Abstract
A novel label-free biosensor concept based on surface plasmon-enhanced diffraction by micro- patterned interfaces was applied to the study of hybridization reactions of target DNA oligonucleotides (15mers and 75mers) from solution to probe DNA oligonucleotides attached via streptavidin to the sensor surface. The self-referencing and quadratic signal amplification mechanism of the sensor allowed highly sensitive detection of the hybridization process. Association and dissociation processes of DNA targets could be recorded in real time and used for the quantification of their binding affinities, which differ considerably with a single base pair mismatch. An equilibrium titration approach was also applied in order to obtain the binding affinities for 15mer targets, yielding similar affinity values. The hybridization efficiencies were found to be higher for the 15mers than for the 75mers, although the latter contained the same recognition sequences. The hybridization efficiency was shown to depend on the probe density and reached nearly 100% for the 15mer fully complementary targets at a probe density of ∼1.2 × 1012 molecules/cm2. Using the assay as an end-point determination method, the lowest detectable coverage of a 15mer oligonucleotide was at least ∼1.1 × 1011 molecules/cm2. The diffraction sensing concept offers a completely novel way to integrate a reference channel in large-scale, label-free screening applications, to improve the stability and to enhance the sensitivity of microarray read-out systems.
INTRODUCTION
The detection and analysis of genetic material has drawn unprecedented research efforts during the past decades due to the increasing interest arising from both application and fundamental research concerns. Many methods for the label-free detection of DNA binding through base pairing have been reported based on optical (1–5), electrochemical (6), piezoelectric (7) and nanomechanical (8) techniques. The basis of operation for a DNA sensor is coupling between a specific base sequence within a DNA target analyte and the complementary oligonucleotide sequence immobilized on the solid surface of a transducer substrate. This DNA hybridization can be detected as a physical signal and can be monitored in situ and in real-time.
Due to the small size (mass) of a typical oligonucleotide, its binding to the surface is usually not sufficient to generate a significant optical contrast. Hence, it is experimentally a major challenge for label-free optical sensors to conduct a thorough investigation of this interaction. A few commercial optical biosensors have realized label-free DNA sensing with the aid of 3-dimensional surface matrices used to enhance the DNA surface coverage (1,5). Only a few reports (3,4) were based on planar functional surfaces and additional signal amplifications were often required for successful investigations (9,10).
Recently, we introduced a novel biosensor, a surface plasmon diffraction sensor (SPDS), based on surface plasmon-enhanced diffraction phenomena at periodic spatial structures (11,12) as a highly sensitive and robust sensing technique. The surface grating structure can diffract the incident light by superimposing discrete momenta m·g (with |g| = 2π/Λ being the magnitude of the grating vector and m being the order of diffraction) generated by the grating constant Λ (cf. Fig. 1A). The high optical intensity of the surface plasmon field greatly enhances the diffraction efficiency of the incident light and allows a very sensitive probing of surface heterogeneities. Modulation of the grating amplitude upon a biological interaction event induces a quadratically amplified change in the diffraction intensity (11). The temperature fluctuations and other bulk effects are automatically compensated during the sensing process, due to the inherently ‘self-referencing’ property of SPDS (12). Therefore, we consider SPDS to be an attractive candidate for detecting and characterizing oligonucleotide hybridization processes.
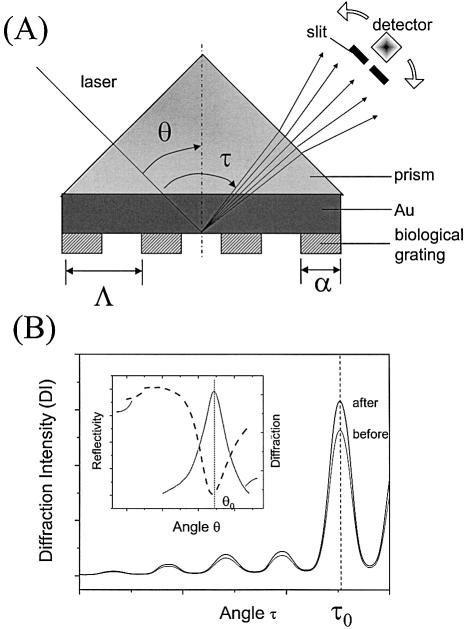
Figure 1.
(A) Schematic geometry for the SPDS. The periodic functional pattern is generated by micro-contact printing (see text for details). θ is the laser incident angle, τ the diffraction angle, α = 42 µm the width of the functional stripes of the surface pattern and Λ = 100 µm the grating constant. (B) Typical diffraction angular scans before and after, for example, hybridization of the target DNA. The inset shows schematically the strong dependence of the monitored diffraction intensity on the surface plasmon coupling angle.
In order to have a highly functional surface and to obtain correct kinetic and thermodynamic parameters of oligonucleotide interactions, it is of vital importance to carefully engineer the functional surface matrices, in addition to instrumental development of the optical DNA sensors. A major aim is to overcome hybridization barriers from, for example, steric hindrance and/or electrostatic repulsion. One successful example, a planar functional layer fabricated by the attachment of thiolated DNA oligonucleotides to the sensor surface via gold–thiol bonds has been shown by SPR (3) and neutron reflectivity (13) studies to be nearly 100% functional. We here employ another type of functional matrix based on a well-developed biotin–streptavidin supramolecular architecture that has already been used extensively in DNA hybridization studies by surface plasmon fluorescence spectroscopy (SPFS) (14,15). The streptavidin monolayer is formed on a mixed self-assembled monolayer (SAM) exposing 5–10% biotin functionalities. The remaining biotin-binding pockets (∼1–2) in the surface-attached streptavidin allow for subsequent attachment of biotinylated DNA probes, with the size of the streptavidin providing a natural limitation of the probe surface density for the next interaction step, target hybridization. This functional multilayer system has been working quite efficiently for SPFS characterization with extraordinarily high sensitivity and a number of different modes of operation. However, due to the distance-dependent fluorescence yield in SPFS and the lack of label-free information on oligonucleotide binding, many details of the hybridization process, e.g. hybridization efficiency, remain unknown.
In this paper, we examine the interactions of four different oligonucleotide DNA targets of different length and base sequence with surface-tethered probe DNA oligonucleotides by SPDS. The measured rate constants are used to calculate affinity constants, which are then compared with values obtained from equilibrium titration experiments. Meanwhile, we provide a preliminary assessment of the detection limit of SPDS, based on the titration experiments. Finally, we address the question of hybridization efficiency (HE) as a function of the probe DNA coverage.
MATERIALS AND METHODS
Materials
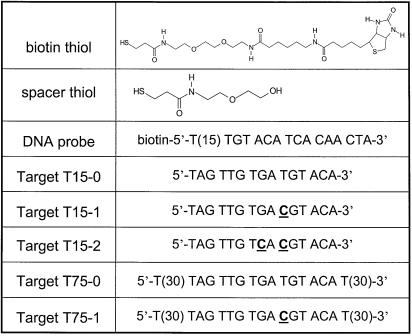
Biotin thiol and spacer thiol were synthesized and kindly provided by Roche Diagnostics. Their molecular structures are shown in Table 1. The biotinylated DNA probe and DNA targets (T15-0, T15-1, T15-2, T75-0 and T75-1) were purchased from MWG-Biotech, and are also listed in Table 1. The 75mer targets (T75-0 and T75-1) consist of the same recognition sequences as the 15mer targets (T15-0 and T15-1), however, they have two 30mer poly(T) flanks. T15-0 and T75-0 are fully complementary to the DNA probe, i.e. they define a zero mismatch (MM0) situation, while a one base mismatch (MM1) was designed in the sequences of T15-1 and T75-1, respectively. A two base mismatch (MM2) was designed for the T15-2 target. Streptavidin (SA) was also kindly provided by Roche Diagnostics. HBS-EP buffer (degassed 10 mM HEPES buffer saline, pH 7.4, 150 mM NaCl, 3 mM EDTA, 0.005% v/v surfactant P-20; Biacore, Uppsala, Sweden) was used for preparation of all of the protein/DNA solutions.
Table 1. Thiols and DNA oligonucleotide sequences used in this study.
Micro-contact printing (µCP) for surface patterning
Strip-like polydimethylsiloxane (PDMS) stamps were fabricated on photo-resist masters using Sylgard 184 silicon elastomer (Dow corning), with a periodicity of Λ = 100 µm and the width of the embossed strip being α = 42 µm. Before printing, the stamp was inked for 5 min in an ethanolic solution of a mixture of biotin thiol and spacer thiol (molar ratio 1:9) with a net concentration of 0.5 mM. Excess thiol solution was removed and the stamp dried in a stream of nitrogen. The stamp was then brought into contact with the freshly evaporated Au (50 nm, ε ≈ –12 + i × 1.3) substrate (LASFN9, n = 1.85 at 633 nm) for 1 min. After rinsing with copious amounts of ethanol, the Au substrate was exposed to an ethanolic solution of the pure spacer thiol (2 mM) for 10 min in order to passivate the non-derivatized areas. The patterning process was completed by rinsing the slide with ethanol and drying it with nitrogen.
Instrumental
The experimental set-up used was essentially based on a Kretschmann surface plasmon resonance spectroscopy (SPR or SPS) instrument and is schematically presented in Figure 1A. A linearly p-polarized HeNe laser (λ = 633 nm, 5 mV) modulated by a frequency chopper was reflected off the Au-coated base of the coupling prism and the reflected/diffracted intensity was measured through a 1 mm slit by a photo-diode detector connected to a lock-in amplifier. The sample arm (including the 90° LASFN9 prism, Au slide and flow cell) and the detector arm were controlled by two coaxial goniometers, respectively, enabling an independent tuning of their angular positions (θ and τ, respectively).
Typical angular diffraction scans are shown schematically in Figure 1B, representing the diffraction patterns before and after binding of, for example, a DNA target. The proportional increase in the diffraction intensity (DI) for every diffraction order indicates an optical thickness increase occurring at the surface of the functional regions. The intensity of each diffraction order depends strongly on the laser incident angle θ, which determines the surface plasmon coupling efficiency, as shown in the inset of Figure 1B. The maximum diffraction intensity of the monitored diffraction order (τ0) appears at the SPR minimum angle θ0. Another reason for having the laser incident angle near θ0 is the fact that at this minimum angle the surface plasmon coupling efficiency remains mostly constant even at small shifts of the minimum angle as a consequence of analyte binding. Therefore, the angular detuning effect is minimized (16). Thus, in kinetic diffraction mode, the laser incident angle was tuned to θ0, the detector rotated to the diffraction order at τ0 and the DI was monitored as a function of time.
A peristaltic pump delivered the sample solutions at a flow rate of 3 ml/min, in order to alleviate the mass transport effect for a correct assessment of the binding kinetics (16). The solutions were circulated in a sealed tubing loop after being manually exchanged, a procedure that is especially advantageous if equilibrium titration experiments are conducted (17).
RESULTS AND DISCUSSION
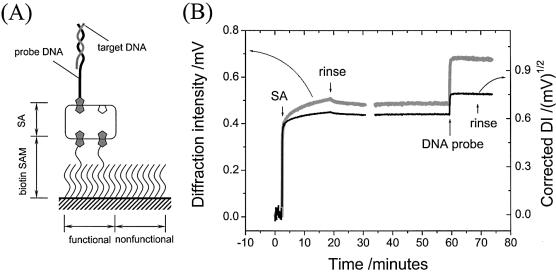
The schematic drawing of the multilayer architecture employed, composed of SAM/streptavidin/probe/target, is shown in Figure 2A. The processes were recorded as sequential increases in the diffraction signal as shown in Figure 2B. One can see from the raw experimental data (gray curve) that the initial DI induced by the patterned biotin SAM surface was quite weak and almost close to zero. This indicates that the optical difference generated by a difference in the SAM composition was insignificant. However, exposing the patterned surface to a 1 µM SA solution led to a rapid increase in the diffraction signal followed by a second slower increase phase. The second phase is considered to be associated with a non-specific aggregation of SA molecules because a pure buffer rinse could gradually wash away the signal accumulated in that phase. After a long rinse, the baseline remained at ∼0.48 mV and was extremely stable. The SPR minimum shift upon SA binding to the patterned surface was measured to be Δθ0 = ∼0.2°. Taking into account the fraction (42%) of functional area relative to the whole surface area, this angle shift on the patterned area is consistent with previous results (Δθ0 = ∼0.45°, corresponding to a thickness of d = ∼3.8 nm assuming n = 1.45 for the proteins) (14) obtained on a surface homogeneously functionalized by a mixed biotin/spacer thiol solution (1:9), confirming the formation of an identical SA coverage (∼2.2×1012 molecules/cm2). The injection of a 1 µM DNA probe solution induced another quick jump in the baseline to a higher level of ∼0.68 mV. Only a minor signal decrease was observed upon exchange of the DNA probe solution for pure buffer, indicating a strong and highly specific binding of the probe oligonucleotides via a biotin–SA linkage.
Figure 2.
(A) Schematic diagram of the multilayer architecture built on the functional pattern surface. (B) Experimental (gray curve) and corrected (black) kinetic curves for SA and DNA probe binding (both from a 1 µM solution).
For further quantitative analysis, the experimental curve was corrected (cf. black curve in Figure 2B), considering the quadratic relationship between DI (Id) and the amplitude of the index of refraction grating (nd), represented by the following equation (18):
Id ∝ I0(π·nd/λ)2 1
Here, nd is the amplitude of the biological grating represented by the optical thickness and I0 and λ are the intensity and wavelength of the light source, respectively. One should notice that the background intensity Ib due to random surface scattering should be subtracted. Ib could be obtained from a measurement on an unpatterned gold surface and was typically found to be ∼0.01 mV. Therefore, Δ√(Id – Ib) gives the increment in the corrected diffraction signal that is considered to be the response linear to the optical thickness of each layer, which in return is a linear function of the mass concentration of the bound biomolecules (19). Taking into consideration that proteins and oligonucleotides have similar refractive indices (n) and do not differ considerably with respect to their SPR response, Δ√(Id – Ib) divided by the corresponding molecular weight (Mw) provides the relative molar surface concentration, and can be used to calculate the stoichiometry between interacting molecules.
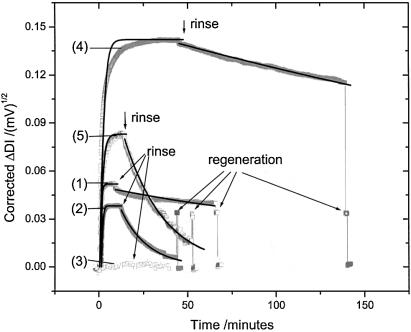
The subsequent association/dissociation measurements of the various DNA targets (T15-0, T15-1, T15-2, T75-0 and T75-1) performed sequentially on the same sensor chip are presented as corrected signals in Figure 3. The working concentration of each target solution was 1 µM to ensure sufficiently high mass transport rates for a correct kinetic evaluation. After reaching equilibrium, the target solution was exchanged against pure buffer in order to dissociate the bound hybrids and to rinse the target strands away. A 1 min pulse injection of 10 mM NaOH/water solution completely regenerated the probe surface (cf. Fig. 3). The same sensor chip could be regenerated at least 30 times and could be used for up to 48 h without significant loss of functionality (recovery >90%).
Figure 3.
Kinetic curves of hybridization (from 1 µM solutions) of different targets, i.e. T15-0 (curve 1), T15-1 (curve 2), T15-2 (curve 3), T75-0 (curve 4), T75-1 (curve 5) to the probe surface. The signals are corrected according to the quadratic dependence of DI on the optical contrast (cf. text). A 1 min pulse injection of 10 mM NaOH solution was used to regenerate the probe surface with ∼100% recovery. Signal exponential fits derived from a 1:1 Langmuir model were applied to derive the association/dissociation phases of the binding curves. The corresponding kinetic constants obtained were used for the KA calculations.
The results of an in-depth analysis of the hybridizations studies, as well as of binding of the SA and probe are listed in Table 2. Firstly, the binding stoichiometry between probe DNA and streptavidin was ∼1:0.75. This means that on average 1.3 probe strands were immobilized on each bound SA molecule. Since the surface concentration for the SA monolayer was ∼2.2 × 1012 molecules/cm2, i.e. each SA molecule occupies an area of ∼45 nm2, the surface concentration of the probe is ∼2.9 × 1012 molecules/cm2, which is close to the so-called ‘high’ probe density reported by Georgiadis and co-workers (3). The HE was also calculated for each target. High HEs (84 and 62%) were calculated for the T15-0 and T15-1 targets. However, substantially lower HEs (46 and 27%) were found for the T75-0 and T75-1 targets. We infer that the two extra poly(T) flanks of the 75mers play a major role in decreasing the HE, owing to steric/electrostatic hindrance. Also, the longer extension of the hybridized 75mers away from the surface may slightly lower their contribution to the optical thickness change sensed by the surface plasmon evanescent field, which decays exponentially into the solution with a depth of Lz ≈ 150 nm.
Table 2. Molecular weight (Mw), corrected response, interaction stoichiometry (hybridization efficiency for probe/target interactions), interaction rate constants kon and koff and affinity constant KA for the interactions shown in Figures 2 and 3.
| Name | MW (kDa) | Responsea | Stoichiometryb | koff (s–1) | kon (M–1 s–1) | KA (M–1) |
|---|---|---|---|---|---|---|
| Streptavidin | ∼60 | 5.1 | 0.75 | |||
| Biotinylated probe | 9.488 | 1.0 | ||||
| T15-MM0 | 4.622 | 0.42 | 0.84 | 1.32 × 10–4 | 6.58 × 104 | 4.98 × 108 |
| T15-MM1 | 4.607 | 0.31 | 0.62 | 1.11 × 10–3 | 2.42 × 104 | 2.18 × 107 |
| T75-MM0 | 22.873 | 1.14 | 0.46 | 3.52 × 10–5 | 9.21 × 103 | 2.62 × 108 |
| T75-MM1 | 22.858 | 0.67 | 0.27 | 7.41 × 10–4 | 1.08 × 104 | 1.46 × 107 |
aDefined as the increment in corrected diffraction intensity (see text for details) normalized to the response of the biotinylated probe.
bDefined as stoichiometry = (streptavidin or DNA target response × Mw of biotinylated probe)/(biotinylated probe response × Mw of streptavidin or DNA target).
The association/dissociation rate constants (kon and koff) of the target oligonucleotides were determined by fitting the working curves to a 1:1 Langmuir model, assuming pseudo-first order association/dissociation kinetics (cf. Fig. 3) (20). Within the Langmuir model we then obtained the affinity constant KA, which is simply the ratio of the two rate constants:
KA = kon/koff 2
At first glance, the one base mismatch induced an apparent difference in the binding curves between T15-0 and T15-1 and T75-0 and T75-1, especially in the dissociation phases. The KA values obtained differed by more than an order of magnitude between the 15mers (4.98 × 108 and 2.18 × 107 M–1 for T15-0 and T15-1, respectively) and the 75mers (2.62 × 108 and 1.46 × 107 M–1 for T75-0 and T75-1, respectively). A two mismatch sequence, T15-2, was also tested, however, it yielded a negligible binding signal. This demonstrates that the hybridization signals obtained were highly specific and the sensor was sensitive to a single base pair mismatch. The affinity parameters of T15-0 and T75-0 and of T15-1 and T75-1, respectively, were close, since they contained the same recognition sequences. It is also worth noticing that the pseudo-first order fitting did not completely match the association behaviors of the 75mer targets. This again reflects the influence of their bulky poly(T) flanks. Firstly, extra time/energy might be needed to change their conformation to form the surface double helix. Secondly, bound 75mers could influence the surface recognition sites, influencing the subsequent binding events. Therefore, the Langmuir 1:1 model did not quite apply for the binding of 75mers, since interfacial steric/electrostatic cross-talk existed. However, the fits still qualitatively reflect the decrease in hybridization affinity on introducing a single base pair mismatch.
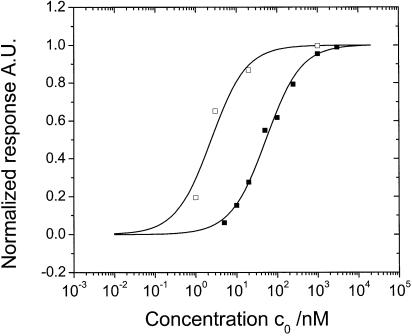
The affinity constants KA of the 15mers were also determined by recording equilibrium binding to the probe surface at different bulk target concentrations c0. The total span of the target concentration was from 1 nM to 3 µM. The normalized equilibrium response was plotted against the corresponding concentrations c0, as shown in Figure 4. A non-linear steady-state fit, based on the Langmuir 1:1 model, allows for the determination of KA, according to:
Figure 4.
Stepwise titration of the 15mer target [T15-0 (hollow square) and T15-1 (filled square), respectively] solutions with increasing concentration in the circulation loop connected to the flow cell. Each concentration was applied until equilibrium was attained. The equilibrium signals are normalized to the saturation responses (corresponding to a target surface concentration of ∼2.4 × 1012 and ∼1.8 × 1012 molecules/cm2 for the T15-0 and T15-1 targets, respectively) of the corresponding target at its maximum concentration. Langmuir fits (solid lines) to the isotherms yield the affinity constants, KA = 4.17 × 108 M–1 and KA = 1.92 × 107 M–1 for T15-0 and T15-1, respectively.
Γ = (KAc0)/(1 + KAc0) 3
with Γ being the normalized response (surface coverage) and c0 the bulk concentration. The affinity constants for T15-0 and T15-1 were 4.17 × 108 and 1.92 × 107 M–1, respectively, in good agreement with the affinity constants obtained from the single association/dissociation study. This implies that the Langmuir model can be applied for parameterization of the hybridization processes of the 15mers.
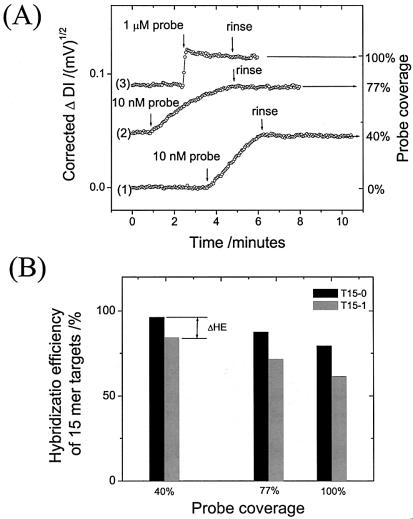
It has been demonstrated (3,21,22) that probe density plays a very important role in target surface hybridization behaviors, i.e. in the binding kinetics and HE. In order to conduct this study with our matrix, we controlled the probe density in our system by bringing a dilute probe concentration into contact with the SA functionalized surface and evaluated the corresponding HEs for 15mers according to the definition in Table 2. Under constant flow conditions, binding of the probe (from a 10 nM solution) was completely controlled by the mass transport rate, due to the low bulk concentration of probe molecules. Thus, binding was greatly slowed down and linear in time before nearly saturating the surface sites, which facilitated easy control of the probe density. Based on the known interaction stoichiometry between SA and probe, the increasing signal could be immediately stopped at any desired probe density level by exchanging the probe solution for pure buffer. As can be seen in Figure 5A, we thus controlled the probe density at three levels of coverage, i.e. 40, 77 and 100%. In order to ensure 100% probe coverage for the third (final) level, a 1 µM probe solution was applied. These levels of coverage were calculated to be equivalent to a probe density of ∼1.2 × 1012, ∼2.2 × 1012 and ∼2.9 × 1012 molecules/cm2, respectively.
Figure 5.
Influence of probe density on the hybridization efficiency of 15mer targets. (A) Control of probe coverage by a sequential loading strategy at low probe concentration. Three steps were applied, with injections of a 10 nM (curve 1), a 10 nM (curve 2) and a 1 µM (curve 3) probe solution followed by a buffer rinse, respectively. The corresponding probe coverages were calculated on the right axis, assuming the 1 µM probe solution rendered 100% probe coverage. (B) Hybridization efficiency as a function of probe coverage for the T15-0 and T15-1 targets (in 2 µM solutions), respectively.
The hybridization experiments using 15mer oligonucleotide targets at a concentration of 2 µM were performed at each level of probe density. A high target concentration was used to ensure (almost) saturated occupation of the available hybridization sites (cf. Fig. 4). The end-point hybridization signals upon association equilibria were corrected, considering the quadratic effect of the diffraction sensor (cf. equation 1). Based on their corresponding molecular weights, the HEs were quantified and are plotted in Figure 5B. One can see that the HE of both targets increased with decreasing probe coverage. This agrees with previous reports using electrochemical (21) and SPR (22) approaches and can be explained by alleviation of the static and/or electrostatic barrier on dilute probe surfaces. For the 40% probe surface density, i.e. at a probe coverage of ∼1.2 × 1012 molecules/cm2, the HE value of T15-0 reached ∼96%, indicating that the surface was (nearly) totally functional, which can be attributed to the good orientation and moderate density built on the SAM/SA supra-architecture. This result is in agreement with previous observations, where HE reached ∼100% at a probe density of 1.5 × 1012 molecules/cm2 for a MM0 25mer and 18mer targets (3). An interesting observation is that the highest HE of the T15-1 target was only ∼85% even at the lowest probe density studied, although operating at a saturation concentration, which was also in agreement with the result in the same report (3). This implies a reduced availability of sites for MM1 target hybridization by a higher hybridization barrier due to the internally mismatched base. On the other hand, the difference in HE (ΔHE, cf. Fig. 5B) between the MM0 and MM1 targets could be only slightly alleviated by lowering the probe density, which may indicate a certain heterogeneity of the probe distribution. One could speculate that this arises from the partial multiple loading of one streptavidin molecule by probe strands, as already indicated by the average stoichiometry between probe and SA (∼1.3) (cf. Table 2).
As a preliminary assessment of the limit of detection (LOD) of this novel DNA sensor, we refer back to the concentration titration experiments (cf. Fig. 4). The saturation response of the titration curves for the T15-0 and T15-1 targets corresponds to coverages of ∼2.4 × 1012 and ∼1.8 × 1012 molecules/cm2, respectively. For T15-1, the 5 nM solution gave an equilibrium signal at 6% of its saturation coverage, which could be easily resolved above the baseline fluctuation. Therefore, SPDS can detect at least 1.1 × 1011 molecules/cm2 of the 15mer oligonucleotide, equivalent to a mass concentration of ∼800 pg/cm2. This preliminary LOD level is already comparable with one of the best performing label-free SPR sensing techniques using near-infrared imaging by Corn’s group, where the LOD was reported to be 10 nM for an 18mer oligonucleotide, corresponding to ∼1011 molecules/cm2 (4). However, the sensitivity is still approximately a factor of 6 poorer than the SPFS assay incorporating a fluorescently labeled DNA target, where the LOD of 2 × 1010 molecules/cm2 was estimated (14). We expect that the LOD of SPDS can be greatly improved with the aid of mass amplification strategies, by which Keating et al. amplified the SPR response by 1000-fold using Au nanoparticals (10).
CONCLUSIONS
SPDS has been successfully applied for direct and rapid detection of oligonucleotides based on an efficient SAM/SA/probe architecture. It is also a tool well-qualified to discriminate single base pair mismatches in oligonucleotides by monitoring their kinetic behaviors in real time. The affinity constants obtained for the 15mers using both kinetics measurements and equilibrium titration were generally in agreement with previously reported data using fluorophore-labeled 15mer targets studied by SPFS (14), although the influence of the dye labeling on the kinetic behavior remains to be evaluated in detail. The strong dependence of the HE was also studied by controlling the probe density on the sensor surface. Substantial improvements in HE were achieved when lowering the probe density, although the absolute amount of hybridized target decreased. A 3-dimensional surface matrix may favor both the amount and efficiency of hybridization in practical applications.
High quality interaction assays can be offered by SPDS, attributed to its self-referencing property. For example, the binding curves in Figure 5A are free of any artifacts due to sample exchange, which exists in many optical biosensors and significantly influences the precise quantification of small binding signals. The LOD was ∼800 pg/mm2 from a rather preliminary assessment, which already compares favorably to many (commercially) available label-free methods (5). Owing to the mature µCP technique and subsequent functionalization chemistry, the sensor surface can be highly reproducible and reusable. More importantly, the concept of diffraction detection offers a novel way to integrate reference channels in the microarray fabrication, which is expected to improve the stability and sensitivity of large-scale label-free screening applications. Further investigations are under way to understand the hybridization behavior of long chain DNA, e.g. PCR products.
Acknowledgments
ACKNOWLEDGEMENTS
The authors would like to thank Roche Diagnostics for providing the thiols and the streptavidin. The authors would also like to thank Mr Lau King Hang Aaron (Institute of Materials Research and Engineering, Singapore) for providing the photo-resist masters fabricated by photolithography. The authors thank Dr Neal R.Armstrong (University of Arizona, AZ), Dr Akira Baba (University of Texas, Houston, TX) and Shengjun Tian (MPIP, Germany) for helpful discussions.
REFERENCES
- 1.Persson B., Stenhag,K., Nilsson,P., Larsson,A., Uhlen,M. and Nygren,P. (1997) Analysis of oligonucleotide probe affinities using surface plasmon resonance: a means for mutational scanning. Anal. Biochem., 246, 34–44. [DOI] [PubMed] [Google Scholar]
- 2.Sauer M., Brecht,A., Charisse,K., Maier,M., Gerster,M., Stemmler,I., Gauglitz,G. and Bayer,E. (1999) Interaction of chemically modified antisense oligonucleotides with sense DNA: a label-free interaction study with reflectometric interference spectroscopy. Anal. Chem., 71, 2850–2857. [DOI] [PubMed] [Google Scholar]
- 3.Peterson A.W., Wolf,L.K. and Georgiadis,R.M. (2002) Hybridization of mismatched or partially matched DNA at surfaces. J. Am. Chem. Soc., 124, 14601–14607. [DOI] [PubMed] [Google Scholar]
- 4.Nelson B.P., Grimsrud,T.E., Liles,M.R., Goodman,R.M. and Corn,R.M. (2001) Surface plasmon resonance imaging measurements of DNA and RNA hybridization adsorption onto DNA microarrays. Anal. Chem., 73, 1–7. [DOI] [PubMed] [Google Scholar]
- 5.Watts H.J., Yeung,D. and Parkes,H. (1995) Real-time detection and quantification of DNA hybridization by an optical biosensor. Anal. Chem., 67, 4283–4289. [DOI] [PubMed] [Google Scholar]
- 6.Wang J. (2002) Electrochemical nucleic acid biosensors. Anal. Chim. Acta, 469, 63–71. [Google Scholar]
- 7.Hook F., Ray,A., Norden,B. and Kasemo,B. (2001) Characterization of PNA and DNA immobilization and subsequent hybridization with DNA using acoustic-shear-wave attenuation measurements. Langmuir, 17, 8305–8312. [Google Scholar]
- 8.Fritz J., Baller,M.K., Lang,H.P.,Rothuizen,H., Vettiger,P., Meyer,E., Guntherodt,H.J., Gerber,C. and Gimzewski,J.K. (2000) Translating biomolecular recognition into nanomechanics. Science, 288, 316–318. [DOI] [PubMed] [Google Scholar]
- 9.Jordan C.E., Frutos,A.G., Thiel,A.J. and Corn,R.M. (1997) Surface plasmon resonance imaging measurements of DNA hybridization adsorption and streptavidin/DNA multilayer formation at chemically modified gold surfaces. Anal. Chem., 69, 4939–4947. [DOI] [PubMed] [Google Scholar]
- 10.He L., Musick,M.D., Nicewarner,S.R., Salinas,F.G., Benkovic,S.J., Natan,M.J. and Keating,C.D. (2000) Colloidal Au-enhanced surface plasmon resonance for ultrasensitive detection of DNA hybridization. J. Am. Chem. Soc., 122, 9071–9077. [Google Scholar]
- 11.Yu F., Tian,S., Yao,D. and Knoll,W. (2004) Surface plasmon enhanced diffraction for label-free bio-sensing. Anal. Chem., in press. [DOI] [PubMed] [Google Scholar]
- 12.Yu F. and Knoll,W. (2004) Immuno-sensor with self-referencing based on surface plasmon diffraction. Anal. Chem., 76, 1971–1975. [DOI] [PubMed] [Google Scholar]
- 13.Levicky R., Herne,T.M., Tarlov,M.J. and Satija,S.K. (1998) Using self-assembly to control the structure of DNA monolayers on Gold: a neutron reflectivity study. J. Am. Chem. Soc., 120, 9787–9792. [Google Scholar]
- 14.Liebermann T., Knoll,W., Sluka,P. and Herrmann,R. (2000) Complement hybridization from solution to surface-attached probe-oligonucleotides observed by surface-plasmon-field-enhanced fluorescence spectroscopy. Colloids Surf. A—Physicochem. Eng. Asp., 169, 337–350. [Google Scholar]
- 15.Neumann T., Johansson,M.-L., Kambhampati,D. and Knoll,W. (2002) Surface-plasmon fluorescence spectroscopy. Adv. Funct. Mater., 12, 575–586. [Google Scholar]
- 16.Yu F., Yao,D. and Knoll,W. (2003) Surface plasmon field-enhanced fluorescence spectroscopy studies of the interaction between an antibody and its surface-coupled antigen. Anal. Chem., 75, 2610–2617. [DOI] [PubMed] [Google Scholar]
- 17.Schuck P., Millar,D.B. and Kortt,A.A. (1998) Determination of binding constants by equilibrium titration with circulating sample in a surface plasmon resonance biosensor. Anal. Biochem., 265, 79–91. [DOI] [PubMed] [Google Scholar]
- 18.Nelson K.A., Casalegno,R., Miller,R.J.D. and Fayer,M.D. (1982) Laser-induced excited-state and ultrasonic wave gratings—amplitude and phase grating contributions to diffraction. J. Chem. Phys., 77, 1144–1152. [Google Scholar]
- 19.Stenberg E., Persson,B., Roos,H. and Urbaniczky,C. (1991) Quantitative-determination of surface concentration of protein with surface-plasmon resonance using radiolabeled proteins. J. Colloid. Interface Sci., 143, 513–526. [Google Scholar]
- 20.Ward L.D. and Winzor,D.J. (2000) Relative merits of optical biosensors based on flow-cell and cuvette designs. Anal. Biochem., 285, 179–193. [DOI] [PubMed] [Google Scholar]
- 21.Steel A.B., Herne,T.M. and Tarlov,M.J. (1998) Electrochemical quantitation of DNA immobilized on gold. Anal. Chem., 70, 4670–4677. [DOI] [PubMed] [Google Scholar]
- 22.Peterson A.W., Heaton,R.J. and Georgiadis,R.M. (2001) The effect of surface probe density on DNA hybridization. Nucleic Acids Res., 29, 5163–5168. [DOI] [PMC free article] [PubMed] [Google Scholar]