Abstract
The OECD has developed test guidelines (TG) to identify agents with genotoxic effects. The in vivo alkaline single cell gel electrophoresis (SCGE) assay is currently being prepared to become such a TG. The performance of a combined SCGE/Pig-a gene mutation study was evaluated with the prototypical genotoxicant benzo[a]pyrene (BaP) at an exposure level known to induce germ cell mutation. We aimed to better understand (i) the strengths and weaknesses of the two methods applied in blood and their potential to predict germ cell mutagenicity, and (ii) the involvement of reactive oxygen species (ROS) following in vivo BaP-exposure. To explore the involvement of ROS on BaP genotoxicity, we utilized a mouse model deficient in a DNA glycosylase. Specifically, C57BL/6 mice (Ogg1+/+ and Ogg1−/−) were treated for three consecutive days with 50 mg BaP/kg/day. DNA damage in nucleated blood cells was measured four hours after the last treatment with the SCGE assay, with and without Formamidopyrimidine DNA glycosylase (Fpg). Pig-a mutant phenotype blood erythrocytes were analysed two and four weeks after treatment. BaP-induced DNA lesions were not significantly increased in either version of the SCGE assay. The phenotypic mutation frequencies for immature and mature erythrocytes were significantly increased after two weeks. These effects were not affected by genotype, suggesting oxidative damage may have a minor role in BaP genotoxicity, at least in the acute exposure situation studied here. While both assays are promising tools for risk assessment, these results highlight the necessity of understanding the limitations regarding each assay’s ability to detect chemicals’ genotoxic potential.
Keywords: hazard identification, risk assessment, alkaline Single Cell Gel Electrophoresis (SCGE), Pig-a mutation assay, benzo[a]pyrene (BaP), genotoxicity
1 Introduction
Hazard identification of chemicals is a key component of risk assessment and includes evaluation of agent’s potential to cause germ or somatic cell mutation. This information can lead to abandonment of product development or in other circumstances is used to determine whether risk assessments should assume a linear relationship between dose and carcinogenicity, or whether non-linear model(s) would be more appropriate. The Organisation for Economic Co-operation and Development (OECD) has developed specific test guidelines (TG) for the evaluation of adverse effects for different DNA damage endpoints to be used in human risk assessment. Since test methods are continuously improving, they are constantly evaluated for their ability to complement and extend the current test batteries with the aim to achieve superior predictability, sensitivity, accuracy, feasibility and reduction of costs and use of experimental animals.
In 2012, a draft for an in vivo rodent alkaline Single Cell Gel Electrophoresis (SCGE) OECD TG was initiated which is now passed by the Working Group of the National Coordinators for the TG Programme and currently in the last stages of final OECD approval. The alkaline SCGE is a very sensitive assay detecting DNA lesions such as strand breaks, alkali labile sites (ALS), crosslinks and repair intermediates to predict potential mutagens [1, 2]. Combined with DNA repair enzymes, such as the Formamidopyrimidine DNA glycosylase (Fpg), a broader range of DNA lesions is detected, including oxidised purines such as 8-oxoguanine [3, 4]. This adaption allows the identification of oxidative DNA lesions.
The Pig-a mutation assay is a more recently developed method detecting phenotypic mutations [5] and was one of six topics discussed at the last International Workshop on Genotoxicity Testing (IWGT, November 2013). This phenotype-based assay is based on the cell surface expression of glycophosphatidylinositol-anchored proteins such as CD24 or CD59 and represents a very efficient platform for studying in vivo somatic cell mutations [6]. The main advantages of this assay are the low requirements for time, costs, blood sample volumes, its potential for multiple sampling, integration into existing OECD TG (subchronic and chronic studies) and the high responsiveness to mutagens [7]. A committee entitled “Relevance and Follow-up of Positive Results from In Vivo Genetic Toxicity Testing” (IVGT), initiated by the Health and Environmental Sciences Institute (HESI), recommends the Pig-a assay (in combination with the micronucleus assay (MN)) in repeat-dose toxicity studies “as it would allow for the first time the simple assessment of aneugenicity, clastogenicity, and mutagenicity” [8].
Combining in vivo Pig-a mutation analysis with the SCGE assay is also expected to have merit, especially given the latter’s ability to study multiple tissue compartments and different types of lesions through the use of enzymes such as Fpg. However, data describing combination of Pig-a and SCGE assays are sparse, and to our knowledge non-existent in mice. The experiments described herein sought to address this data gap by combining in vivo Pig-a assay as a phenotypic reporter of gene mutation with the SCGE method to detect primary DNA damage. From a scientific – but not regulatory – point of view, we aimed to better understand (i) the strengths and weaknesses of the two methods applied in blood in a regulatory perspective and their potential to predict germ cell mutagenicity, and (ii) the involvement of reactive oxygen species (ROS) for the genotoxicity of BaP following in vivo exposure.
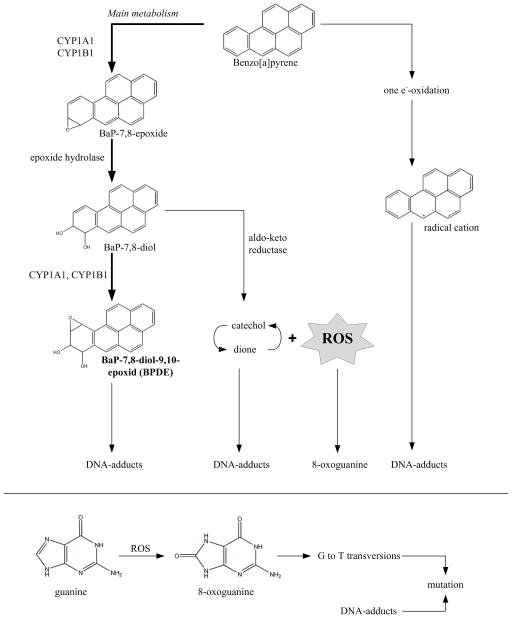
We chose to study the prototypical genotoxic carcinogen benzo[a]pyrene (BaP). We have shown previously that BaP causes germ cell mutagenicity [9] and we wanted to investigate if blood can be used as a proxy when using same dose levels. BaP is rapidly distributed to all tissues [10] and mainly metabolised by cytochrome P450s and epoxide hydrolase to BaP-7,8-diol (IARC, 2012) (Figure 1). This key metabolite is further oxidized to BaP-7,8-diol-9,10-epoxide (BPDE), the major metabolite causing carcinogenesis. Alternatively, BaP-7,8-diol can be oxidized by aldo-keto-reductases (AKRs) entering a futile cycle forming catechol and dione via an o-quinone [11] with concomitant ROS generation. BPDE, catechol and dione intermediates can form DNA adducts inducing G→T transversions, the signature mutation of BaP exposure as shown for CHO cells [12]. In addition, the futile cycle will produce ROS [13] as a side product of the redox reaction [14] (Figure 1). ROS will lead to induction of oxidative DNA lesions such as the pre-mutagenic 8-oxoguanine which, similar to BPDE, leads to G→T transversions [15]. Despite this knowledge, it is still unknown to what extent ROS is induced by BaP-exposure and whether ROS contributes to the mutagenicity of BaP in vivo [13].
Figure 1. Metabolism of Benzo[a]pyrene (BaP).
The metabolism primarily occurs via cytochrome P450s (CYP1A1 and CYP1B1), peroxidases (epoxide hydrolase) and aldo-keto reductase. The major metabolite which is thought to cause carcinogenesis is BaP-7,8-diol-9,10-epoxide (BPDE) which is metabolised via BaP-7,8-epoxide and BaP-7,8-diol. BPDE forms DNA-adducts which can induce G→T transversions. In addition, many metabolites (such as catechol and dione within a futile cycle) form reactive oxygen species (ROS) thus adding to oxidative stress and contributing to the formation of oxidative DNA lesions such as 8-oxoguanine, a pre-mutagenic DNA lesion. Radical cations are formed in a minor quantity.
Mice were treated with a dose of BaP that we have previously shown to be mutagenic to mouse germ cells [9]. Consistent with 3Rs principles, harvest and analysis of blood cells was timed to accommodate SCGE and Pig-a endpoints. Furthermore, to evaluate the extent to which radical oxygen species (ROS) are responsible for BaP-associated genotoxicity, the SCGE assay was conducted with Fpg, and mice with and without deficiencies in the repair of 8-oxoguanine (Ogg1−/− versus Ogg+/−) were used. The resulting data are discussed in terms of the merits and limitations of combining disparate genotoxicity endpoints in blood, and also the extent to which ROS is involved in DNA damage caused by acute BaP exposure.
2 Materials and Methods
2.1 Reagents
Benzo[a]pyrene (BaP, CAS 50-32-8, cat.no. B1760), corn oil (CAS 8001-30-7, cat. No. C8267), and N-ethyl-N-nitrosourea (ENU, CAS 759-73-9, cat.no. N3385) were purchased from Sigma Aldrich Norway AS, Oslo, Norway. Lympholyte®-Mammal cell separation reagent was purchased from CedarLane, Burlington, NC, USA. Anti-PE MicroBeads, LS+ Positive Selection Columns and QuadroMACS™ Separator were purchased from Miltenyi Biotec GmbH, Bergisch Gladbach, Germany. CountBright™ Absolute Counting Beads were from Invitrogen, Life Technologies™, Carlsbad, CA, USA. Heat-inactivated fetal bovine serum (FCS) was from PAA Laboratories, Pasching, Austria. Anticoagulant Solution, Buffered Salt Solution, Nucleic Acid Dye Solution (SYTO®13), Anti-CD24-PE and Anti-CD61-PE were from the Prototype In Vivo Mouse MutaFlow® Kit, Litron Laboratories, Rochester, NY, USA. Low melting point agarose (NuSieve®GTG®Agarose) and Gelbond® films were purchased from Lonza, Rockland, ME, USA. SYBR®Gold Nucleic Acid Gel Stain (10,000 × concentrate in DMSO) was from Life Technologies™, Carlsbad, CA, USA.
2.2 Animals, treatments, and blood harvest
Wild type C57BL/6 mice (Ogg1+/+) with BigBlue™ background were originally purchased from Stratagene (La Jolla, CA, USA). Ogg1 gene knockout mice (deficient of the mouse 8-oxoguanine-DNA-Glycosylase, Ogg1−/−, mixed BL6/SV129) [16] were generously provided by University of Oslo, Norway. Mice were back-crossed in-house with Ogg1+/+ BigBlue™ C57BL/6 mice for nine generations, to achieve isogenic strains with similar genetic background (C57BL/6). Homozygote genotypes (Ogg1+/+ and Ogg1−/−) were bred in-house. Animals were kept at a 12 h light/dark circle, controlled temperature (20 – 24 °C) and humidity (55 ± 10%), following the European convention (2006), appendix A [17]. Up to three mice were kept in IVC plastic cages (Innovive, San Diego, CA, USA) with Nestpack bedding (Datesand Ltd., Manchester, UK). Water (tap water) and food (SDS RM1, Special Diet Services, Essex, UK) were available ad libitum. The experiments were approved by the National Experimental Animal Board of Norway. A total of 26 animals were used in this study. Rodents, 10 – 11 weeks old at start of treatment, had a mean weight of 22.4 ± 1.9 g.
Mice were randomly assigned to treatment groups: BaP (8 mice per genotype), or corn oil (control group, 3 mice per genotype). BaP stock solution in corn oil (7.5 mg/ml) was freshly made every day. Mice were ip injected on three consecutive days (day 0, 1 and 2) with 50 mg/kg bw/day (injection volume for BaP varied between 120 – 170 μl, while the injection volume of vehicle corn oil was 160 μl), giving a total dose of 150 mg BaP/kg bw. The dose was chosen in accordance to previous experiments were BaP induced mutations in liver and male germ cells [9]. ENU was used as positive control and the stock solution was freshly prepared for each treatment: four wild type mice were ip injected with 22 mg/kg/day on three consecutive days with ENU or NaCl (vehicle for ENU, negative control), respectively, giving a total dose of 66 mg ENU/kg bw. The injection volume for ENU/NaCl was 200 μl. One mouse died during the last injection (NaCl).
For the Pig-a assay, blood samples were taken one week prior to treatment (day −5), and at day 16 and day 34 from the saphenous vein using a 21-G needle and heparinised capillary tubes (Fisherbrand®, Pittsburgh, PA, USA). For SCGE, blood was drawn four hours after the last injection (day 2) as suggested in the proposed OECD test guideline for the rodent alkaline SCGE in 2012 [18]. The study design is shown in Figure 2. 60 μl (Pig-a) or 30 μl (SCGE) of free-flowing blood was added to 100 μl anticoagulant (supplied in the Prototype In Vivo Mouse MutaFlow® Kit) and mixed well. Diluted samples for SCGE were kept on ice and processed immediately while Pig-a samples were kept at room temperature and processed within two hours.
Figure 2. Study design.

Eight C57BL/6 mice of two different genotypes (Ogg1+/+ and Ogg1−/−) were treated with BaP on three consecutive days (total of 150 mg/kg bw, ip). In addition, three mice of each genotype were injected with corn oil as vehicel control. Blood for the Pig-a mutation assay was drawn one week prior BaP-treatment (day −5), and two weeks (day 16) and four weeks (day 34) after the last chemical treatment. Blood for the SCGE was drawn four hours after the last injections of BaP or vehicle control.
2.3 Single Cell Gel Electrophoresis (SCGE)
The high throughput SCGE was established in our lab as described [19] and was performed with minor modifications.
The homogenous blood/anticoagulant suspension (130 μl) was diluted 1:10 in 0.75% low melting point agarose at 37 °C; triplicates (3 × 4 μl) from each individual were immediately applied on a cold GelBond® film. Lysis was performed overnight in lysis buffer (2.5 M NaCl, 0.1 M Na2EDTA, 0.01 M Tris, 0.2 M NaOH, 0.038 M N-Laurylsarcosine, 10% DMSO, 1% TritonX-100, pH10). Films were transferred to enzyme buffer (0.04 M Hepes, 0.1 M KCl, 0.5 mM Na2EDTA, pH7.6) for 60 min at 4 °C. For enzyme treatment, 0.02 μg/ml Formamidopyrimidine DNA glycosylase (crude Fpg extract, a bacterial homologue to the eukaryotic Ogg1 detecting a wide range of oxidative DNA lesions [20]), and 0.2 mg/ml BSA were added to fresh pre-warmed (37 °C) enzyme buffer. The Fpg concentration was optimized based on previous titration experiments with a photoactivated drug (Ro12-9786) as recommended [21]. Films were incubated in this buffer (+/− Fpg) for 60 min at 37 °C. For unwinding, the films were immersed in electrophoresis solution (0.3 M NaOH, 0.001 M Na2EDTA, pH > 13) for 40 min, followed by 20 min alkaline electrophoresis at 8 °C, 25 V and 0.8 V/cm over the platform in fresh electrophoresis solution, with circulation as described [19]. After electrophoresis, films were washed in water (1 min) and in neutralisation solution (0.322 M Tris-HCl, 0.078 M Tris-base, pH7.5) for 2 × 5 min, and dried after soaking in ethanol (5 min in 70% and 90 min in 96% EtOH). Prior to analysis, cells were stained by immersing the films in SYBR® Gold Nucleic Acid Gel Stain in TE-buffer (1 mM Na2EDTA, 10 mM Tris-HCl, pH8). Fifty randomly chosen cells per replicate were scored using 20 × magnification in an Olympus BX51 microscope (light source: Olympus BH2-RFL-T3, Olympus optical Co., Ltd.; camera: A312f-VIS, BASLER, Ahrensburg, Germany) with the Comet IV analysis software (Perceptive Instruments Ltd., Bury St. Edmunds, UK).
2.4 Pig-a mutation assay
The Pig-a gene mutation assay was performed as described in the Prototype Mouse MutaFlow® Kit instruction manual. Blood/anticoagulant mixture (160 μl) was added to 3 ml Lympholyte®-Mammal and centrifuged (20 min, 800 × g, RT) to isolate leuko- and platelet-depleted red blood cells (RBC) and reticulocytes (RET, immature red blood cells). After rinsing the pellet carefully three times with PBS, RBCs were incubated with anti-CD24-PE (labelling wild type (wt) erythrocytes)/anti-CD61-PE (labelling platelets) and anti-PE magnetic particles (binding to the wt-erythrocytes and platelets). Incubations lasted for 20 min and were separated by washing/centrifugation steps. A small fraction of each sample was taken and added to a Nucleic Acid Dye/Counting Bead Solution and incubated at 37 °C for 30 min. This “pre-column” sample provides information about the cell-to-bead-ratio. The majority of the sample was transferred to a magnetic column (Miltenyi Biotec) to selectively remove CD24 cells and CD61 positive platelets. The eluate (CD24-negative cells, i.e. RBCCD24- and RETCD24-) was concentrated in a further spinning step, washed and subsequently incubated with 300 μl Nucleic Acid Dye/Counting Bead Solution at 37 °C for 30 min. This so-called “post-column” sample gives information about the mutant phenotype cell-to-bead-ratio. Cells were analysed using a flow cytometer (BD LSRII, FACSDiva Software, BD Bioscience, San Jose, CA, USA). The ratios from pre- and post-columns are used to calculate the mutation frequency as described previously for rats [22]. The total numbers of reticulocyte and erythrocyte equivalents are derived from the cell-to-bead ratios in the pre-column sample and the number of counting beads observed in the post-column sample. At least 100 × 106 RBCs and 3 × 106 RETs per sample were recorded.
It should be noted that the Prototype MutaFlow® protocol as used herein (v120301) has been modified since this study was completed. Since that time, post-column samples are no longer incubated at 37 °C for 30 min, a step that is now known to lyse mouse erythrocytes, especially RNA-negative erythrocytes. Thus, the reliability of the RBCCD24- values presented herein likely underestimate mutagen-induced effects.
2.5 Statistical Analysis
The raw data of the SCGE were processed using the Comet Assay Spreadsheet Generator Version 1.3.1 (Perceptive Instruments Ltd., Bury St. Edmunds, UK). Subsequently, the % tail intensity (TI, i.e. % DNA in tail) per individual was calculated as suggested by Bright and coworkers [23]: 50 comets of one replicate were summarized as median % TI, and three technical replicates were summarized as mean % TI, giving one data point for each animal.
The raw data of the Pig-a assay was processed and calculated as described by Dertinger and co-workers [24] using Microsoft Excel 2010. Prior to statistical analysis the mutant phenotype cell frequencies of RBCCD24- and RETCD24- were transformed. An offset of 0.1 was added to all RETCD24- values to allow log10-transformation.
Statistical analysis was performed using SigmaPlot 12.0 (Systat Software, San Jose, CA, USA) for both Pig-a data and SCGE data. To find significant differences between different time points (day 16 and day 34 compared to control day −5) as well as BaP treatment (compared to vehicle corn oil) a multiple comparison (Dunnett’s t-test) within a one-way analysis of variance (ANOVA) was performed. Significance was evaluated at a 5% level (p < 0.05).
3 Results
3.1 Single Cell Gel Electrophoresis (SCGE)
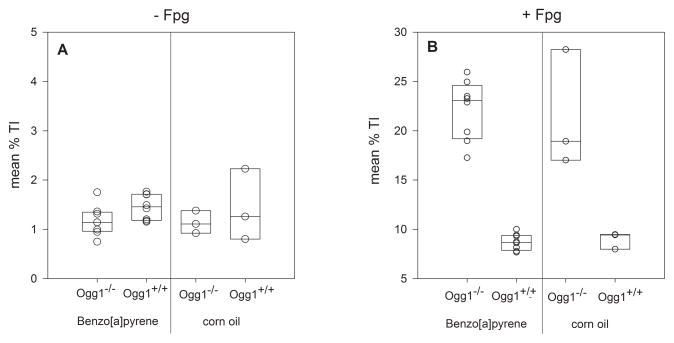
Neither treatment (BaP or corn oil) or genotype (Ogg1−/− or Ogg1+/+) was associated with significant increases in DNA lesions (strand breaks and ALS) in the alkaline SCGE in peripheral blood leukocytes four hours after last treatment (Figure 3A, Table I).
Figure 3. DNA lesions in mouse blood cells measured by the alkaline SCGE without (A) and with Fpg (B) four hours after treatment with BaP.
Each circle represents data for one mouse as mean (n = 8 for BaP, n = 3 for corn oil), based on three technical replicates per mouse. Mouse genotypes and treatments are indicated on the horizontal axis. The box plot shows the median and the first and third quartile.
Table I.
Results of the alkaline SCGE without and with Fpg-treatment
| [% TI]a
|
||||||
|---|---|---|---|---|---|---|
| Enzyme treatment | genotype | n | treatment | mean | median | SD |
| none | Ogg1−/− | 8 | BaP | 1.17 | 1.14 | 0.31 |
| Ogg1+/+ | 8 | BaP | 1.45 | 1.45 | 0.26 | |
| Ogg1−/− | 3 | corn oil | 1.14 | 1.11 | 0.23 | |
| Ogg1+/+ | 3 | corn oil | 1.43 | 1.26 | 0.73 | |
| Ogg1+/+ | 2 | ENUb | 17.14 | |||
| Ogg1+/+ | 1 | NaClc | 1.65 | |||
|
| ||||||
| Fpg | Ogg1−/− | 8 | BaP | 22.08 | 23.08 | 3.05 |
| Ogg1+/+ | 8 | BaP | 8.71 | 8.68 | 0.83 | |
| Ogg1−/− | 3 | corn oil | 21.39 | 18.93 | 5.99 | |
| Ogg1+/+ | 3 | corn oil | 8.96 | 9.42 | 0.85 | |
| Ogg1+/+ | 2 | ENUb | 37.98 | |||
| Ogg1+/+ | 1 | NaClc | 8.84 | |||
Mean, median and standard deviation (SD) is given for each group of eight (BaP) and three (corn oil) mice, respectively.
Positive control
Negative control (vehicle of ENU)
When including the repair-enzyme Fpg in the SCGE to reveal oxidative DNA lesions, more than a 2-fold increase of % TI was observed in Ogg1−/− mice compared to Ogg1+/+ (Figure 3B, Table I). The increased level of oxidative DNA lesions to 20% TI with Fpg in Ogg1−/− mice is in agreement with previous experiments in our laboratory [25] and was independent of the treatment (BaP or corn oil). The median % TI data was slightly higher in Ogg1−/− mice treated with BaP compared to Ogg1−/− mice treated with corn oil (23.08% TI and 18.93% TI), but this difference was not statistically significant (Table I). Consequently, BaP did not cause significantly increased levels of DNA strand breaks, alkali labile sites or oxidative DNA lesions in peripheral blood leukocytes compared to corn oil, at four hours after the last treatment in either Ogg1−/− or in Ogg1+/+ mice.
3.2 Pig-a mutation assay
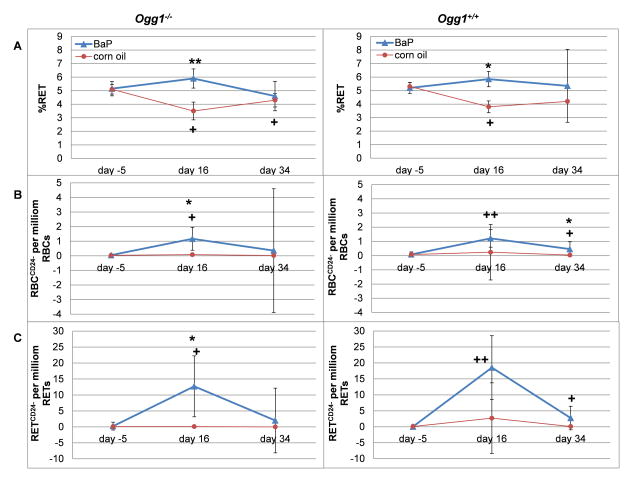
The frequency of mutant phenotype reticulocytes and erythrocytes (RETCD24- and RBCCD24-, respectively) was measured to examine the genotoxic effects of BaP (Figure 4). In general, BaP exposure resulted in increased mean frequencies of mutant phenotype cells at all post-exposure time points with a peak at day 16. This was observed for both Ogg1−/− and Ogg1+/+ genotypes.
Figure 4. Pig-a mutant frequencies and percent reticulocytes in blood cells of mice treated with BaP.
Data is given as median (SD) for the percentage of reticulocytes (A), and the mutation frequencies of red blood cells (B) and reticulocytes (C). Data was compared to each other by Dunnett’s multiple comparison within a one way ANOVA. Significant differences to the vehicle control (corn oil) of the same genotype and sampling day are indicated by asterisks: p ≤ 0.05 (*) and p ≤ 0.001 (**). Significant differences between day 16 and day 34, respectively, to day −5 are marked with crosses: p ≤ 0.05 (+) and p ≤ 0.001 (++).
In RETCD24- the increase at day 16 was statistically significant compared to pre-treatment (p = 0.002 and p < 0.001 for Ogg1−/− and Ogg1+/+ mice, respectively) (Figure 4C, Table II). At day 34 only Ogg1+/+ mice showed a significant increase in RETCD24- relative to the untreated group (p = 0.004).
Table II.
Results of the Pig-a assay
| %RET
| |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| genotype | treatment | n | day −5
|
n | day 16
|
n | day 34
|
||||||||||
| mean | median | SD | mean | median | SD | p-value vehiclec | p-value day −5d | mean | median | SD | p-value vehiclec | p-value day −5d | |||||
| Ogg1−/− | BaP | 8 | 5.29 | 5.15 | 0.53 | 8 | 6.04 | 5.90 | 0.71 | <0.001 | 7 | 4.99 | 4.60 | 1.09 | |||
| Ogg1+/+ | BaP | 8 | 5.19 | 5.20 | 0.41 | 8 | 5.75 | 5.85 | 0.57 | 0.001 | 6 | 6.08 | 5.35 | 2.70 | |||
| Ogg1−/− | corn oil | 3 | 5.07 | 5.10 | 0.35 | 3 | 3.60 | 3.50 | 0.66 | 0.023 | 3 | 4.30 | 4.30 | 0.50 | 0.006 | ||
| Ogg1+/+ | corn oil | 3 | 5.37 | 5.30 | 0.31 | 3 | 4.00 | 3.80 | 0.44 | 0.003 | 3 | 4.17 | 4.20 | 0.06 | |||
| Ogg1+/+ | ENUa | 3 | 4.25 | 2 | 7.40 | 2 | 4.65 | ||||||||||
| Ogg1+/+ | NaClb | 2 | 5.05 | 1 | 3.80 | 1 | 3.60 | ||||||||||
| mutation frequency (RBCCD24-) per million RBCs
| |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| genotype | treatment | n | day −5
|
n | day 16
|
n | day 34
|
||||||||||
| mean | median | SD | mean | median | SD | p-value vehiclec | p-value day −5d | mean | median | SD | p-value vehiclec | p-value day −5d | |||||
| Ogg1−/− | BaP | 4 | 0.08 | 0.05 | 0.09 | 8 | 1.25 | 1.17 | 0.79 | <0.001 | 0.003 | 8 | 2.14 | 0.36 | 4.24 | ||
| Ogg1+/+ | BaP | 8 | 0.13 | 0.09 | 0.18 | 8 | 1.14 | 1.22 | 0.62 | <0.001 | 8 | 0.59 | 0.46 | 0.51 | 0.004 | 0.005 | |
| Ogg1−/− | corn oil | 2 | 0.03 | 0.03 | 0.01 | 3 | 0.09 | 0.08 | 0.04 | 3 | 0.02 | 0.02 | 0.02 | ||||
| Ogg1+/+ | corn oil | 3 | 0.10 | 0.08 | 0.07 | 3 | 1.26 | 0.24 | 1.95 | 3 | 0.03 | 0.04 | 0.01 | ||||
| Ogg1+/+ | ENUa | 2 | 0.14 | 2 | 13.39 | 2 | 3.64 | ||||||||||
| Ogg1+/+ | NaClb | 2 | 0.08 | 1 | 0.09 | 1 | 0.03 | ||||||||||
| mutation frequency (RETCD24-) per million RETs
| |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| genotype | treatment | n | day −5
|
n | day 16
|
n | day 34
|
||||||||||
| mean | median | SD | mean | median | SD | p-value vehiclec | p-value day −5d | mean | median | SD | p-value vehiclec | p-value day −5d | |||||
| Ogg1−/− | BaP | 4 | 0.78 | 0.25 | 1.22 | 8 | 13.64 | 12.70 | 9.56 | <0.001 | 0.002 | 8 | 5.76 | 2.00 | 10.14 | ||
| Ogg1+/+ | BaP | 8 | 0.29 | 0.05 | 0.58 | 8 | 17.43 | 18.55 | 10.00 | <0.001 | 8 | 3.50 | 2.75 | 3.70 | 0.004 | ||
| Ogg1−/− | corn oil | 2 | 0.10 | 0.10 | 0.00 | 3 | 0.27 | 0.10 | 0.29 | 3 | 0.13 | 0.00 | 0.23 | ||||
| Ogg1+/+ | corn oil | 3 | 0.17 | 0.10 | 0.21 | 3 | 7.70 | 2.70 | 11.08 | 3 | 0.27 | 0.10 | 0.38 | ||||
| Ogg1+/+ | ENUa | 2 | 0.40 | 2 | 125.80 | 2 | 66.10 | ||||||||||
| Ogg1+/+ | NaClb | 2 | 0.10 | 1 | 0.00 | 1 | 0.10 | ||||||||||
Note. Data is based on ≥ 3 million RETs and ≥ 100 million RBCs equivalents per mouse
Positive control
Negative control (vehicle of ENU)
Significant difference between BaP treatment compared to vehicle (corn oil) of the same day (one way ANOVA, Dunnett’s t-test)
Significant difference between treatment on day 16 (and day 34) compared to pre-treatment (day −5) (one way ANOVA, Dunnett’s t-test)
RBCCD24- frequencies confirmed the observations from the population of RETs with significantly increased frequency of mutant phenotype cells at day 16 (p = 0.003 and p < 0.001 for Ogg1−/− and Ogg1+/+, respectively, compared to day −5). This effect was also observed at day 34 in Ogg1+/+ mice (p = 0.005) (Figure 4B, Table II). Ogg1−/− mice, on the other hand, did not show a significantly increased mutation frequency at day 34 (compared to day −5).
Treatment with corn oil had no significant impact on RETCD24- or RBCCD24-frequencies.
One Ogg1+/+ mouse gave a relatively high RETCD24- frequency at day 16 that was not recapitulated at the later time point (day 34) (Table II). Since there was no obvious technical reason to exclude this individual data point, data are presented as median in order to provide a better overall indication of group values (Figure 4).
The percentage of RETs (% RET) is the ratio of newly formed RNA-positive erythrocytes relative to all erythrocytes, and is used as a measure of bone marrow cytotoxicity. BaP treated mice exhibited increased % RET at day 16, which thereafter decreased at day 34 (Figure 4A, Table II). Relative to pre-dose blood samples, % RET in mice treated with corn oil was initially reduced at the first time point post-treatment (day 16), whereas they returned to pre-dose levels at day 34.
In general, Ogg1−/− was not statistically different from Ogg1+/+ regarding the time course of mutation frequency.
4 Discussion
In this work we focused on two genotoxic methods to measure mutagenicity in somatic cells. We used (i) the conventional alkaline SCGE (as proposed in the OECD TG, i.e. without Fpg, as well as with Fpg to quantify oxidative DNA lesions) and (ii) the more recently developed Pig-a mutation assay. We wanted to evaluate the merits and limitations of these assays when studying the mutagenic potential of chemicals such as BaP in blood. Also, by choosing a dose that has previously been shown to mutate germ cells, we wanted to assess the ability of hematopoietic cells to serve as a surrogate for germ cell effects.
From the perspective of hazard identification, the alkaline SCGE is increasingly used as an in vivo follow-up test to obtain supportive evidence of an agent’s genotoxic potential after positive in vitro genotoxicity tests. The SCGE detects DNA lesions (single and double strand breaks) as well as ALS, i.e. potentially pre-mutagenic DNA lesions. The Pig-a assay on the other hand is a phenotypic reporter of gene mutation. BaP was used herein as a reference mutagenic agent known to be a germ cell mutagen [26]. BaP is rapidly distributed to all tissues and metabolised in blood, liver, skin and kidney with t1/2 of 4.2 – 6.1 hours in rats [10]. Metabolites of BaP such as BPDE form bulky DNA adducts [27] and are considered to be highly carcinogenic [28]. The level of DNA adducts increases between three and twelve hours in human hepatoma cells [29], and are also measurable in mouse liver four days after exposure [9]. Further, BPDE DNA adducts are also induced in the bone marrow [30] (28 day repeat dose study).
Despite the high sensitivity of SCGE at low doses [19], BaP was not observed to affect this endpoint in blood cells (single strand breaks and ALS). This is in contrast to previous findings in a germ cell mutation assay using the same dose level [9]. Our negative SCGE results in leukocytes are in agreement with other reports that examined even higher dose levels of BaP [31]. The SCGE results in blood are likely explained by the bulky nature of BPDE DNA adducts which are repaired via nucleotide excision repair (NER). It is known that such lesions are generally not detected by SCGE while strand breaks which occur as repair intermediates during the NER process are measurable; they exist during short periods and at very low levels.
On the other hand, the Pig-a assay did record an increased frequency of mutant phenotype cells two weeks after BaP treatment. Both RBCCD24- and RETCD24- were increased confirming that BaP was distributed to the bone marrow. This is in accordance with previous BaP studies using Pig-a [30, 32, 33] and other mutation assays (lacZ mutation assay [30], cII mutation assay [9]). Somewhat surprisingly, RBCCD24- frequencies did not increase at day 34 as it would have been expected after the increased RETCD24- frequency was noted two weeks prior to this. One possible explanation is that acute BaP exposure affected a narrow population of erythrocyte precursors, and this short-duration effect led to a modest increase in RBCCD24-. Alternatively, as explained above, a simpler and perhaps more likely explanation is that the alpha version of the Mouse MutaFlow Pig-a Kit underestimated RBCCD24-frequencies, and this reduced the sensitivity of the endpoint. In general, the mutation frequencies for BaP were low compared to the very potent ENU used as positive control (Table II), confirming the sensitivity of the Pig-a mutation assay. The ratio of reticulocytes (%RET) is also slightly increased after BaP treatment, indicating that BaP either leads selectively to cytotoxicity of RBCs or that BaP stimulates cell division in the bone marrow. It has been shown that BaP stimulates cell proliferation [32, 33]. Repeated bleeding of a small blood volume is not expected to influence haematopoiesis and hence, does not appreciably influence the frequency of Pig-a mutant phenotype cells (personal communication, B. Heflich).
As noted above, in order to specifically investigate the contribution of ROS to BaP-induced genotoxicity, we designed the current study with non-standard elements. Firstly, Fpg was included in the alkaline SCGE protocol to examine the ability of BaP to induce oxidative DNA lesions caused by the side products within the BaP metabolism [13] (Figure 1). Secondly, the repair deficient Ogg1−/− mouse model was used. These mice potentially accumulate oxidative DNA lesions since their repair enzyme recognizing 8-oxoguanine (Ogg1) is non-functional. In fact, a slight increase in oxidative lesions was measured in BaP treated Ogg1−/− mice compared to corn oil treated Ogg1−/− mice (Figure 3B, Table I). However, this increase was insignificant. ROS seem therefore to have a minor role in causing oxidative DNA lesions at this time point (four hours after last exposure).
Overall, it appears that BaP-induced DNA lesions and mutant phenotype cells may be independent of Ogg1. This is in contrast to results from in vitro studies which do show an increased formation of oxidative lesions and DNA strand breaks after exposure to BaP [34]. Reasons for the lack of detectable oxidative DNA lesions in our study could be that radicals and ROS are produced in minor quantities, that induced lesions are repaired via alternative back-up DNA repair systems other than Ogg1, or that the recommended sampling time point (taking from the OECD TG draft) is premature with respect to ROS generation within the BaP metabolism. Another possibility relates to the fact that in vitro systems are highly oxygenated (approximately 21%) compared to in vivo systems (2 – 13%), and this difference could potentially explain the disparate results. Finally, the natural antioxidant defence mechanisms present in vivo may not fully recapitulated in vitro.
For regulatory purposes it is important to understand the limitations of a test assay. The alkaline SCGE may lead to false negative results when an agent gives rise to bulky DNA lesions or alkali-stabile base modifications as suggested in this study. Whereas the Pig-a assay is currently limited to blood cells, the SCGE assay is theoretically applicable in any tissue from which it is possible to isolate single cells or nuclei, including reproductive tissues such as testicle and enables thus direct assessment of potential germ cell mutagens [25, 35]. Therefore, experiments must be designed according to the assumed type(s) and tissue locations of chemicals’ genotoxic effects and interpreted with great care. The use of test batteries is a practical solution because it allows one to consider multiple endpoints and tissue compartments.
The Pig-a assay appears to have great potential since it reveals chemicals that cause gene mutation whereas the SCGE detects pre-mutagenic DNA lesions across tissues of interest. Both assays provide many practical advantages: only minute volumes (30 μl and 60 μl for SCGE and Pig-a mutation assay, respectively) of blood are necessary. Blood is one of few biological samples that can be taken repeatedly without sacrificing animals, thus one reduces the amount of animals following the 3Rs-concept. Hence, the development of mutagenicity can be followed over time, allowing the implementation of both assays in subchronic and chronic studies. Sampling can be done prior to evident toxicity, allowing results from animals which are otherwise not considered useful due to systemic toxicity. To measure mutagenicity in very small blood volumes also opens the possibility to use blood cells as biomarker for exposure for genotoxic substances [36]. The Pig-a mutation assay is discussed to be included in already adapted repeated dose studies such as the repeated dose 28-day oral toxicity study in rodents (OECD 407) [32, 37] since RETCD24- and RBCCD24-accumulate with repeated dosing. A cross-species potential (including humans) is clear for the SCGE and has also been shown for the Pig-a mutation assay [38–40]. Both assays give results shortly after exposure to mutagenic agents, compared to traditional endpoints such as carcinogenicity which takes years in rodents and decades in humans to develop. Most importantly, both are in vivo assays, therefore not only allowing the investigation of effects of the chemical and its metabolites, but also the identification of DNA repair pathways for a compound. Furthermore, the assays are easily transferable between laboratories [41]. Other OECD TGs that are used to evaluate in vivo genotoxicity of chemicals are the Micronuclei assay (OECD TG 474) and the in vivo transgenic rodent (TGR) cell gene mutation assay (OECD TG 488). As with the SCGE, the TGR can be applied to any tissue. However, such experiments require transgenic rodents carrying reporter genes, whereas the Pig-a assay can be performed in any rodent. The MN assay identifies aneugens and clastogens but not subtler yet still important gene mutation effects. Thus, a combination of Pig-a and MN assay would appear to represent a powerful battery as pointed out previously [5] and its inclusion in future experiments similar to this study should be considered. The SCGE, on the other hand, is very sensitive even at very low doses [19] and reaches the highest scores with respect to sensitivity and specificity for predicting known carcinogens in rodents compared to the MN and TGR assays [36].
5 Conclusion
Taken together, the data presented herein reinforce the view that combination and integrated study designs represent important developments that address the 3Rs. SCGE/Pig-a is one such combination that appears to have merit, both for its ability to cover different types of DNA damage and tissue compartments. As demonstrated here, the use of enzymes such as Fpg as well as the availability of rodents with specific genotypes can contribute to the elucidation of specific modes of genotoxic action. On a cautionary note, it is also clear from these data that the choice and combination of in vivo methods should be done on basis of the agent’s chemical nature, and that a detailed understanding of the applicability of each assay as well as its restrictions is vital in order to interpret data correctly.
ip injection of C57BL/6 male mice on three consecutive days with 50 mg BaP/kg bw
BaP does not trigger ALS, SSB or oxidative DNA lesions in the SCGE
BaP induce a significant increase of the Pig-a mutation frequency
SCGE and Pig-a mutation assay are useful tools in risk assessment
Acknowledgments
Ogg1 gene knockout mice were generously provided by Dr. A. Klungland, University of Oslo, Oslo University Hospital, Oslo, Norway. The study was supported by the Norwegian Research Council (grant no. 190794) and partly funded by a grant from the National Institute of Health/National Institute of Environmental Health Sciences (NIEHS; grant no. R44ES018017).
ABBREVIATIONS
- Fpg
Formamidopyrimidine
- DNA
glycosylase
- OECD
Organisation for Economic Cooperation and Development
- TG
test guideline
- MN
micronucleus
- ENU
N-ethyl-N-nitrosourea
- RBC
red blood cells
- RET
reticulocytes
- NER
nucleotide excision repair
- Ogg1
8-oxoguanine
Footnotes
The contents are solely the responsibility of the authors, and do not necessarily represent the official views of the NIEHS.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kirkland D, Speit G. Evaluation of the ability of a battery of three in vitro genotoxicity tests to discriminate rodent carcinogens and non-carcinogens III. Appropriate follow-up testing in vivo. Mutat Res. 2008;654:114–132. doi: 10.1016/j.mrgentox.2008.05.002. [DOI] [PubMed] [Google Scholar]
- 2.Singh NP, McCoy MT, Tice RR, Schneider EL. A simple technique for quantitation of low levels of DNA damage in individual cells. Exp Cell Res. 1988;175:184–191. doi: 10.1016/0014-4827(88)90265-0. [DOI] [PubMed] [Google Scholar]
- 3.Boiteux S, Gajewski E, Laval J, Dizdaroglu M. Substrate specificity of the Escherichia coli Fpg protein (formamidopyrimidine-DNA glycosylase): excision of purine lesions in DNA produced by ionizing radiation or photosensitization. Biochemistry. 1992;31:106–110. doi: 10.1021/bi00116a016. [DOI] [PubMed] [Google Scholar]
- 4.Tchou J, Kasai H, Shibutani S, Chung MH, Laval J, Grollman AP, Nishimura S. 8-oxoguanine (8-hydroxyguanine) DNA glycosylase and its substrate specificity. Proc Natl Acad Sci U S A. 1991;88:4690–4694. doi: 10.1073/pnas.88.11.4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dobrovolsky VN, Miura D, Heflich RH, Dertinger SD. The in vivo Pig-a gene mutation assay, a potential tool for regulatory safety assessment. Environmental and molecular mutagenesis. 2010;51:825–835. doi: 10.1002/em.20627. [DOI] [PubMed] [Google Scholar]
- 6.Araten DJ, Nafa K, Pakdeesuwan K, Luzzatto L. Clonal populations of hematopoietic cells with paroxysmal nocturnal hemoglobinuria genotype and phenotype are present in normal individuals. Proc Natl Acad Sci U S A. 1999;96:5209–5214. doi: 10.1073/pnas.96.9.5209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dertinger SD, Heflich RH. In vivo assessment of Pig-a gene mutation-recent developments and assay validation. Environ Mol Mutagen. 2011;52:681–684. doi: 10.1002/em.20685. [DOI] [PubMed] [Google Scholar]
- 8.Schuler M, Gollapudi BB, Thybaud V, Kim JH. Need and potential value of the Pig-ain vivo mutation assay-a HESI perspective. Environmental and molecular mutagenesis. 2011;52:685–689. doi: 10.1002/em.20687. [DOI] [PubMed] [Google Scholar]
- 9.Olsen AK, Andreassen A, Singh R, Wiger R, Duale N, Farmer PB, Brunborg G. Environmental exposure of the mouse germ line: DNA adducts in spermatozoa and formation of de novo mutations during spermatogenesis. PLoS One. 2010;5:e11349. doi: 10.1371/journal.pone.0011349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marie C, Bouchard M, Heredia-Ortiz R, Viau C, Maitre A. A toxicokinetic study to elucidate 3-hydroxybenzo(a)pyrene atypical urinary excretion profile following intravenous injection of benzo(a)pyrene in rats. J Appl Toxicol. 2010;30:402–410. doi: 10.1002/jat.1511. [DOI] [PubMed] [Google Scholar]
- 11.Smithgall TE, Harvey RG, Penning TM. Spectroscopic identification of ortho-quinones as the products of polycyclic aromatic trans-dihydrodiol oxidation catalyzed by dihydrodiol dehydrogenase. A potential route of proximate carcinogen metabolism. J Biol Chem. 1988;263:1814–1820. [PubMed] [Google Scholar]
- 12.Zhan DJ, Heflich RH, Fu PP. Molecular characterization of mutation and comparison of mutation profiles in the hprt gene of Chinese hamster ovary cells treated with benzo[a]pyrene trans-7,8-diol-anti-9,10-epoxide, 1-nitrobenzo[a]pyrene trans-7,8-diol-anti-9,10-epoxide, and 3-nitrobenzo[a]pyrene trans-7,8- diol-anti-9,10-epoxide. Environmental and molecular mutagenesis. 1996;27:19–29. doi: 10.1002/(SICI)1098-2280(1996)27:1<19::AID-EM3>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 13.Penning TM, Ohnishi ST, Ohnishi T, Harvey RG. Generation of reactive oxygen species during the enzymatic oxidation of polycyclic aromatic hydrocarbon trans-dihydrodiols catalyzed by dihydrodiol dehydrogenase. Chem Res Toxicol. 1996;9:84–92. doi: 10.1021/tx950055s. [DOI] [PubMed] [Google Scholar]
- 14.Miller KP, Ramos KS. Impact of cellular metabolism on the biological effects of benzo[a]pyrene and related hydrocarbons. Drug Metab Rev. 2001;33:1–35. doi: 10.1081/dmr-100000138. [DOI] [PubMed] [Google Scholar]
- 15.Nakabeppu Y, Sakumi K, Sakamoto K, Tsuchimoto D, Tsuzuki T, Nakatsu Y. Mutagenesis and carcinogenesis caused by the oxidation of nucleic acids. Biological chemistry. 2006;387:373–379. doi: 10.1515/BC.2006.050. [DOI] [PubMed] [Google Scholar]
- 16.Klungland A, Rosewell I, Hollenbach S, Larsen E, Daly G, Epe B, Seeberg E, Lindahl T, Barnes DE. Accumulation of premutagenic DNA lesions in mice defective in removal of oxidative base damage. Proc Natl Acad Sci U S A. 1999;96:13300–13305. doi: 10.1073/pnas.96.23.13300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.European Convention. European Convention for the Protection of Vertebrate Animals Used for Experimental and Other Scientific Purposes, Appendix A. European Convention; 2006. [Google Scholar]
- 18.OECD. Draft OECD Guideline for the Testing of Chemicals. Rodent alkaline single cell gel electrophoresis (Comet) assay. 2012 [Google Scholar]
- 19.Gutzkow KB, Langleite TM, Meier S, Graupner A, Collins AR, Brunborg G. High-throughput comet assay using 96 minigels. Mutagenesis. 2013;28:333–340. doi: 10.1093/mutage/get012. [DOI] [PubMed] [Google Scholar]
- 20.Boiteux S, O’Connor TR, Lederer F, Gouyette A, Laval J. Homogeneous Escherichia coli FPG protein. A DNA glycosylase which excises imidazole ring-opened purines and nicks DNA at apurinic/apyrimidinic sites. J Biol Chem. 1990;265:3916–3922. [PubMed] [Google Scholar]
- 21.Collins AR, Oscoz AA, Brunborg G, Gaivao I, Giovannelli L, Kruszewski M, Smith CC, Stetina R. The comet assay: topical issues. Mutagenesis. 2008;23:143–151. doi: 10.1093/mutage/gem051. [DOI] [PubMed] [Google Scholar]
- 22.Dertinger SD, Bryce SM, Phonethepswath S, Avlasevich SL. When pigs fly: immunomagnetic separation facilitates rapid determination of Pig-a mutant frequency by flow cytometric analysis. Mutat Res. 2011;721:163–170. doi: 10.1016/j.mrgentox.2011.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bright J, Aylott M, Bate S, Geys H, Jarvis P, Saul J, Vonk R. Recommendations on the statistical analysis of the Comet assay. Pharm Stat. 2011;10:485–493. doi: 10.1002/pst.530. [DOI] [PubMed] [Google Scholar]
- 24.Dertinger SD, Phonethepswath S, Avlasevich SL, Torous DK, Mereness J, Bryce SM, Bemis JC, Bell S, Weller P, MacGregor JT. Efficient monitoring of in vivo pig-a gene mutation and chromosomal damage: summary of 7 published studies and results from 11 new reference compounds. Toxicol Sci. 2012;130:328–348. doi: 10.1093/toxsci/kfs258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hansen SH, Olsen AK, Soderlund EJ, Brunborg G. In vitro investigations of glycidamide-induced DNA lesions in mouse male germ cells and in mouse and human lymphocytes. Mutat Res. 2010;696:55–61. doi: 10.1016/j.mrgentox.2009.12.012. [DOI] [PubMed] [Google Scholar]
- 26.Demarini DM. Declaring the existence of human germ-cell mutagens. Environmental and molecular mutagenesis. 2012;53:166–172. doi: 10.1002/em.21685. [DOI] [PubMed] [Google Scholar]
- 27.Verma N, Pink M, Rettenmeier AW, Schmitz-Spanke S. Review on proteomic analyses of benzo[a]pyrene toxicity. Proteomics. 2012;12:1731–1755. doi: 10.1002/pmic.201100466. [DOI] [PubMed] [Google Scholar]
- 28.Osborne MR, Crosby NT. Cambridge monographs on cancer research. Cambridge University Press; Cambridge, England: 1987. Benzopyrenes. [Google Scholar]
- 29.van Delft JH, Mathijs K, Staal YC, van Herwijnen MH, Brauers KJ, Boorsma A, Kleinjans JC. Time series analysis of benzo[A]pyrene-induced transcriptome changes suggests that a network of transcription factors regulates the effects on functional gene sets. Toxicol Sci. 2010;117:381–392. doi: 10.1093/toxsci/kfq214. [DOI] [PubMed] [Google Scholar]
- 30.Lemieux CL, Douglas GR, Gingerich J, Phonethepswath S, Torous DK, Dertinger SD, Phillips DH, Arlt VM, White PA. Simultaneous measurement of benzo[a]pyrene-induced Pig-a and lacZ mutations, micronuclei and DNA adducts in Muta Mouse. Environ Mol Mutagen. 2011;52:756–765. doi: 10.1002/em.20688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bowen DE, Whitwell JH, Lillford L, Henderson D, Kidd D, Mc GS, Pearce G, Beevers C, Kirkland DJ. Evaluation of a multi-endpoint assay in rats, combining the bone-marrow micronucleus test, the Comet assay and the flow-cytometric peripheral blood micronucleus test. Mutat Res. 2011;722:7–19. doi: 10.1016/j.mrgentox.2011.02.009. [DOI] [PubMed] [Google Scholar]
- 32.Dertinger SD, Phonethepswath S, Franklin D, Weller P, Torous DK, Bryce SM, Avlasevich S, Bemis JC, Hyrien O, Palis J, MacGregor JT. Integration of mutation and chromosomal damage endpoints into 28-day repeat dose toxicology studies. Toxicol Sci. 2010;115:401–411. doi: 10.1093/toxsci/kfq070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bhalli JA, Shaddock JG, Pearce MG, Dobrovolsky VN, Cao X, Heflich RH, Vohr HW. Report on stage III Pig-a mutation assays using benzo[a]pyrene. Environ Mol Mutagen. 2011;52:731–737. doi: 10.1002/em.20675. [DOI] [PubMed] [Google Scholar]
- 34.Park JH, Mangal D, Tacka KA, Quinn AM, Harvey RG, Blair IA, Penning TM. Evidence for the aldo-keto reductase pathway of polycyclic aromatic trans-dihydrodiol activation in human lung A549 cells. Proc Natl Acad Sci USA. 2008;105:6846–6851. doi: 10.1073/pnas.0802776105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Olsen AK, Duale N, Bjoras M, Larsen CT, Wiger R, Holme JA, Seeberg EC, Brunborg G. Limited repair of 8-hydroxy-7,8-dihydroguanine residues in human testicular cells. Nucleic Acids Res. 2003;31:1351–1363. doi: 10.1093/nar/gkg216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.EFSA Scientific Committee. Scientific opinion on genotoxicity testing strategies applicable to food and feed safety assessment. EFSA Journal. 2011:69. [Google Scholar]
- 37.Miura D, Dobrovolsky VN, Kimoto T, Kasahara Y, Heflich RH. Accumulation and persistence of Pig-A mutant peripheral red blood cells following treatment of rats with single and split doses of N-ethyl-N-nitrosourea. Mutat Res. 2009;677:86–92. doi: 10.1016/j.mrgentox.2009.05.014. [DOI] [PubMed] [Google Scholar]
- 38.Phonethepswath S, Bryce SM, Bemis JC, Dertinger SD. Erythrocyte-based Pig-a gene mutation assay: demonstration of cross-species potential. Mutat Res. 2008;657:122–126. doi: 10.1016/j.mrgentox.2008.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dobrovolsky VN, Elespuru RK, Bigger CA, Robison TW, Heflich RH. Monitoring humans for somatic mutation in the endogenous PIG-a gene using red blood cells. Environmental and molecular mutagenesis. 2011;52:784–794. doi: 10.1002/em.20667. [DOI] [PubMed] [Google Scholar]
- 40.Dobrovolsky VN, Shaddock JG, Mittelstaedt RA, Manjanatha MG, Miura D, Uchikawa M, Mattison DR, Morris SM. Evaluation of Macaca mulatta as a model for genotoxicity studies. Mutation research. 2009;673:21–28. doi: 10.1016/j.mrgentox.2008.11.006. [DOI] [PubMed] [Google Scholar]
- 41.Dertinger SD, Phonethepswath S, Weller P, Nicolette J, Murray J, Sonders P, Vohr HW, Shi J, Krsmanovic L, Gleason C, Custer L, Henwood A, Sweder K, Stankowski LF, Jr, Roberts DJ, Giddings A, Kenny J, Lynch AM, Defrain C, Nesslany F, van der Leede BJ, Van DT, Schuermans A, Tanaka K, Hiwata Y, Tajima O, Wilde E, Elhajouji A, Gunther WC, Thiffeault CJ, Shutsky TJ, Fiedler RD, Kimoto T, Bhalli JA, Heflich RH, MacGregor JT. International Pig-a gene mutation assay trial: evaluation of transferability across 14 laboratories. Environ Mol Mutagen. 2011;52:690–698. doi: 10.1002/em.20672. [DOI] [PubMed] [Google Scholar]