Abstract
Major efforts are underway to systematically define the somatic and germline genetic variations causally associated with disease. Genome-wide genetic analysis of actual clinical samples is, however, limited by the paucity of genomic DNA available. Here we have tested the fidelity and genome representation of φ29 polymerase-based genome amplification (φ29MDA) using direct sequencing and high density oligonucleotide arrays probing >10 000 SNP alleles. Genome representation was comprehensive and estimated to be 99.82% complete, although six regions encompassing a maximum of 5.62 Mb failed to amplify. There was no degradation in the accuracy of SNP genotyping and, in direct sequencing experiments sampling 500 000 bp, the estimated error rate (9.5 × 10–6) was the same as in paired unamplified samples. The detection of cancer-associated loss of heterozygosity and copy number changes, including homozygous deletion and gene amplification, were similarly robust. These results suggest that φ29MDA yields high fidelity, near-complete genome representation suitable for high resolution genetic analysis.
INTRODUCTION
Understanding the genomic alterations of disease-derived cells is of vital importance for the development of more rational diagnostics and treatment strategies. In principle, the completed sequence of the human genome allows for the interrogation of virtually every single base for variations between the canonical and disease genome sequences. However, genetic analysis of clinical samples is often limited, in practice, by the amount of genomic DNA available. New technologies allowing in vitro production of numerous copies of the entire genome are currently available. However, the extent to which such methods result in a representative copy of the initial genome and the fidelity with which such representative copies are produced remains largely uncharacterized.
Whole genome amplification by multiple displacement amplification is based on the use of φ29 DNA polymerase and random primers (henceforth referred to as φ29MDA) (1–4). φ29 polymerase combines high processivity with a strand displacement ability leading to the synthesis of DNA fragments >70 kb (5) and favoring uniform representation of sequences. In addition, the enzyme possesses 3′→5′ exonuclease activity resulting in error rates thought to be between 10–5 and 10–6 (6).
In this study, we have used φ29MDA to amplify genomic DNA from both normal and cancer cells. We then tested the accuracy and genome-wide coverage of φ29MDA through both direct sequencing of ∼500 000 bp of DNA and the use of high density oligonucleotide arrays interrogating >10 000 single nucleotide polymorphisms (SNPs). Genome-wide amplification achieved an estimated 99.82% coverage of the human genome. SNP call concordance was comparable to that of unamplified replicates and, in the context of large-scale exon resequencing, highly accurate sequence data were obtained from φ29MDA samples when compared to samples amplified by standard methods. Finally, our ability to detect several types of chromosomal aberrations, including loss of heterozygosity (LOH), homozygous deletions and gene amplification remained robust compared to non-amplified samples. In summary, our results show that φ29MDA affords highly accurate and comprehensive whole genome amplification suitable for high resolution genetic analysis.
MATERIALS AND METHODS
DNA samples
Genomic DNA samples from cell lines were obtained from ATCC (Manassas, VA). Of the 16 samples used, six were breast cancer cell/normal pairs, namely cell lines HCC1008 (renamed HCC1007 for consistency), HCC1143, HCC1599, HCC1937, HCC2218 and HCC38 and their normal counterparts generated by EBV-induced transformation of peripheral blood lymphocytes obtained from the same patients (HCC1007 BL, HCC1143 BL, HCC1599 BL, HCC1937 BL, HCC2218 BL and HCC38 BL). Samples HCC2157 BL and HCC1954 BL were unmatched. Two normal reference DNA sets from blood samples were also obtained from Affymetrix (Santa Clara, CA). For exon resequencing experiments genomic DNA from 20 samples of lung adenocarcinoma was used. DNA was quantified using a PicoGreen® dsDNA quantitation kit (Molecular Probes Inc., Eugene, OR).
φ29 polymerase multiple strand displacement whole genome amplification
Whole genome amplification reagents (REPLI-g™ 625S; Molecular Staging Inc., New Haven, CT) were used as follows. Briefly, 10 ng template DNA was mixed with 4× master mix (containing the reaction buffer and hexamer primers) and the DNA polymerase in a 50 µl final volume reaction. Completed reactions were transferred to a 96-well plate and incubated for 16 h at 30°C, followed by incubation at 65°C for 3 min to inactivate the enzyme. For five samples (HCC1007BL, HCC1007, HCC1143BL, HCC1143 and HCC1599BL), template DNA was denatured using alkaline denaturation prior to the φ29MDA reaction (version 2 φ29MDA). In all cases, 10 µg amplified DNA was purified using a QIAquick PCR purification kit (Qiagen Inc., Valencia, CA). Purified DNA was quantified and 250 ng was used for SNP array analysis and 5 ng for PCR followed by sequencing.
Single nucleotide polymorphism array experiments
10K Mapping Arrays and hybridization reagents were obtained from Affymetrix (7,8). Aliquots of 250 ng genomic or whole genome amplification DNA was restricted with XbaI. A single double-stranded linker was ligated to the XbaI fragments using T4 DNA ligase. XbaI fragments were then amplified by single primer PCR as per the manufacturer’s protocol. PCR products were purified from free primers and nucleotides by differential precipitation in 2-propanol/sodium perchlorate. The PCR products were quantified spectrophotometrically and assayed for the appropriate size distribution on a DNA analyzer (Agilent 2100 Bioanalyzer). The purified PCR products were fragmented with DNase I and the resulting DNA was labeled with a single biotin at each free 3′-OH using terminal deoxynucleotidyl transferase and a dideoxy biotinylated nucleoside triphosphate. The biotinylated fragments were added to a hybridization solution [12× MES (1.22 M), 100% DMSO, 50× Denhardt’s solution, 0.5 M EDTA, 10 mg/ml herring sperm DNA, 3 nM Oligo B2, 1 mg/ml human Cot-1, 1% Tween-20, 5 M TMACL] containing a biotinylated control oligonucleotide (for quality control) and then hybridized to a 10K SNP microarray chip overnight at 48°C. The arrays were then washed six times with non-stringent buffer A (6× SSPE, 0.01% Tween-20) and stringent buffer B (0.6× SSPE, 0.01% Tween-20). Bound DNA was then detected by incubation with streptavidin followed by biotinylated anti-streptavidin, followed by phycoerythrin-conjugated streptavidin (SAPE). Bound fluorescent antibody was detected using a confocal laser scanner (570 nm) and the positions and intensities of the fluorescence emissions were captured.
Single nucleotide polymorphism data visualization and analysis
dChipSNP (9,10), a bioinformatics platform developed for SNP array data analysis, was used to automate the process of making LOH calls based on paired normal/tumor SNP calls and displaying the SNP data according to their chromosomal locations, along with cytogenetic band information. Figures 1B and 3B and C were generated based on ‘Chromosome View’ in dChipSNP. The SNP call rates of unamplified samples were compared to those of the paired φ29MDA samples using the one sample Wilcoxon rank sum test. The SNP or LOH call concordance rate between two samples was calculated as the proportion of concordant markers among all markers excluding those in which either one or both of the two samples was NO_CALL. We used the two sample Wilcoxon rank sum test to compare the SNP call concordance rate between unamplified samples and the two types of φ29MDA samples (obtained from non-denatured and denatured DNA, respectively) to the concordance rate between unamplified replicates and also to compare the SNP call concordance rates between the two types of φ29MDA samples. To assess the quality of φ29MDA on the entire genome, all the arrays were first normalized using the invariant set method and then for each SNP marker a signal intensity index was calculated using a model-based approach (11). The fold changes in the signal intensity index between unamplified samples and φ29MDA samples were calculated for all SNP markers and statistical significance was assessed using a permutation test with multiple comparison adjustment by the maxT procedure (12).
Figure 1.
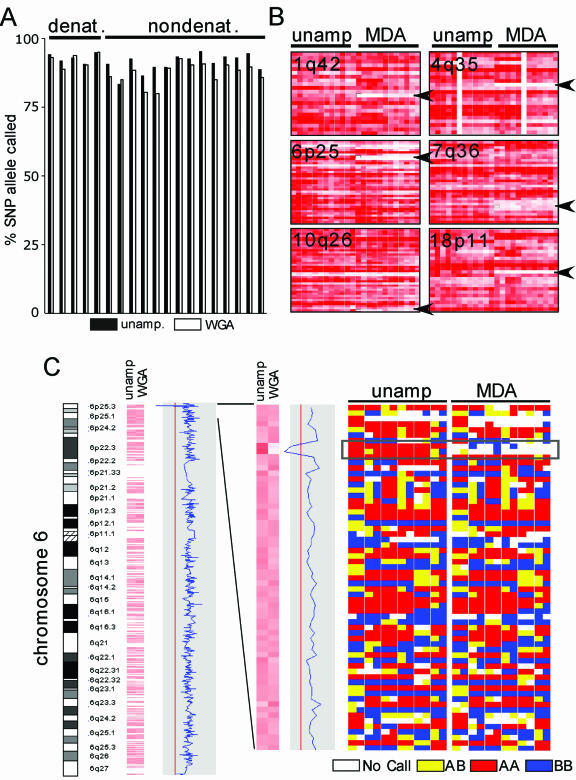
Genome coverage following whole genome amplification. (A) The SNP call rate before and after whole genome amplification. Probes were prepared from 250 ng DNA from samples of breast cancer cell line or paired normal cell line DNA either before (unamplified) or after (φ29MDA) amplification and hybridized to Affymetrix 10K SNP arrays. In addition, 14 samples were amplified without denaturation prior to whole genome amplification, while five samples were amplified after denaturation (as indicated). (B) Regions with loss of representation after φ29MDA. For each indicated region, the specific SNP allele hybridization intensity normalized on a scale of 0 (absent and white) to 6 (red) is shown for unamplified or amplified samples. Each row represents a single SNP allele, while each column represents a single DNA sample. Columns 1–5 correspond to denatured DNA samples (HCC1007BL, HCC1007, HCC1143BL, HCC1143 and HCC1599BL). (C) The absence of amplification of a region in the chromosome cytoband 6p25.3. Using dChip-Signal, the mean signal intensity for each SNP allele on the 10K array was compared between samples before (left) and after (right) whole genome amplification. Signal intensity is shown normalized on a scale of 0 (white) to 6 (red). An enlargement of this region is shown in the middle panel. The right panel indicates the calls for each specific SNP allele where white is no call, yellow is AB, blue is BB and red is AA. The region indicated by the dotted line shows absent or incorrect calls in samples after MDA, corresponding to the region where signal loss was detected in dChip-Signal. Columns 1–5 correspond to denatured DNA samples.
Figure 3.
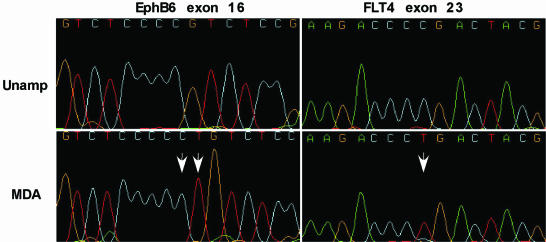
Rare homozygous sequence variations in φ29MDA samples. DNA sequence traces for unamplified (upper) and φ29MDA (lower) samples showing a homozygous 2 bp (CT) insertion and a homozygous C→T substitution (white arrows) detected by NQS analysis. All samples were non-denatured.
Copy number analysis
Based on the signal intensities, the copy number of each SNP locus in cancer cell lines samples was estimated by hidden Markov model analysis of the signal intensity variation along each chromosome, compared to a set of normal references (13). In addition to the unamplified normal samples described above, over 50 additional samples from normal tissue were included as unamplified normal references (generously provided by E.Hochberg and J.Ritz) to improve the accuracy of copy number estimates. For the φ29MDA DNA samples, φ29MDA DNA obtained from immortalized lymphocytes (described above) was used as normal references. Copy number estimates were binned in the categories 0, 1, 2, 3, 4 and >4 before calculating copy number discordance rates.
PCR and sequencing
One hundred tyrosine kinase exons and flanking intronic sequences were amplified using specific primers in a 384-well format nested PCR set-up (J. G. Paez, J. C. Lee, M. Meverson and W. R. Sellars, in preparation). The nested primers were tagged with M13 tails. PCR was performed using 5 ng of both amplified and unamplified DNA. PCR products were purified using SPRI (solid phase reversible immobilization) chemistry followed by bidirectional dye terminator fluorescent sequencing with universal M13 primers. Sequencing fragments were detected via capillary electrophoresis using an ABI Prism 3700 DNA Analyzer (Applied Biosystems, Foster City, CA). PCR and sequencing was performed at Agencourt Bioscience Corp. (Beverly, MA).
Sequence data analysis
Forty DNA samples consisting of 20 generated by φ29MDA and 20 corresponding unamplified matched DNA were sequenced against 100 amplicons. In all, 4000 forward (F) and 4000 reverse (R) chromatogram reads (2000 sets) were analyzed in batch by Mutation Surveyor (Soft Genetics Inc. Version 2.03, release 9/25/2003). High quality sequence variations found in both the forward and reverse directions were exported into a flat text file for comparison between φ29MDA and unamplified DNA variation calls. A Perl script was written to analyze the text files and data in which any of the four possible reads (φ29MDA F&R, unamplified F&R) did not pass the quality standards for Mutation Surveyor analyses were omitted. Filtering the data resulted in 1295 of the original 2000 sets of reads passing, representing ∼500 000 bases analyzed (F&R = 1 base). To analyze the sequencing data using the Neighborhood Quality Standard (NQS) (14,15), pair-wise alignments for each unamplified/φ29MDA sample pair were generated for each amplicon and differences were detected using the NQS. Briefly, a base met NQS if the Phred quality (16) was ≥30 and the 5 bases on each side had Phred quality scores ≥25. Sample pairs with fewer than 50 NQS bases were discarded. In samples potentially containing deletions, average quality from neighboring bases was used as a surrogate for base quality. The number of passing sample pairs (≥50 NQS bases compared) was 1585, leading to a total of 404 335 NQS bases (F&R = 1 base).
RESULTS
Genome representation after φ29 polymerase-based whole genome amplification
Genomic DNA from seven breast cancer cell lines and seven EBV-immortalized lymphoblastoid cell lines was used for these studies. Twelve of these samples represent six pairs comprising a cancer-derived cell line and a lymphoblastoid cell line representing the paired normal genome. Aliquots of 10 ng of each genomic DNA sample were subjected to φ29MDA, yielding an average of 25 µg total DNA/sample. In addition, genomic DNA from five samples was denatured by treatment with alkali prior to φ29MDA. To ascertain the extent to which φ29MDA resulted in whole genome representation, we determined the rate of detection for ∼10 000 SNPs alleles in each DNA sample, using high density oligonucleotide arrays. Here, 250 ng φ29MDA DNA or unamplified DNA was used to prepare probes for the Affymetrix 10K SNP array. High quality array data was obtained on all samples as judged by the performance of the spiked controls. As a first measure of genome representation we compared the number of SNPs successfully detected or called (the call rate) in the φ29MDA samples versus unamplified samples by pair-wise comparison (Fig. 1A). While all φ29MDA samples performed acceptably, in samples prepared by φ29MDA without a preceding denaturation step the mean call rate was 88.92%, compared to a mean of 92.45% in the unamplified samples. The call rate improved substantially with denaturing of genomic DNA prior to φ29MDA, such that the call rates were similar between φ29MDA and unamplified samples (92.06 versus 92.93%, P = 0.24).
We next determined whether this small decrease in the mean call rate reflected random loss of genomic representation differing in each φ29MDA sample preparation or alternatively reflected non-random loss of amplification of specific regions of the genome. Recent data from our group has shown that the oligonucleotide SNP arrays can be used not only for the robust specification of allele calls, but can also detect copy number changes (13). Thus, as a second approach to the question of genome representation after φ29MDA, we normalized oligonucleotide signal intensity across all arrays using the invariant probe set method (11) and determined whether specific regions of the genome were under-represented in producing hybridizing fragments, as indicated by consistent loss of the hybridization signal. Here, the mean signal intensity for each SNP allele in the amplified samples was compared to the mean signal intensity of the unamplified samples by plotting the log2 ratio of the mean φ29MDA to unamplified signal (as shown for chromosome 6 in Fig. 1C). Regions of statistically significant signal loss were then sought. Fourteen regions of consistent signal loss were detected in the φ29MDA amplified samples (Table 1). Conversely, no regions of loss were found in unamplified samples when compared to the amplified samples. When the specific SNP alleles in each region were examined in detail, six regions (1q42, 4q35, 6p25, 7q36, 10q26 and 18p11) showed both significantly reduced oligonucleotide signal intensity and consistent no-call designations in the φ29MDA samples (Fig. 1B and Table 1, shaded in gray). An example of one such region located on chromosome 6p25.3 is shown in Figure 1C, where the mean signal intensity of φ29MDA and unamplified samples, as well as the specific allele calls, are shown. The remaining eight regions had low signal intensity, suggesting inadequate amplification, however, there was apparently sufficient amplification to allow accurate detection of the specific SNP alleles. The six regions of low intensity and poor SNP calls represent a maximum of 5.64 Mb (measured from the first flanking retained SNP markers). For the currently assembled human genome size of 3070 Mb (17) these data give an estimated genome coverage of (3070 – 5.64 Mb/3070 Mb) × 100 = 99.82%.
Table 1. Under-represented regions after whole genome amplification.
| Cytobanda | Locationb | Markerc | SNP calld (%) |
|---|---|---|---|
| 1q42.3 | 231.859845–232.716009 | 273247C | 0 |
| 2p25.3 | 2.981319–3.533432 | 510038A | 100 |
| 4q35.2 | 189.005358–189.512271 | 53705A | 0 |
| 6p25.3 | 1.308549–1.308710 (min) | 47914C, 47913A | 0 |
| 1.267150–1.461644 (max) | |||
| 7p13 | 43.671625–45.002746 | 446257A | 100 |
| 7q36.3 | 154.928147–155.709859 | 696854A | 0 |
| 8p11.1 | 40.228919–46.972292 | 1135786A | 100 |
| 9q34.11 | 122.379760–125.318769 | 423667A | 100 |
| 10q26.3 | 132.676552-qter | 725725A | 0 |
| 16q22.1 | 66.320884–69.686706 | 56839C | 100 |
| 18p11.21 | 11.601378–12.539851 | 64448A | 0 |
| 20p11.23 | 20.264205–20.826382 | 48858G | 100 |
| 20q13.33 | 60.531791–62.575257 | 108798A | 100 |
| 22q13.1 | 37.019787–39.869701 | 242417C | 100 |
aCytoband is based on the hg15 golden path assembly (NCBI build 33) (http://genome.ucsc.edu/).
bChromosomal location in Mb.
cSNP probe set name.
dFor the six φ29MDA samples, six (100%) or zero (0%) SNP calls were obtained.
These regions of loss are not physically constrained to particular chromosomal segments such as telomeres, thus we examined each region to determine whether there were any obvious clues as to why such regions might be more difficult to amplify. While each region contains repetitive elements of the LINE, SINE and MER1 families, it is unclear whether this bears causally on the lack of amplification for these small regions.
Fidelity of whole genome amplification for detecting SNP alleles
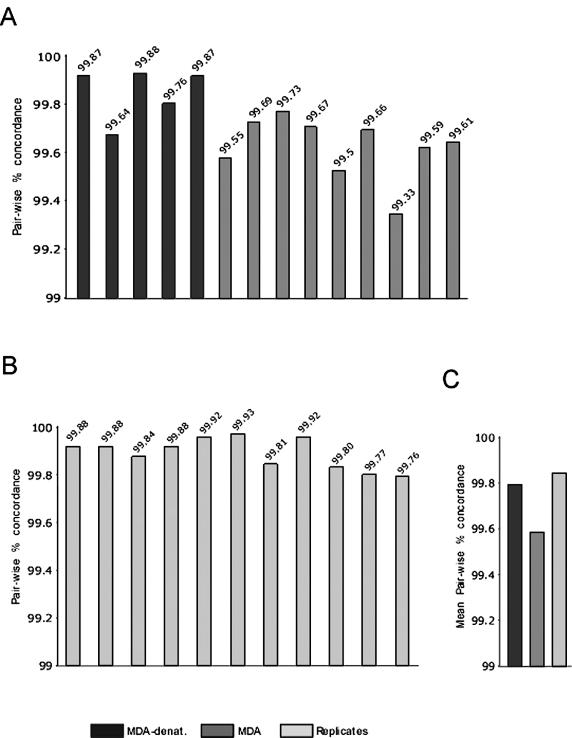
In order to ascertain whether φ29MDA gives robust SNP allele calls when used in a large-scale SNP genotyping application, we next determined the SNP call concordance between φ29MDA amplified and unamplified pairs. Here, only SNPs giving ‘calls’ in both the unamplified and the φ29MDA samples were considered. To assess the experimental variability, SNP concordance was measured in 10 pairs of replicate samples. Concordance was 99.85% among paired replicates, 99.59% (P = 0.000004) when non-denatured φ29MDA samples were compared to the unamplified samples and 99.80% (P = 0.12) when alkaline denatured φ29MDA samples were compared to unamplified samples (Fig. 2A–C).
Figure 2.
SNP allele concordance. (A) SNP allele concordance between pairs of φ29MDA amplified and unamplified samples (black and dark gray). (B) SNP allele concordance between pairs of unamplified replicates (light gray). (C) Mean concordance in the same pair-wise comparisons as shown in (A) and (B).
Examination of the discrepancies between non-denatured φ29MDA and unamplified samples showed that 63% were heterozygotes called as homozygotes. Approximately 30% of all calls were heterozygotes and hence the heterozygote drop-out rate is ∼0.73% for this method (data not shown). It is thought that the inclusion of an alkaline denaturation step may reduce stochastic effects and serve to ensure even priming of maternal and paternal alleles. Indeed, the heterozygote drop-out rate decreased by 24-fold from 0.73 to <0.03% with inclusion of the alkaline denaturation step. In no case were homozygote conversions detected.
As the rate of discordant calls found in the denatured samples was nearly identical to that found in the replicate data, we conclude that under these conditions the SNP error rate after φ29MDA is within the experimental error for this assay and thus the estimated upper bound of the φ29MDA error rate is <0.2%. These data indicate that highly reliable SNP genotyping can be obtained after φ29MDA genome amplification.
Direct sequencing comparison
In order to test more robustly the fidelity of φ29MDA, 20 samples of DNA obtained from lung adenocarcinoma were subjected to φ29MDA. Next, all 20 φ29MDA amplified and the corresponding unamplified genomic DNA samples were used in nested PCR amplification reactions to generate sequencing templates for 100 tyrosine kinase exons. All resulting amplicons were then used in fluorescent dye terminator sequencing reactions with standard M13 forward and reverse primers. A total of 1295 paired φ29MDA and unamplified sequencing reads were available for analysis by Mutation Surveyor, representing nearly 500 000 paired bases for each method. Single direction sequences where the opposite read was not available were excluded. In this dataset, variants from the canonical sequence were detected using Mutation Surveyor followed by manual review. There were 234 variants detected by both methods, four variants detected in unamplified samples not found in the φ29MDA samples and five variants found in the φ29MDA samples that were not found in the unamplified samples (Table 2). From these data we estimate that the error rate of PCR-based amplification and sequencing is 7.6 × 10–6 and that of φ29MDA followed by PCR-based sequencing is 9.5 × 10–6. These error rates are not different statistically.
Table 2. Direct sequence comparison.
| No. of variations | Error (×10–6) | Concordance | |
|---|---|---|---|
| Both | 234 | ||
| Unamplified | 4 | 7.6 | 0.9999924 |
| Whole genome amplified | 5 | 9.5 | 0.9999905 |
We also approached the question of fidelity using the NQS (14,15) to detect homozygous changes between unamplified and amplified samples. Because NQS excludes heterozygous base calls, it produces a lower bound on the discrepancy rate between amplified and unamplified sample pairs. In 404 335 bases that passed the NQS quality criteria, five differences were detected: four substitutions and one two-base insertion. The overall concordance rate was therefore 1 – (5/404335) = 0.999985. In all five discrepancies, the φ29MDA amplified sample differed from the canonical sequence. Illustrative of two of these variations are the dinucleotide insertion and the homozygous base pair substitution shown in Figure 3. Thus a lower bound for error introduced by φ29MDA estimated by NQS is 1.24 × 10–5.
These error rates are relatively low in the context of PCR-based sequencing efforts, nonetheless, the possibility of a few φ29MDA-induced alterations including homozygous changes detected by the NQS method indicates that sequencing reconfirmation of candidate variants from unamplified DNA samples or from independently amplified samples may be warranted, as is currently the case for alterations detected in PCR amplified samples.
Detection of large-scale genetic alterations after whole genome amplification
Methods for robust whole genome amplification may enable genome-wide analysis of the somatic genetic alterations from samples that are limited to a few hundred cells. Samples where there is an obvious need for such amplification include needle aspiration biopsy specimens, samples of preneoplastic lesions, circulating cancer cells and foci of cancer isolated by laser capture microdissection (LCM). Cancer genomes harbor numerous large-scale genetic alterations, including translocations, homozygous deletions, chromosomal and gene amplifications and regions undergoing LOH resulting from either hemizygous deletion or gene conversion. To determine whether such alterations are reproducibly detected after φ29MDA, the SNP array data for six paired cancer/normal samples were analyzed for LOH and for copy number differences (e.g. amplifications and deletions) before and after φ29MDA.
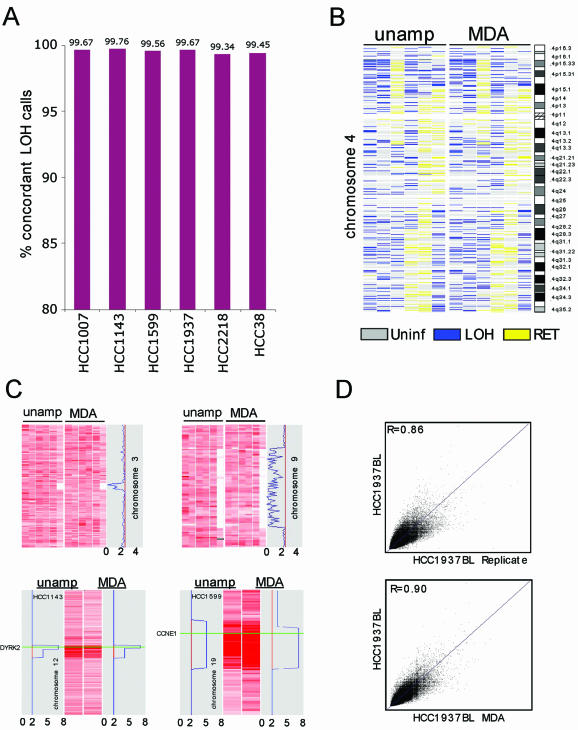
To determine the LOH concordance rate, each SNP allele found to be heterozygous in the normal and undergoing LOH (reduction to a homozygous state) in the cancer was compared in amplified and unamplified pairs. Here, the mean pair-wise concordance rate was 99.58% and was highly consistent across all six pairs (Fig. 4A). Regions of LOH typically encompass many such heterozygote SNPs, thus this degree of LOH concordance leads to highly reproducible LOH maps, as shown for chromosome 4 (Fig. 4B).
Figure 4.
Detection of LOH, homozygous deletions and gene amplification after φ29MDA. (A) Concordance of LOH detection in cancer/normal samples before and after φ29MDA. HCC1007 and HCC1143 tumor/normal pairs and HCC1599 BL were denatured prior to amplification. (B) LOH map of chromosome 4 representative of whole genome LOH maps from six breast cancer/normal pairs. Informative SNP loci showing retention of heterozygosity are shown in yellow, uninformative SNPs (i.e. homozygous or no-call in normal) in gray and SNP alleles undergoing LOH in blue. The first two columns correspond to denatured tumor/normal DNA samples. For the third column, only the normal DNA was denatured. The remaining three columns correspond to all non-denatured tumor/normal pairs. (C) Detection of homozygous deletions and gene amplification. SNP signal intensity is displayed as rows where intensity is normalized on a scale of 0 (white) to 6 (dark red). The upper panels show two representative regions of homozygous deletion on chromosomes 3 and 9 in six breast cancer/normal pairs. The first two columns correspond to tumor samples denatured prior amplification. The lower panel shows representative regions of amplification on chromosomes 12 and 19. Only the individual cancer cell line samples showing gain are shown. Copy number loss or gain is indicated by the blue line in the gray box. HCC1143 was denatured prior amplification whereas HCC1599 was not. (D) Representative SNP hybridization intensity concordance plots. Hybridization intensities from unamplified DNA are plotted against corresponding intensities from a second replicate hybridization of unamplified DNA (top) and from whole genome amplified DNA (bottom). The best fit linear correlation is represented in blue. Correlation coefficients (top left corners), representing signal intensity concordance rates, are similar in both cases. These data are representative examples of all pair-wise MDA/unamplified and unamplified/replicate comparisons examined in this manner. The mean R values for these comparisons are presented in the text.
Recent data from our group has shown that homozygous deletion events and copy number changes are robustly detected using signal intensity measures derived from the SNP oligonucleotide probe sets (13). We therefore compared the signal intensity for each SNP locus for the test samples to a set of normal controls. A hidden Markov model was constructed to estimate the most likely copy number for each locus in the test sample along each chromosome (11). To determine whether homozygous deletions are likewise detected after φ29MDA, we applied this method to the data from our cell lines. In unamplified samples, 11 homozygous deletions were detected. Eleven of 11 were also detected in the samples after φ29MDA. Representative examples of homozygous deletions on chromosomes 3 and 9 are shown in Figure 4C (upper panels). The deletion at 3p15 in the HCC 38 cell line has been reported previously (18).
To determine the concordance of copy number estimates between φ29MDA and unamplified samples, the pair-wise relationship of hybridization intensity for each replicate sample or φ29MDA sample was compared. The mean signal intensity concordance in replicate samples was R = 0.896 ± 0.0276 with R values ranging from 0.788 to 0.943, while the mean signal intensity concordance of the φ29MDA and unamplified sample pairs was 0.894 ± 0.0459, with a range from 0.835 to 0.925. Representative concordance plots are shown in Figure 4D.
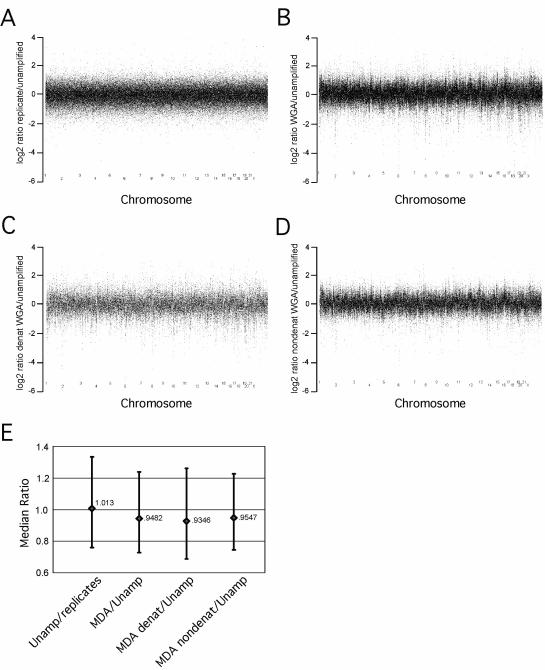
To further ascertain the range of variation in genome representation, signal intensity ratios were calculated where, at each SNP position, the intensity in the unamplified sample was divided by either a replicate pair or by the corresponding φ29MDA amplified sample (Fig. 5A and B). In addition, the mean intensity ratio and the 25th and 75th percentile boundaries for the intensity ratios for all markers were determined and plotted separately (Fig. 5E). The comparison of unamplified replicates versus φ29MDA shows a slight systematic under-representation as the mean intensity ratio is <1 for the comparison of unamplified samples to amplified samples (see Fig. 5E). In addition, there is a clear structure within the φ29MDA plots not apparent in the comparison of replicated unamplified samples (compare Fig. 5B and A). The nature of this additional structure within these data is not clear, however, it appears to be topological. This structure does not appear to contribute to the overall variation as there is no difference between replicates and φ29MDA amplified samples with respect to the extent of overall variation (Fig. 5E). The addition of a denaturing step prior to amplification appears to have little or no effect in this structure nor was there an alteration in variation (Fig. 5C and D).
Figure 5.
Variation in genome amplification after φ29MDA. All the arrays were normalized using the common baseline array HCC1937 BL_250303 by the invariant set method. Then the model-based signal intensities for all SNPs were calculated by the PM/MM difference model. The log2 signal ratios of all SNPs were calculated and plotted separately for each of the: (A) 11 unamplified replicate pairs; (B) 14 φ29MDA/unamplified pairs; (C) five denatured φ29MDA/unamplified pairs; (D) nine non-denatured φ29MDA/unamplified pairs. The three plots were adjusted to have the same scale for better comparison, thus a few extreme outliers are not shown in some plots. (E) The median ratio for each of the sets of comparisons as well as the range of values encompassing the 25–75th percentile values are shown.
For the purposes of estimation of relative copy number in cancer samples, standardization using φ29MDA amplified non-tumor samples as the copy-normal controls appears to overcome these problems. To evaluate how well copy number estimates were preserved after φ29MDA, copy number estimates were binned in six categories: 0, 1, 2, 3, 4 and >4. Using these binned estimates, the copy number concordance for unamplified φ29MDA pairs was 87%, comparable to that obtained for unamplified replicates. The majority of copy number discrepancies were single copy disagreements, while <1% were discrepant by more than one copy number difference (Table 3). This allowed us to compare the detection of regions of copy number amplification between φ29MDA and unamplified samples. In this set of cell lines eight regions of high copy number amplification were identified in the unamplified cancer samples and were confirmed by quantitative real-time PCR. All eight were also detected in φ29MDA samples with comparable copy number estimates (see Table 4).
Table 3. Distribution of copy number differences.
| Percent concordant | Percent with copy number differences of | ||||
|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | ||
| Unamplified replicatesa | 88 | 12 | 0.03 | ||
| Unamplified vs MDAb | 87 | 13 | 0.2 | 0.03 | 0.001 |
Copy numbers were estimated at each SNP locus by comparing the signal intensities observed in each sample to the signal intensities observed in a set of germline DNA samples (see Materials and Methods).
aComparison between repeated runs of unamplified DNA (six samples).
bComparison between whole genome amplified and corresponding unamplified DNA (18 samples).
Table 4. Regions of high level genome amplification detected in amplified or unamplified cancer cell line genomes.
| Cytobanda | Candidate geneb | Cell line | Quantitative numberc | Copy no. predictiond | |
|---|---|---|---|---|---|
| Unamplified | φ29MDA | ||||
| 11q13.3 | CCND1 | HCC1143 | 25.44 | 7 | 6 |
| 12q14.3 | DYRK2 | HCC1143 | 10.05 | 9 | 6 |
| 19q12 | CCNE1 | HCC1599 | 13.48 | 7 | 6 |
| 8q24.1 | MYC | HCC38 | 3.35 | 3 | 3 |
| 8q24.1 | MYC | HCC1937 | 3.69 | 3 | 4 |
| 8q24.1 | MYC | HCC1143 | 4.04 | 4 | 3 |
| 8q24.1 | MYC | HCC1599 | 4.69 | 4 | 4 |
| 8q24.1 | MYC | HCC2218 | 3.70 | 4 | 4 |
aCytoband is based on the hg15 golden path assembly (NCBI build 33) (http://genome.ucsc.edu/).
bThe gene is either within or very close to the predicted region.
cRepresents copy number estimated by quantitative real-time PCR.
dCopy numbers as predicted by hidden Markov modeling from SNP array intensity data analysis of unamplified and MDA DNA. Only cell line HCC1143 was denatured prior to amplification.
DISCUSSION
Large-scale sequencing, SNP genotyping and cancer genomic studies will, over the next few years, allow definition of the germline and somatic genetic alterations causally linked to oncogenesis. Currently, such studies require substantial quantities of high quality DNA. Indeed, a number of efforts directed at cancer somatic genetics have been initiated with cell lines because of the quantity of DNA required (19). Techniques such as φ29MDA that allow the robust amplification of entire genomes from a limited number of genome copies hold the potential to allow large-scale studies to be conducted from primary sample sources. Sources of interest include not only surgically resected specimens, but also genomes isolated from needle biopsies, buccal swabs, circulating cancer cells, LCM cells and ultimately from single tumor cells. In order for amplified DNA to be utilized in high resolution genetic analysis, uniform and accurate genome-wide representation are required.
Genome coverage
Here we report the first genome-scale survey to determine the extent of coverage by φ29MDA using high density SNP arrays. The results presented herein rely on sampling >10 000 SNP markers spaced across the genome with a mean intermarker distance of 210 kb. The 10K SNP array, however, contains no probes for markers on 13p, 14p, 15p, 21p, 22p or Y and poor representation of centromeric and telomeric regions of other chromosomes. This array representation further varies between chromosomes, being sparsest for chromosomes 17, 19, 20 and 22. The median intermarker distance, which leaves out these areas, is 105 kb. Thus, while this sampling was not perfectly distributed across the genome, it is likely that the genome coverage results reported herein can be extrapolated to these uncovered regions. In this study, call rate and allele genotype were determined for φ29MDA and paired unamplified samples. Overall, φ29MDA samples rendered call rates comparable to, but slightly lower than (although not reaching statistical significance), those of unamplified DNA. While genome coverage was estimated to be 99.82%, we identified six regions that were consistently non-represented in whole genome amplification DNA. In addition, eight additional regions were under-represented. Although we obtained correct calls for SNPs in these eight regions, signal intensity was consistently lower across all samples, so this reduction in representation might impair other genetic analysis techniques, e.g. gene copy number determination. For most applications this extent of genome coverage will suffice, however, if specific susceptibility genes or susceptibility loci fall within such under- or unamplified regions, important genetic contributors to disease might be missed. Prior studies assessing the genome coverage of φ29MDA (4,20,21) interrogated a total of 100 loci distributed across all chromosomes and noted robust coverage of these specific loci, though under-representation of LINE1 elements was also noted. Whether this contributes to the failure to amplify the six regions detected in our studies is not yet clear.
Application to large-scale SNP genotyping efforts
In this survey of 10 000 SNP genotypes there was high concordance between φ29MDA and unamplified DNA. This is in keeping with previous reports on a limited number of SNPs where concordance was 100 (4,20) and 99.7% (21). In our study there was no statistical difference between φ29MDA and unamplified samples in SNP call concordance. Indeed, previous data as well as our own support the notion that preservation of SNP heterozygotes after φ29MDA amplification is quite robust. Based on our data on genome representation, if SNPs fall within regions that are poorly amplified there will be a loss of detection for these SNPs. Thus, as higher density SNP maps become available, the number of indeterminate SNP calls (or ‘missing SNP data’) may grow. The overall SNP error rate after φ29MDA is within that expected from experimental error of this assay, therefore, we conclude that φ29MDA DNA can be used for genome-wide SNP mapping studies. Preliminary results using φ29MDA to perform similar genotyping studies on DNA obtained after LCM demonstrate call and concordance rates similar to those obtained with cell line DNA (data not shown).
Application to high throughput sequencing efforts
The fidelity of a PCR-based sequencing pipeline is constrained by the least accurate step. In our case, the limiting step is PCR amplification using a proofreading-deficient Taq polymerase. Direct sequencing of a pool of PCR products tends to minimize, although not eliminate, random errors introduced by the polymerase during PCR. Thus, the quality of initial template DNA introduced into the PCR reaction is critical. φ29 polymerase has been reported to possess an intrinsic proofreading activity ensuring accurate DNA replication (6). To investigate the fidelity of MDA in our system, we resequenced 100 tyrosine kinase amplicons in 20 pairs of whole genome amplification or unamplified samples. Although, we observed virtually identical error rates (∼1 × 10–5) for both φ29MDA and unamplified samples, we did find a small excess of homozygous changes in φ29MDA samples. This may be due to the fact that the resequencing study was done without the denaturing step prior to MDA. Thus, in keeping with our SNP concordance results, we hypothesize that the inclusion of a denaturation step before amplification may partially correct the allele bias. The rate of error introduced by φ29MDA appears to be quite low, although, nonetheless, confirmational sequencing of candidate variants from unamplified DNA samples or from independently amplified samples is warranted.
Application to the detection of LOH and copy number changes
Large-scale genetic alterations, including changes in gene copy number such as deletions and amplification or LOH, are hallmarks of the perturbed cancer cell genome (22). As expected from the very high SNP concordance rate, the concordance of LOH between φ29MDA and unamplified cancer samples is also very high (99.58%), allowing the construction of robust LOH maps (23) for the study of the genetic alterations of cancer cell genomes.
With respect to copy number alterations, our results indicate that MDA results in a change in the copy number structure but does not increase the mean variation in signal intensity (Fig. 5). While this manuscript was in preparation, Lage et al. (24) reported significant over- and under-representation of clusters of yeast ORFs evaluated by array-CGH. Most of the under-represented loci map to the ends of yeast chromosomes. Similar results were obtained on human cDNA arrays. Whether these alterations are similar to those seen in our data remains to be determined. Additional studies using quantitative PCR have shown that loci representation in φ29MDA DNA, as a percentage relative to an unamplified genomic DNA standard, ranged between 50 and 300%, a maximum 6-fold bias between any two loci (20). Despite these changes in genome representation, for the purpose of detecting copy number changes in cancer samples we found that normalization against φ29MDA non-tumor samples led to copy number estimates that were relatively well preserved after φ29MDA, with a copy number concordance of 87% between φ29MDA and unamplified samples. In addition, copy number variation was typically confined to within a single copy number unit and variation outside of that bound was unusual. There are three reasons that may explain high copy number concordance rates in our data despite the signal intensity variations seen here and in other studies. First, the SNP probes tend to be under-represented in telomeric and centromeric regions, where increased copy number variability was noted. Second, many of the signal intensity changes between unamplified and φ29MDA amplified samples appear to be systematic rather than random errors and are corrected by normalizing against a φ29MDA amplified control. This finding was also noted by Lage et al. in their work with MDA using the Bst polymerase (24). Finally, signal intensity changes in individual loci do not necessarily translate into copy number errors when applying our method for determining copy number based on hidden Markov models. This method utilizes the signal intensities of neighboring loci when estimating the copy number for a given locus, therefore averaging over random errors. Importantly, cancer-associated high level amplification and homozygous deletions were readily and reliably detected in φ29MDA samples.
In summary, our results show that φ29MDA DNA provides a highly accurate and comprehensive representation of the unamplified genome, suitable for high resolution genetic analysis, including SNP genotyping, gene copy number detection and direct sequencing. Thus, it is now reasonable to expect that such technologies may be widely applied on a genome scale to primary clinical samples.
Acknowledgments
ACKNOWLEDGEMENTS
The authors thank Maura Berkeley and Dr Edward A. Fox at the Dana-Farber Cancer Institute microarray facility for valuable assistance with the 10K mapping arrays and Dr Michael Egholm, Dr Levi Garraway, Dr Heidi Greulich and Kevin McKernan for their critical reading of the manuscript.
REFERENCES
- 1.Alsmadi O.A., Bornarth,C.J., Song,W., Wisniewski,M., Du,J., Brockman,J.P., Faruqi,A.F., Hosono,S., Sun,Z., Du,Y. et al. (2003) High accuracy genotyping directly from genomic DNA using a rolling circle amplification based assay. BMC Genomics, 4, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dean F.B., Nelson,J.R., Giesler,T.L. and Lasken,R.S. (2001) Rapid amplification of plasmid and phage DNA using phi 29 DNA polymerase and multiply-primed rolling circle amplification. Genome Res., 11, 1095–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nelson J.R., Cai,Y.C., Giesler,T.L., Farchaus,J.W., Sundaram,S.T., Ortiz-Rivera,M., Hosta,L.P., Hewitt,P.L., Mamone,J.A., Palaniappan,C. et al. (2002) TempliPhi, phi29 DNA polymerase based rolling circle amplification of templates for DNA sequencing. Biotechniques, (suppl.), 44–47. [PubMed] [Google Scholar]
- 4.Dean F.B., Hosono,S., Fang,L., Wu,X., Faruqi,A.F., Bray-Ward,P., Sun,Z., Zong,Q., Du,Y., Du,J. et al. (2002) Comprehensive human genome amplification using multiple displacement amplification. Proc. Natl Acad. Sci. USA, 99, 5261–5266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blanco L., Bernad,A., Lazaro,J.M., Martin,G., Garmendia,C. and Salas,M. (1989) Highly efficient DNA synthesis by the phage phi 29 DNA polymerase. Symmetrical mode of DNA replication. J. Biol. Chem., 264, 8935–8940. [PubMed] [Google Scholar]
- 6.Esteban J.A., Salas,M. and Blanco,L. (1993) Fidelity of phi 29 DNA polymerase. Comparison between protein-primed initiation and DNA polymerization. J. Biol. Chem., 268, 2719–2726. [PubMed] [Google Scholar]
- 7.Kennedy G.C., Matsuzaki,H., Dong,S., Liu,W.M., Huang,J., Liu,G., Su,X., Cao,M., Chen,W., Zhang,J. et al. (2003) Large-scale genotyping of complex DNA. Nat. Biotechnol., 21, 1233–1237. [DOI] [PubMed] [Google Scholar]
- 8.Janne P.A., Li,C., Zhao,X., Girard,L., Chen,T.H., Minna,J., Christiani,D., Johnson,B.E. and Meyerson,M. (2004) High-resolution single nucleotide polymorphism array and clustering analysis of loss of heterozygosity in human lung cancer cell lines. Oncogene, 23, 2716–2726. [DOI] [PubMed] [Google Scholar]
- 9.Lieberfarb M.E., Lin,M., Lechpammer,M., Li,C., Tanenbaum,D.M., Febbo,P.G., Wright,R.L., Shim,J., Kantoff,P.W., Loda,M. et al. (2003) Genome-wide loss of heterozygosity analysis from laser capture microdissected prostate cancer using single nucleotide polymorphic allele (SNP) arrays and a novel bioinformatics platform dChipSNP. Cancer Res., 63, 4781–4785. [PubMed] [Google Scholar]
- 10.Lin M., Wei,L.J., Sellers,W.R., Lieberfarb,M., Wong,W.H. and Li,C. (2004) dChipSNP: significance curve and clustering of SNP-array-based loss-of-heterozygosity data. Bioinformatics, doi: 10.1093/bioinformatics/bth069. [DOI] [PubMed] [Google Scholar]
- 11.Li C. and Wong,W.H. (2001) Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc. Natl Acad. Sci. USA, 98, 31–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Westfall P.H. and Young,S.S. (1993) Resampling-based Multiple Testing: Examples and Methods for P-value Adjustment. Wiley, New York, NY. [Google Scholar]
- 13.Zhao X., Li,C., Paez,J.G., Chin,K., Jänne,P.A., Chen,T.H., Girard,L., Minna,J., Christiani,D., Leo,C. et al. (2004) An integrated view of copy number and allelic alterations in the cancer genome using single nucleotide polymorphism arrays. Cancer Res., 64, 3060–3071. [DOI] [PubMed] [Google Scholar]
- 14.Altshuler D., Pollara,V.J., Cowles,C.R., Van Etten,W.J., Baldwin,J., Linton,L. and Lander,E.S. (2000) An SNP map of the human genome generated by reduced representation shotgun sequencing. Nature, 407, 513–516. [DOI] [PubMed] [Google Scholar]
- 15.Mullikin J.C., Hunt,S.E., Cole,C.G., Mortimore,B.J., Rice,C.M., Burton,J., Matthews,L.H., Pavitt,R., Plumb,R.W., Sims,S.K. et al. (2000) An SNP map of human chromosome 22. Nature, 407, 516–520. [DOI] [PubMed] [Google Scholar]
- 16.Ewing B. and Green,P. (1998) Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res., 8, 186–194. [PubMed] [Google Scholar]
- 17.UCSC (2003) UCSC genome browser. NCBI build 34, July 2003 freeze. Available at http://genome.ucsc.edu.
- 18.Sundaresan V., Chung,G., Heppell-Parton,A., Xiong,J., Grundy,C., Roberts,I., James,L., Cahn,A., Bench,A., Douglas,J. et al. (1998) Homozygous deletions at 3p12 in breast and lung cancer. Oncogene, 17, 1723–1729. [DOI] [PubMed] [Google Scholar]
- 19.Davies H., Bignell,G.R., Cox,C., Stephens,P., Edkins,S., Clegg,S., Teague,J., Woffendin,H., Garnett,M.J., Bottomley,W. et al. (2002) Mutations of the BRAF gene in human cancer. Nature, 417, 949–954. [DOI] [PubMed] [Google Scholar]
- 20.Hosono S., Faruqi,A.F., Dean,F.B., Du,Y., Sun,Z., Wu,X., Du,J., Kingsmore,S.F., Egholm,M. and Lasken,R.S. (2003) Unbiased whole-genome amplification directly from clinical samples. Genome Res., 13, 954–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lovmar L., Fredriksson,M., Liljedahl,U., Sigurdsson,S. and Syvanen,A.C. (2003) Quantitative evaluation by minisequencing and microarrays reveals accurate multiplexed SNP genotyping of whole genome amplified DNA. Nucleic Acids Res., 31, e129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hanahan D. and Weinberg,R.A. (2000) The hallmarks of cancer. Cell, 100, 57–70. [DOI] [PubMed] [Google Scholar]
- 23.Lindblad-Toh K., Tanenbaum,D.M., Daly,M.J., Winchester,E., Lui,W.O., Villapakkam,A., Stanton,S.E., Larsson,C., Hudson,T.J., Johnson,B.E. et al. (2000) Loss-of-heterozygosity analysis of small-cell lung carcinomas using single-nucleotide polymorphism arrays. Nat. Biotechnol., 18, 1001–1005. [DOI] [PubMed] [Google Scholar]
- 24.Lage J.M., Leamon,J.H., Pejovic,T., Hamann,S., Lacey,M., Dillon,D., Segraves,R., Vossbrinck,B., Gonzalez,A., Pinkel,D. et al. (2003) Whole genome analysis of genetic alterations in small DNA samples using hyperbranched strand displacement amplification and array-CGH. Genome Res., 13, 294–307. [DOI] [PMC free article] [PubMed] [Google Scholar]