernández-López,M. (2002) Identification and characterization of bacterial class E group II introns. Gene, 299, 245–250. [DOI] [PubMed] [Google Scholar]
- 22.Ferat J.L., Le Gouar,M. and Michel,F. (2003) A group II intron has invaded the genus Azotobacter and is inserted within the termination codon of the essential groEL gene. Mol. Microbiol., 49, 1407–1423. [DOI] [PubMed] [Google Scholar]
- 23.Gorbalenya A.E. (1994) Self-splicing group I and group II introns encode homologous (putative) DNA endonucleases of a new family. Protein Sci., 3, 1117–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shub D.A., Goodrich-Blair,H. and Eddy,S.R. (1994) Amino acid sequence motif of group I endonucleases is conserved in open reading frames of group II introns. Trends Biochem. Sci., 19, 402–404. [DOI] [PubMed] [Google Scholar]
- 25.Aizawa Y., Xiang,Q., Lambowitz,A.M. and Pyle,A.M. (2003) The pathway for DNA recognition and RNA integration by a group II intron retrotransposon. Mol. Cell, 11, 795–805. [DOI] [PubMed] [Google Scholar]
- 26.Eskes R., Yang,J., Lambowitz,A.M. and Perlman,P.S. (1997) Mobility of yeast mitochondrial group II introns: engineering a new site specificity and retrohoming via full reverse splicing. Cell, 88, 865–874. [DOI] [PubMed] [Google Scholar]
- 27.Cousineau B., Smith,D., Lawrence-Cavanagh,S., Mueller,J.E., Yang,J., Mills,D., Manias,D., Dunny,G., Lambowitz,A.M. and Belfort,M. (1998) Retrohoming of a bacterial group II intron: mobility via complete reverse splicing, independent of homologous recombination. Cell, 94, 451–462. [DOI] [PubMed] [Google Scholar]
- 28.Eskes R., Liu,L., Ma,H., Chao,M.Y., Dickson,L., Lambowitz,A.M. and Perlman,P.S. (2000) Multiple homing pathways used by yeast mitochondrial group II introns. Mol. Cell. Biol., 20, 8432–8446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martínez-Abarca F., García-Rodríguez,F.M. and Toro,N. (2000) Homing of a bacterial group II intron with an intron-encoded protein lacking a recognizable endonuclease domain. Mol. Microbiol., 35, 1405–1412. [DOI] [PubMed] [Google Scholar]
- 30.San Filippo J. and Lambowitz,A.M. (2002) Characterization of the C-terminal DNA-binding/DNA endonuclease region of a group II intron-encoded reverse transcriptase. J. Mol. Biol., 324, 933–951. [DOI] [PubMed] [Google Scholar]
- 31.Guo H., Zimmerly,S., Perlman,P.S. and Lambowitz,A.M. (1997) Group II intron endonucleases use both RNA and protein subunits for recognition of specific sequences in double-stranded DNA. EMBO J., 16, 6835–6848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiménez-Zurdo J.I., García-Rodríguez,F.M., Barrientos-Durán,A. and Toro,N. (2003) DNA-target requirements for homing in vivo of a bacterial group II intron encoding a protein lacking the DNA endonuclease domain. J. Mol. Biol., 326, 413–423. [DOI] [PubMed] [Google Scholar]
- 33.Muñoz-Adelantado E., San Filippo,J., Martínez-Abarca,F., García-Rodríguez,F.M., Lambowitz,A.M. and Toro,N. (2003) Mobility of the Sinorhizobium meliloti group II intron RmInt1 occurs by reverse splicing into DNA, but requires an unknown reverse transcriptase priming mechanism. J. Mol. Biol., 327, 931–943. [DOI] [PubMed] [Google Scholar]
- 34.Zhong J. and Lambowitz,A.M. (2003) Group II intron mobility using nascent strands at DNA replication forks to prime reverse transcription. EMBO J., 22, 4555–4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Villadas P.J., Velázquez,E., Martínez-Molina,E. and Toro,N. (1995) Identification of nodule dominant Rhizobium meliloti strains carrying pRmeGR4b-type plasmid within indigenous soil populations by PCR using primers derived from specific DNA sequences. FEMS Microbiol. Ecol., 17, 161–168. [Google Scholar]
- 36.Robertsen B.K., Aiman,P., Darvill,A.G., McNeil,M. and Albershein,P. (1981) The structure of acidic extracellular polysaccharides secreted by Rhizobium leguminosarum and Rhizobium trifolii. Plant Physiol., 67, 389–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 38.Blatny J.M., Brautaset,T., Winther-Larsen,H.C., Haugan,K. and Valla,S. (1997) Construction and use of a versatile set of broad-host-range cloning and expression vectors based on the RK2 replicon. Appl. Environ. Microbiol., 63, 370–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marsh J.L., Erfle,M. and Wykes,E.J. (1984) The pIC plasmid and phage vectors with versatile cloning sites for recombinant selection by insertional inactivation. Gene, 32, 481–484. [DOI] [PubMed] [Google Scholar]
- 40.Macián F., Pérez-Roger,I. and Armengol,M.E. (1994) An improved vector system for constructing transcriptional lacZ fusions: analysis of regulation of the dnaA, dnaN, recF asnd gyrB genes of Escherichia coli. Gene, 145, 17–24. [DOI] [PubMed] [Google Scholar]
- 41.Clackson T., Güssow,D. and Jones,P.T. (1993) General applications of PCR to gene cloning and manipulation. In Mcpherson,M.J., Quirce,P. and Taylor,G.R. (eds), PCR A Practical Approach. Oxford University Press, Oxford, pp. 202–204. [Google Scholar]
- 42.Belfort M., Chandry,P.S. and Pedersen-Lane,J. (1987) Genetic delineation of functional components of the group I intron in the phage T4 td gene. Cold Spring Harbor Symp. Quant. Biol., 52, 181–192. [DOI] [PubMed] [Google Scholar]
- 43.Ditta G., Stanfield,S., Corbin,D. and Helinski,D.R. (1980) Broad host range DNA cloning system for Gram-negative bacteria. Construction of gene bank of Rhizobium meliloti. Proc. Natl Acad. Sci. USA, 77, 7347–7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Toro N., Martínez-Abarca,F., Fernández-Lopez,M. and Muñoz-Adelantado,E. (2003) Diversity of group II introns in the genome of Sinorhizobium meliloti strain 1021: splicing and mobility of RmInt1. Mol. Genet. Genomics, 268, 628–636. [DOI] [PubMed] [Google Scholar]
- 45.Knonieczny I. (2003) Strategies for helicase recruitment and loading in bacteria. EMBO Rep., 4, 37–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kovach M.E., Phillips,R.W., Elzer,P.H., Roop.,R.M.II. and Peterson,K.M. (1994) pBBR1MCS: a broad-host-range cloning vector. Biotechniques, 16, 800–802. [PubMed] [Google Scholar]
- 47.Antoine R. and Locht,C. (1992) Isolation and molecular characterization of a novel broad-host-range plasmid from Bordetella bronchiseptica with sequence similarities to plasmids from gram-positive organisms. Mol. Microbiol., 6, 1785–1799. [DOI] [PubMed] [Google Scholar]
- 48.Wang H. and Lambowitz,A.M. (1993) The Mauriceville plasmid reverse transcriptase can initiate cDNA synthesis de novo and may be related to reverse transcriptase and DNA polymerase progenitor. Cell, 75, 1071–1081. [DOI] [PubMed] [Google Scholar]