ABSTRACT
Obligate symbioses with bacteria allow insects to feed on otherwise unsuitable diets. Some symbionts have extremely reduced genomes and have lost many genes considered to be essential in other bacteria. To understand how symbiont genome degeneration proceeds, we compared the genomes of symbionts in two leafhopper species, Homalodisca vitripennis (glassy-winged sharpshooter [GWSS]) and Graphocephala atropunctata (blue-green sharpshooter [BGSS]) (Hemiptera: Cicadellidae). Each host species is associated with the anciently acquired “Candidatus Sulcia muelleri” (Bacteroidetes) and the more recently acquired “Candidatus Baumannia cicadellinicola” (Gammaproteobacteria). BGSS “Ca. Baumannia” retains 89 genes that are absent from GWSS “Ca. Baumannia”; these underlie central cellular functions, including cell envelope biogenesis, cellular replication, and stress response. In contrast, “Ca. Sulcia” strains differ by only a few genes. Although GWSS “Ca. Baumannia” cells are spherical or pleomorphic (a convergent trait of obligate symbionts), electron microscopy reveals that BGSS “Ca. Baumannia” maintains a rod shape, possibly due to its retention of genes involved in cell envelope biogenesis and integrity. Phylogenomic results suggest that “Ca. Baumannia” is derived from the clade consisting of Sodalis and relatives, a group that has evolved symbiotic associations with numerous insect hosts. Finally, the rates of synonymous and nonsynonymous substitutions are higher in “Ca. Baumannia” than in “Ca. Sulcia,” which may be due to a lower mutation rate in the latter. Taken together, our results suggest that the two “Ca. Baumannia” genomes represent different stages of genome reduction in which many essential functions are being lost and likely compensated by hosts. “Ca. Sulcia” exhibits much greater genome stability and slower sequence evolution, although the mechanisms underlying these differences are poorly understood.
IMPORTANCE
In obligate animal-bacterial symbioses, bacteria experience extreme patterns of genome evolution, including massive gene loss and rapid evolution. However, little is known about this process, particularly in systems with complementary bacterial partners. To understand whether genome evolution impacts symbiont types equally and whether lineages follow the same evolutionary path, we sequenced the genomes of two coresident symbiotic bacteria from a plant sap-feeding insect and compared them to the symbionts from a related host species. We found that the older symbiont has a highly reduced genome with low rates of mutation and gene loss. In contrast, the younger symbiont has a larger genome that exhibits higher mutation rates and varies dramatically in the retention of genes related to cell wall biogenesis, cellular replication, and stress response. We conclude that while symbiotic bacteria evolve toward tiny genomes, this process is shaped by different selection intensities that may reflect the different ages and metabolic roles of symbiont types.
INTRODUCTION
Obligate mutualisms with bacteria provide insects with the capability to feed on nutritionally deficient diets. Such symbionts are often housed within specialized host cells (bacteriocytes) and transmitted to offspring through transovariolar mechanisms (1). This maternally transmitted, symbiotic lifestyle imposes a reduced effective population size that results in increased fixation of deleterious mutations, including inactivation of nonessential genes (2, 3). As a result, the genomes of some bacterial symbionts are among the smallest known, with several being <200 kb (4–8). Despite this extreme reduction, genes are retained to support nutritional requirements of the host, such as the synthesis of B vitamins or essential amino acids (EAA), depending on the insect’s diet (9–13). Among the lost genes, some correspond to bacterial functions considered to be essential, and the mechanisms compensating for these losses remain poorly understood (reviewed in reference 14).
The genome sizes vary among related symbiont strains from different host species (reviewed in reference 15). In the symbiont genus “Candidatus Blattabacterium,” found in cockroaches and primitive termites, the genome sizes vary by >50 kb due mainly to differential losses of vitamin and EAA pathways. The host switch from omnivory to wood feeding was accompanied by the acquisition of a stable gut microbiota hypothesized to provide functions previously assumed by “Ca. Blattabacterium,” potentially enabling further gene loss (16, 17). In some cases, the acquisition of a cosymbiont enables an ancestral symbiont to lose genes that encode redundant functions. For example, in the aphid Cinara cedri, Buchnera aphidicola has a genome ~225 kb smaller than that of other Buchnera species, apparently the consequence of acquiring an additional symbiont, Serratia symbiotica (5, 18). Even more extreme genome reduction has occurred in the mealybug symbiont, “Candidatus Tremblaya princeps,” which has adopted its own intracellular bacterium (8, 19, 20). Finally, even when symbiont lineages inhabit related hosts with similar feeding habits, their genome sizes vary due to gene losses underlying essential bacterial functions (e.g., information processing, energy metabolism, and cell envelope synthesis) (15). Examples include Buchnera aphidicola in aphids, “Candidatus Carsonella ruddi” in psyllids, “Ca. Blattabacterium” species in cockroaches, and “Candidatus Portiera aleyrodidarum” in whiteflies (4, 16, 21–28). Ongoing gene losses likely change host-symbiont interactions in a lineage-specific fashion, but these patterns remain largely unexplored.
Members of the plant sap-feeding insect suborder Auchenorrhyncha (Hemiptera) have relied on at least two obligate bacterial symbionts for EAA synthesis for over 260 million years. This insect clade is broadly associated with “Candidatus Sulcia muelleri” (Bacteroidetes), which synthesizes a set of seven or eight EAAs, and one of several coprimary symbionts, which synthesize the two or three remaining EAAs (reviewed in reference 13). “Ca. Sulcia muelleri” is a highly conserved symbiont, hypothesized to be present in a shared ancestor of the Auchenorrhyncha and retained in most lineages (29, 30). It retains a central role in EAA metabolism and a conserved genomic architecture (reviewed in reference 31) but shows variations in genome size, mostly affecting capabilities for tryptophan biosynthesis, transcription/translation, and energy metabolism (7, 13). The ancestral Auchenorrhyncha also possessed a second symbiont, in the Betaproteobacteria, named “Candidatus Nasuia deltocephalinicola” in leafhoppers and “Candidatus Zinderia insecticola” in spittlebugs; this symbiont codiversified with “Ca. Sulcia muelleri” and hosts since its origin (7, 30, 32) but was lost and replaced in numerous host lineages. It was replaced by “Candidatus Hodgkinia cicadicola” (Alphaproteobacteria) in cicadas, a Sodalis-like symbiont (Gammaproteobacteria) in the spittlebug subfamily Philaenini, and by “Candidatus Baumannia cicadellinicola” (Gammaproteobacteria) in leafhoppers (6, 31, 33). The acquisition of “Ca. Baumannia cicadellinicola” may have permitted leafhoppers to transition from phloem to xylem sap (11, 34). The genome of “Ca. Baumannia cicadellinicola” from Homalodisca vitripennis (glassy-winged sharpshooter [GWSS], formerly named Homalodisca coagulata) is one of the largest genomes sequenced from obligate symbionts of insects (11).
To address the question of the genomic stability of obligate symbionts living together within the same host, we sequenced complete “Ca. Baumannia cicadellinicola” and “Ca. Sulcia muelleri” genomes from Graphocephala atropunctata (Cicadellinae), commonly known as the blue-green sharpshooter (BGSS). BGSS was selected because it is distantly related (tribe: Cicadellini) to GWSS (tribe: Proconiini) (34). (These symbiont strains are referred to hereinafter as S-BGSS and S-GWSS for the two strains of “Ca. Sulcia muelleri” and as B-BGSS and B-GWSS for the two strains of “Ca. Baumannia cicadellinicola.”) Both insect species vector Pierce’s disease (Xylella fastidiosa) in the United States, causing millions of dollars in agricultural losses (35). A comparative genomic approach targeting the differences between the symbiotic systems of these two leafhopper species offers the opportunity to (i) discern the patterns of genome evolution in a symbiont lineage that may be at an intermediate stage of genome streamlining and (ii) elucidate the evolutionary path to highly reduced genomes.
RESULTS AND DISCUSSION
Symbiont genome characteristics for BGSS.
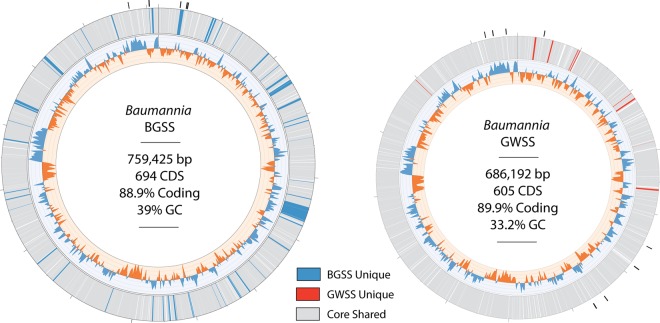
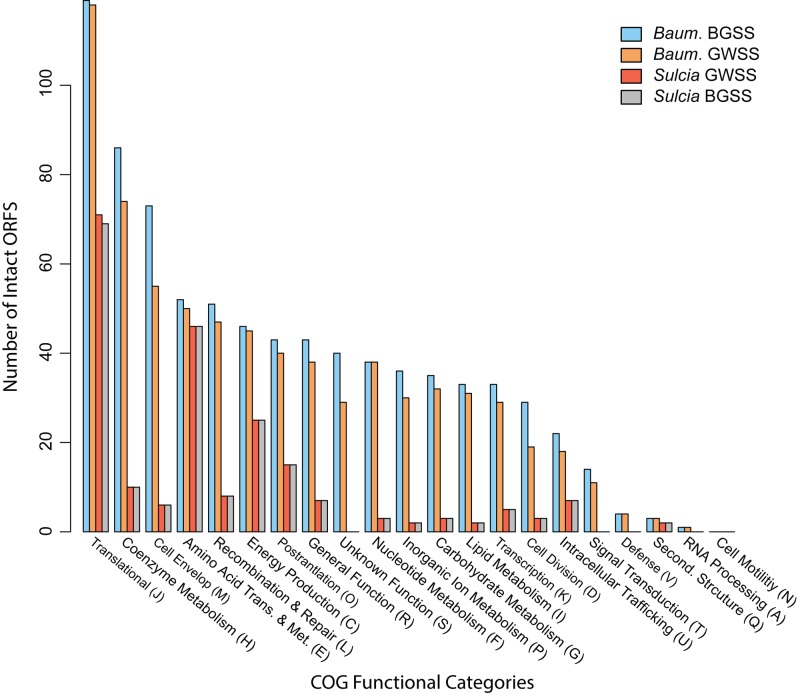
The genome sizes for S-BGSS and B-BGSS are 244,618 and 759,425 bp, respectively (Fig. 1). Both symbionts have low GC content, which is typical in obligate symbioses: the GC content is 17% for S-BGSS and 39% for B-BGSS. B-BGSS contains 694 predicted protein-coding sequences (CDS), of which 20 encode hypothetical proteins. It contains two ribosomal cassettes, 39 tRNAs, and 10 CDS that are truncated or pseudogenized with uncertain function (see Table S2 in the supplemental material). S-BGSS has 225 CDS, with 10 encoding hypothetical proteins, a single ribosomal cassette, 31 tRNAs, and no identifiable pseudogenes (Table S3). Figure 2 shows clusters of orthologous groups (COG) assignments for genes in both symbiont genomes from BGSS and GWSS. For B-BGSS, 69 genes could not be assigned.
FIG 1 .
Comparison of “Ca. Baumannia cicadellinicola” genomes from two different sharpshooter leafhopper host species, Graphocephala atropunctata (blue-green sharpshooter [BGSS]) and the previously sequenced Homalodisca vitripennis (glassy-winged sharpshooter [GWSS]) (11). Genes are color-coded as core shared genes or unique to BGSS or GWSS “Ca. Baumannia” as indicated in the key. The genomes are completely syntenic except for large contiguous gene deletions. The graph on the inner track shows genomewide GC skew, and the outer track shows predicted genes for each genome. Genes are color-coded according to whether they are unique to a genome (e.g., blue for unique BGSS genes) or whether they are shared between genomes (grey). Black tick marks around the outer track show inferred origins of replication.
FIG 2 .
Bar chart showing distribution of functional categories of clusters of orthologous groups (COGs) for predicted protein-coding genes for “Ca. Baumannia cicadellinicola” and “Ca. Sulcia muelleri” from BGSS and GWSS. Bars are color coded according to genome (see key). ORFS, open reading frames.
S-BGSS genome and function.
To date, three sequenced genomes are available for “Ca. Sulcia” in sharpshooter leafhoppers, including S-BGSS (this study), S-GWSS (12), and “Ca. Sulcia muelleri” from Draeculacephala minerva (36). The three genomes are perfectly syntenic and vary in content by only a few genes involved in the transcriptional and translational machinery (Fig. 2). S-BGSS has lost the phenylalanyl-tRNA synthetase gene (pheST), and “Ca. Sulcia muelleri” from D. minerva has lost a ribosomal subunit gene (rplJ) and the 16S methyltransferase gene (rsmD). Otherwise, the identical metabolic functions of “Ca. Sulcia muelleri” within sharpshooters have recently been reviewed (12, 36). Genes that encode tRNA synthetases are routinely purged from symbionts with tiny genomes, including the other “Ca. Sulcia muelleri” (7, 8, 27). Similarly, some ribosomal proteins are often lost from symbiont genomes, as are other rRNA processing and methylation genes (reviewed in reference 15). The extreme genomic conservation of sharpshooter “Ca. Sulcia muelleri” is striking given that its hosts diverged over 70 million years ago. This stands in stark contrast to its companion symbiont, “Ca. Baumannia cicadellinicola” (reviewed below).
Functional overview of “Ca. Baumannia cicadellinicola” strains.
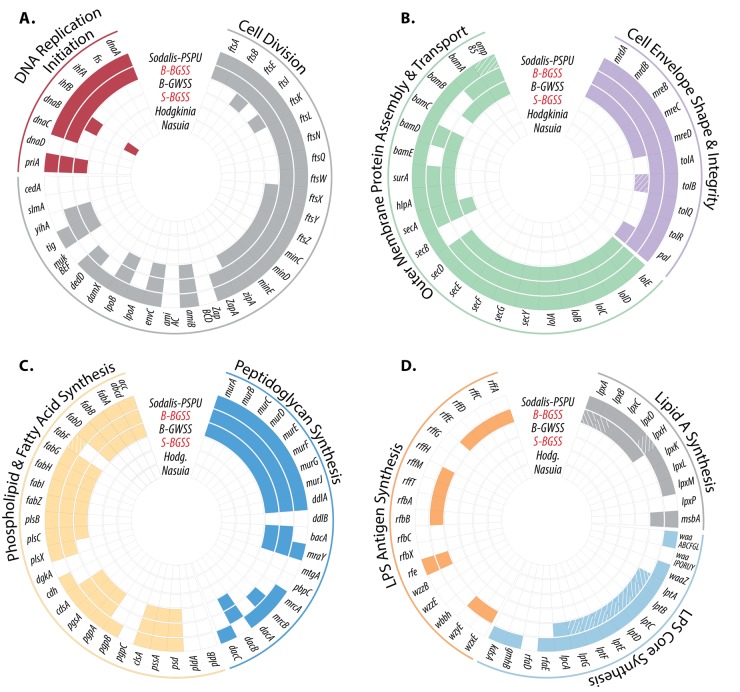
The B-BGSS genome is 73 kb larger than the previously sequenced genome of B-GWSS (686 kb) and retains 89 additional genes (Fig. 1 and 3). Aside from deleted regions, the “Ca. Baumannia cicadellinicola” genomes are perfectly syntenic. Unique B-BGSS genes are distributed in nearly every COG category but are enriched for functions related to cell envelope synthesis (category M; n = 18), coenzyme and inorganic metabolite synthesis and transport (categories H and P; n = 18), and cell cycle and division (category D; n = 10) (Fig. 2; see Table S2 in the supplemental material). Thus, B-BGSS appears to retain more autonomy involving capabilities in these categories (Fig. 3A to D). For example, B-BGSS retains the complete pathway for an additional ubiquinone coenzyme involved in the electron transport chain, transcriptional regulation genes (e.g., metJ, hha, cpdA, etc.), and transporters (e.g., iron [feoABC], glutamate/aspartate [gltP], and sialic acid [nanT] transporter genes). In the sections below, we compare and contrast the major metabolic differences implied by the “Ca. Baumannia cicadellinicola” genomes.
FIG 3 .
Schematics showing the presence and absence of genes involved in central cellular processes for the obligate symbionts of the Auchenorrhyncha. Colored boxes indicate gene presence, white boxes show gene absence, and hatched boxes show predicted pseudogenes. “Ca. Baumannia cicadellinicola” (B) and “Ca. Sulcia muelleri” (S) symbionts are abbreviated according to their hosts (BGSS and GWSS). Hodgkinia or Hodg. is “Candidatus Hodgkinia cicadicola,” and Nasuia is “Candidatus Nasuia deltocephalinicola.” A single “Ca. Sulcia muelleri” strain is shown as representative for the sharpshooter strains (S-BGSS), which have lost most of these major pathways in all auchenorrhynchan hosts. Genomes are arranged by size from smallest (inner ring) to largest (outer ring). The genomes sequenced in this study are highlighted in red.
B-GWSS retains only 15 unique genes but no major pathways that are absent from B-BGSS. These differences include genes for proteins involved in translational machinery (epmAB), outer membrane (OM) synthesis and integrity (lipoprotein genes bamBE and lpp, respectively), a tricarboxylic acid (TCA) cycle enzyme (succinyl-CoA synthetase β-subunit gene [sucC]), and methionine synthesis (metAB) and three other putative genes with uncertain functions (e.g., genes for two membrane-bound proteins and endonuclease). Thus, the capabilities of B-GWSS are largely a subset of those of B-BGSS.
B-BGSS additionally contains a plasmid (pB-BGSS) that is 6,564 bp in length, with GC content of 32.0% (see Fig. S1 in the supplemental material). No plasmid was reported for the B-GWSS genome sequence. pB-BGSS carries five genes that are shared with several plasmids sequenced from B. aphidicola in aphids (37, 38), including genes encoding two replication initiation protein variants (RepA1-2), two Hsp20 small heat shock proteins (IbpA), and a highly conserved integral membrane protein (YqhA). The functional presence of two replication initiation proteins is unclear, as is the overall role of yqhA. IbpA is known to stabilize denatured proteins for refolding under heat stress (39). pB-BGSS encodes an additional protein, QmcA, which can suppress the activities of FtsH and HtpX (40), which are encoded on the main chromosome. FtsH and HtpX are involved in the proper folding and degradation of aberrant proteins in the membrane or misfolded proteins that result from heat stress, respectively (reviewed in reference 41). Thus, it appears that pB-BGSS may play a regulatory role in mitigating protein malfunction in “Ca. Baumannia cicadellinicola,” possibly during stress.
Essential amino acid synthesis.
One of the most notable gene losses in B-BGSS relative to the genome of B-GWSS involves the first two steps of methionine biosynthesis—one of the few inferred requirements of “Ca. Baumannia cicadellinicola” by the leafhopper host. B-BGSS has lost metA, and metB appears to have been recently pseudogenized, as it exists as several fragments with interspersed deleted regions (verified with Sanger sequencing). Together, metAB are responsible for the initiating steps in methionine synthesis that catalyze the reaction leading from homoserine to cystathionine. In the absence of these genes, it is unclear how methionine synthesis is initiated. Some other symbionts, including B. aphidicola and “Ca. Tremblaya princeps,” retain only metE (8, 42), underlying the final step in methionine synthesis. Recent genomic and transcriptomic data have revealed that the host might be able to initiate the first few steps of methionine synthesis from sulfate (43, 44). In both aphid and mealybug bacteriocytes, genes that can accomplish this task are highly expressed (e.g., cystathionine γ-lyase) (see references 19 and 44). These findings add to evidence that insect hosts are likely to be intimately involved in these pathways. Whether the host can supplement methionine pathway losses remains untested for “Ca. Baumannia cicadellinicola,” but this difference provides evidence for a greater level of host-symbiont complementation in BGSS than in GWSS with respect to amino acid synthesis.
Bacterial morphology and cell envelope synthesis.
The cell shape differs markedly between the two “Ca. Baumannia cicadellinicola” symbionts: B-BGSS retains a rod-shaped cell resembling those of other Gammaproteobacteria, whereas B-GWSS occurs as spheroid cells (Fig. 4B and C) (29). The basis for the difference is not clear. Although both genomes retain genes encoding rod shape-determining proteins (Fig. 3B, MrdAB and MreBCD), B-BGSS retains more genetic pathways involving cell envelope integrity and cellular division, which are discussed further below. In other ancient obligate symbionts with highly reduced genomes, altered and bizarre cellular morphologies are often observed, including spheroid (B. aphidicola), pleomorphic (“Ca. Zinderia,” “Ca. Nasuia,” “Ca. Tremblaya princeps,” and “Candidatus Moranella endobia”), and large straplike (“Ca. Carsonella” species, “Ca. Hodgkinia” species, and “Ca. Sulcia” species) shapes (1, 8, 13, 14, 29, 45, 46). This morphological degeneration is correlated with the loss of genes that underlie the production of components of the outer and inner membranes and cell wall. For example, the obligate enlarged and amorphous cells of symbionts from mealybugs, psyllids, and most members of the Auchenorrhyncha no longer encode genes for the synthesis of peptidoglycan and phospholipids, nor do they have genes for synthesizing any membrane-associated lipids (4, 8, 12, 13). The larger genome of B. aphidicola in pea aphids, on the other hand, encodes genes for synthesizing cell wall components but not for the essential phospholipid membrane (42). In these cases, the host must provide a membrane, but how this functional transition evolved remains an open question.
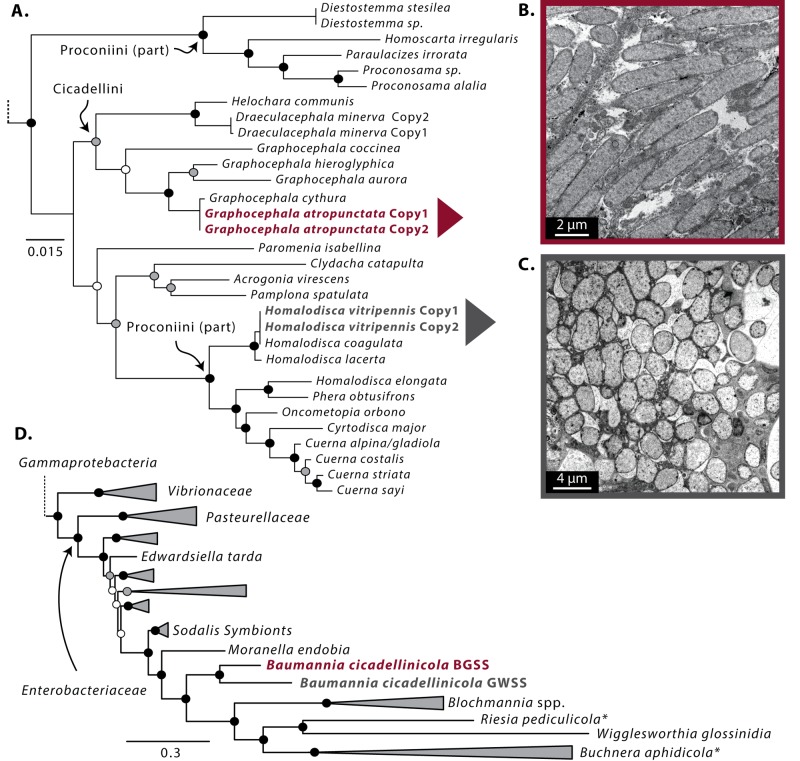
FIG 4 .
Phylogenetic relationships and transmission electron micrographs (TEM) of “Ca. Baumannia cicadellinicola” strains. (A to C) Phylogenetic relationships of “Ca. Baumannia cicadellinicola” from across sharpshooter leafhoppers inferred with 16S rRNA sequences from previous studies (33, 34). Colored arrows show the focal “Ca. Baumannia cicadellinicola” strains and corresponding TEM images for B-BGSS (B) and B-GWSS (C). (D) Phylogenomic placement of the investigated “Ca. Baumannia cicadellinicola” genomes within the Enterobacteriaceae (Gammaproteobacteria) and as sister to the Sodalis-like clade (n = 4, see Fig. S3 for specific Sodalis species). Asterisks next to symbiont names show possible artifacts of long-branch attraction (66). Riesia pediculicola is “Candidatus Riesia pediculicola.”
Both B-GWSS and B-BGSS retain considerable machinery for cell envelope biogenesis, as their genomes encode the capabilities to synthesize fatty acids (acc and fab clusters), phospholipid membrane (plsBC), and cell wall components (mur cluster) (Fig. 3). However, B-BGSS retains additional capabilities to synthesize part of an outer membrane (OM). The OM is characteristic of free-living Gram-negative bacteria; it provides protection from the environment, and it is the etiological agent of septic shock in animals (reviewed in references 47 and 48). The OM is formed by an inner phospholipid leaflet and an outer lipopolysaccharide (LPS) leaflet that comprises a basal lipid A layer, lipopolysaccharide core, and surface antigens. B-BGSS but not B-GWSS encodes pathways for the partial synthesis and assembly of lipid A (lpx cluster and msbA), but both have lost nearly all genes for the synthesis of the core LPS chains (waa and lpt clusters, respectively). B-BGSS but not B-GWSS also contains a partial O antigen synthesis pathway (rfbAB) and a nearly complete set of genes for the assembly of an enterobacterial common antigen (ECA; rff cluster, wzyE elongation factor, and wzxE flippase). Bacterial antigens are typically anchored to the LPS core. Finally, both B-GWSS and B-BGSS contain genes coding for the transport and assembly of integral OM proteins (e.g., hlpA, encoding a periplasmic transporter, protein-folding surA, and OMP85 family genes [tamA]). B-GWSS may still embed exogenous proteins in its OM, but the molecular origin of the OM is unknown.
The B-BGSS genome encodes additional pathways that contribute to the structure and integrity of the cell envelope. It has a nearly complete Tol-Pal system that anchors the outer and inner membranes to the peptidoglycan cell wall (Fig. 3B) (49–51). Mutants with knockouts of the Tol genes in Erwinia chyrsanthemi (Enterobacteriaceae) can have altered bacterial cell shape (52). Thus, it is plausible that the rod shape observed in B-BGSS but not B-GWSS is linked to the retention of the Tol-Pal system in B-BGSS.
Our results suggest that the capability of “Ca. Baumannia” strains to synthesize a cell envelope is in flux. As symbionts lose the ability to produce their own cell envelope, the host presumably compensates, which may yield more direct control over the symbiont cell population size, metabolite exchange, and symbiont localization. This suggests that this ability is being ceded to the host, as appears to have already occurred in other ancient symbioses in insects (7, 12, 13, 26, 27, 53). The B-GWSS and B-BGSS genomes may provide clues to the earlier steps during this process. For example, apparent loss of the ability to synthesize the core region of LPS and the retention of the ECA are shared features between B-BGSS and the other recently established Sodalis-like endosymbionts of grain weevils and Philaenine spittlebugs (31, 54). Possibly there is early selection to denude LPS molecules because they are endotoxic and elicit host immune responses, as is observed in the grain weevil-Sodalis symbiosis (54, 55). The host may use the remaining surface molecules to recognize and control symbiont localization to the bacteriocyte.
Symbiont replication and division.
The genome of B-BGSS retains several genetic pathways and genes that are involved in accomplishing cellular replication and division. Broadly, cellular replication requires initiation, chromosome replication, cell envelope expansion, and cell division (reviewed in reference 51 ). B-BGSS retains the capability to initiate and complete chromosome replication; in contrast, this ability has been stripped from B-GWSS (Fig. 3A) (11). These genes include dnaA, which is responsible for initiating DNA replication, and fis and ihfAB, which increase the precision and stability of chromosomal replication (56, 57). All three genes function at the origin of replication (oriC), which appears to be absent from the genomes of both B-GWSS and B-BGSS, as there is no obvious GC skew (Fig. 1). Genome scans with Ori-Finder identify five oriC regions under liberal settings, but none are identified under more strict defaults. However, genomic Blast searches against the DoriC database did not reveal any oriC orthologs. To date, the only other symbiont associated with the Auchenorrhyncha that contains recognizable genes for initiating its own DNA replication is the relatively young Sodalis-like symbiont in spittlebugs (Fig. 3A) (31). Thus, the loss of these abilities is an eventual outcome in obligate symbioses.
The propagation and establishment of obligate symbionts in successive host insect generations requires symbiont cell replication and division. However, this remains one of the major essential functions typically lost from bacterial symbionts with small genomes. In this respect, B-BGSS retains vastly more genetic capabilities and autonomy than does B-GWSS (Fig. 3A). Specifically, during cell elongation and septation (splitting), a series of divisome proteins in the Fts cluster and other auxiliary proteins consecutively bind to form the septal ring at the division site (58). While both the B-GWSS and the B-BGSS genome retain the genes for the FtsZ and MinCDE proteins that initialize and localize septal ring formation (59), only B-BGSS encodes nearly all of the divisome proteins. B-GWSS has lost the genes for two early-stage proteins (FtsA and ZapA) and the entire downstream half of the pathway (Fig. 3A, FtsQBLWIN). Many of these genes are universally conserved in bacteria, and experimental evidence demonstrates that the loss of any is highly detrimental to cellular division in Escherichia coli (60). It is unlikely that B-GWSS is able to initiate and carry out cellular division on its own and, in contrast to the capabilities of B-BGSS, some other mediating source is likely required.
Bacterial cell growth requires the peptidoglycan sacculus extension, structural maintenance, and cleavage. While both the B-GWSS and the B-BGSS genome are genetically depauperate in these categories, only B-BGSS clearly retains some genes that can accomplish these tasks. B-BGSS retains the bifunctional transglycosylase/transpeptidase, mrcB, that is essential in cell growth and is one of the main peptidoglycan sacculus synthesis genes (reviewed in reference 61). Thus, it is unclear how B-GWSS accomplishes cell wall growth. Structural maintenance and integrity of the cell envelope during growth are at least partially maintained by the tol-pal pathway, which is also involved in outer membrane constriction during cell division (62). As discussed above, it is missing from the B-GWSS genome. Experimental evidence suggests that the tol-pal pathway is not essential in cell growth and viability because it is partially redundant with lpoB-mrcB functions (63). However, the latter genes are missing from B-GWSS as well, and their deletion usually results in severe cell division defects and lethality. The final cell division step is to cleave budding cells. Both the B-GWSS and the B-BGSS genome retain the gene for only one of three enzymes that cleave peptidoglycan sacculus, AmiB. Combinatorial knockouts of the missing amiA and amiC result in a high percentage of undivided E. coli cells (64). This morphological defect has not been observed in either B-GWSS or B-BGSS, indicating that either AmiB or an exogenous source efficiently cleaves cells. As an example of the latter case, the mealybug has horizontally acquired amiD from other bacteria, which is capable of cleaving peptidoglycan and is one of the most highly expressed genes in the bacteriocyte (19).
Stress response.
Among the increased capabilities of B-BGSS is the regulation of metabolic functions under stress conditions and nutrient limitation. In particular, B-BGSS retains the spoT gene that is required for operating the stringent response (SR). The SR suppresses the transcription of proteins involved in metabolic processes, chromosome replication, cellular growth, and division when bacteria are starved for amino acids and other required elements (e.g., iron and carbon) (reviewed in reference 65). This is accomplished by modulating the levels of the alarmone, ppGpp, which in abundance acts as a transcriptional down-regulator. During normal environmental and growth conditions, spoT operates to degrade ppGpp, which is constitutively synthesized by relA and spoT depending on the nutritive conditions. Notably, B-GWSS retains relA, which indicates that it can produce ppGpp, but the regulation of its function is unknown. Possibly the host insect plays a role in controlling the levels of ppGpp. This in turn would provide the opportunity to control symbiont metabolism and growth. The retention of spoT-relA in B-BGSS, in contrast to other reduced-genome symbionts, provides further evidence that this symbiont retains an unusual degree of autonomy over its own metabolism. Conceivably, the maintenance of the SR can be beneficial to the host and symbiont, as it would render B-BGSS less metabolically active and taxing to the host during times of nutritional stress.
Origins of “Ca. Baumannia cicadellinicola.”
To ascertain the potential origins of “Ca. Baumannia cicadellinicola” strains, we reconstructed their phylogenetic relationships within sharpshooters and across Gammaproteobacteria. The phylogenetic relationships for “Ca. Baumannia” strains inferred with 16S rRNA sequences support the placement of B-BGSS with other “Ca. Baumannia cicadellinicola” strains previously sequenced from other Graphocephala lineages in the tribe Cicadellini (Fig. 4A, bootstrap support [BS] = 58 to 100) (34). The previously sequenced B-GWSS lineage is placed within the tribe Proconiini (BS = 100), which is paraphyletic according to symbiont molecular data. These results largely corroborate the previous findings of Takiya et al. (34), who also demonstrated the one-to-one codiversification of “Ca. Baumannia” and sharpshooters.
Our phylogenomic results support the placement of “Ca. Baumannia cicadellinicola” in the Enterobacteriaceae (Gammaproteobacteria) (Fig. 4D, BS = 100). There is strong support for a monophyletic clade that includes “Ca. Baumannia cicadellinicola” and some other insect symbionts together with the Sodalis cluster of symbionts (Fig. 4D, BS = 100). However, this result should be treated with caution, as variations in rates of evolution and biases in base substitution patterns may confound the results. For example, the result that symbionts from disparate insect groups, such as ants and sharpshooters, derive from a single ancestor could be specious due to the high levels of divergence of these symbiont lineages and inadequate sampling of free-living diversity in the same group. Furthermore, previous work by Husnik et al. (66), using an amino acid recoding approach aimed at reducing the effects of biased molecular evolution (e.g., Dayhoff6 models) in a Bayesian framework, suggested that some taxa in the monophyletic symbiont clade resolved here (and reported in other papers, e.g., references 67 and 68) derive from independent origins; i.e., B. aphidicola in aphids from a Pantoea/Erwinia-like ancestor and “Candidatus Riesia pediculicola” from an Arsenophonus-like ancestor (Fig. 4D, asterisks). Representatives from both bacterial lineages were included in this study, but recoding efforts did not change the tree topology.
A previous phylogenomic study also suggested that “Ca. Baumannia cicadellinicola” is derived from Sodalis-like ancestors (66). Sodalis species are common insect symbionts found in a diversity of insect orders that sometimes have replaced more ancient symbionts (67, 69). The acquisition and replacement of obligate symbionts by Sodalis-like bacteria has been reported in giant scales (Llaveia spp.), weevils (Sitophilus spp.), and spittlebugs (tribe Philaenini) (31, 66, 70). The long-term coprimary symbionts of some psyllids and mealybugs also appear to be derived from a Sodalis-like ancestor (8, 27).
Molecular evolution in sharpshooter symbionts.
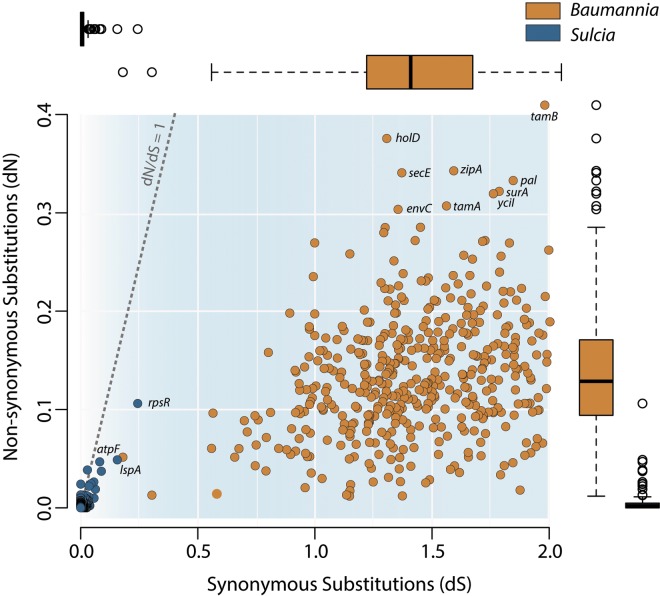
We analyzed the genomewide rates of synonymous (dS) and nonsynonymous (dN) substitutions for the two symbionts (Fig. 5; see Table S4 in the supplemental material). Because “Ca. Baumannia cicadellinicola” and “Ca. Sulcia muelleri” in these hosts diverged in synchrony with their host lineage and with each other (34), the time of divergence for B-GWSS and B-BGSS is the same as that for S-GWSS and S-BGSS, implying that differences in pairwise divergences are due to differences in rates of nucleotide base substitution. In fact, the rates are higher in “Ca. Baumannia”: the average pairwise identities in each symbiont’s genomes are 77.0% and 98.6% for “Ca. Baumannia cicadellinicola” and “Ca. Sulcia muelleri,” respectively. The average dS rates are higher in “Ca. Baumannia cicadellinicola” (1.41) than in “Ca. Sulcia muelleri” (0.02). Of these, 128 “Ca. Baumannia cicadellinicola” genes are in saturation for dS (>2.0). The average dN is also higher in “Ca. Baumannia cicadellinicola” (0.13) than in “Ca. Sulcia muelleri” (0.005). Compared with “Ca. Baumannia cicadellinicola,” all genes have extremely depressed rates of molecular evolution (Fig. 5; Table S4). Higher rates in “Ca. Baumannia cicadellinicola” than in “Ca. Sulcia muelleri” were noted previously for 16S rRNA sequences (34); our analysis shows that this is a genomewide difference.
FIG 5 .
Distribution of genomewide synonymous (dS) and nonsynonymous (dN) rates of molecular evolution in “Ca. Sulcia muelleri” (blue) and “Ca. Baumannia cicadellinicola” (orange). Rates are inferred from pairwise comparisons of all shared orthologs between the symbiont strains obtained from BGSS and GWSS. Adjacent box plots show median, quartiles, and maximum and minimum distributions for dS and dN values on the x- and y-axes, respectively. Box plots are color-coded according to genome.
The average dN/dS ratios in “Ca. Baumannia cicadellinicola” (0.10) and “Ca. Sulcia muelleri” (0.30) genes are less than 1, indicating that most or all genes are under purifying selection. No genes have dN/dS ratios above 1 in “Ca. Baumannia cicadellinicola.” In “Ca. Sulcia muelleri,” the genes have extremely low dN and dS values that often approximate zero, so the dN/dS values do not give strong evidence of either positive or purifying selection (Fig. 5; see Table S4 in the supplemental material).
The disparity in the rates of molecular evolution between symbionts is particularly enigmatic considering that “Ca. Baumannia cicadellinicola” retains more genes involved in DNA replication and repair than does “Ca. Sulcia muelleri” (DNA polymerase complex, complete mutSHL, recA, and uvrD). In B. aphidicola, accelerated rates of evolution are thought to reflect the loss of DNA repair enzymes (71). However, given that “Ca. Sulcia muelleri” has lower rates of evolution and fewer repair enzymes than does “Ca. Baumannia cicadellinicola,” this hypothesis does not readily explain these results. An alternative explanation that could account for the lower dS in “Ca. Sulcia muelleri” is its lower number of tRNA genes, which potentially enforces more constraints on codon usage (“Ca. Baumannia cicadellinicola” has 39 tRNA genes, and “Ca. Sulcia muelleri” has 31 tRNA genes). Indeed, “Ca. Sulcia muelleri” does have stronger codon usage bias, reflecting its stronger bias in genomic base composition (as has been suggested for “Ca. Buchnera aphidicola” in aphids [72]), and potentially, this relates to tRNA retention in the genome and abundance in cells. However, a similar disparity in substitution rates is observed in the rRNA genes: the “Ca. Baumannia” 23S and 16S percent identities are 92% and 94%, respectively, while “Ca. Sulcia” 23S and 16S are both >98% identical. Thus, the elevated rates for both protein and rRNA genes in “Ca. Baumannia cicadellinicola” may reflect mechanisms associated with the fidelity of DNA replication.
Conclusion.
The majority of symbionts of the Auchenorrhyncha have been stripped of most independent capabilities, regardless of the ecology and diet type of the host. Thus, it appears that in heritable, obligate nutritional mutualisms, all bacterial players are slated for extreme genome reduction. The genomic differences between “Ca. Baumannia cicadellinicola” and “Ca. Sulcia muelleri” strains indicate that these symbionts are at different phases in this process. In “Ca. Sulcia muelleri,” one of the oldest known bacterial symbionts, the rates of gene loss and the rates of molecular evolution are low. In “Ca. Baumannia cicadellinicola,” on the other hand, gene loss is ongoing and affects cell envelope synthesis, cell replication, and gene expression and transport. Furthermore, sequenced “Ca. Baumannia cicadellinicola” strains retain only partial metabolic pathways and many pseudogenes, suggesting a relatively recent inactivation of some functions. The piecemeal loss of these capabilities must render symbiotic bacteria increasingly dependent on the host. In contrast, “Ca. Sulcia muelleri” may have already attained a stage of extreme dependence precluding further change without dire consequences to the symbiosis. As this process proceeds, the host genome may evolve in tandem, to fill the metabolic gaps produced by the degenerating genomes of its obligate symbionts. Indeed, emerging studies focusing on the role of the host in sustaining symbioses with degenerate partners have demonstrated host complementation of impaired symbiont metabolisms (8, 19, 44, 73).
MATERIALS AND METHODS
Material preparation and genome sequencing.
We obtained Graphocephala atropunctata females from stock colonies maintained at the University of California, Berkeley, by Rodrigo Almeida. Bacteriomes were isolated from approximately 20 insects by dissecting in 95% ethanol, and purified genomic DNA (gDNA) was extracted using Qiagen blood and tissue kits. Total gDNA was prepared and sequenced using 454 GS FLX technologies according to the manufacturer’s protocol.
We obtained 226,183 reads, totaling 54,196,507 nucleotide bases and containing sequences from both symbionts and host. The initial assembly, performed with Newbler version 1.1.02.15, produced 575 contigs over 500 bases with an N50 of 30,660 and a maximum contig size of 229,604. The S-BGSS genome initially assembled into 16 contigs with an average depth of coverage of 14.7×; the contigs were joined using PCR and Sanger sequencing to give a single circular contig. The initial annotation of S-BGSS revealed 44 potential genes that contained inactivating substitutions or frame shifts. Because 454 sequencing is known to introduce shifts in lengths of homopolymers, we suspected that many of these genes were actually intact. These genes were first checked by manually comparing the region to the finished S-GWSS genome. If the S-BGSS gene was in-frame with the S-GWSS gene with the exception of a homopolymer region, the homopolymer length in S-BGSS was adjusted to match that of S-GWSS. PCR and Sanger sequencing were performed on 23 of these suspected errors, and the results verified that these genes were in-frame. In total, 27 of 44 genes were adjusted based on these data.
The B-BGSS genome initially assembled into 11 contigs with an average depth of coverage of 17.5×. These were joined using PCR and Sanger sequencing to yield a single circular contig. As for S-BGSS, indel errors were checked for 42 genes, of which 12 were verified by PCR and Sanger sequencing and 30 were adjusted. Several representative genes of the major pathways that appeared to be disrupted were further verified with PCR and sequencing, including metB, lpxAB, and lptABCD.
Genome annotation.
Initial annotations for the S-BGSS and B-BGSS genomes were done using the IMG-ER suite (74). The annotations were finalized by manually checking predicted genes with Glimmer3 (75). Predicted protein identities were validated using HMMER3 (76). Origins of replication for B-BGSS were inferred using Ori-Finder and blast searches against the DoriC database (77, 78). Functional characterization and comparisons between the symbiont genomes of GWSS and BGSS were done by assigning predicted proteins to clusters of orthologous groups (COG) (79). Hand-curated genomes were produced with Geneious, version 5.5.8, and visualized using Circos, version 0.61 (80).
TEM.
“Ca. Baumannia” cell morphology was observed with transmission electron microscopy (TEM). Preliminary dissections were conducted to confirm the structure of bacteriomes and location of bacterial symbionts, following the method of Moran et al. (33). Collected tissue was preserved in 4% glutaraldehyde and 2% formaldehyde (EM grade). Dissected bacteriome tissues were embedded in 1% agarose and fixed in an aldehyde mixture containing 4% glutaraldehyde and 2% formaldehyde in 0.1 M cacodylate with 2 mM Ca and 4 mM Mg. The following steps were done with a microwave-vacuum procedure (Pelco BioWave Pro), following the manufacturer’s protocol. Samples were postfixed in reduced osmium tetroxide (2% osmium tetroxide and 2% potassium ferrocyanide in 0.1 M cacodylate buffer), washed with distilled water 5 times for 10 min each, and then dehydrated in 50%, 70%, and 95% ethyl alcohol (EtOH) rinses and 2 times in a 100% EtOH rinse, with 2% uranyl acetate in the 70% step. After two rinses in 100% acetone, the specimens were infiltrated with epoxy resin (Hard-Plus 812; Electron Microscopy Sciences) by soaking in resin-acetone mixtures of 30% and 60%, followed by two steps in 100% resin. Samples were placed in silica molds and incubated for 48 h at 60°C. Thin sections 60 to 70 nm thick were picked up on copper mesh grids and imaged with an FEI Tecnai transmission electron microscope.
Phylogenetics and molecular evolution of sharpshooter symbionts.
Two approaches were taken to determine the phylogenetic relationships of B-BGSS. First, single-locus 16S rRNA phylogenies were reconstructed to infer its placement among previously sequenced “Ca. Baumannia cicadellinicola” strains (see Table S1 for full taxon sampling) (11, 33, 34). The sequences were aligned using Muscle (81) and reconstructed with maximum likelihood (ML) using RAxML, version 7.4.4, as described below (82).
Second, a phylogenomics approach with amino acid sequences was used to determine the placement of “Ca. Baumannia cicadellinicola” within the Gammaproteobacteria, using 176 sequenced genomes of Gammaproteobacteria. Conserved orthologous genes were identified using the Phylo_amphora2 suite, which implements HMMER3 markerscan (E value cutoff of 0.01) (83). Four outgroups were selected from the Betaproteobacteria and Alphaproteobacteria. Matrices were aligned with MAFFT, version 7.017, with the L-INS-I model (84). Redundant taxa and alignments with <50% pairwise identity or with >10% missing taxa were removed. This resulted in an alignment of 64 proteins (17,041 columns) for 180 taxa. Prottest3 was used to assign each gene alignment a unique amino acid likelihood model (85). Phylogenetic trees were reconstructed using ML implemented in RAxML. Genes were concatenated into a single matrix, partitioned by loci, and assigned an independent general time reversible (GTR)-based substitution model. ML searches were done for 100 bootstrap partitions and a final ML tree search. In an attempt to reduce the effect of compositional heterogeneity and long-branch attraction (LBA), ML searches were also run with a recoded amino acid matrix using the Dayhoff6 model (86). While this has been effective at breaking LBA and symbiont clustering in other analyses (66), it did not alter the tree topology in our ML analyses and it reduced the bootstrap support at internal nodes (data not shown).
To determine the dynamics of molecular evolution in these two symbiont types, we estimated the genomewide rates of synonymous (dS) and nonsynonymous (dN) substitutions. Orthologous genes were extracted and aligned using a codon-based model in TranslatorX (87). Pairwise dN/dS values were estimated with the CODEML module in PHYML (settings: Mode = −2, CodonFreq = 2) (88). Genes for which dS was near saturation (dS > 2) or for which dN/dS was incalculable (e.g., dS = 0) were removed from subsequent analyses. All data matrices and batch analyses were run and parsed with custom Python scripts.
Nucleotide sequence accession numbers.
Fully annotated genomes were submitted to GenBank under accession numbers CP008985 and CP008986 for B-BGSS and S-BGSS, respectively.
SUPPLEMENTAL MATERIAL
“Ca. Baumannia” BGSS plasmid (pB-BGSS). Download
Complete phylogenetic tree for “Ca. Baumannia cicadellinicola” strains across sharpshooters using the 16S rRNA loci. Download
Complete phylogenomic tree for Gammaproteobacteria. Download
Phylogenetic taxon sampling.
Predicted genes in the BGSS “Ca. Baumannia” genome.
Predicted genes in the BGSS “Ca. Sulcia” genome.
Molecular characteristics and dN and dS mutation rates for all pairwise comparisons between “Ca. Baumannia cicadellinicola” and “Ca. Sulcia muelleri” strains.
ACKNOWLEDGMENTS
We thank Celia Del Cid, Fabien Labroussaa, and Rodrigo Almeida for their generous contribution of sharpshooter specimens. We thank Alejandro Caro-Qunitero and Allison Joyce for assistance with various aspects of this work. We also thank Erica Di Pierro and Stephen Trent for their helpful insights on symbiont membranes.
G.M.B. is funded by a NIFA fellowship from the United States Department of Agriculture (2013-2013-03420). J.P.M. was funded by the University of Arizona’s Center for Insect Science through an NIH training grant (1K 12 GM00708). This work was funded by the National Science Foundation (IOS 1347116 to N.A.M. and G.M.B.)
Footnotes
Citation Bennett GM, McCutcheon JP, MacDonald BR, Romanovicz D, Moran NA. 2014. Differential genome evolution between companion symbionts in an insect-bacterial symbiosis. mBio 5(5):e01697-14. doi:10.1128/mBio.01697-14.
REFERENCES
- 1. Buchner P. 1965. Endosymbiosis of animals with plant microorganisms. John Wiley & Sons, New York, NY. [Google Scholar]
- 2. Moran NA. 1996. Accelerated evolution and Muller’s ratchet in endosymbiotic bacteria. Proc. Natl. Acad. Sci. U. S. A. 93:2873–2878. 10.1073/pnas.93.7.2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Moran NA, Baumann P. 2000. Bacterial endosymbionts in animals. Curr. Opin. Microbiol. 3:270–275. 10.1016/S1369-5274(00)00088-6 [DOI] [PubMed] [Google Scholar]
- 4. Nakabachi A, Yamashita A, Toh H, Ishikawa H, Dunbar HE, Moran NA, Hattori M. 2006. The 160-kilobase genome of the bacterial endosymbiont Carsonella. Science 314:267. 10.1126/science.1134196 [DOI] [PubMed] [Google Scholar]
- 5. Pérez-Brocal V, Gil R, Ramos S, Lamelas A, Postigo M, Michelena JM, Silva FJ, Moya A, Latorre A. 2006. A small microbial genome: the end of a long symbiotic relationship? Science 314:312–313. 10.1126/science.1130441 [DOI] [PubMed] [Google Scholar]
- 6. McCutcheon JP, McDonald BR, Moran NA. 2009. Origin of an alternative genetic code in the extremely small and GC-rich genome of a bacterial symbiont. PLoS Genet. 5:e1000565. 10.1371/journal.pgen.1000565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bennett GM, Moran NA. 2013. Small, smaller, smallest: the origins and evolution of ancient dual symbioses in a phloem-feeding insect. Genome Biol. Evol. 5:1675–1688. 10.1093/gbe/evt118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. McCutcheon JP, von Dohlen CD. 2011. An interdependent metabolic patchwork in the nested symbiosis of mealybugs. Curr. Biol. 21:1366–1372. 10.1016/j.cub.2011.06.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Akman L, Yamashita A, Watanabe H, Oshima K, Shiba T, Hattori M, Aksoy S. 2002. Genome sequence of the endocellular obligate symbiont of tsetse flies, Wigglesworthia glossinidia. Nat. Genet. 32:402–407. 10.1038/ng986 [DOI] [PubMed] [Google Scholar]
- 10. Kirkness EF, Haas BJ, Sun W, Braig HR, Perotti MA, Clark JM, Lee SH, Robertson HM, Kennedy RC, Elhaik E, Gerlach D, Kriventseva EV, Elsik CG, Graur D, Hill CA, Veenstra JA, Walenz B, Tubío JM, Ribeiro JM, Rozas J, Johnston JS, Reese JT, Popadic A, Tojo M, Raoult D, Reed DL, Tomoyasu Y, Kraus E, Krause E, Mittapalli O, Margam VM, Li HM, Meyer JM, Johnson RM, Romero-Severson J, Vanzee JP, Alvarez-Ponce D, Vieira FG, Aguadé M, Guirao-Rico S, Anzola JM, Yoon KS, Strycharz JP, Unger MF, Christley S, Lobo NF, Seufferheld MJ, Wang N, Dasch GA, Struchiner CJ, Madey G, Hannick LI, Bidwell S, Joardar V, Caler E, Shao R, Barker SC, Cameron S, Bruggner RV, Regier A, Johnson J, Viswanathan L, Utterback TR, Sutton GG, Lawson D, Waterhouse RM, Venter JC, Strausberg RL, Berenbaum MR, Collins FH, Zdobnov EM, Pittendrigh BR. 2010. Genome sequences of the human body louse and its primary endosymbiont provide insights into the permanent parasitic lifestyle. Proc. Natl. Acad. Sci. U. S. A. 107:12168–12173. 10.1073/pnas.1003379107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wu D, Daugherty SC, Van Aken SE, Pai GH, Watkins KL, Khouri H, Tallon LJ, Zaborsky JM, Dunbar HE, Tran PL, Moran NA, Eisen JA. 2006. Metabolic complementarity and genomics of the dual bacterial symbiosis of sharpshooters. PLoS Biol. 4:e188. 10.1371/journal.pbio.0040188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. McCutcheon JP, Moran NA. 2007. Parallel genomic evolution and metabolic interdependence in an ancient symbiosis. Proc. Natl. Acad. Sci. U. S. A. 104:19392–19397. 10.1073/pnas.0708855104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. McCutcheon JP, Moran NA. 2010. Functional convergence in reduced genomes of bacterial symbionts spanning 200 million years of evolution. Genome Biol. Evol. 3:708–718. 10.1093/gbe/evq055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. McCutcheon JP, Moran NA. 2011. Extreme genome reduction in symbiotic bacteria. Nat. Rev. Microbiol. 10:13–26. 10.1038/nrmicro2670 [DOI] [PubMed] [Google Scholar]
- 15. Moran NA, Bennett GM. 2 June 2014. The tiniest tiny genomes. Annu. Rev. Microbiol. 64:195–215. 10.1146/annurev-micro-091213-112901 [DOI] [PubMed] [Google Scholar]
- 16. Neef A, Latorre A, Peretó J, Silva FJ, Pignatelli M, Moya A. 2011. Genome economization in the endosymbiont of the wood roach Cryptocercus punctulatus due to drastic loss of amino acid synthesis capabilities. Genome Biol. Evol. 3:1437–1448. 10.1093/gbe/evr118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sabree ZL, Huang CY, Okusu A, Moran NA, Normark BB. 2013. The nutrient supplying capabilities of Uzinura, an endosymbiont of armoured scale insects. Environ. Microbiol. 15:1988–1999. 10.1111/1462-2920.12058 [DOI] [PubMed] [Google Scholar]
- 18. Lamelas A, Gosalbes MJ, Manzano-Marín A, Peretó J, Moya A, Latorre A. 2011. Serratia symbiotica from the aphid Cinara cedri: a missing link from facultative to obligate insect endosymbiont. PLoS Genet. 7:e1002357. 10.1371/journal.pgen.1002357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Husnik F, Nikoh N, Koga R, Ross L, Duncan RP, Fujie M, Tanaka M, Satoh N, Bachtrog D, Wilson AC, von Dohlen CD, Fukatsu T, McCutcheon JP. 2013. Horizontal gene transfer from diverse bacteria to an insect genome enables a tripartite nested mealybug symbiosis. Cell 153:1567–1578. 10.1016/j.cell.2013.05.040 [DOI] [PubMed] [Google Scholar]
- 20. López-Madrigal S, Latorre A, Porcar M, Moya A, Gil R. 2011. Complete genome sequence of “Candidatus Tremblaya princeps” strain PCVAL, an intriguing translational machine below the living-cell status. J. Bacteriol. 193:5587–5588. 10.1128/JB.05749-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Degnan PH, Ochman H, Moran NA. 2011. Sequence conservation and functional constraint on intergenic spacers in reduced genomes of the obligate symbiont Buchnera. PLoS Genet. 7:e1002252. 10.1371/journal.pgen.1002252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nakabachi A, Ueoka R, Oshima K, Teta R, Mangoni A, Gurgui M, Oldham NJ, van Echten-Deckert G, Okamura K, Yamamoto K, Inoue H, Ohkuma M, Hongoh Y, Miyagishima SY, Hattori M, Piel J, Fukatsu T. 2013. Defensive bacteriome symbiont with a drastically reduced genome. Curr. Biol. 23:1478–1484. 10.1016/j.cub.2013.06.027 [DOI] [PubMed] [Google Scholar]
- 23. Sabree ZL, Huang CY, Arakawa G, Tokuda G, Lo N, Watanabe H, Moran NA. 2012. Genome shrinkage and loss of nutrient-providing potential in the obligate symbiont of the primitive termite Mastotermes darwiniensis. Appl. Environ. Microbiol. 78:204–210. 10.1128/AEM.06540-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jiang ZF, Xia F, Johnson KW, Bartom E, Tuteja JH, Stevens R, Grossman RL, Brumin M, White KP, Ghanim M. 2012. Genome sequences of the primary endosymbiont “Candidatus Portiera aleyrodidarum” in the whitefly Bemisia tabaci B and Q biotypes. J. Bacteriol. 194:6678–6679. 10.1128/JB.01841-12 PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Santos-Garcia D, Farnier PA, Beitia F, Zchori-Fein E, Vavre F, Mouton L, Moya A, Latorre A, Silva FJ. 2012. Complete genome sequence of “Candidatus Portiera aleyrodidarum” BT-QVLC, an obligate symbiont that supplies amino acids and carotenoids to Bemisia tabaci. J. Bacteriol. 194:6654–6655. 10.1128/JB.01793-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sloan DB, Moran NA. 2012. Endosymbiotic bacteria as a source of carotenoids in whiteflies. Biol. Lett. 8:986–989. 10.1098/rsbl.2012.0664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sloan DB, Moran NA. 2012. Genome reduction and co-evolution between the primary and secondary bacterial symbionts of psyllids. Mol. Biol. Evol. 29:3781–3792. 10.1093/molbev/mss180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Patiño-Navarrete R, Moya A, Latorre A, Peretó J. 2013. Comparative genomics of Blattabacterium cuenoti: the frozen legacy of an ancient endosymbiont genome. Genome Biol. Evol. 5:351–361. 10.1093/gbe/evt011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Moran NA, Tran P, Gerardo NM. 2005. Symbiosis and insect diversification: an ancient symbiont of sap-feeding insects from the bacterial phylum Bacteroidetes. Appl. Environ. Microbiol. 71:8802–8810. 10.1128/AEM.71.12.8802-8810.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Urban JM, Cryan JR. 2012. Two ancient bacterial endosymbionts have coevolved with the planthoppers (Insecta: Hemiptera: Fulgoroidea). BMC Evol. Biol. 12:87. 10.1186/1471-2148-12-87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Koga R, Moran NA. 2014. Swapping symbionts in spittlebugs: evolutionary replacement of a reduced genome symbiont. ISME J. 8:1237–1246. 10.1038/ismej.2013.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Koga R, Bennett GM, Cryan JR, Moran NA. 2013. Evolutionary replacement of obligate symbionts in an ancient and diverse insect lineage. Environ. Microbiol. 15:2073–2081. 10.1111/1462-2920.12121 [DOI] [PubMed] [Google Scholar]
- 33. Moran NA, Dale C, Dunbar H, Smith WA, Ochman H. 2003. Intracellular symbionts of sharpshooters (Insecta: Hemiptera: Cicadellinae) form a distinct clade with a small genome. Environ. Microbiol. 5:116–126. 10.1046/j.1462-2920.2003.00391.x [DOI] [PubMed] [Google Scholar]
- 34. Takiya DM, Tran PL, Dietrich CH, Moran NA. 2006. Co-cladogenesis spanning three phyla: leafhoppers (Insecta: Hemiptera: Cicadellidae) and their dual bacterial symbionts. Mol. Ecol. 15:4175–4191. 10.1111/j.1365-294X.2006.03071.x [DOI] [PubMed] [Google Scholar]
- 35. Purcell AH. 1976. Seasonal changes in host plant preference of the blue-green sharpshooter Hordnia circellata (Homoptera: Cicadellidae). Pan-Pac. Entomol. 52:33–37 [Google Scholar]
- 36. Woyke T, Tighe D, Mavromatis K, Clum A, Copeland A, Schackwitz W, Lapidus A, Wu D, McCutcheon JP, McDonald BR, Moran NA, Bristow J, Cheng JF. 2010. One bacterial cell, one complete genome. PLoS One 5:e10314. 10.1371/journal.pone.0010314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gil R, Sabater-Muñoz B, Perez-Brocal V, Silva FJ, Latorre A. 2006. Plasmids in the aphid endosymbiont Buchnera aphidicola with the smallest genomes. A puzzling evolutionary story. Gene 370:17–25. 10.1016/j.gene.2005.10.043 [DOI] [PubMed] [Google Scholar]
- 38. van Ham RC, Moya A, Latorre A. 1997. Putative evolutionary origin of plasmids carrying the genes involved in leucine biosynthesis in Buchnera aphidicola (endosymbiont of aphids). J. Bacteriol. 179:4768–4777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Matuszewska M, Kuczyńska-Wiśnik D, Laskowska E, Liberek K. 2005. The small heat shock protein IbpA of Escherichia coli cooperates with IbpB in stabilization of thermally aggregated proteins in a disaggregation competent state. J. Biol. Chem. 280:12292–12298. 10.1074/jbc.M412706200 [DOI] [PubMed] [Google Scholar]
- 40. Chiba S, Ito K, Akiyama Y. 2006. The Escherichia coli plasma membrane contains two PHB (prohibitin homology) domain protein complexes of opposite orientations. Mol. Microbiol. 60:448–457. 10.1111/j.1365-2958.2006.05104.x [DOI] [PubMed] [Google Scholar]
- 41. Akiyama Y. 2009. Quality control of cytoplasmic membrane proteins in Escherichia coli. J. Biochem. 146:449–454. 10.1093/jb/mvp071 [DOI] [PubMed] [Google Scholar]
- 42. Shigenobu S, Watanabe H, Hattori M, Sakaki Y, Ishikawa H. 2000. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature 407:81–86. 10.1038/35024074 [DOI] [PubMed] [Google Scholar]
- 43. Wilson AC, Ashton PD, Calevro F, Charles H, Colella S, Febvay G, Jander G, Kushlan PF, Macdonald SJ, Schwartz JF, Thomas GH, Douglas AE. 2010. Genomic insight into the amino acid relations of the pea aphid, Acyrthosiphon pisum, with its symbiotic bacterium Buchnera aphidicola. Insect Mol. Biol. 19(Suppl 2):249–258. 10.1111/j.1365-2583.2009.00942.x [DOI] [PubMed] [Google Scholar]
- 44. Hansen AK, Moran NA. 2011. Aphid genome expression reveals host-symbiont cooperation in the production of amino acids. Proc. Natl. Acad. Sci. U. S. A. 108:2849–2854. 10.1073/pnas.1013465108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Douglas AE. 1988. Experimental studies on the mycetome symbiosis in the leafhopper Euscelis incisus. J. Insect Physiol. 34:1–10. 10.1016/0022-1910(88)90033-9 [DOI] [Google Scholar]
- 46. Charles H, Balmand S, Lamelas A, Cottret L, Pérez-Brocal V, Burdin B, Latorre A, Febvay G, Colella S, Calevro F, Rahbé Y. 2011. A genomic reappraisal of symbiotic function in the aphid/Buchnera symbiosis: reduced transporter sets and variable membrane organisations. PLoS One 6:e29096. 10.1371/journal.pone.0029096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bos MP, Robert V, Tommassen J. 2007. Biogenesis of the gram-negative bacterial outer membrane. Annu. Rev. Microbiol. 61:191–214. 10.1146/annurev.micro.61.080706.093245 [DOI] [PubMed] [Google Scholar]
- 48. Raetz CR, Whitfield C. 2002. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71:635–700. 10.1146/annurev.biochem.71.110601.135414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Walburger A, Lazdunski C, Corda Y. 2002. The Tol/pal system function requires an interaction between the C-terminal domain of TolA and the N-terminal domain of TolB. Mol. Microbiol. 44:695–708. 10.1046/j.1365-2958.2002.02895.x [DOI] [PubMed] [Google Scholar]
- 50. Cascales E, Lloubès R. 2004. Deletion analyses of the peptidoglycan-associated lipoprotein Pal reveals three independent binding sequences including a TolA box. Mol. Microbiol. 51:873–885. 10.1046/j.1365-2958.2003.03881.x [DOI] [PubMed] [Google Scholar]
- 51. Typas A, Banzhaf M, Gross CA, Vollmer W. 2011. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat. Rev. Microbiol. 10:123–136. 10.1038/nrmicro2677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Dubuisson JF, Vianney A, Hugouvieux-Cotte-Pattat N, Lazzaroni JC. 2005. Tol-pal proteins are critical cell envelope components of Erwinia chrysanthemi affecting cell morphology and virulence. Microbiology 151(Pt 10):3337–3347. 10.1099/mic.0.28237-0 [DOI] [PubMed] [Google Scholar]
- 53. McCutcheon JP, McDonald BR, Moran NA. 2009. Convergent evolution of metabolic roles in bacterial co-symbionts of insects. Proc. Natl. Acad. Sci. U. S. A. 106:15394–15399. 10.1073/pnas.0906424106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Oakeson KF, Gil R, Clayton AL, Dunn DM, von Niederhausern AC, Hamil C, Aoyagi A, Duval B, Baca A, Silva FJ, Vallier A, Jackson DG, Latorre A, Weiss RB, Heddi A, Moya A, Dale C. 2014. Genome degeneration and adaptation in a nascent stage of symbiosis. Genome Biol. Evol. 6:76–93. 10.1093/gbe/evt210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Vigneron A, Charif D, Vincent-Monégat C, Vallier A, Gavory F, Wincker P, Heddi A. 2012. Host gene response to endosymbiont and pathogen in the cereal weevil Sitophilus oryzae. BMC Microbiol. 12(Suppl 1):S14. 10.1186/1471-2180-12-S1-S14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ryan VT, Grimwade JE, Camara JE, Crooke E, Leonard AC. 2004. Escherichia coli prereplication complex assembly is regulated by dynamic interplay among Fis, IHF and DnaA. Mol. Microbiol. 51:1347–1359. 10.1046/j.1365-2958.2003.03906.x [DOI] [PubMed] [Google Scholar]
- 57. Fuller RS, Kornberg A. 1983. Purified dnaA protein in initiation of replication at the Escherichia coli chromosomal origin of replication. Proc. Natl. Acad. Sci. U. S. A. 80:5817–5821. 10.1073/pnas.80.19.5817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sánchez-Pulido L, Devos D, Genevrois S, Valencia M. 2003. POTRA: a conserved domain in the FtsQ family and a class of β-barrel outer membrane proteins. Trends Biochem. Sci. 28:523–526. 10.1016/j.tibs.2003.08.003 [DOI] [PubMed] [Google Scholar]
- 59. Lutkenhaus J, Pichoff S, Du S. 2012. Bacterial cytokinesis: from Z ring to divisome. Cytoskeleton 69:778–790. 10.1002/cm.21054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Vicente M, Rico AI, Martínez-Arteaga R, Mingorance J. 2006. Septum enlightenment: assembly of bacterial division proteins. J. Bacteriol. 188:19–27. 10.1128/JB.188.1.19-27.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Höltje JV. 1998. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol. Mol. Biol. Rev. 62:181–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Gerding MA, Ogata Y, Pecora ND, Niki H, de Boer PA. 2007. The trans-envelope Tol-Pal complex is part of the cell division machinery and required for proper outer-membrane invagination during cell constriction in E. coli. Mol. Microbiol. 63:1008–1025. 10.1111/j.1365-2958.2006.05571.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Typas A, Banzhaf M, van den Berg van Saparoea B, Verheul J, Biboy J, Nichols RJ, Zietek M, Beilharz K, Kannenberg K, von Rechenberg M, Breukink E, den Blaauwen T, Gross CA, Vollmer W. 2010. Regulation of peptidoglycan synthesis by outer-membrane proteins. Cell 143:1097–1109. 10.1016/j.cell.2010.11.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Heidrich C, Templin MF, Ursinus A, Merdanovic M, Berger J, Schwarz H, De Pedro MA, Höltje JV. 2001. Involvement of N-acetylmuramyl-l-alanine amidases in cell separation and antibiotic-induced autolysis of Escherichia coli. Mol. Microbiol. 41:167–178. 10.1046/j.1365-2958.2001.02499.x [DOI] [PubMed] [Google Scholar]
- 65. Boutte CC, Crosson S. 2013. Bacterial lifestyle shapes stringent response activation. Trends Microbiol. 21:174–180. 10.1016/j.tim.2013.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Husník F, Chrudimský T, Hypša V. 2011. Multiple origins of endosymbiosis within the Enterobacteriaceae (γ-Proteobacteria): convergence of complex phylogenetic approaches. BMC Biol. 9:87. 10.1186/1741-7007-9-87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Clayton AL, Oakeson KF, Gutin M, Pontes A, Dunn DM, Niederhausern AC, Weiss RB, Fisher M, Dale C. 2012. A novel human-infection-derived bacterium provides insights into the evolutionary origins of mutualistic insect–bacterial symbioses. PLoS Genet. 8:e1002990. 10.1371/journal.pgen.1002990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Rosas-Pérez T, Rosenblueth M, Rincón-Rosales R, Mora J, Martínez-Romero E. 2014. Genome sequence of “Candidatus Walczuchella monophlebidarum” the flavobacterial endosymbiont of Llaveia axin axin (Hemiptera: Coccoidea: Monophlebidae). Genome Biol. Evol. 6:714–726. 10.1093/gbe/evu049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Snyder AK, McMillen CM, Wallenhorst P, Rio RV. 2011. The phylogeny of Sodalis-like symbionts as reconstructed using surface-encoding loci. FEMS Microbiol. Lett. 317:143–151. 10.1111/j.1574-6968.2011.02221.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Conord C, Despres L, Vallier A, Balmand S, Miquel C, Zundel S, Lemperiere G, Heddi A. 2008. Long-term evolutionary stability of bacterial endosymbiosis in Curculionoidea: additional evidence of symbiont replacement in the Dryophthoridae family. Mol. Biol. Evol. 25:859–868. 10.1093/molbev/msn027 [DOI] [PubMed] [Google Scholar]
- 71. Moran NA, McLaughlin HJ, Sorek R. 2009. The dynamics and time scale of ongoing genomic erosion in symbiotic bacteria. Science 323:379–382. 10.1126/science.1167140 [DOI] [PubMed] [Google Scholar]
- 72. Rispe C, Delmotte F, van Ham RC, Moya A. 2004. Mutational and selective pressures on codon and amino acid usage in Buchnera, endosymbiotic bacteria of aphids. Genome Res. 14:44–53. 10.1101/gr.1358104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Sloan DB, Nakabachi A, Richards S, Qu J, Murali SC, Gibbs RA, Moran NA. 2014. Parallel histories of horizontal gene transfer facilitated extreme reduction of endosymbiont genomes in sap-feeding insects. Mol. Biol. Evol. 31:857–871. 10.1093/molbev/msu004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Markowitz VM, Mavromatis K, Ivanova NN, Chen IM, Chu K, Kyrpides NC. 2009. IMG ER: a system for microbial genome annotation expert review and curation. Bioinformatics 25:2271–2278. 10.1093/bioinformatics/btp393 [DOI] [PubMed] [Google Scholar]
- 75. Delcher AL, Bratke KA, Powers EC, Salzberg SL. 2007. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 23:673–679. 10.1093/bioinformatics/btm009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Finn RD, Clements J, Eddy SR. 2011. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39:W29–W37. 10.1093/nar/gkr367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Gao F, Zhang CT. 2008. Ori-finder: a web-based system for finding oriCs in unannotated bacterial genomes. BMC Bioinformatics 9:79. 10.1186/1471-2105-9-79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Gao F, Luo H, Zhang CT. 2013. DoriC 5.0: an updated database of oriC regions in both bacterial and archaeal genomes. Nucleic Acids Res. 41:D90–D93. 10.1093/nar/gkt090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Tatusov RL, Koonin EV, Lipman DJ. 1997. A genomic perspective on protein families. Science 278:631–637. 10.1126/science.278.5338.631 [DOI] [PubMed] [Google Scholar]
- 80. Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. 2009. Circos: an information aesthetic for comparative genomics. Genome Res. 19:1639–1645. 10.1101/gr.092759.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690. 10.1093/bioinformatics/btl446 [DOI] [PubMed] [Google Scholar]
- 83. Wang Z, Wu M. 2013. A phylum-level bacterial phylogenetic marker database. Mol. Biol. Evol. 30:1258–1262. 10.1093/molbev/mst059 [DOI] [PubMed] [Google Scholar]
- 84. Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30:772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Darriba D, Taboada GL, Doallo R, Posada D. 2014. High-performance computing selection of models of DNA substitution for multicore clusters. Int. J. High Perform. Comput. Appl. 28:112–125. 10.1177/1094342013495095 [DOI] [Google Scholar]
- 86. Dayhoff MO, Schwartz RM, Orcutt BC. 1978. A model of evolutionary change in proteins, chapter 22, p 345–352 In Dayhoff MO, Atlas of protein sequence and structure. National Biomedical Research Foundation, Silver Spring, MD [Google Scholar]
- 87. Abascal F, Zardoya R, Telford MJ. 2010. TranslatorX: multiple alignment of nucleotide sequences guided by amino acid translations. Nucleic Acids Res. 38:W7–W13. 10.1093/nar/gkq291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Yang Z, Nielsen R. 2000. Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol. Biol. Evol. 17:32–43. 10.1093/oxfordjournals.molbev.a026236 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
“Ca. Baumannia” BGSS plasmid (pB-BGSS). Download
Complete phylogenetic tree for “Ca. Baumannia cicadellinicola” strains across sharpshooters using the 16S rRNA loci. Download
Complete phylogenomic tree for Gammaproteobacteria. Download
Phylogenetic taxon sampling.
Predicted genes in the BGSS “Ca. Baumannia” genome.
Predicted genes in the BGSS “Ca. Sulcia” genome.
Molecular characteristics and dN and dS mutation rates for all pairwise comparisons between “Ca. Baumannia cicadellinicola” and “Ca. Sulcia muelleri” strains.