ABSTRACT
How sublethal levels of antibiotics and heavy metals select for clinically important multidrug resistance plasmids is largely unknown. Carriage of plasmids generally confers substantial fitness costs, implying that for the plasmid-carrying bacteria to be maintained in the population, the plasmid cost needs to be balanced by a selective pressure conferred by, for example, antibiotics or heavy metals. We studied the effects of low levels of antibiotics and heavy metals on the selective maintenance of a 220-kbp extended-spectrum β-lactamase (ESBL) plasmid identified in a hospital outbreak of Klebsiella pneumoniae and Escherichia coli. The concentrations of antibiotics and heavy metals required to maintain plasmid-carrying bacteria, the minimal selective concentrations (MSCs), were in all cases below (almost up to 140-fold) the MIC of the plasmid-free susceptible bacteria. This finding indicates that the very low antibiotic and heavy metal levels found in polluted environments and in treated humans and animals might be sufficiently high to maintain multiresistance plasmids. When resistance genes were moved from the plasmid to the chromosome, the MSC decreased, showing that MSC for a specific resistance conditionally depends on genetic context. This finding suggests that a cost-free resistance could be maintained in a population by an infinitesimally low concentration of antibiotic. By studying the effect of combinations of several compounds, it was observed that for certain combinations of drugs each new compound added lowered the minimal selective concentration of the others. This combination effect could be a significant factor in the selection of multidrug resistance plasmids/bacterial clones in complex multidrug environments.
IMPORTANCE
Antibiotic resistance is in many pathogenic bacteria caused by genes that are carried on large conjugative plasmids. These plasmids typically contain multiple antibiotic resistance genes as well as genes that confer resistance to biocides and heavy metals. In this report, we show that very low concentrations of single antibiotics and heavy metals or combinations of compounds can select for a large plasmid that carries resistance to aminoglycosides, β-lactams, tetracycline, macrolides, trimethoprim, sulfonamide, silver, copper, and arsenic. Our findings suggest that the low levels of antibiotics and heavy metals present in polluted external environments and in treated animals and humans could allow for selection and enrichment of bacteria with multiresistance plasmids and thereby contribute to the emergence, maintenance, and transmission of antibiotic-resistant disease-causing bacteria.
INTRODUCTION
Earlier studies showed that concentrations of antibiotics up to 230-fold below the MIC for susceptible bacteria (MICsusc) can both enrich preexisting resistance in a population and select for de novo resistant mutants (1, 2). Previous work focused on resistance caused by mutations or resistance genes located on the chromosome, and it remains unclear if the same selective effect of very low antibiotic concentrations (<<MIC) can be applied to clinically identified multidrug resistance plasmids (3–7). These types of plasmids generally confer resistance to a wide range of different antibiotics, biocides, and heavy metals, and they often carry a significant fitness cost in the absence of selection (8–12). Despite these costs, resistance plasmids are widespread and they are one of the most common causes of β-lactam resistance in Gram-negative bacteria caused by extended-spectrum β-lactamases (ESBLs), a serious problem in hospitals around the world (3, 13, 14). Here, we studied the 220-kbp multidrug resistance plasmid pUUH239.2, isolated in 2005 from a large nosocomial outbreak at the Uppsala University Hospital (15–18). Besides the genes that confer resistance to antibiotics (β-lactams, tetracyclines, aminoglycosides, macrolides, sulfonamides, trimethoprim, and ciprofloxacin) and biocides, the plasmid also carries genes conferring resistance to silver, copper, and arsenic. Because of the genetic linkage between the resistances, exposure to any of these drugs, or any combination of them, could coselect for the maintenance of the whole plasmid and all its associated resistances.
Metal resistance genes are often found on plasmids together with antibiotic resistance genes (16, 19–23), and metals may therefore be an additional factor that indirectly selects for antibiotic resistance. In support of this notion, observational studies show that antibiotic-resistant bacteria are enriched at locations contaminated with metals (21, 24, 25). Arsenic and copper are both naturally present due to geologic sources (26–28), and in addition, anthropogenic activities significantly contribute to their release and accumulation in the environment, particularly in certain agricultural locations (25, 29–33). For example, copper has extensively been used as a swine fodder additive (34, 35) and arsenic is a component in swine and poultry feed (29, 32, 33), in pesticides applied to cotton fields (36), and in wood-treating agents (31).
Apart from heavy metals and biocides, antibiotics are released into the environment from a number of different sources. For example, human urine and feces from antibiotic users can contain considerable amounts of active residues, which ultimately will reach sewage water and sludge (which subsequently may be spread on farmland to recycle nutrients), where they can exert a selective pressure (37). Similarly, large amounts of antibiotics are used in animal production, and depending on farming practices, urine and feces are collected, treated, and distributed to land and water (37–40). Finally, in certain settings direct discharge of antibiotics in connection with pharmaceutical industries can be very high (41–43), which also may result in pollution of surrounding groundwaters (44). Thus, agricultural areas, industry runoff, and sewage waters provide ample opportunity for various metals and antibiotics to mix and enter streams, lakes, and soil (25, 45). Little is known about the interactions taking place between substances in these mixtures and whether their effects are combined with regard to selective ability of resistance genes/mutations (25, 46).
Here, we examined how sub-MICs of several classes of antibiotics and heavy metals singly and in combination select for a multidrug resistance plasmid. Results show that the concentrations of single antibiotics and heavy metals needed to maintain the plasmid were in all cases well below the MIC of the plasmid-free susceptible bacteria and that for combinations of several compounds even lower concentrations of each individual drug were needed to maintain the plasmid. Such combination effects could be a significant factor in the selection of multidrug resistance plasmids in complex, natural multidrug-containing environments.
RESULTS
Experimental setup.
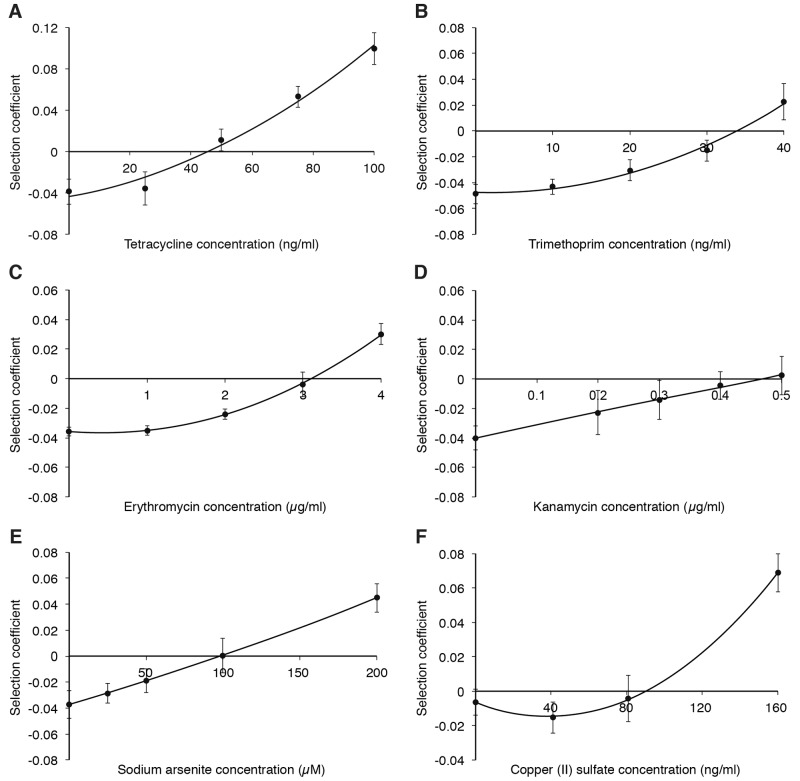
To be able to determine the minimal selective concentrations (MSCs) of the different antibiotics, isogenic strains of Escherichia coli, with or without the plasmid pUUH239.2, were competed against each other. The MSC is the drug concentration where the fitness cost of the resistance plasmid is balanced by the selective effect of the added drug (1, 2). The two strains were genetically tagged with blue or yellow fluorescent protein genes (mTagBFP2 [47] or SYFP2 [48]), to allow tracking of the two populations over time using flow cytometry. To correct for the difference in cost between the two fluorescent markers, control competitions were performed between two plasmid-free strains carrying the respective blue and yellow variants of the fluorescent proteins. For all competition experiments, the selection coefficients were adjusted to correct for the small differences in fitness observed between the fluorescent markers (see Table S1 in the supplemental material). To avoid conjugation of the plasmid from the resistant to the susceptible strain during the competition assays, the traJ gene was deleted from the plasmid. This gene, encoding the TraJ regulatory protein, is essential for conjugation since it controls the expression of the transfer genes of the plasmid (49–53). The competitions were performed by serial dilutions in batch cultures for up to 40 generations in the presence of the antibiotics tetracycline, trimethoprim (a dihydrofolate reductase inhibitor), erythromycin (a macrolide), and kanamycin (an aminoglycoside), as well as the heavy metals arsenic and copper. The data in Fig. 1 show the selection coefficients from competition experiments plotted as functions of antibiotic concentrations. The point where the curve intercepts the y axis represents the fitness cost of the plasmid, whereas the intercept on the x axis, s = 0, represents the MSC, the concentration where the cost of carrying the plasmid is balanced by the selection pressure of the drug.
FIG 1 .
Selection coefficients as function of antibiotic concentrations during competition experiments between strains carrying the pUUH239.2 plasmid and susceptible strains at low levels of antibiotics and heavy metals. Competitions in the presence of tetracycline (A), trimethoprim (B), erythromycin (C), kanamycin (D), arsenite (E), and copper (F). Data can be found in Table S3 in the supplemental material. Standard deviations are indicated.
Antibiotics and heavy metals at sub-MIC levels select for the multidrug resistance plasmid pUUH239.2.
As for many plasmids, the multidrug resistance plasmid pUUH239.2 has a considerable fitness cost (4% reduction in exponential growth rate) (16), which means that some selective pressure needs to favor the plasmid-carrying hosts to allow for the plasmid to persist in the population. To determine the levels of antibiotics and metals that are necessary for plasmid selection, competition assays were conducted. Figure 1A to F shows the selection coefficient of the plasmid strain as a function of the concentration of the relevant antibiotic or metal. These results demonstrate that the MSCs were significantly below the MICs of the susceptible strain: for tetracycline (Fig. 1A), the MSC value was 1/17 of the MICsusc; for trimethoprim (Fig. 1B), the value was 1/6; for erythromycin (Fig. 1C), the value was 1/4; and for kanamycin (Fig. 1D), the value was 2/3. These values represent absolute antibiotic concentrations of 45 ng/ml (tetracycline), 33 ng/ml (trimethoprim), 3 µg/ml (erythromycin), and 470 ng/ml (kanamycin). For heavy metals, a similar pattern was observed. Thus, the MSC of arsenite was close to 140-fold lower than the MICsusc (Fig. 1E). This MSC was also observed when the plasmid-carrying strain initially was present at a lower fraction (1:10 and 1:100) than the competing plasmid-free strain (see Fig. S1 in the supplemental material). Similarly for copper(II) sulfate, the MSC was more than 10-fold lower than the MICsusc (Fig. 1F). In summary, these results demonstrate that the concentrations of the antibiotics as well as those of arsenite and copper(II) required for plasmid selection lie well below (2- to almost 140-fold) the concentration necessary to prevent growth of a plasmid-free strain.
Combinations of antibiotics and heavy metals can have synergistic effects on selection.
In many environments such as wastewater and sewage treatment plants, as well as in agricultural and industrial areas, bacteria often encounter a complex mix of low levels of many different antibiotics, biocides, and heavy metals (25, 54–58), and it is therefore important to investigate how these substances interact in terms of their combined selection potential. To study this combination effect, competitions were performed with several different combinations of low levels of (i) two different antibiotics (trimethoprim and erythromycin), (ii) one antibiotic and one heavy metal (tetracycline and arsenic), or (iii) two different antibiotics (tetracycline and trimethoprim) and a heavy metal (arsenic). In competitions i and iii, we used three concentrations close to the MSC of each compound and then performed competitions at all possible combinations of the three concentrations. For trimethoprim and erythromycin, this resulted in 9 different combinations (Fig. 2A), while for tetracycline, trimethoprim, and arsenic, 27 different combinations were investigated (Fig. 2C). The concentrations were normalized to the MSC of each compound, and the normalized concentrations were then summarized for each combination. If we assume that the selection effects of the compounds are additive, we would expect that a total normalized concentration value of 1 should give a selection coefficient of zero. In competition ii, the concentration of arsenic was kept constant while the concentration of tetracycline was varied.
FIG 2 .
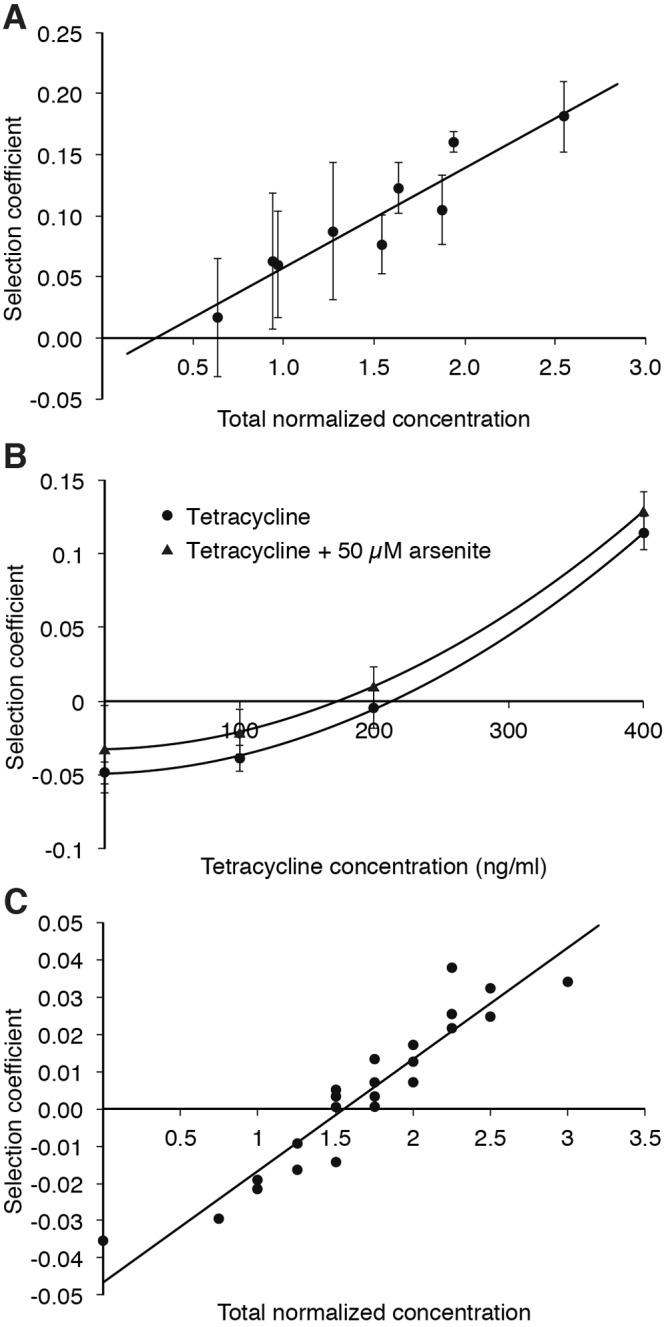
Selection coefficients as a function of antibiotic and heavy metal concentrations during competition experiments between resistant strains carrying the pUUH239.2 plasmid and susceptible plasmid-free strains. (A) Competitions in the presence of combinations of trimethoprim and erythromycin. The total normalized concentration is the sum of the two normalized concentrations, where the concentration of each compound was first normalized to its respective MSC. Data can be found in Table S4 in the supplemental material. (B) Competitions in the presence of combinations of arsenic and tetracycline. These competitions were performed in LB medium, explaining why the selection coefficients of tetracycline in this experiment were different from the values in Fig. 1A, where Mueller-Hinton medium was used. Data can be found in Table S5. The differences in MSC with and without arsenic were significant (P value = 0.0001 in a paired t test). (C) Competitions in the presence of combinations of arsenic, tetracycline, and trimethoprim. The total normalized concentration is the sum of all normalized concentrations, where the concentration of each compound was first normalized to its respective MSC. Data can be found in Table S6. Standard deviations are indicated.
The results show that for competition i, these two antibiotics have a synergistic effect at low levels, and when combined, they cause an enrichment of resistance even when the total normalized concentration is below 1 (Fig. 2A; intercept on x axis is <1 MSC).
For combination ii, arsenite at 0.5× MSC was combined with increasing amounts of tetracycline to assess the effect of this combination. The antibiotic and metal together conferred a combination effect where the presence of each substance adds to the total selection coefficient. The presence of the metal resulted in an increase of the selection coefficient by an average of 1.6 ± 0.1 percentage points and a subsequent 20% decrease of the MSC of tetracycline (Fig. 2B). In the third test (iii), the selective potential of a combination of three compounds, arsenite, tetracycline, and trimethoprim, was examined by combining three different concentrations (0.25×, 0.5×, and 1× MSC) of each substance. By plotting the selection coefficient as a function of total normalized concentration, a trend is apparent where the combined effect of each drug leads to a higher selection coefficient (Fig. 2C). However, in contrast to the erythromycin and trimethoprim combination, the selective effect was less than the sum of the MSCs (Fig. 2C, intercept on x axis is >1 MSC).
MSC is conditional and depends on the fitness cost of resistance.
To further elucidate the dependence of MSC on the fitness cost of resistance, three of the resistance cassettes, tetRA (tetracycline resistance), dhfr (trimethoprim resistance), and the mph operon (erythromycin resistance), were transferred from the pUUH239.2 plasmid to the Escherichia coli chromosome by genetic recombineering (59). The results of the competitions between these strains and isogenic susceptible strains are shown in Fig. 3. These data demonstrate that the MSC is directly dependent on the fitness cost of the resistance. Thus, when the fitness cost conferred by the rest of the plasmid is eliminated, the selection coefficient curve is shifted upwards, thereby reducing the MSC value. For tetracycline, the MSC value was 1/25 of the MICsusc (Fig. 3A); for trimethoprim, the value was less than 1/100 (Fig. 3B); and for erythromycin, the value was less than 1/60 (Fig. 3C). These values represent absolute antibiotic concentrations of 30 ng/ml (tetracycline), less than 2 ng/ml (trimethoprim), and less than 200 ng/ml (erythromycin) for the chromosomally carried resistance compared to 45 ng/ml (tetracycline), 33 ng/ml (trimethoprim), and 3,000 ng/ml (erythromycin) when the corresponding resistance was present on the plasmid. As these resistance genes on the chromosome do not confer any significant fitness costs, the MSC values approach zero, making the MSC values for trimethoprim and erythromycin uncertain in this low range.
FIG 3 .
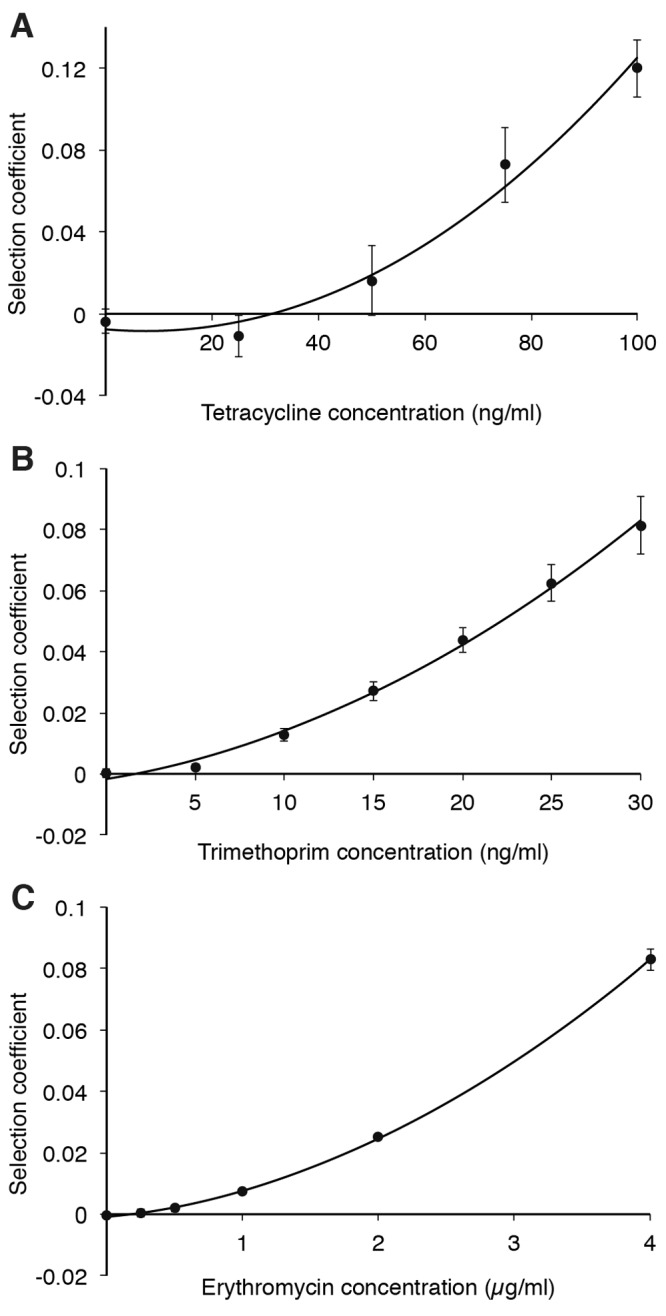
Selection coefficients as function of antibiotic concentrations during competition experiments between strains carrying chromosomally carried resistance genes and susceptible strains at low levels of antibiotics. Competitions in the presence of tetracycline (A), trimethoprim (B), and erythromycin (C). Data can be found in Table S7 in the supplemental material. Standard deviations are indicated.
DISCUSSION
In this study, we investigated the selective effect of four clinically relevant antibiotics as well as two heavy metals on the maintenance of the ESBL plasmid pUUH239.2. This plasmid confers resistance not only to β-lactams but also to aminoglycosides, tetracycline, trimethoprim, sulfonamides, and erythromycin, as well as biocides such as quaternary ammonium compounds and the heavy metals copper, silver, and arsenic. Results show that selection of the multidrug resistance plasmid can occur at concentrations far below the MIC of a susceptible strain and that this general pattern is observed for many different classes of antibiotics and heavy metals. To exclude the possibility that any genetic changes might occur in the plasmid or chromosome during the course of the competition experiments (40 generations of growth), we performed whole-genome sequencing (WGS) of 12 independent lineages of plasmid-carrying bacteria that were grown for 40 generations in the presence of low levels of the different antibiotics and heavy metals. Results showed that none of the 12 lineages acquired any mutations in the chromosome or plasmid (see Table S9 in the supplemental material).
The strongest selection in this study was observed with arsenic, where the level required for selection, the MSC, is close to 140-fold lower than the MIC of the susceptible wild-type strain. The MSC value of arsenite determined here corresponds to 7 mg/liter arsenic, a level that has been found in many types of environments. For example, it often occurs in certain sites that have been contaminated by mining (28, 60, 61). Furthermore, there are large areas around the world where the groundwater naturally contains high levels of inorganic arsenic, such as in Bangladesh (28, 62, 63), where millions of people get their drinking water from wells containing up to 300 µg/liter arsenic (64). Arsenic compounds have also been used in large quantities as growth promotion agents in animal feed, mainly as roxarsone, the organoarsenic compound 3-nitro-4-hydroxyphenylarsonic acid (65). The level of roxarsone in feed can be as high as 45.4 mg/kg, which corresponds to a molar amount of arsenic approximately 3-fold above the MSC for arsenite determined here (7 mg/liter). Thus, bacteria present in animal intestines might be exposed to arsenic levels that are sufficiently high to selectively maintain bacteria carrying a multidrug resistance plasmid, similar to the one studied here. Furthermore, most of the roxarsone is excreted unchanged from the animals, and litter from poultry houses using roxarsone-containing feed has been shown to contain approximately 30 mg/kg of arsenic. It is estimated that the broiler manure produced in the United States alone during the year 2000 contained roxarsone levels equivalent to 2.5 × 105 kg of arsenic, eventually ending up contaminating soil and water when manure from the animals was spread onto agricultural fields as fertilizer (29). It is likely that also other metals to which bacteria are known to carry resistance genes, such as silver, copper, mercury, and cadmium, would constitute a similar selective force. This is problematic since metals of various kinds are widely applied in industries, agriculture, construction, health care, and other areas from which they are released into the environment (66–75).
The combined effect of mixtures of substances such as metals and antibiotics that are present in the environment is an additional complicating factor regarding selection (46, 76). The fact that combined effects have been demonstrated here is of great concern since it shows that even very small amounts of several compounds, where each individual compound is not sufficiently high to confer selection, could in combination add up and result in selection of resistance. The combined effects of the compounds tested here vary from synergistic to less than additive. Thus, the combination of the antibiotics erythromycin and trimethoprim shows a synergistic effect, where the presence of one of the drugs lowers the MSC of the other. Likewise, combining various concentrations of arsenite, tetracycline, and trimethoprim results in stronger selection, but here, the effect is less than additive. The difference between these two mixtures of drugs may be the result of the specific mechanism of action of each substance interfering with the other, and other combinations of metals and antibiotics with different mechanisms of action could therefore result in either greater or lesser combination effects. However, based on these experiments it seems that each new compound added to the mixture will lower the MSC of the others. Because of the high number of compounds present in many environments, this combination effect could be a contributing factor to the persistence of multidrug resistance plasmids, even though each individual compound is present at levels below the minimal selective concentration.
Finally, our data show that the MSC for a specific resistance is conditional and depends on the fitness costs of the resistance mutation/gene. Thus, when the cost associated with the resistance was reduced (here achieved by moving the resistance genes from the plasmid to the chromosome), the MSC was also reduced correspondingly. For the three antibiotics examined, the reduction in MSC associated with the transfer of the resistance from the plasmid to the chromosome was 2- to 15-fold. An important implication from this finding is that a cost-free resistance could in principle be maintained in a population by an infinitesimally low concentration of antibiotic, provided that there are no threshold effects with regard to the inhibitory effect of antibiotics. As the initial cost of resistance mutations can often be efficiently compensated by second-site mutations without loss of resistance, it is expected that cost-free resistances are common in natural settings (77).
MATERIALS AND METHODS
Strain construction.
All strains in this study were derived from the wild-type E. coli MG1655 strain and are listed in Table 1. Single-cell tracking of the different strains during the competition assays was enabled by integration of chromosomal copies of either a blue (mTagBFP2) (47) or yellow (SYFP2) (48) fluorescent protein gene. Both genes were codon optimized for expression in E. coli and synthesized by DNA 2.0. The two genes were inserted into galK using the λ red recombineering system as previously described (59) and moved by P1 transduction into the wild-type strain. In strains DA26735 to DA26738, the fluorescent genes are expressed by the promoter PLlacO (78), while in the remaining fluorescent strains, they are expressed from the weaker promoter J23101 (79) (GenBank accession numbers KM018299 to KM018302). The strains carrying the plasmid pUUH239.2 (GenBank accession number CP002474.1) were constructed by conjugation from a clinical isolate into a strain carrying the λ red recombineering system (59) on the chromosome. To eliminate the risk of plasmid conjugation during the competitions, the gene controlling the transfer operon of the plasmid, traJ (49–53), was replaced with a chloramphenicol resistance marker using recombineering. The pUUH239.2 ΔtraJ::cat plasmid was then conjugated into the strains carrying chromosomally integrated fluorescent markers by overexpression of a cloned copy of traJ in trans from a second plasmid (GenBank accession number KM018297) in the donor strain.
TABLE 1 .
Strains and genotypes
| Strain | Genotype | Source or reference |
|---|---|---|
| DA5438 | E. coli MG1655 wild type (parent) | Strain collection |
| DA25916 | /pUUH239.2 | This study |
| DA26735 | ΔlacIZYA::FRT galK::mTagBFP2-amp | This study |
| DA26736 | ΔlacIZYA::FRT galK::SYFP2-amp | This study |
| DA26737 | ΔlacIZYA::FRT galK::mTagBFP2-amp/pUUH239.2 ΔtraJ::cat | This study |
| DA26738 | ΔlacIZYA::FRT galK::SYFP2-amp/pUUH239.2 ΔtraJ::cat | This study |
| DA28200 | galK::SYFP2-FRT | This study |
| DA28202 | galK::mTagBFP2-FRT | This study |
| DA28893 | galK::SYFP2-FRT ΔbglGFB::FRT | This study |
| DA28895 | galK::mTagBFP2-FRT ΔbglGFB::FRT | This study |
| DA28702 | galK::SYFP2-FRT ΔbglGFB::dhfr | This study |
| DA28704 | galK::mTagBFP2-FRT ΔbglGFB::dhfr | This study |
| DA28706 | galK::SYFP2-FRT ΔbglGFB::mph | This study |
| DA28708 | galK::mTagBFP2-FRT ΔbglGFB::mph | This study |
| DA28759 | galK::SYFP2-FRT ΔbglGFB::tetRA | This study |
| DA28761 | galK::mTagBFP2-FRT ΔbglGFB::tetRA | This study |
The strains carrying chromosomally integrated resistance genes from pUUH239.2 were constructed by first replacing the genes bglGFB with a chloramphenicol marker flanked by FLP recombination target (FRT) sites as well as terminators to ensure transcriptional insulation from the surrounding genes. In a second recombineering step, the chloramphenicol marker was replaced with resistance genes amplified from the pUUH239.2 plasmid (dhfr, tetRA, and the mph operon). The chromosomally integrated pUUH239.2 resistance genes were then moved into the fluorescent strains by P1 transduction. The ΔbglGFB::cat marker was also moved into the fluorescent strains using P1 transduction, and the chloramphenicol marker was removed using FLP recombinase to generate the ΔbglGFB::FRT strains DA28893 and DA28895. Primers used are listed in Table S2 in the supplemental material.
MIC measurements.
Susceptibilities of the strains to different antibiotics (Table 2) were determined using Etests according to the manufacturer’s instructions (bioMérieux, Marcy l’Étoile, France). Susceptibility to copper and arsenic was determined using broth microdilution with an initial bacterial inoculum of 106 cells and an incubation time of 18 to 20 h at 37°C without shaking. The MIC of sodium arsenite was determined in LB medium and defined as the lowest concentration where no visible growth was observed. MIC determinations of copper(II) sulfate were conducted in minimal medium (M9 glucose) with limited access to oxygen.
TABLE 2 .
MICs, MSCs, and relative fitness values for the different strains useda
| Strain | Resistance | Fitness | Arsenite |
Cu(II) sulfate |
Tet |
Trm |
Ery |
Kan |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MSC (µM) | MIC (µM) | MSC (ng/ml) | MIC (ng/ml) | MSC (ng/ml) | MIC (ng/ml) | MSC (ng/ml) | MIC (ng/ml) | MSC (µg/ml) | MIC (µg/ml) | MSC (µg/ml) | MIC (µg/ml) | |||
| DA5438 | None (wt) | 1.0 | NA | 12,500 | NA | 1,300 | NA | 750 | NA | 190 | NA | 12 | NA | 0.75 |
| DA25916 | pUUH239.2 | 0.96 | 90 | 25,000 | 90 | >80,000 | 45 | 24,000 | 33 | >32,000 | 3 | 96 | 0.47 | 24 |
| DA28759 | tetRA | 0.99 | NA | NA | NA | NA | 30 | 48,000 | NA | NA | NA | NA | NA | NA |
| DA28702 | dhfr | 1.0 | NA | NA | NA | NA | NA | NA | <2 | >32,000 | NA | NA | NA | NA |
| DA28706 | mph | 1.0 | NA | NA | NA | NA | NA | NA | NA | NA | <0.2 | >256 | NA | NA |
Abbreviations: MSC, minimal selective concentration; Tet, tetracycline; Trm, trimethoprim; Ery, erythromycin; Kan, kanamycin; NA, not applicable; wt, wild type.
Competition assays.
Competition experiments were performed between a susceptible strain expressing mTagBFP2 or SYFP2 and an isogenic resistant strain expressing the other marker. Overnight cultures were mixed 1:1 and serially passaged with a 1,000-fold dilution every 24 h, resulting in 10 generations of growth per serial passage. The ratios between the two competing strains were measured by counting 105 cells at each serial passage using flow cytometry (BD FACSAria IIu). The selection coefficients were calculated using the regression model s = {ln[R(t)/R(0)]}/[t], as previously described (80), where R is the ratio of resistant to susceptible strain. Control experiments were performed to determine the relative cost of expressing the mTagBFP2 marker compared to the SYFP2 marker, and this difference in cost was used for compensation of the calculated selection coefficients (see Table S1 in the supplemental material). Competitions were terminated if nonlinear slopes were detected in plots of the ratio of resistant to susceptible bacteria over time. Nonlinearity indicates that a beneficial adaptive mutation occurred in one of the populations (periodic selection). This phenomenon is impossible to completely avoid (81) since medium adaptation mutations will always occur at some frequency (82). In our experiments, only 4 out of 525 competitions were discarded (see Tables S3 to S8) and the adaptive mutations appearing in these competitions were not further investigated. The competitions using arsenic and using arsenic plus tetracycline were performed in Luria-Bertani broth, the competitions using copper were performed in minimal medium (M9 plus 0.2% glucose) with limited access to oxygen, and the remaining competitions were performed in Mueller-Hinton medium (similar results were obtained in these two media).
WGS.
To investigate whether the bacteria undergo any genetic changes during the competition experiments, 12 independent cultures of bacteria carrying the plasmid (strain DA26737) were analyzed using whole-genome sequencing (WGS) before and after being passaged in the presence of low levels (2× MSC) of all the different antibiotics and heavy metals. Two independent cultures per antimicrobial agent were passaged for 40 generations and analyzed (see Table S9 in the supplemental material). Genomic DNA was prepared from 3 ml of overnight cultures using the Genomic-tip 100/G columns and the Genomic DNA buffer set (Qiagen, Netherlands), according to the manufacturer’s instructions. The genomic DNA was then sequenced using the Illumina sequencing technology with 500-bp paired-end libraries by the BGI sequencing facility (Hong Kong). Mutations were identified by assembling the sequencing reads to the reference genome sequence in CLC Genomics Workbench 7.5 (CLC Bio, Denmark). Illumina reads of each strain were individually mapped onto the reference genome of the wild-type MG1655 and the reference pUUH239.2 plasmid sequenced earlier in our laboratory. The WGS data show that no mutations or rearrangements (83) were found in the plasmid or chromosome during this time frame.
Nucleotide sequence accession numbers.
Nucleotide sequence accession numbers are as follows: Klebsiella pneumoniae plasmid pUUH239.2, CP002474.1; conjugation activation plasmid pMH3CIq-PT5lac-traJ for pUUH239.2, KM018297; synthetic fluorescent protein expression cassette cat-J23101-mTagBFP2, KM018299; synthetic fluorescent protein expression cassette cat-J23101-SYFP2, KM018300; synthetic fluorescent protein expression cassette amp-PLlacO-mTagBFP2, KM018302; synthetic fluorescent protein expression cassette amp-PLlacO-SYFP2, KM018301.
SUPPLEMENTAL MATERIAL
Competitions with low initial fractions of resistant bacteria. Competition experiments at different concentrations of arsenite and different starting fractions of resistant bacteria carrying the pUUH239.2 plasmid. (A) Initial ratio of susceptible to resistant mutants, 10:1. (B) Initial ratio of susceptible to resistant mutants, 100:1. Data can be found in Table S8. Standard deviations are indicated. Download
Costs of fluorescence markers.
Primers used in this study.
Data for Fig. 1
Data for Fig. 2A
Data for Fig. 2B
Data for Fig. 2C
Data for Fig. 3
Data for Fig. S1
Summary of whole-genome sequencing data.
ACKNOWLEDGMENTS
This work was supported by grants from the Swedish Research Council, FORMAS, and the European Commission (project EvoTAR) to D.I.A.
Footnotes
Citation Gullberg E, Albrecht LM, Karlsson C, Sandegren L, Andersson DI. 2014. Selection of a multidrug resistance plasmid by sublethal levels of antibiotics and heavy metals. mBio 5(5):e01918-14. doi:10.1128/mBio.01918-14.
REFERENCES
- 1. Gullberg E, Cao S, Berg OG, Ilbäck C, Sandegren L, Hughes D, Andersson DI. 2011. Selection of resistant bacteria at very low antibiotic concentrations. PLoS Pathog. 7:e1002158. 10.1371/journal.ppat.1002158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Liu A, Fong A, Becket E, Yuan J, Tamae C, Medrano L, Maiz M, Wahba C, Lee C, Lee K, Tran KP, Yang H, Hoffman RM, Salih A, Miller JH. 2011. Selective advantage of resistant strains at trace levels of antibiotics: a simple and ultrasensitive color test for detection of antibiotics and genotoxic agents. Antimicrob. Agents Chemother. 55:1204–1210. 10.1128/AAC.01182-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hawkey PM, Jones AM. 2009. The changing epidemiology of resistance. J. Antimicrob. Chemother. 64(Suppl 1):i3–i10. 10.1093/jac/dkp256 [DOI] [PubMed] [Google Scholar]
- 4. Smet A, Martel A, Persoons D, Dewulf J, Heyndrickx M, Cloeckaert A, Praud K, Claeys G, Catry B, Herman L, Haesebrouck F, Butaye P. 2009. Comparative analysis of extended-spectrum-{beta}-lactamase-carrying plasmids from different members of Enterobacteriaceae isolated from poultry, pigs and humans: evidence for a shared {beta}-lactam resistance gene pool? J. Antimicrob. Chemother. 63:1286–1288. 10.1093/jac/dkp101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kang HY, Kim J, Seol SY, Lee YC, Lee JC, Cho DT. 2009. Characterization of conjugative plasmids carrying antibiotic resistance genes encoding 16S rRNA methylase, extended-spectrum beta-lactamase, and/or plasmid-mediated AmpC beta-lactamase. J. Microbiol. 47:68–75. 10.1007/s12275-008-0158-3 [DOI] [PubMed] [Google Scholar]
- 6. Carattoli A. 2009. Resistance plasmid families in Enterobacteriaceae. Antimicrob. Agents Chemother. 53:2227–2238. 10.1128/AAC.01707-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Amos GC, Hawkey PM, Gaze WH, Wellington EM. 2014. Waste water effluent contributes to the dissemination of CTX-M-15 in the natural environment. J. Antimicrob. Chemother. 69:1785–1791. 10.1093/jac/dku079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bouma JE, Lenski RE. 1988. Evolution of a bacteria/plasmid association. Nature 335:351–352. 10.1038/335351a0 [DOI] [PubMed] [Google Scholar]
- 9. Modi RI, Adams J. 1991. Coevolution in bacterial-plasmid populations. Evolution 45:656–667 [DOI] [PubMed] [Google Scholar]
- 10. Dahlberg C, Chao L. 2003. Amelioration of the cost of conjugative plasmid carriage in Escherichia coli K12. Genetics 165:1641–1649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dionisio F. 2005. Plasmids survive despite their cost and male-specific phages due to heterogeneity of bacterial populations. Evol. Ecol. Res. 7:1089–1107 [Google Scholar]
- 12. Humphrey B, Thomson NR, Thomas CM, Brooks K, Sanders M, Delsol AA, Roe JM, Bennett PM, Enne VI. 2012. Fitness of Escherichia coli strains carrying expressed and partially silent IncN and IncP1 plasmids. BMC Microbiol. 12:53. 10.1186/1471-2180-12-53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pitout JD. 2010. Infections with extended-spectrum beta-lactamase-producing Enterobacteriaceae: changing epidemiology and drug treatment choices. Drugs 70:313–333. 10.2165/11533040-000000000-00000 [DOI] [PubMed] [Google Scholar]
- 14. D’Andrea MM, Arena F, Pallecchi L, Rossolini GM. 2013. CTX-M-type β-lactamases: a successful story of antibiotic resistance. Int. J. Med. Microbiol. 303:305–317. 10.1016/j.ijmm.2013.02.008 [DOI] [PubMed] [Google Scholar]
- 15. Lytsy B, Sandegren L, Tano E, Torell E, Andersson DI, Melhus Å. 2008. The first major extended-spectrum beta-lactamase outbreak in Scandinavia was caused by clonal spread of a multiresistant Klebsiella pneumoniae producing CTX-M-15. APMIS 116:302–308. 10.1111/j.1600-0463.2008.00922.x [DOI] [PubMed] [Google Scholar]
- 16. Sandegren L, Linkevicius M, Lytsy B, Melhus Å, Andersson DI. 2012. Transfer of an Escherichia coli ST131 multiresistance cassette has created a Klebsiella pneumoniae-specific plasmid associated with a major nosocomial outbreak. J. Antimicrob. Chemother. 67:74–83. 10.1093/jac/dkr405 [DOI] [PubMed] [Google Scholar]
- 17. Adler M, Anjum M, Andersson DI, Sandegren L. 2013. Influence of acquired β-lactamases on the evolution of spontaneous carbapenem resistance in Escherichia coli. J. Antimicrob. Chemother. 68:51–59. 10.1093/jac/dks368 [DOI] [PubMed] [Google Scholar]
- 18. Tängdén T, Adler M, Cars O, Sandegren L, Löwdin E. 2013. Frequent emergence of porin-deficient subpopulations with reduced carbapenem susceptibility in ESBL-producing Escherichia coli during exposure to ertapenem in an in vitro pharmacokinetic model. J. Antimicrob. Chemother. 68:1319–1326. 10.1093/jac/dkt044 [DOI] [PubMed] [Google Scholar]
- 19. Foster TJ. 1983. Plasmid-determined resistance to antimicrobial drugs and toxic metal ions in bacteria. Microbiol. Rev. 47:361–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ghosh A, Singh A, Ramteke PW, Singh VP. 2000. Characterization of large plasmids encoding resistance to toxic heavy metals in Salmonella abortus equi. Biochem. Biophys. Res. Commun. 272:6–11. 10.1006/bbrc.2000.2727 [DOI] [PubMed] [Google Scholar]
- 21. Stepanauskas R, Glenn TC, Jagoe CH, Tuckfield RC, Lindell AH, King CJ, McArthur JV. 2006. Coselection for microbial resistance to metals and antibiotics in freshwater microcosms. Environ. Microbiol. 8:1510–1514. 10.1111/j.1462-2920.2006.01091.x [DOI] [PubMed] [Google Scholar]
- 22. Baker-Austin C, Wright MS, Stepanauskas R, McArthur JV. 2006. Co-selection of antibiotic and metal resistance. Trends Microbiol. 14:176–182. 10.1016/j.tim.2006.02.006 [DOI] [PubMed] [Google Scholar]
- 23. Stokes HW, Gillings MR. 2011. Gene flow, mobile genetic elements and the recruitment of antibiotic resistance genes into gram-negative pathogens. FEMS Microbiol. Rev. 35:790–819. 10.1111/j.1574-6976.2011.00273.x [DOI] [PubMed] [Google Scholar]
- 24. Hölzel CS, Müller C, Harms KS, Mikolajewski S, Schäfer S, Schwaiger K, Bauer J. 2012. Heavy metals in liquid pig manure in light of bacterial antimicrobial resistance. Environ. Res. 113:21–27. 10.1016/j.envres.2012.01.002 [DOI] [PubMed] [Google Scholar]
- 25. Zhu YG, Johnson TA, Su JQ, Qiao M, Guo GX, Stedtfeld RD, Hashsham SA, Tiedje JM. 2013. Diverse and abundant antibiotic resistance genes in Chinese swine farms. Proc. Natl. Acad. Sci. U. S. A. 110:3435–3440. 10.1073/pnas.1302581110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cullen WR, Reimer KJ. 1989. Arsenic speciation in the environment. Chem. Rev. 89:713–764. 10.1021/cr00094a002 [DOI] [Google Scholar]
- 27. Flemming CA, Trevors JT. 1989. Copper toxicity and chemistry in the environment: a review. Water Air Soil Pollut. 44:143–158. 10.1007/BF00228784 [DOI] [Google Scholar]
- 28. Smedley PL, Kinniburgh DG. 2002. A review of the source, behaviour and distribution of arsenic in natural waters. Appl. Geochem. 17:517–568. 10.1016/S0883-2927(02)00018-5 [DOI] [Google Scholar]
- 29. Garbarino JR, Bednar AJ, Rutherford DW, Beyer RS, Wershaw RL. 2003. Environmental fate of Roxarsone in poultry litter. I. Degradation of Roxarsone during composting. Environ. Sci. Technol. 37:1509–1514. 10.1021/es026219q [DOI] [PubMed] [Google Scholar]
- 30. Bolan NS, Khan MA, Donaldson J, Adriano DC, Matthew C. 2003. Distribution and bioavailability of copper in farm effluent. Sci. Total Environ. 309:225–236. 10.1016/S0048-9697(03)00052-4 [DOI] [PubMed] [Google Scholar]
- 31. Belluck DA, Benjamin SL, Baveye P, Sampson J, Johnson B. 2003. Widespread arsenic contamination of soils in residential areas and public spaces: an emerging regulatory or medical crisis? Int. J. Toxicol. 22:109–128. 10.1080/10915810305087 [DOI] [PubMed] [Google Scholar]
- 32. Li Y-X, Chen T-B. 2005. Concentrations of additive arsenic in Beijing pig feeds and the residues in pig manure. Resour. Conserv. Recycl. 45:356–367. 10.1016/j.resconrec.2005.03.002 [DOI] [Google Scholar]
- 33. Silbergeld EK, Nachman K. 2008. The environmental and public health risks associated with arsenical use in animal feeds. Ann. N. Y. Acad. Sci. 1140:346–357. 10.1196/annals.1454.049 [DOI] [PubMed] [Google Scholar]
- 34. Apgar GA, Kornegay ET, Lindemann MD, Notter DR. 1995. Evaluation of copper sulfate and a copper lysine complex as growth promoters for weanling swine. J. Anim. Sci. 73:2640–2646 [DOI] [PubMed] [Google Scholar]
- 35. Hasman H, Kempf I, Chidaine B, Cariolet R, Ersbøll AK, Houe H, Bruun Hansen HC, Aarestrup FM. 2006. Copper resistance in Enterococcus faecium, mediated by the tcrB gene, is selected by supplementation of pig feed with copper sulfate. Appl. Environ. Microbiol. 72:5784–5789. 10.1128/AEM.02979-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Baker RS, Barrentine WL, Bowman DH, Hawthorne WL, Pettiet JV. 1976. Crop response and arsenic uptake following soil incorporation of MSMA. Weed Sci. 24:322–326 [Google Scholar]
- 37. Kümmerer K. 2009. Antibiotics in the aquatic environment—a review—part I. Chemosphere 75:417–434. 10.1016/j.chemosphere.2008.11.086 [DOI] [PubMed] [Google Scholar]
- 38. Sarmah AK, Meyer MT, Boxall AB. 2006. A global perspective on the use, sales, exposure pathways, occurrence, fate and effects of veterinary antibiotics (VAs) in the environment. Chemosphere 65:725–759. 10.1016/j.chemosphere.2006.03.026 [DOI] [PubMed] [Google Scholar]
- 39. Kümmerer K. 2009. Antibiotics in the aquatic environment—a review—part II. Chemosphere 75:435–441. 10.1016/j.chemosphere.2008.12.006 [DOI] [PubMed] [Google Scholar]
- 40. Pruden A, Larsson DG, Amézquita A, Collignon P, Brandt KK, Graham DW, Lazorchak JM, Suzuki S, Silley P, Snape JR, Topp E, Zhang T, Zhu YG. 2013. Management options for reducing the release of antibiotics and antibiotic resistance genes to the environment. Environ. Health Perspect. 121:878–885. 10.1289/ehp.1206446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Larsson DG, de Pedro C, Paxeus N. 2007. Effluent from drug manufactures contains extremely high levels of pharmaceuticals. J. Hazard. Mater. 148:751–755. 10.1016/j.jhazmat.2007.07.008 [DOI] [PubMed] [Google Scholar]
- 42. Li D, Yang M, Hu J, Ren L, Zhang Y, Li K. 2008. Determination and fate of oxytetracycline and related compounds in oxytetracycline production wastewater and the receiving river. Environ. Toxicol. Chem. 27:80–86. 10.1897/07-080.1 [DOI] [PubMed] [Google Scholar]
- 43. Sim WJ, Lee JW, Lee ES, Shin SK, Hwang SR, Oh JE. 2011. Occurrence and distribution of pharmaceuticals in wastewater from households, livestock farms, hospitals and pharmaceutical manufactures. Chemosphere 82:179–186. 10.1016/j.chemosphere.2010.10.026 [DOI] [PubMed] [Google Scholar]
- 44. Fick J, Söderström H, Lindberg RH, Phan C, Tysklind M, Larsson DG. 2009. Contamination of surface, ground, and drinking water from pharmaceutical production. Environ. Toxicol. Chem. 28:2522–2527. 10.1897/09-073.1 [DOI] [PubMed] [Google Scholar]
- 45. Finley RL, Collignon P, Larsson DG, McEwen SA, Li XZ, Gaze WH, Reid-Smith R, Timinouni M, Graham DW, Topp E. 2013. The scourge of antibiotic resistance: the important role of the environment. Clin. Infect. Dis. 57:704–710. 10.1093/cid/cit355 [DOI] [PubMed] [Google Scholar]
- 46. Lindberg RH, Björklund K, Rendahl P, Johansson MI, Tysklind M, Andersson BA. 2007. Environmental risk assessment of antibiotics in the Swedish environment with emphasis on sewage treatment plants. Water Res. 41:613–619. 10.1016/j.watres.2006.11.014 [DOI] [PubMed] [Google Scholar]
- 47. Subach OM, Cranfill PJ, Davidson MW, Verkhusha VV. 2011. An enhanced monomeric blue fluorescent protein with the high chemical stability of the chromophore. PLoS One 6:e28674. 10.1371/journal.pone.0028674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kremers GJ, Goedhart J, van Munster EB, Gadella TW. 2006. Cyan and yellow super fluorescent proteins with improved brightness, protein folding, and FRET Förster radius. Biochemistry 45:6570–6580. 10.1021/bi0516273 [DOI] [PubMed] [Google Scholar]
- 49. Achtman M, Manning PA, Kusecek B, Schwuchow S, Willetts N. 1980. A genetic analysis of F sex factor cistrons needed for surface exclusion in Escherichia coli. J. Mol. Biol. 138:779–795. 10.1016/0022-2836(80)90065-0 [DOI] [PubMed] [Google Scholar]
- 50. Willetts N, Skurray R. 1980. The conjugation system of F-like plasmids. Annu. Rev. Genet. 14:41–76. 10.1146/annurev.ge.14.120180.000353 [DOI] [PubMed] [Google Scholar]
- 51. Silverman PM, Wickersham E, Harris R. 1991. Regulation of the F plasmid traY promoter in Escherichia coli by host and plasmid factors. J. Mol. Biol. 218:119–128. 10.1016/0022-2836(91)90878-A [DOI] [PubMed] [Google Scholar]
- 52. Frost LS, Ippen-Ihler K, Skurray RA. 1994. Analysis of the sequence and gene products of the transfer region of the F sex factor. Microbiol. Mol. Biol. Rev. 58:162–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Will WR, Frost LS. 2006. Characterization of the opposing roles of H-NS and TraJ in transcriptional regulation of the F-plasmid tra operon. J. Bacteriol. 188:507–514. 10.1128/JB.188.2.507-514.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Pan X, Qiang Z, Ben W, Chen M. 2011. Residual veterinary antibiotics in swine manure from concentrated animal feeding operations in Shandong Province, China. Chemosphere 84:695–700. 10.1016/j.chemosphere.2011.03.022 [DOI] [PubMed] [Google Scholar]
- 55. Harrison EZ, Oakes SR, Hysell M, Hay A. 2006. Organic chemicals in sewage sludges. Sci. Total Environ. 367:481–497. 10.1016/j.scitotenv.2006.04.002 [DOI] [PubMed] [Google Scholar]
- 56. Kim K-R, Owens G, Kwon S-I, So K-H, Lee D-B, Ok YS. 2011. Occurrence and environmental fate of veterinary antibiotics in the terrestrial environment. Water Air Soil Pollut. 214:163–174. 10.1007/s11270-010-0412-2 [DOI] [Google Scholar]
- 57. Lapworth DJ, Baran N, Stuart ME, Ward RS. 2012. Emerging organic contaminants in groundwater: a review of sources, fate and occurrence. Environ. Pollut. 163:287–303. 10.1016/j.envpol.2011.12.034 [DOI] [PubMed] [Google Scholar]
- 58. Olofsson U, Bignert A, Haglund P. 2012. Time-trends of metals and organic contaminants in sewage sludge. Water Res. 46:4841–4851. 10.1016/j.watres.2012.05.048 [DOI] [PubMed] [Google Scholar]
- 59. Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645. 10.1073/pnas.120163297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wilson FH, Hawkins DB. 1978. Arsenic in streams, stream sediments, and ground water, Fairbanks area, Alaska. Environ. Geol. 2:195–202. 10.1007/BF02380485 [DOI] [Google Scholar]
- 61. Welch AH, Westjohn DB, Helsel DR, Wanty RB. 2000. Arsenic in ground water of the United States: occurrence and geochemistry. Ground Water 38:589–604. 10.1111/j.1745-6584.2000.tb00251.x [DOI] [Google Scholar]
- 62. Nickson R, McArthur J, Burgess W, Ahmed KM, Ravenscroft P, Rahman M. 1998. Arsenic poisoning of Bangladesh groundwater. Nature 395:338. 10.1038/26387 [DOI] [PubMed] [Google Scholar]
- 63. Harvey CF, Swartz CH, Badruzzaman AB, Keon-Blute N, Yu W, Ali MA, Jay J, Beckie R, Niedan V, Brabander D, Oates PM, Ashfaque KN, Islam S, Hemond HF, Ahmed MF. 2002. Arsenic mobility and groundwater extraction in Bangladesh. Science 298:1602–1606. 10.1126/science.1076978 [DOI] [PubMed] [Google Scholar]
- 64. Smith AH, Lingas EO, Rahman M. 2000. Contamination of drinking-water by arsenic in Bangladesh: a public health emergency. Bull. World Health Organ. 78:1093–1103 [PMC free article] [PubMed] [Google Scholar]
- 65. Wershaw RL, Garbarino JR, Burkhardt MR. 1999. Roxarsone in natural water systems. Effects of animal feeding operations on water resources and the environment. In Proceedings of the technical meeting, Fort Collins, Colorado, August 30–September 1, 1999. U.S. Geological Survey, Fort Collins, CO [Google Scholar]
- 66. Nriagu JO, Pacyna JM. 1988. Quantitative assessment of worldwide contamination of air, water and soils by trace metals. Nature 333:134–139. 10.1038/333134a0 [DOI] [PubMed] [Google Scholar]
- 67. Kelly J, Thornton I, Simpson PR. 1996. Urban geochemistry: a study of the influence of anthropogenic activity on the heavy metal content of soils in traditionally industrial and non-industrial areas of Britain. Appl. Geochem. 11:363–370. 10.1016/0883-2927(95)00084-4 [DOI] [Google Scholar]
- 68. Glover RD, Miller JM, Hutchison JE. 2011. Generation of metal nanoparticles from silver and copper objects: nanoparticle dynamics on surfaces and potential sources of nanoparticles in the environment. ACS Nano 5:8950–8957. 10.1021/nn2031319 [DOI] [PubMed] [Google Scholar]
- 69. Li X, Poon C-S, Liu PS. 2001. Heavy metal contamination of urban soils and street dusts in Hong Kong. Appl. Geochem. 16:1361–1368. 10.1016/S0883-2927(01)00045-2 [DOI] [Google Scholar]
- 70. Hylander LD, Goodsite ME. 2006. Environmental costs of mercury pollution. Sci. Total Environ. 368:352–370. 10.1016/j.scitotenv.2005.11.029 [DOI] [PubMed] [Google Scholar]
- 71. Wong CS, Li X, Thornton I. 2006. Urban environmental geochemistry of trace metals. Environ. Pollut. 142:1–16. 10.1016/j.envpol.2005.09.004 [DOI] [PubMed] [Google Scholar]
- 72. Bird G, Macklin MG, Brewer PA, Zaharia S, Balteanu D, Driga B, Serban M. 2009. Heavy metals in potable groundwater of mining-affected river catchments, northwestern Romania. Environ. Geochem. Health 31:741–758. 10.1007/s10653-009-9259-0 [DOI] [PubMed] [Google Scholar]
- 73. Fabrega J, Luoma SN, Tyler CR, Galloway TS, Lead JR. 2011. Silver nanoparticles: behaviour and effects in the aquatic environment. Environ. Int. 37:517–531. 10.1016/j.envint.2010.10.012 [DOI] [PubMed] [Google Scholar]
- 74. Wei B, Yang L. 2010. A review of heavy metal contaminations in urban soils, urban road dusts and agricultural soils from China. Microchem. J. 94:99–107. 10.1016/j.microc.2009.09.014 [DOI] [Google Scholar]
- 75. Luo XS, Yu S, Zhu YG, Li XD. 2012. Trace metal contamination in urban soils of China. Sci. Total Environ. 421-422:17–30. 10.1016/j.scitotenv.2011.04.020 [DOI] [PubMed] [Google Scholar]
- 76. Eguchi K, Nagase H, Ozawa M, Endoh YS, Goto K, Hirata K, Miyamoto K, Yoshimura H. 2004. Evaluation of antimicrobial agents for veterinary use in the ecotoxicity test using microalgae. Chemosphere 57:1733–1738. 10.1016/j.chemosphere.2004.07.017 [DOI] [PubMed] [Google Scholar]
- 77. Andersson DI, Hughes D. 2010. Antibiotic resistance and its cost: is it possible to reverse resistance? Nat. Rev. Microbiol. 8:260–271 [DOI] [PubMed] [Google Scholar]
- 78. Lutz R, Bujard H. 1997. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 25:1203–1210. 10.1093/nar/25.6.1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Kelly JR, Rubin AJ, Davis JH, Ajo-Franklin CM, Cumbers J, Czar MJ, de Mora K, Glieberman AL, Monie DD, Endy D. 2009. Measuring the activity of BioBrick promoters using an in vivo reference standard. J. Biol. Eng. 3:4. 10.1186/1754-1611-3-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Dykhuizen DE. 1990. Experimental studies of natural selection in bacteria. Annu. Rev. Ecol. Syst. 21:373–398. 10.1146/annurev.es.21.110190.002105 [DOI] [Google Scholar]
- 81. Atwood KC, Schneider LK, Ryan FJ. 1951. Periodic selection in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 37:146–155. 10.1073/pnas.37.3.146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Conrad TM, Joyce AR, Applebee MK, Barrett CL, Xie B, Gao Y, Palsson BØ. 2009. Whole-genome resequencing of Escherichia coli K-12 MG1655 undergoing short-term laboratory evolution in lactate minimal media reveals flexible selection of adaptive mutations. Genome Biol. 10:R118. 10.1186/gb-2009-10-10-r118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Adler M, Anjum M, Berg OG, Andersson DI, Sandegren L. 2014. High fitness costs and instability of gene duplications reduce rates of evolution of new genes by duplication-divergence mechanisms. Mol. Biol. Evol. 31:1526–1535. 10.1093/molbev/msu111 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Competitions with low initial fractions of resistant bacteria. Competition experiments at different concentrations of arsenite and different starting fractions of resistant bacteria carrying the pUUH239.2 plasmid. (A) Initial ratio of susceptible to resistant mutants, 10:1. (B) Initial ratio of susceptible to resistant mutants, 100:1. Data can be found in Table S8. Standard deviations are indicated. Download
Costs of fluorescence markers.
Primers used in this study.
Data for Fig. 1
Data for Fig. 2A
Data for Fig. 2B
Data for Fig. 2C
Data for Fig. 3
Data for Fig. S1
Summary of whole-genome sequencing data.