ABSTRACT
Pairing high-throughput sequencing technologies with high-throughput mutagenesis enables genome-wide investigations of pathogenic organisms. Knowledge of the specific functions of protein domains encoded by the genome of the hepatitis C virus (HCV), a major human pathogen that contributes to liver disease worldwide, remains limited to insight from small-scale studies. To enhance the capabilities of HCV researchers, we have obtained a high-resolution functional map of the entire viral genome by combining transposon-based insertional mutagenesis with next-generation sequencing. We generated a library of 8,398 mutagenized HCV clones, each containing one 15-nucleotide sequence inserted at a unique genomic position. We passaged this library in hepatic cells, recovered virus pools, and simultaneously assayed the abundance of mutant viruses in each pool by next-generation sequencing. To illustrate the validity of the functional profile, we compared the genetic footprints of viral proteins with previously solved protein structures. Moreover, we show the utility of these genetic footprints in the identification of candidate regions for epitope tag insertion. In a second application, we screened the genetic footprints for phenotypes that reflected defects in later steps of the viral life cycle. We confirmed that viruses with insertions in a region of the nonstructural protein NS4B had a defect in infectivity while maintaining genome replication. Overall, our genome-wide HCV mutant library and the genetic footprints obtained by high-resolution profiling represent valuable new resources for the research community that can direct the attention of investigators toward unidentified roles of individual protein domains.
IMPORTANCE
Our insertional mutagenesis library provides a resource that illustrates the effects of relatively small insertions on local protein structure and HCV viability. We have also generated complementary resources, including a website (http://hangfei.bol.ucla.edu) and a panel of epitope-tagged mutant viruses that should enhance the research capabilities of investigators studying HCV. Researchers can now detect epitope-tagged viral proteins by established antibodies, which will allow biochemical studies of HCV proteins for which antibodies are not readily available. Furthermore, researchers can now quickly look up genotype-phenotype relationships and base further mechanistic studies on the residue-by-residue information from the functional profile. More broadly, this approach offers a general strategy for the systematic functional characterization of viruses on the genome scale.
INTRODUCTION
In this work, we combined transposon insertion mutagenesis with next-generation sequencing (NGS) to functionally characterize a viral genome in a systematic manner. NGS techniques have boosted deposition of new sequences in public databases. In contrast, increases in our knowledge of viral gene function lag behind and rely on testing individual mutant viruses. For genome-scale studies, this approach would consume extensive time and labor. Thus, virologists could benefit from rapid and high-throughput methods that uncover relationships between sequenced genes and encoded functions. The microbiology field has already integrated NGS with traditional transposon mutagenesis for a systems-level analysis of microorganisms (1). In brief, these strategies recover representative transposon insertion libraries grown under defined selection conditions; sequencing of insertion site junctions then determines the relative frequencies of each insertion mutant at various points during the selection. In this way, one experiment can report the contribution of thousands of bacterial genome positions to the outcome of any selection. The first additions of transposons to virologists’ tool kits demonstrated the usefulness of insertional mutagenesis for the functional profiling of small viral genomes (2). We and others have increased the profiling throughput by establishing a massively parallel profiling platform based on capillary electrophoresis; this resulted in genome-wide functional profiles for hepatitis C virus (HCV), Venezuelan equine encephalitis virus (VEEV), and norovirus (3–5). In their studies of VEEV, Beitzel et al. improved the sensitivity and precision of genetic footprinting through Roche 454 massively parallel sequencing (4). Recently, Heaton et al. took advantage of the increased sequencing capacity of Illumina technology in a genome-wide mutagenesis study of influenza virus (6), but in-depth NGS-based analyses of mutagenized positive-stranded RNA viruses such as HCV have been missing.
To increase the resolution of genetic footprinting, we profiled HCV insertion mutant pools by Illumina sequencing. We chose this virus because of HCV’s relevance to public health and the rich body of existing literature, which includes experimentally determined protein structures. An estimated 130 to 170 million people worldwide suffer from HCV infection (7). Over time, infection can lead to chronic inflammation and liver cancer. HCV research lacked infectious model systems to study the viral life cycle until 2005, when several groups reported successful propagation of HCV in cell culture (8, 9). In host hepatocytes, translation of the compact RNA genome (9.6 kb) produces ten viral proteins (10); ascribing functions to these proteins remains an active area of investigation. The core protein and envelope glycoproteins E1 and E2 make up the structural components of the virus (11). Replication of the viral genome depends on the nonstructural proteins NS3 to NS5B (12), as these proteins form the viral replication complex. Virus particles do not appear to incorporate the p7 protein (13). In a replicon system, genome replication takes places without p7 and NS2 (12, 14). However, more and more studies have been implicating both p7 and NS2 in virion assembly (15). Similarly, the nonstructural protein NS4B, which appears to play many roles in genome replication (16), also appears to play a role in postreplication steps (17–19).
In this study, we established a resource that illustrates the effect of 5-amino-acid (aa) insertions on HCV viability. Insertions are spread randomly across the entire HCV genome, with a single insertion for every mutant, covering 99% of HCV codons. We addressed whether the genetic footprints mirror the biological functions and known structures of HCV proteins. We then used the resource to guide us in the construction of infectious epitope-tagged viruses, to screen the data set for insertions that affect later steps of the virus life cycle, and to identify a critical region in NS4B. This systems-level analysis of the HCV genome provides a comprehensive resource that can benefit the entire research community by triggering mechanistic studies based on the profile’s residue-by-residue information.
RESULTS
Combining random transposon mutagenesis with NGS yields a comprehensive data set for profiling nearly every position of the HCV genome.
How disruptions of local protein domains affect HCV fitness remains an important question for virologists. More broadly, how disruptions introduced by genomic mutations affect protein function is of general interest to the biological community. To create a comprehensive resource that addresses these questions we combined transposon insertion sequencing with genetic footprinting (see Fig. S1 in the supplemental material). First, we generated a library of HCV mutants harboring one 15-nt insertion at one random location by Mu transposon insertion mutagenesis and subsequent removal of the transposon fragment (see Fig. S1A in the supplemental material). Following in vitro transcription of the DNA library, we delivered the resulting HCV RNA library into cultured Huh-7.5.1 cells for genetic selection. We then recovered the total cell-associated RNA at 96 h posttransfection (pool 1 [P1]) and after an additional passage of the conditioned cell culture medium (pool 2 [P2]). The original mutagenized RNA library represented the unselected input pool (pool 0 [P0]).
To analyze the genetic footprints of the libraries from pools 0, 1, and 2, we enriched insertion sites and digitally counted each insertion mutant site by NGS (see Fig. S1C and D in the supplemental material). Our experiment produced 6.4, 3.0, and 4.5 million reads for pools 0, 1, and 2, respectively; the initial library from pool 0 contained 7,978 unique insertion positions in genomic regions coding for HCV proteins (see Dataset S1 in the supplemental material). Thus, our experiment covered 88% of the entire 9,102 nucleotides of protein-encoding space. The insertions changed 99% of the HCV codons and 98% of encoded amino acid positions. In conclusion, leveraging random transposon mutagenesis and high-throughput insertion site sequencing yielded a data set large enough to assay nearly every encoded amino acid position.
Selection profiles of individual HCV genes vary according to the functions of encoded proteins as structural or replication complex proteins.
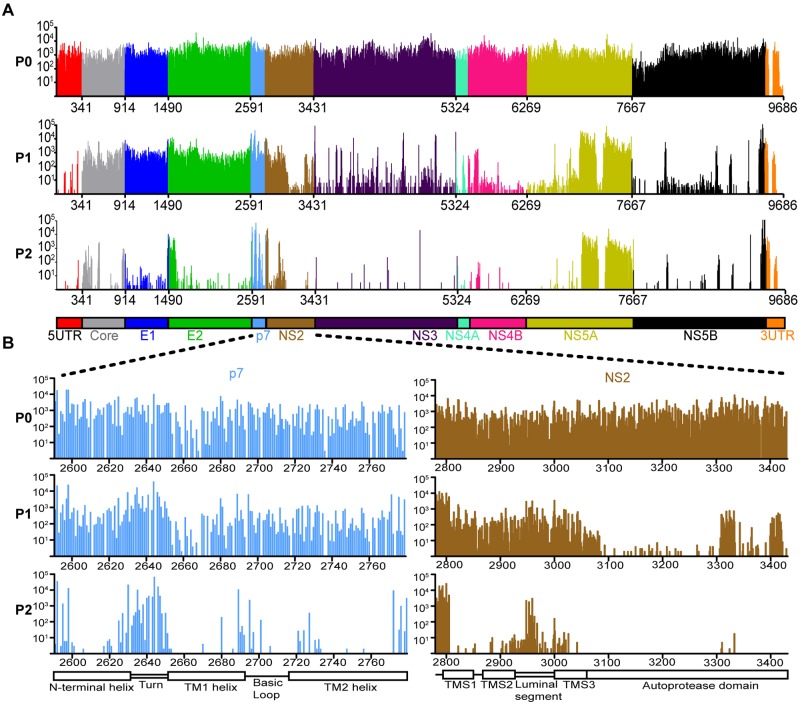
To visualize the genetic footprints of this comprehensive insertion library, we generated a global insertion map based on the sequencing data (Fig. 1A). The profile’s resolution also allowed us to align these footprints with the domain organization of encoded proteins p7 and NS2 (Fig. 1B). We have uploaded higher-resolution maps of the remaining eight HCV proteins to our HCV resource website (http://hangfei.bol.ucla.edu). For each insertion position, we determined the number of sequencing reads in pools 0, 1, and 2. We also calculated the percentage of total insertion positions detected in the respective protein region in pools 1 and 2 relative to the number of insertion positions detected in the input pool 0 (Table 1). In pools 1 and 2, we detected 55% and 17%, respectively, of the original pool 0 insertion mutants (Table 1). This overall decrease in pools 1 and 2, also seen in Fig. 1, suggested that passaging in cell culture indeed exerted a selection pressure on mutant viruses. This selection pressure was particularly strong for mutant viruses containing insertions at positions located in the NS2 autoprotease domain, NS3, NS4A, NS4B, and NS5B (Fig. 1 and Table 1). The trend toward insertions in NS3-NS5B reflected the role of proteins forming the replication complex, required early on in pool 1 to initiate genome replication. In contrast, we detected 95 to 98% of insertions in core, E1, E2, p7, and the NS2 transmembrane domain in pool 1 but only 15 to 45% in pool 2 (Table 1). The trend toward insertions in core, E1, and E2 reflected the roles of structural proteins during the HCV life cycle: they are dispensable for genome replication in pool 1 but essential for virus propagation in pool 2. Note how insertions in NS2 and NS5A displayed a mixed selection pattern; in pool 2 we detected only 5% of mutants in certain regions of NS2 and NS5A (NS2 autoprotease domain and NS5A RNA binding domain), whereas mutants in other regions (NS2 transmembrane domain and rest of NS5A) persisted at 27% and 76%, respectively (Table 1).
FIG 1 .
Functional map of the entire HCV genome. (A) Digital counting of each insertion mutant in sequenced libraries. (Top) Bar graph of raw sequencing reads from all 15-nt insertion mutants at each passage (input [P0], pool 1 [P1], and pool 2 [P2]). Each bar represents a unique mutant, and bar heights indicate corresponding sequencing reads. (Bottom) Cartoon of HCV genome organization. (B) (Top) Closer look at the genetic footprint of insertion sites located in the p7 and NS2 genes. (Bottom) Cartoon of p7 and NS2 protein domain organization. TM1 helix and TM2 helix, transmembrane helix 1 and 2; TMS1, TMS2, and TMS3, transmembrane segments 1, 2, and 3.
TABLE 1 .
Gene-by-gene summary of the high-resolution functional profilea
| Gene region | Size (nt) | No. (%b) of insertion positions detected in: |
Protein function | ||
|---|---|---|---|---|---|
| Pool 0 | Pool 1 | Pool 2 | |||
| Core | 574 | 497 | 472 (95) | 144 (27) | Virion capsid |
| E1 | 576 | 498 | 479 (96) | 123 (33) | Virion envelope |
| E2 | 1100 | 983 | 938 (95) | 158 (15) | Virion envelope |
| p7 | 191 | 176 | 172 (98) | 81 (45) | Viroporin, assembly |
| NS2 TM domain | 302 | 273 | 261 (96) | 105 (27) | Scaffold, assembly |
| NS2 protease domain | 348 | 309 | 128 (41) | 18 (5) | Autoprotease |
| NS3 | 1895 | 1657 | 512 (30) | 79 (3) | RC, protease, helicase |
| NS4A | 161 | 142 | 48 (32) | 11 (4) | RC, NS3 cofactor |
| NS4B | 784 | 695 | 241 (34) | 42 (5) | RC, membranous web |
| NS5A domain I | 638 | 542 | 175 (31) | 39 (5) | Binding to RNA/membranes |
| NS5A rest | 760 | 664 | 575 (87) | 508 (76) | Regulatory, assembly |
| NS5B | 1,710 | 1,481 | 361 (22) | 127 (5) | RC complex, polymerase |
| NS5B C terminus | 64 | 61 | 49 (80) | 39 (64) | Membrane tail |
| Total | 9,102 | 7,978 | 4,411 (55) | 1,474 (17) | |
RC, replication complex; TM, transmembrane.
Percentage of the total number of insertion positions detected in pool 0.
In summary, the functional map of the HCV genome revealed very distinct patterns of selection. The separation of insertion mutants based on shared selection patterns largely overlapped with separation based on the genetically encoded function, namely, as structural components (core, E1, and E2) and replication-complex components (NS3, NS4A, NS4B, and NS5B). This congruence supported the validity of the functional profile.
Functional annotation of transmembrane protein structures supports the biological relevance of previously solved protein structures.
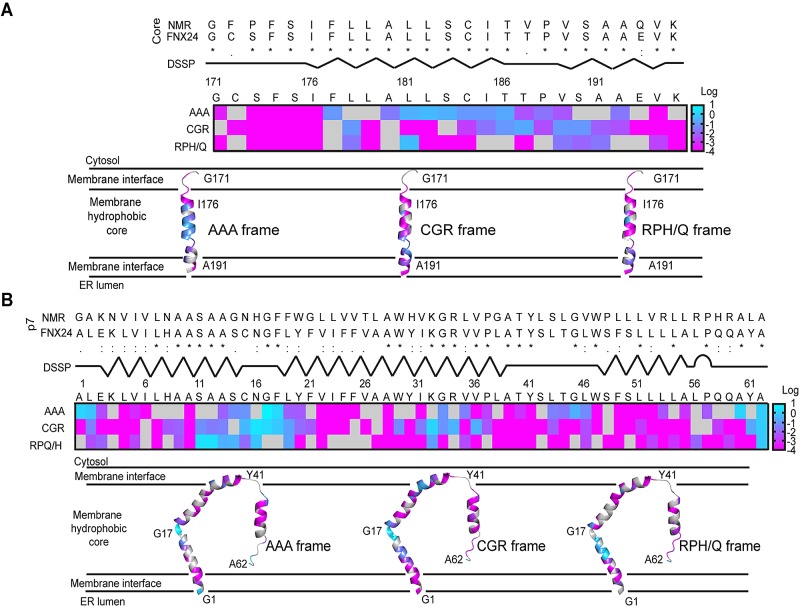
To determine whether we could match our screen data with established functional HCV regions, we next focused our analysis on protein domains with known nuclear magnetic resonance (NMR) structures. Structures of entire membrane proteins are difficult to obtain; thus, genetic footprints in membrane segments of the HCV genome can provide useful insight into structure-function relationships. Note that the actual encoded amino acid insertion represents one of three possible motifs depending on an insertion’s location with respect to the overall HCV reading frame. This results in 5-aa insertions each containing one of the following sets of three constant amino acids: AAA, CGR, and RPH/Q (note that the other two inserted amino acids depend on the insertion location). First, we examined the C-terminal signal peptide region of the core protein. As a measure of fitness, we calculated the ratio of normalized reads in pool 2 and reads in pool 1 (P2/P1). We then displayed the log of the P2/P1 ratios for the AAA, CGR, and RPH/Q motifs as a heat map and color-coded a previously solved structure (20) (Fig. 2A). P2/P1 ratios for insertions in the membrane hydrophobic core, namely, the helix extending from aa 175 to 186, were highest for insertions encoding alanine residues (Fig. 2A, cyan/blue colors for the AAA frame). In contrast, insertions of proline or glycine were less tolerated. Whereas alanine is known to have high helix-forming propensities, the propensities of proline and glycine are poor (21). Thus, the observed genetic footprints are consistent with incorporation of alanine insertions into the local alpha-helical structure. On the other hand, insertion of disruptive proline and glycine residues would compromise helical structures, thereby impairing viral fitness.
FIG 2 .
Functional annotation of protein structures of HCV transmembrane regions. (A) Heat map of P2/P1 values in the C-terminal region of HCV core protein. Each box provides the phenotype of mutants containing an insertion after the indicated amino acid. Gray shows a lack of the respective insertion mutant in pool 0. Mutants are separated by their AAA, CGR and RPH/Q amino acid motifs. The color bar corresponds to a range of P2/P1 values [log(P2/P1) = 1 to −4]. Dictionary of secondary structure of proteins (DSSP) provides an overview of helices (zigzag lines) and unstructured loops (straight lines). (Top) Alignment of core amino acid sequences of HCV strains from this study (strain FNX24) and deposited structure (PDB code 2LIF). (Bottom) Ribbon diagrams of annotated structure, along with a tentative model of the peptide’s orientation within the endoplasmic reticulum (ER). (B) Heat map of P2/P1 values in the p7 protein. DSSP secondary structure indicates a turn (arc) toward the C-terminal end of the protein. PDB code 2M6X.
Second, we matched a published NMR structure of the p7 protein (22) to our genetic footprints (Fig. 2B). Tolerated insertion mutants clustered within a region connecting the N-terminal helix and first transmembrane helix (TM1). Moreover, amino acid residues preceding transmembrane helix 2 (TM2) tolerated insertion of the AAA motif (Fig. 2B, stretch of four blue boxes [LTGL]). Areas of insertion toleration in the p7 protein localized mostly to turns and loops. Although the p7 protein is rather small (63 aa), six p7 monomers can self-assemble to form larger ion channel complexes (22). Annotation of the entire viroporin structure allowed us to visualize an area tolerating all three insertion motifs in the middle part of the ion channel (see Fig. S2A in the supplemental material [side view, yellow dashed area]). Most of the tolerated insertions localized in a region forming an unstructured loop between the N-terminal helix and TM1 (aa 15 to 18). Thus, the functional information from the genetic footprints allowed us to define the boundaries of an area of toleration for 5-aa insertions within the structural landscape of the oligomerized p7 ion channel. Finally, we explored the footprint of an additional HCV transmembrane protein, NS2. When we overlaid the previously solved structure of NS2’s membrane-bound portion (23, 24) with our functional profile, we located tolerated regions proximal to a short helix in transmembrane segment 1 and in a short loop connecting transmembrane segments 2 and 3 (see Fig. S2B in the supplemental material). To summarize, our genetic footprints were consistent with solved structures of HCV transmembrane proteins and allowed us to match tolerated insertion regions with the protein’s structure. Note that our screen determined the phenotypes of insertion mutants in an infectious cell culture system that recapitulated the entire virus life cycle. Thus, our functional profile supported the biological relevance of previously solved structures, which were based on fragments of the core and NS2 proteins.
Identification of nonessential regions aids in the design of viable epitope-tagged viruses.
Our functional annotation of existing protein structures had already suggested that the genetic footprints may identify regions of insertion toleration. Thus, we wanted to use our footprints to direct the systematic insertion of short protein sequences into the HCV genome. Genetic insertion of epitope tags allows researchers to purify tagged viral proteins and associated complexes from infected cells using well-characterized tag-specific antibodies. Moreover, these antibodies can aid in visualizing the subcellular localization of tagged proteins. To identify candidate regions for epitope-tag insertion and produce a map of hot spots tolerating insertions, we filtered the functional profile based on calculated ratios between normalized reads of pools 2 and 0 (P2/P0), as well as pools 1 and 0 (P1/P0), using the following criteria: hot spot regions had to tolerate 5-aa insertions regardless of the insertion reading frame at a P2/P0 ratio cutoff of >0.01 (Fig. 3A). Moreover, to remove any insertional mutant viruses with defects in genome replication, we disregarded any mutant with P1/P0 ratios of ≤0.1. This screen revealed hot spots in core, E2, p7, NS2, and NS5A (Fig. 3A). After considering the structural organization of these proteins, we picked specific locations for the insertion of epitope tags (Fig. 3B). Because the lengths and amino acid sequences of inserted epitopes differed from those of the screen’s 5-aa insertions, we inserted a panel of tags with various sizes and sequences (see Table S1 in the supplemental material). This variety increased the likelihood of producing viable epitope-tagged mutants. After constructing the individual epitope-tagged mutants, we measured their fitness in infectivity assays. Most epitope-tagged mutants remained viable (Fig. 3B). Viruses with a 100-fold reduction in infectivity still produced robust levels of 1,000 infectious particles per ml. Moreover, we confirmed that the tags remained functional: antibodies against the respective tag immunostained transfected cells viewed by fluorescence microscopy (see Fig. S3 in the supplemental material). The subcellular localization of tagged proteins was consistent with previous studies (13, 24, 25) and detected only in infected cells (see Fig. S3 in the supplemental material).
FIG 3 .
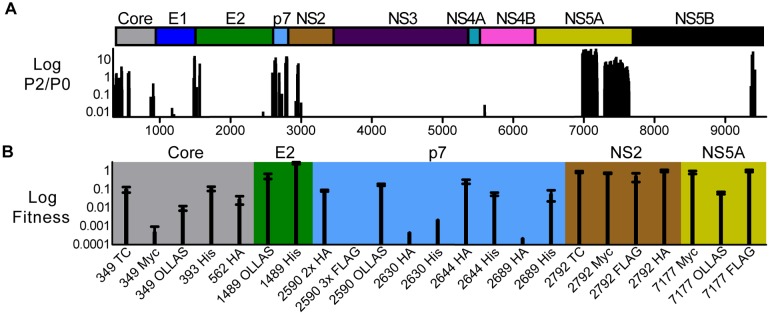
Screening data set for hot spots tolerating small insertions. (A) Insertion mutants’ P2/P1 ratios across the HCV genome. Only data points that fall within a cluster of ≥3 consecutive insertion mutants with nonzero P2/P1 ratios were plotted. We discarded all insertion mutants with P1/P0 values of <0.1 and P2/P0 values of <0.1. At the top is a cartoon depicting HCV genome organization. (B) Engineering infectious epitope-tagged HCV. The graph shows the fitness of cloned epitope-tagged viruses. Fitness is the ratio of mutant virus infectivity to the wild-type infectivity, as determined by a limiting dilution assay of supernatants taken from transfected Huh-7.5.1 cells. Numbers (349 to 7177) refer to the genome position after which the sequence encoding the respective epitope tag was inserted. TC, tetracysteine.
Epitope-tagged proteins offer added flexibility for multicolor fluorescence microscopy due to the wide availability of anti-tag antibodies. Multilabel microscopy often relies on primary antibodies raised in different species. Species-specific fluorochrome-conjugated secondary antibodies then allow the detection of multiple proteins in the same sample. Using the 7177-FLAG virus, which generates FLAG-tagged NS5A protein, we visualized the spatial distribution of three viral proteins simultaneously using primary antibodies against FLAG, HCV core, and E2 combined with species-specific anti-rabbit, anti-mouse, and anti-human antibodies (see Fig. S4 in the supplemental material). In addition, staining of infected cells with the lipophilic dye BODIPY 493/503 provided a cellular marker that revealed the location of lipid droplets (LDs). We observed small areas of overlapping signals between the three viral proteins in the vicinity of LDs. Thus, our epitope-tagged virus facilitated a four-color microscopy approach, which can visualize putative viral assembly sites.
Taken together, this work confirmed that we could tap our HCV genetic footprint resource to guide the successful placement of epitope tags for the production of replication-competent reporter viruses, which have a wide range of applications, such as fluorescence microscopy.
Analysis of mutant viruses’ genetic footprints uncovers new functional regions encoded by the HCV genome.
In the analyses described above, the data set’s genetic footprints agreed with known functions and protein structures of HCV proteins. Next, we wanted to test whether this resource could produce biological information beyond a viral protein’s known functions. Based on our finding that the selection profiles varied according to protein function, we predicted that the values of P1/P0 and P2/P0 could indicate whether an insertion affected an early or late step of the HCV life cycle. Because insertions in core, E1, and E2 had average P1/P0 ratios of 1.2, we hypothesized that insertions with P1/P0 ratios within the broad range of 0.65 to 1.85 would be competent for genome replication. In contrast, P2/P0 ratios of >0.01 would be consistent with a defect at a later step of the life cycle and an inability to produce infectious viral particles.
To find new nonstructural-protein functions not related to traditional roles in genome replication, we applied the aforementioned criteria (along with a minimum of 20 for input P0 reads) and generated a candidate list of 13 mutants with insertions in nonstructural proteins (Table 2). Because insertions in NS4B dominated this list (8 out of 13 mutants), we focused our validation efforts on NS4B. Individually cloned NS4B mutants with insertions at positions 5571, 5584, 5597, 5607, and 5615 showed a defect in spreading in cell culture, as evidenced by immunofluorescence studies examining the number of HCV-positive cells after transfection of viral genome RNA (Fig. 4A). Moreover, the insertions reduced infectivity to 1 to 6% of the levels seen for cells transfected with a wild-type genome at 3 days following transfection (Fig. 4B). We observed similar defects in infectious virus production at 1 and 5 days (Fig. 4B). Both our immunofluorescence (Fig. 4B) and Western blotting (data not shown) suggested that the insertions in NS4B reduced NS5A protein levels only slightly, which indicated that viral RNA genomes still replicated robustly. Thus, we conclude that the decreases in infectious virus particle production may be due to additional defects in steps after genome replication, such as viral assembly or egress. In summary, we identified a new region in the NS4B protein that may to be essential for steps following genome replication. Importantly, we showed that our data set could serve as a resource to screen for insertion mutants that reveal protein regions with previously unrecognized biological functions.
TABLE 2 .
Screening of functional profile produces a candidate list of nonstructural proteins with roles in steps following genome replicationa
| Genome position | HCV protein | Amino acid sequence | Count for: |
P1/P0 | P2/P0 | ||
|---|---|---|---|---|---|---|---|
| P0 | P1 | P2 | |||||
| 5594 | NS4B | QAQDIQLRPQQPAMQAS | 20 | 24 | 0 | 1.2 | 0 |
| 5571 | NS4B | GLLQQASAAAASKQAQD | 211 | 264 | 2 | 1.2 | 0.007 |
| 5574 | NS4B | LLQQASNAAASKQAQDI | 24 | 26 | 0 | 1.1 | 0 |
| 5583 | NS4B | QASKQAHAAAAQDIQPA | 73 | 87 | 0 | 1.2 | 0 |
| 5584 | NS4B | ASKQAQCGRTQDIQPAM | 699 | 471 | 3 | 0.7 | 0.004 |
| 5596 | NS4B | AQDIQPCGRKPAMQASW | 39 | 42 | 0.2 | 1.1 | 0.006 |
| 5597 | NS4B | AQDIQPVRPQPAMQASW | 190 | 134 | 0 | 0.7 | 0 |
| 5607 | NS4B | IQPAMQAAAAQASWPKV | 535 | 521 | 2 | 1.0 | 0.003 |
| 6847 | NS5A | LPCEPECGRTEPDADVL | 174 | 122 | 0.9 | 0.7 | 0.005 |
| 7290 | NS5A | VAGCALPAAALPPPKKA | 35 | 38 | 0 | 1.1 | 0 |
| 7671 | NS5B | TTVCCSIAAASMSYSWT | 82 | 112 | 0 | 1.4 | 0 |
| 8979 | NS5B | ILMVQDTAAADTLDQNL | 36 | 41 | 0.2 | 1.1 | 0.006 |
| 8991 | NS5B | QDTLDQNAAAQNLNFEM | 31 | 31 | 0 | 1.0 | 0 |
Filtering criteria: P0 > 20; 0.65 < P1/P0 < 1.85; P2/P0 ≤ 0.01; only HCV proteins NS3 to -5B; P0, P1, and P2 total reads normalized to 1,000,000.
FIG 4 .
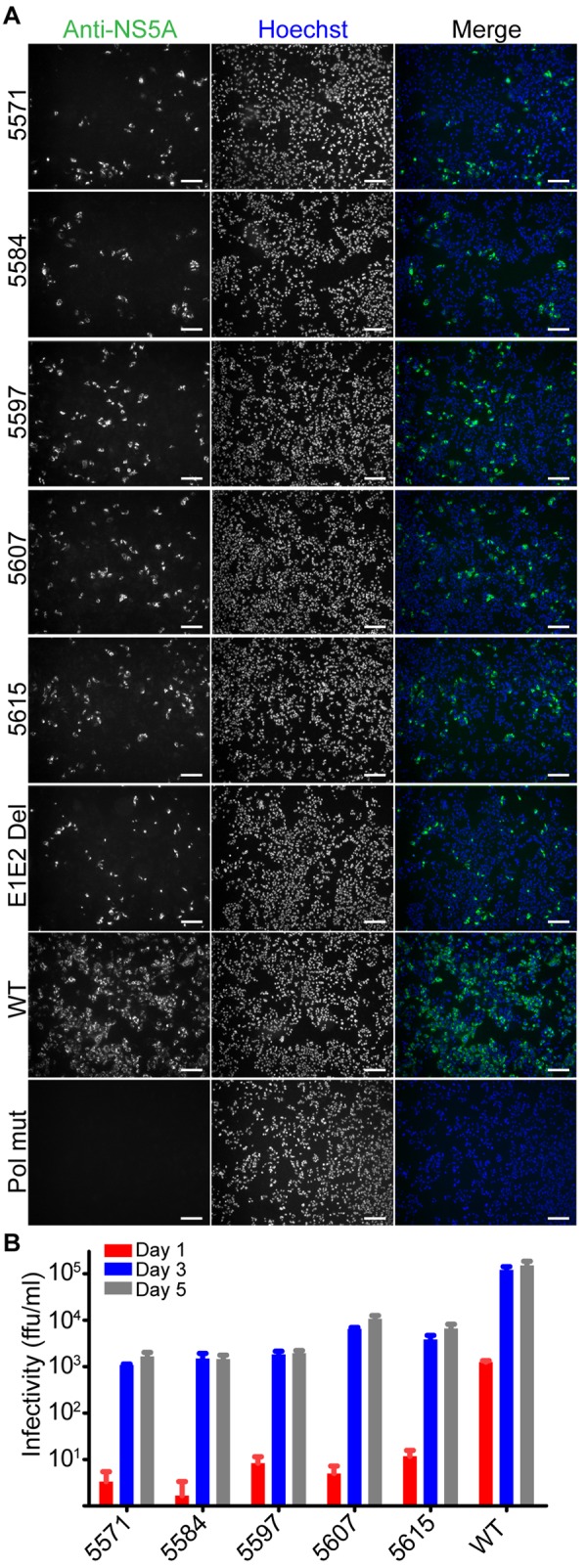
Validation of NS4B’s role in later steps of the viral life cycle. (A) Huh-7.5.1 cells were transfected with individually cloned mutants containing insertions identified in Table 2. Controls were FNX24 parental virus (wild-type), an E1E2 deletion mutant (most of the E1 and E2 coding regions were deleted, making this mutant replication competent but assembly deficient), and a Pol mutant (the mutant’s polymerase motif contains a GDD-to-GNN mutation, making the mutant genome replication defective). Three days after transfection of mutant RNA genomes, cells were fixed and processed for an immunofluorescence assay using an anti-NS5A antibody. The Hoechst dye provides a nuclear counterstain. Scale bar: 20 µm. (B) Infectivity of cell culture supernatants taken from cells transfected with NS4B insertion mutants. We collected cell supernatants at multiple time points after transfection of RNA genomes and determined supernatant infectivity by limiting dilution assay.
DISCUSSION
In this study, high-throughput sequencing of transposon insertion sites in the complete genome of HCV provided us with a resource that captured biological functions of HCV proteins, aided in the design of reporter viruses, and led to the identification of new functional regions involved in later steps of the viral life cycle. Note that the current functional profile of the HCV genome improved upon our previously published profile (3) in several ways (see Text S1 in the supplemental material). We could clearly distinguish selection patterns for insertions in genes encoding the structural proteins from those in genes encoding replication complex proteins. In addition, the genetic footprints also reflected biological functions mediated by different domains of viral proteins. For example, a closer look at the NS2 profile (Fig. 1B) and NS5A profile (Fig. 1A) revealed different selection patterns for functionally distinct protein domains (see Text S2 in the supplemental material). At the same time, when examining pool 1 and 2 counts of single insertion mutants, we cannot rule out the possibility that the electroporation and sequencing library preparation steps introduce a certain level of biological noise in pools 1 and 2. We suggest that when insertion positions are being selected for follow-up experiments, further statistical analyses and data trimming should be used to filter out this noise. Nonetheless, validation of screening results with individual mutants showed consistent phenotypes for select insertion mutants of p7, NS3, NS4B, NS5A, and NS5B (data not shown). Overall, the agreement between this study’s genetic footprints and known biological functions of HCV proteins support the validity of this resource.
To further validate this resource, we surveyed the profile from a structural angle, focusing on NMR structures of individual membrane segments of HCV proteins. Our genetic footprints match the structures in several ways. When we annotated protein structures with the fitness values of insertional mutants, we observed insertion toleration in external surface loops, unstructured linker regions and a short turn separating two transmembrane helices. These areas appear to accommodate additional amino acid residues more readily, as seen in our previously published functional profile of NS5B (3). Moreover, toleration in unstructured loops seems to be a general theme for viral proteins, as evidenced by insertional mutagenesis studies of poliovirus and norovirus (5, 26, 27). Our functional annotation of transmembrane proteins p7 and NS2 also highlights this method’s promise in complementing the structural biology of membrane proteins. Difficulties in the crystallization of membrane proteins and challenges in NMR structure determinations of larger proteins (28) often prevent the determination of high-resolution structures, which is the first step in studying structure-function relationships. Therefore, we predict that the resource described in this paper, which examines the phenotypes of insertions in membrane proteins within the context of their native biological environment, can support the biological relevance of solved membrane protein structures and advance our comprehension of structure-function relationships.
After we examined the validity of the functional profile, we wanted to show applications of this resource. First, we created maps of nonessential areas of the HCV genome. These maps guided the construction of epitope-tagged viruses. This information can guide the exact placement of exogenous sequences to produce viable tagged viruses. Previously, transposon-mediated random insertion mutagenesis in the coding region of NS5A and subsequent selection of viable HCV replicons led to the identification of two sites near the NS5A C terminus tolerating insertion of heterologous sequences (29). This approach led to the generation of replicons and, later, infectious viruses encoding green fluorescent protein (GFP); these reagents have become essential tools to directly visualize functional HCV replication complexes in host cells (29–31). In a similar way, we wanted to take advantage of our genetic footprints in the entire HCV genome to identify hot spots for insertion toleration. These insertion hot spots would then provide candidate regions for insertion of epitope tags. We identified the N termini of E2, p7, and NS2 as prime candidates to insert small peptides with limited fitness cost. Note that independent studies have engineered individual epitope-tagged viruses at similar locations (13, 24, 32–36). We expanded on these results by revealing that these N termini tolerate various epitope tags, in addition to the originally published tags. Our hot spot analysis for the core and p7 proteins also yielded two unpublished internal locations for the design of viable epitope-tagged viruses. We also found that insertion into internal sites was more sensitive to the size and amino acid identity of the actual inserted tag. An alternative approach (see Text S3 in the supplemental material), similar to the one described by Moradpour et al., may be necessary to develop epitope-tagged viruses for certain protein domains, i.e., E1, NS3, NS4A, NS4B, and NS5B, whose functions are easily disrupted by insertions (29). Nevertheless, the genetic footprinting approach outlined in this study can assist the rational design of epitope-tagged viruses.
We also show the usefulness of an epitope-tagged reporter virus in multiplexed immunofluorescence assays, which revealed areas of colocalization between the HCV structural proteins core/E2 and the nonstructural protein NS5A. These areas were close to LDs, which are cellular fat storage organelles that play critical roles in the HCV assembly process (37, 38). In previous confocal microscopy experiments, the structural proteins E2 and core colocalized frequently adjacent to small lipid droplets (39). Furthermore, core and NS5A signals overlap on LDs or on the endoplasmic reticulum at various stages of virion assembly (37, 40). To our knowledge, our four-color microscopy images are the first to confirm colocalization between these three viral proteins (core, E2, and NS5A) around lipid droplets within the same cell. Additional experiments that combine the use of epitope-tagged viruses and high-resolution microscopy approaches will be needed to further characterize these putative assembly sites. Toward this end, the recent application of three-color 3D superresolution microscopy to the study of a FLAG-tagged E2 virus has shown great promise in the analysis of assembly sites (41).
The second application of this resource took advantage of the method’s semiquantitative measures of viral fitness. By limiting our searches to particular P1/P0 and P2/P0 ratios, we generated a candidate list of areas in nonstructural proteins potentially mediating later steps of the viral life cycle. Insertions in a short stretch of the NS4B gene dominated this list. This protein is a key player in the formation of the HCV replication complex. In addition, NS4B appears to play some role in postreplication of the HCV life cycle (17–19, 42). Insertions from the candidate list fall between a predicted amphipathic alpha helix (43, 44) and a structurally resolved alpha helix at the N terminus of the protein (44). Although a clear role of this region in mediating later steps in the HCV life cycle, such as particle assembly, has not yet been reported, a recent study suggests that particular residues in the N terminus could play minor roles in HCV assembly (45). Additional studies will need to investigate the exact mechanism of action underlying the role of this NS4B region in later steps of the viral life cycle. In summary, we envision our footprinting resource as an aid to preliminary identification of new functional candidate regions, triggering detailed individual studies that address possible mechanisms of action.
In addition to the sequencing files, linked to the BioProject entry PRJNA237836 at the NCBI BioProject database, we provide this resource in a downloadable spreadsheet format (Dataset S1). We have also designed a website to quickly screen for desired phenotypes. In this way, the entire research community can retrieve the sequenced phenotypes of insertional mutant viruses. Anyone can then reconstruct the corresponding insertion mutant or introduce variations of mutations at the profiled amino acid location for follow-up studies. These types of studies will be critical for examining how the mutation of a select amino acid residue affects HCV protein function. Although this study used genome-wide transposon insertion sequencing to assess the basic requirements for HCV viability in cell culture, the approach lends itself to analysis of requirements for growth or survival under any selective condition. For example, studies comparing genetic footprints in the presence and absence of an antiviral agent such as interferon are ongoing in our laboratory. Moreover, analogous experiments could examine the effect of knocking down or overexpressing cellular host factors. Fitness analyses of HCV mutants in these cell lines could shed light on detailed interactions of HCV with the overexpressed/knocked-down factors. Finally, transposon insertion sequencing experiments should strive to select mutagenized libraries in additional model systems. Examples of these systems would include primary hepatocytes as well as stem cell-derived hepatocyte-like cells (46). Ultimately, applying the transposon insertion sequencing approach to animal models of HCV infection would be truly exciting. The emergence of new small animal models and improvement of existing models (47) make high-throughput functional studies of viruses using in vivo models a realistic goal in the near future.
MATERIALS AND METHODS
Cells.
The Huh-7.5.1 cell line was kindly provided by Francis Chisari (Scripps Research Institute, La Jolla, CA). The cells were cultured in supplemented medium as described previously (3).
Plasmids and antibodies.
We synthesized pFNX-HCV based on the chimeric sequence of the genotype 2a J6/JFH1 virus. Construction of this clone, which also included a mutation in the endogenous NotI site, and the generation of envelope deletion and polymerase mutant controls were described previously (48). We constructed individual mutant viruses with 15-nt insertions or epitope tags by site-directed PCR mutagenesis using the primers listed in Table S1 in the supplemental material, which also lists antibodies and epitope tag sequences used.
Transposon mutagenesis.
The plasmid carrying FNX-HCV was modified by in vitro bacteriophage Mu transposon mutagenesis (Thermo Scientific, Waltham, MA), followed by incubation on plates containing kanamycin and ampicillin to select for bacteria transformed with transposon insertion-containing plasmids. We isolated mutant plasmids from a total of one million individual bacterial colonies (100-fold the number of possible insertions in the HCV genome) and pooled and digested plasmids with NotI enzyme (New England Biosciences, Ipswich, MA) to remove the transposon fragment, followed by a self-ligation step, which left 15 nucleotides (nt) (5′-N1N2N3N4N5TGCGGCCGCA-3′, where N1 to N5 are 5 duplicated nucleotides from target DNA) randomly inserted across the virus genome.
In vitro transcription, transfection, and insertion library passage.
A total of 16 µg of the pFNX library DNA served as the template for in vitro transcription using the T7 Ribomax Express kit (Promega, Madison, WI). We transfected 120 µg of DNase-treated RNA into 4.8 × 107 Huh-7.5.1 cells by electroporation as described before (3). Cells were resuspended in 40 ml of complete Dulbecco’s modified Eagle medium (DMEM) and plated in T-75 culture flasks. Medium was replaced at 8 h posttransfection, removed after an incubation period of 96 h, and tested for infectivity. The resulting titer of this reconstituted mutant library was 8.4 × 104 focus-forming units (FFU) per ml. We passaged the library in Huh-7.5.1 cells for another round at a low multiplicity of infection (MOI) (0.2). Total RNA was isolated from the cells using Tri-reagent (Molecular Research Center Inc., Cincinnati, OH). The DNase-treated, purified RNA was used for functional profiling analysis.
Library preparation, Illumina sequencing, and analysis.
Total extracted RNA from P0, P1, and P2 was reverse transcribed into cDNA with Superscript III (Life Technologies, Carlsbad, CA). The cDNA then served as the template for PCR to amplify thirteen overlapping fragments covering the entire virus genome. We then enriched for insertion sites and loaded the final products directly into the flow cell for sequencing. Text S4 in the supplemental material provides additional details on the enrichment method. Sequencing of multiplexed insertion mutant libraries was carried out using the Illumina IIx genome analyzer (Illumina, San Diego, CA). Additional details on the sequencing data analysis are provided in Text S4.
Mapping functional data to protein structures, cloning of individual insertion mutants, viral titration, and immunofluorescence.
Text S4 in the supplemental material further describes how heat maps showing P2/P1 ratios for the three insertion frames were obtained at each amino acid position in defined stretches of core, p7, and NS2 proteins. Moreover, experimental details on the cloning of individual epitope-tagged viruses and NS4B insertion mutant viruses, as well as the functional assays, including virus titration and immunofluorescence, are provided as well.
Accession numbers.
Project metadata can be found under the accession number PRJNA237836 at the NCBI BioProject resource (http://www.ncbi.nlm.nih.gov/bioproject). We have linked this experiment’s sequencing reads to this BioProject through the NCBI Sequence Read Archive.
SUPPLEMENTAL MATERIAL
Comparison of current profile with previously published HCV profile. Download
Examination of NS2 and NS5A functional profiles. Download
Approach for epitope tagging in hard-to-tag areas of the HCV genome. Download
Supplemental materials and methods. Download
HCV footprinting data set. The entire data set is available in this Excel spreadsheet. This file includes information on genomic positions of insertional mutant viruses, amino acid sequences of insertions, raw counts (P0, P1, and P2), normalized counts, and P1/P0, P2/P0, and P1/P0 ratios. Download
Strategy for functional profiling of HCV insertion libraries by massively parallel sequencing. (A) Generation of an HCV insertion library by random insertion mutagenesis of a plasmid carrying the HCV genome (pFNX-HCV). kanR and ampR are kanamycin and ampicillin resistance genes. (B) Passaging the mutagenized HCV RNA insertion library. (C) Library preparation and Illumina sequencing. Download
Mapping tolerated insertions to the structure of p7 viroporin and N terminus of NS2. (A) Surface views of annotated p7 viroporin structure (PDB code 2M6X). Gray indicates a lack of the respective insertion mutant in pool 0. We colored amino acid residues in the p7 structure according to the heat map in Fig. 2B. Functional data for AAA, CGR, and RPH/Q frames are shown separately. Surface transparency was set to 65% for top and bottom views to reveal ribbon representations inside viroporin. Yellow dotted circles indicate area tolerating insertions for three reading frames. (B) Helical segments (zigzag lines), turns (arcs) and unstructured loops (straight lines) were deduced from NMR analyses (PDB codes 2JY0, 2KWT, and 2KWZ) of NS2 peptides (aa 1 to 27, 27 to 59, and 60 to 99). (Bottom) Tentative model of NS2 topology within the ER membrane. Download
Immunofluorescence assay of cells infected with epitope-tagged HCV. Huh-7.5.1 cells were infected with indicated epitope-tagged viruses. Three days after infection, we fixed and processed cells for immunofluorescence using antibodies against the inserted epitope tag and either NS5A or E2 viral proteins. Hoechst dye stains the nuclei of both infected and uninfected cells. Images were acquired by epifluorescence microscopy. Scale bar: 10 µm. Download
Four-color fluorescence microscopy of cells infected with NS5A-FLAG virus. (A) (Right) Overview of an infected cell, with the nucleus marked by a dashed line. We acquired multiplexed images with specific filters for each fluorochrome and pseudocolored each channel as indicated: lipid droplets in gray, core in green, E2 in red, and NS5A-FLAG in blue. Bar, 10 µm. The box indicates a perinuclear area rich in lipid droplets and expressing viral proteins. (Left) Zoom-in of the boxed region. Bar, 1 µm. (B) To better visualize localization of each viral protein with regard to lipid droplets, these zoom-in views show only two channels at a time. Here, we pseudocolored lipid droplets in red and each of the three viral proteins in green (NS5A, E2, and core). Arrows indicate areas containing spatially overlapping signals in the NS5A, E2, and core channels. Download
List of primers and antibodies used in this study.
ACKNOWLEDGMENTS
This work was supported by grants from the National Natural Science Foundation of China (NSFC) (81172314) and the National Institutes of Health (AI078133) (R.S.), grant P30CA016042 (Jonsson Comprehensive Cancer Center), and grant P30AI028697 (UCLA AIDS Institute/CFAR).
We thank F. Chisari for providing the Huh-7.5.1 cell line. We are grateful to S. Foung and C. M. Rice for the human anti-E2 antibody CBH5 and mouse anti-NS5A antibody. We also thank Asim Dasgupta, Samuel French, and Yong-Hoon Kim for their comments during the writing process. Confocal laser scanning microscopy was performed at the Department of Anesthesiology (we thank Yibing Wang, Hongmei Ruan, and Tom Vondriska).
Footnotes
Citation Remenyi R, Qi H, Su S, Chen Z, Wu NC, Arumugaswami V, Truong S, Chu V, Stokelman T, Lo H, Olson CA, Wu T, Chen S, Lin C, Sun R. 2014. A comprehensive functional map of the hepatitis C virus genome provides a resource for probing viral proteins. mBio 5(5):e01469-14. doi:10.1128/mBio.01469-14.
REFERENCES
- 1. van Opijnen T, Camilli A. 2013. Transposon insertion sequencing: a new tool for systems-level analysis of microorganisms. Nat. Rev. Microbiol. 11:435–442. 10.1038/nrmicro3033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kekarainen T, Savilahti H, Valkonen JP. 2002. Functional genomics on potato virus A: virus genome-wide map of sites essential for virus propagation. Genome Res. 12:584–594. 10.1101/gr.220702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Arumugaswami V, Remenyi R, Kanagavel V, Sue EY, Ngoc Ho T, Liu C, Fontanes V, Dasgupta A, Sun R. 2008. High-resolution functional profiling of hepatitis C virus genome. PLoS Pathog. 4:e1000182. 10.1371/journal.ppat.1000182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Beitzel BF, Bakken RR, Smith JM, Schmaljohn CS. 2010. High-resolution functional mapping of the Venezuelan equine encephalitis virus genome by insertional mutagenesis and massively parallel sequencing. PLoS Pathog. 6:e1001146. 10.1371/journal.ppat.1001146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Thorne L, Bailey D, Goodfellow I. 2012. High-resolution functional profiling of the norovirus genome. J. Virol. 86:11441–11456. 10.1128/JVI.00439-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Heaton NS, Sachs D, Chen CJ, Hai R, Palese P. 2013. Genome-wide mutagenesis of influenza virus reveals unique plasticity of the hemagglutinin and NS1 proteins. Proc. Natl. Acad. Sci. U. S. A. 110:20248–20253. 10.1073/pnas.1320524110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Alter MJ. 2007. Epidemiology of hepatitis C virus infection. World J. Gastroenterol. 13:2436–2441. 10.3748/wjg.v13.i17.2436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lindenbach BD, Evans MJ, Syder AJ, Wölk B, Tellinghuisen TL, Liu CC, Maruyama T, Hynes RO, Burton DR, McKeating JA, Rice CM. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623–626. 10.1126/science.1114016 [DOI] [PubMed] [Google Scholar]
- 9. Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, Murthy K, Habermann A, Kräusslich HG, Mizokami M, Bartenschlager R, Liang TJ. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791–796. 10.1038/nm1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lindenbach BD, Rice CM. 2005. Unravelling hepatitis C virus replication from genome to function. Nature 436:933–938. 10.1038/nature04077 [DOI] [PubMed] [Google Scholar]
- 11. Penin F, Dubuisson J, Rey FA, Moradpour D, Pawlotsky JM. 2004. Structural biology of hepatitis C virus. Hepatology 39:5–19. 10.1002/hep.20032 [DOI] [PubMed] [Google Scholar]
- 12. Lohmann V, Körner F, Koch J, Herian U, Theilmann L, Bartenschlager R. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110–113. 10.1126/science.285.5424.110 [DOI] [PubMed] [Google Scholar]
- 13. Vieyres G, Brohm C, Friesland M, Gentzsch J, Wölk B, Roingeard P, Steinmann E, Pietschmann T. 2013. Subcellular localization and function of an epitope-tagged p7 viroporin in hepatitis C virus-producing cells. J. Virol. 87:1664–1678. 10.1128/JVI.02782-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Blight KJ, Kolykhalov AA, Rice CM. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972–1974. 10.1126/science.290.5498.1972 [DOI] [PubMed] [Google Scholar]
- 15. Lindenbach BD. 2013. Virion assembly and release. Curr. Top. Microbiol. Immunol. 369:199–218. 10.1007/978-3-642-27340-7_8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gouttenoire J, Penin F, Moradpour D. 2010. Hepatitis C virus nonstructural protein 4B: a journey into unexplored territory. Rev. Med. Virol. 20:117–129. 10.1002/rmv.640 [DOI] [PubMed] [Google Scholar]
- 17. Han Q, Manna D, Belton K, Cole R, Konan KV. 2013. Modulation of hepatitis C virus genome encapsidation by nonstructural protein 4B. J. Virol. 87:7409–7422. 10.1128/JVI.03523-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jones DM, Patel AH, Targett-Adams P, McLauchlan J. 2009. The hepatitis C virus NS4B protein can trans-complement viral RNA replication and modulates production of infectious virus. J. Virol. 83:2163–2177. 10.1128/JVI.01885-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Paul D, Romero-Brey I, Gouttenoire J, Stoitsova S, Krijnse-Locker J, Moradpour D, Bartenschlager R. 2011. NS4B self-interaction through conserved C-terminal elements is required for the establishment of functional hepatitis C virus replication complexes. J. Virol. 85:6963–6976. 10.1128/JVI.00502-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Oehler V, Filipe A, Montserret R, da Costa D, Brown G, Penin F, McLauchlan J. 2012. Structural analysis of hepatitis C virus core-E1 signal peptide and requirements for cleavage of the genotype 3a signal sequence by signal peptide peptidase. J. Virol. 86:7818–7828. 10.1128/JVI.00457-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pace CN, Scholtz JM. 1998. A helix propensity scale based on experimental studies of peptides and proteins. Biophys. J. 75:422–427. 10.1016/S0006-3495(98)77529-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. OuYang B, Xie S, Berardi MJ, Zhao X, Dev J, Yu W, Sun B, Chou JJ. 2013. Unusual architecture of the p7 channel from hepatitis C virus. Nature 498:521–525. 10.1038/nature12283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jirasko V, Montserret R, Appel N, Janvier A, Eustachi L, Brohm C, Steinmann E, Pietschmann T, Penin F, Bartenschlager R. 2008. Structural and functional characterization of nonstructural protein 2 for its role in hepatitis C virus assembly. J. Biol. Chem. 283:28546–28562. 10.1074/jbc.M803981200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jirasko V, Montserret R, Lee JY, Gouttenoire J, Moradpour D, Penin F, Bartenschlager R. 2010. Structural and functional studies of nonstructural protein 2 of the hepatitis C virus reveal its key role as organizer of virion assembly. PLoS Pathog. 6:e1001233. 10.1371/journal.ppat.1001233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rouillé Y, Helle F, Delgrange D, Roingeard P, Voisset C, Blanchard E, Belouzard S, McKeating J, Patel AH, Maertens G, Wakita T, Wychowski C, Dubuisson J. 2006. Subcellular localization of hepatitis C virus structural proteins in a cell culture system that efficiently replicates the virus. J. Virol. 80:2832–2841. 10.1128/JVI.80.6.2832-2841.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Teterina NL, Lauber C, Jensen KS, Levenson EA, Gorbalenya AE, Ehrenfeld E. 2011. Identification of tolerated insertion sites in poliovirus non-structural proteins. Virology 409:1–11. 10.1016/j.virol.2010.09.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Teterina NL, Pinto Y, Weaver JD, Jensen KS, Ehrenfeld E. 2011. Analysis of poliovirus protein 3A interactions with viral and cellular proteins in infected cells. J. Virol. 85:4284–4296. 10.1128/JVI.02398-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Huang C, Mohanty S. 2010. Challenging the limit: NMR assignment of a 31 kDa helical membrane protein. J. Am. Chem. Soc. 132:3662–3663. 10.1021/ja100078z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Moradpour D, Evans MJ, Gosert R, Yuan Z, Blum HE, Goff SP, Lindenbach BD, Rice CM. 2004. Insertion of green fluorescent protein into nonstructural protein 5A allows direct visualization of functional hepatitis C virus replication complexes. J. Virol. 78:7400–7409. 10.1128/JVI.78.14.7400-7409.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nevo-Yassaf I, Yaffe Y, Asher M, Ravid O, Eizenberg S, Henis YI, Nahmias Y, Hirschberg K, Sklan EH. 2012. Role for TBC1D20 and Rab1 in hepatitis C virus replication via interaction with lipid droplet-bound nonstructural protein 5A. J. Virol. 86:6491–6502. 10.1128/JVI.00496-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wölk B, Büchele B, Moradpour D, Rice CM. 2008. A dynamic view of hepatitis C virus replication complexes. J. Virol. 82:10519–10531. 10.1128/JVI.00640-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ma Y, Anantpadma M, Timpe JM, Shanmugam S, Singh SM, Lemon SM, Yi M. 2011. Hepatitis C virus NS2 protein serves as a scaffold for virus assembly by interacting with both structural and nonstructural proteins. J. Virol. 85:86–97. 10.1128/JVI.01070-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Merz A, Long G, Hiet MS, Brügger B, Chlanda P, Andre P, Wieland F, Krijnse-Locker J, Bartenschlager R. 2011. Biochemical and morphological properties of hepatitis C virus particles and determination of their lipidome. J. Biol. Chem. 286:3018–3032. 10.1074/jbc.M110.175018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Popescu CI, Callens N, Trinel D, Roingeard P, Moradpour D, Descamps V, Duverlie G, Penin F, Héliot L, Rouillé Y, Dubuisson J. 2011. NS2 protein of hepatitis C virus interacts with structural and non-structural proteins towards virus assembly. PLoS Pathog. 7:e1001278. 10.1371/journal.ppat.1001278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Prentoe J, Bukh J. 2011. Hepatitis C virus expressing flag-tagged envelope protein 2 has unaltered infectivity and density, is specifically neutralized by flag antibodies and can be purified by affinity chromatography. Virology 409:148–155. 10.1016/j.virol.2010.10.034 [DOI] [PubMed] [Google Scholar]
- 36. Stapleford KA, Lindenbach BD. 2011. Hepatitis C virus NS2 coordinates virus particle assembly through physical interactions with the E1-E2 glycoprotein and NS3-NS4A enzyme complexes. J. Virol. 85:1706–1717. 10.1128/JVI.02268-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Miyanari Y, Atsuzawa K, Usuda N, Watashi K, Hishiki T, Zayas M, Bartenschlager R, Wakita T, Hijikata M, Shimotohno K. 2007. The lipid droplet is an important organelle for hepatitis C virus production. Nat. Cell Biol. 9:1089–1097. 10.1038/ncb1631 [DOI] [PubMed] [Google Scholar]
- 38. Shavinskaya A, Boulant S, Penin F, McLauchlan J, Bartenschlager R. 2007. The lipid droplet binding domain of hepatitis C virus core protein is a major determinant for efficient virus assembly. J. Biol. Chem. 282:37158–37169. 10.1074/jbc.M707329200 [DOI] [PubMed] [Google Scholar]
- 39. Counihan NA, Rawlinson SM, Lindenbach BD. 2011. Trafficking of hepatitis C virus core protein during virus particle assembly. PLoS Pathog. 7:e1002302. 10.1371/journal.ppat.1002302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Galli A, Scheel TK, Prentoe JC, Mikkelsen LS, Gottwein JM, Bukh J. 2013. Analysis of hepatitis C virus core/NS5A protein co-localization using novel cell culture systems expressing core-NS2 and NS5A of genotypes 1-7. J. Gen. Virol. 94:2221–2235. 10.1099/vir.0.053868-0 [DOI] [PubMed] [Google Scholar]
- 41. Eggert D, Rosch K, Reimer R, Herker E. 2014. Visualization and analysis of hepatitis C virus structural proteins at lipid droplets by super-resolution microscopy. PLoS One 9:e102511. 10.1371/journal.pone.0102511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Han Q, Aligo J, Manna D, Belton K, Chintapalli SV, Hong Y, Patterson RL, van Rossum DB, Konan KV. 2011. Conserved GXXXG- and S/T-like motifs in the transmembrane domains of NS4B protein are required for hepatitis C virus replication. J. Virol. 85:6464–6479. 10.1128/JVI.02298-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Elazar M, Liu P, Rice CM, Glenn JS. 2004. An N-terminal amphipathic helix in hepatitis C virus (HCV) NS4B mediates membrane association, correct localization of replication complex proteins, and HCV RNA replication. J. Virol. 78:11393–11400. 10.1128/JVI.78.20.11393-11400.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gouttenoire J, Montserret R, Kennel A, Penin F, Moradpour D. 2009. An amphipathic alpha-helix at the C terminus of hepatitis C virus nonstructural protein 4B mediates membrane association. J. Virol. 83:11378–11384. 10.1128/JVI.01122-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Blight KJ. 2011. Charged residues in hepatitis C virus NS4B are critical for multiple NS4B functions in RNA replication. J. Virol. 85:8158–8171. 10.1128/JVI.00858-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wilson GK, Stamataki Z. 2012. In vitro systems for the study of hepatitis C virus infection. Int. J. Hepatol. 2012:292591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Billerbeck E, de Jong Y, Dorner M, de la Fuente C, Ploss A. 2013. Animal models for hepatitis C. Curr. Top. Microbiol. Immunol. 369:49–86. 10.1007/978-3-642-27340-7_3 [DOI] [PubMed] [Google Scholar]
- 48. Chu D, Ren S, Hu S, Wang WG, Subramanian A, Contreras D, Kanagavel V, Chung E, Ko J, Amirtham Jacob Appadorai RS, Sinha S, Jalali Z, Hardy DW, French SW, Arumugaswami V. 2013. Systematic analysis of enhancer and critical cis-acting RNA elements in the protein-encoding region of the hepatitis C virus genome. J. Virol. 87:5678–5696. 10.1128/JVI.00840-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Comparison of current profile with previously published HCV profile. Download
Examination of NS2 and NS5A functional profiles. Download
Approach for epitope tagging in hard-to-tag areas of the HCV genome. Download
Supplemental materials and methods. Download
HCV footprinting data set. The entire data set is available in this Excel spreadsheet. This file includes information on genomic positions of insertional mutant viruses, amino acid sequences of insertions, raw counts (P0, P1, and P2), normalized counts, and P1/P0, P2/P0, and P1/P0 ratios. Download
Strategy for functional profiling of HCV insertion libraries by massively parallel sequencing. (A) Generation of an HCV insertion library by random insertion mutagenesis of a plasmid carrying the HCV genome (pFNX-HCV). kanR and ampR are kanamycin and ampicillin resistance genes. (B) Passaging the mutagenized HCV RNA insertion library. (C) Library preparation and Illumina sequencing. Download
Mapping tolerated insertions to the structure of p7 viroporin and N terminus of NS2. (A) Surface views of annotated p7 viroporin structure (PDB code 2M6X). Gray indicates a lack of the respective insertion mutant in pool 0. We colored amino acid residues in the p7 structure according to the heat map in Fig. 2B. Functional data for AAA, CGR, and RPH/Q frames are shown separately. Surface transparency was set to 65% for top and bottom views to reveal ribbon representations inside viroporin. Yellow dotted circles indicate area tolerating insertions for three reading frames. (B) Helical segments (zigzag lines), turns (arcs) and unstructured loops (straight lines) were deduced from NMR analyses (PDB codes 2JY0, 2KWT, and 2KWZ) of NS2 peptides (aa 1 to 27, 27 to 59, and 60 to 99). (Bottom) Tentative model of NS2 topology within the ER membrane. Download
Immunofluorescence assay of cells infected with epitope-tagged HCV. Huh-7.5.1 cells were infected with indicated epitope-tagged viruses. Three days after infection, we fixed and processed cells for immunofluorescence using antibodies against the inserted epitope tag and either NS5A or E2 viral proteins. Hoechst dye stains the nuclei of both infected and uninfected cells. Images were acquired by epifluorescence microscopy. Scale bar: 10 µm. Download
Four-color fluorescence microscopy of cells infected with NS5A-FLAG virus. (A) (Right) Overview of an infected cell, with the nucleus marked by a dashed line. We acquired multiplexed images with specific filters for each fluorochrome and pseudocolored each channel as indicated: lipid droplets in gray, core in green, E2 in red, and NS5A-FLAG in blue. Bar, 10 µm. The box indicates a perinuclear area rich in lipid droplets and expressing viral proteins. (Left) Zoom-in of the boxed region. Bar, 1 µm. (B) To better visualize localization of each viral protein with regard to lipid droplets, these zoom-in views show only two channels at a time. Here, we pseudocolored lipid droplets in red and each of the three viral proteins in green (NS5A, E2, and core). Arrows indicate areas containing spatially overlapping signals in the NS5A, E2, and core channels. Download
List of primers and antibodies used in this study.