Abstract
Because of their stringent sequence specificity, the 3C-like proteases from tobacco etch virus (TEV3) and human rhinovirus are often used for the removal of affinity tags. The latter enzyme is rumored to have greater catalytic activity at 4°C, the temperature at which fusion protein substrates are usually digested. Here, we report that experiments with fusion protein and peptide substrates confirm this conjecture. Whereas the catalytic efficiency of rhinovirus 3C protease is approximately the same at its optimum temperature (30°C) and at 4°C, TEV protease is 10-fold less active at the latter temperature, due primarily to a reduction in kcat.
Keywords: 3C protease, affinity tag removal, fusion protein protease, protease, PreScission protease, tobacco etch virus (TEV) protease
Genetically engineered tags are frequently added to recombinant proteins to improve their yield, help protect them from proteolysis, enhance their solubility, and facilitate their purification [1]. Yet all tags, whether large or small, have the potential to interfere with the structure and biological activity of proteins. Consequently, reliable methods for removing them are needed. Although both chemical and enzymatic approaches have been used to cleave fusion proteins at specific sites, only the natural proteolytic enzymes have the requisite specificity to be broadly useful for this purpose. In recent years, the 3C-like proteases from certain RNA viruses have emerged as the reagents of choice. Two of the most popular are the tobacco etch virus (TEV) and human rhinovirus 3C (R3C) proteases [2].
TEV and R3C proteases recognize and cleave related but distinct sequences [2]. To this day, arguments persist about which enzyme is the superior reagent for cleaving fusion proteins. A noteworthy advantage of TEV protease is that it tolerates a variety of amino acid residues in the P1′ and P2′ positions of its recognition site [3–5], whereas R3C exhibits a strict requirement for glycine and proline residues in these positions, respectively [6]. Consequently, in many cases TEV protease (but not R3C protease) can be used to generate a digestion product with no non-native amino acid residues appended to its N-terminus. Nonetheless, it is common practice to digest fusion proteins overnight at 4°C [7,8], and it has been rumored that R3C protease has significantly greater catalytic activity at this temperature than does TEV protease [2,9]. Remarkably, however, no study has ever been carried out in which the temperature-dependence of the two enzymes has been directly compared.
In the present study, two types of substrates were used to compare the activity of TEV and R3C proteases at different temperatures: kinetic parameters Km and kcat were derived from data obtained with synthetic peptide substrates, whereas fusion proteins were used to monitor the temperature dependence of protease activity under “typical” reaction conditions. The synthetic oligopeptides TENLYFQ↓SGTRR and SLEVLFQ↓GPVRR (Genscript, Piscataway, NJ) were used as substrates to assay the activity of TEV (S219V) [10] and R3C proteases [11], respectively. The arrows indicate the sites of enzyme-mediated hydrolysis. The lyophylized peptides were dissolved in distilled water. The enzymes (1 mg/ml stock solutions) were diluted at least 50-fold with the 2X reaction buffer (50 mM sodium phosphate pH 7.0, 800 mM NaCl, 5 mM DTT, 10% glycerol). Assays were initiated by mixing 20 μl of the diluted protease (50–200 nM final concentration) with 20 μl of substrate solution (0.02–0.72 mM final concentration). Measurements were performed at a minimum of six different substrate concentrations bracketing Km. The reactions were incubated at 4°C or 30°C for 30 min and then stopped by the addition of 160 μl 1% TFA. An aliquot was injected onto a Waters Symmetry 300 C18 reverse-phase chromatography column (4.6 × 250 mm) using an automatic injector. The substrates and cleavage products were separated with an increasing water-acetonitrile gradient (0–100%) in the presence of 0.05% TFA. The kcat values were calculated by assuming 100% activity for the enzymes. Kinetic parameters were determined by fitting the data obtained at less than 20% substrate hydrolysis to the Michaelis-Menten equation, using the Enzyme Kinetics Module of the SigmaPlot program (Systat Software, Inc. Chicago, IL). Standard deviations were calculated according to Boross et al. [12].
The kinetic measurements are reported in Table 1. They revealed that the Km of the two enzymes is similar at 4°C, but at 30°C the Km of R3C is 10-fold higher than that of TEV protease. However, the increase in Km for R3C at the higher temperature is accompanied by a concomitant 10-fold increase in the kcat. Consequently, the catalytic efficiency (kcat/Km) of R3C is about the same at both temperatures. On the other hand, the Km of TEV protease actually decreases (i.e. apparent affinity increases) by 2-fold as the temperature changes from 4°C to 30°C while the kcat rises by about 6-fold. The net result of these changes is that TEV protease exhibits approximately the same catalytic efficiency as R3C at 30°C. At 4°C, although the two enzymes have similar Km values, the kcat (and therefore also the kcat/Km) of TEV protease is 10-fold lower than that of R3C at the same temperature.
Table 1.
Kinetic Parameters for Processing of Peptide Substrates by Rhinovirus 3C and TEV Proteases.
| Enzyme | Temperature (°C) | Km (mM) | kcat (s−1) | kcat/Km |
|---|---|---|---|---|
| TEV | 4 | 0.087 ± 0.012 | 0.036 ± 0.003 | 0.41 ± 0.06 |
| TEV | 30 | 0.037 ± 0.011 | 0.22 ± 0.01 | 6.95 ± 1.79 |
| R3C | 4 | 0.099 ± 0.013 | 0.41 ± 0.02 | 4.14 ± 0.57 |
| R3C | 30 | 0.391 ± 0.071 | 2.38 ± 0.14 | 6.09 ± 1.16 |
The kinetic data are supplemented by results obtained with fusion protein substrates. Two otherwise identical MBP-NusG fusion proteins containing a recognition site for either TEV protease (-ENLYFQ↓S-) or R3C protease (-LEVLFQ↓GP-) in the linker between the two domains were expressed and purified as described [5]. Experiments were performed at a series of temperatures ranging between 4°C and 40°C, as indicated in figure 1. The substrate concentration was 0.5 mg/ml (7 μM), which is typical for the digestion of a fusion protein substrate. The enzyme concentration was either 200 nM (TEV) or 50 nM (R3C). Reactions were initiated by adding the enzymes to reaction buffer (50mM Tris pH 8.0, 0.5 mM EDTA, 1.0 mM DTT) containing the substrates and terminated after various times by the addition of SDS-PAGE sample buffer. The reaction products were separated by SDS-PAGE and stained with Coomassie Brilliant Blue. The band intensities of the substrates and products were quantified by densitometry using an Alpha Imager CCD camera and ImageQuant software (Alpha Innotech, San Leandro, CA). Initial velocities were calculated under conditions of less than 20% substrate hydrolysis.
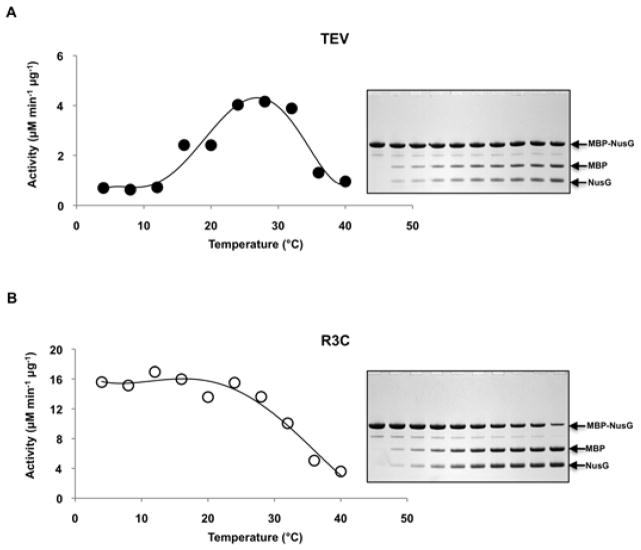
Figure 1.
Temperature Dependence of Initial Reaction Velocity. MBP-NusG fusion protein substrates were digested by TEV [A] or R3C [B] proteases at the indicated temperatures. In the respective insets (right), representative SDS-PAGE gels are shown for uncleaved (Lane 1) and cleaved (Lanes 2-10; 2, 5, 10, 20, 30, 40, 50, 60, and 120 min, respectively) substrates at 4°C.
Figure 1 shows a plot of the initial reaction velocity vs. temperature for TEV and R3C proteases using fusion protein substrates. The R3C protease appeared to have an approximately 4-fold higher maximum cleavage rate on the protein substrate as compared to that of TEV protease, as against a rather constant value of kcat/Km (table 1), likely due to the different sizes of the substrates (polyprotein versus oligopeptide) as well as ionic strength in the reaction. In agreement with the kinetic data obtained from reactions conducted with peptide substrates, the turnover rate for the R3C protease exhibits little change between 4°C and 30°C whereas that of TEV protease increases steadily over this temperature range, reaching its optimum temperature at approximately 30°C. The activity of both enzymes begins to decrease above 30°C. The sharp drop in the activity of TEV protease can be attributed to denaturation of the enzyme [10]. It is unknown whether this is also the case for R3C and is currently under study.
In conclusion, the experiments reported here unequivocally confirm, for the first time, that R3C is 10-fold more active than TEV protease at 4°C, the temperature at which fusion proteins are most often digested. The opposite directional change of the Km values for the proteases between 4°C and 30°C suggests that the enthalpic and entropic contributions to their substrate binding energy may be entirely different. This could be explained in part by more extensive interactions between R3C than TEV protease with their respective substrates, particularly in the S1′/P1′ and S2′/P2′ positions. If so, then it is unlikely that an endoprotease could be engineered to embody the most desirable characteristics of both enzymes: that is, R3C-like kinetics at 4°C along with the relaxed specificity in the “prime” sites that is exhibited by TEV protease.
Acknowledgments
We thank Dr. Ari Geerlof for the rhinovirus 3C protease expression vector. This research was funded by the Intramural Research Program of the Center for Cancer Research, National Cancer Institute, National Institutes of Health and in part by the by the Hungarian Science and Research Fund (OTKA K101591 to JT).
Footnotes
Abbreviations used: DTT, dithiothreitol; EDTA, ethylenediaminetetraacetic acid; MBP, maltose-binding protein; NusG, Aquifex aeolicus N-utilization substance G protein; R3C, rhinovirus 3C; TEV, tobacco etch virus; TFA, trifluoroacetic acid.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Waugh DS. Making the most of affinity tags. Trends Biotechnol. 2005;23:316–320. doi: 10.1016/j.tibtech.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 2.Waugh DS. An overview of enzymatic reagents for the removal of affinity tags. Protein Expression Purif. 2011;80:283–293. doi: 10.1016/j.pep.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dougherty WG, Cary SM, Parks TD. Molecular genetic analysis of a plant virus polyprotein cleavage site: a model. Virology. 1989;171:356–364. doi: 10.1016/0042-6822(89)90603-x. [DOI] [PubMed] [Google Scholar]
- 4.Dougherty WG, Carrington JC, Cary SM, Parks TD. Biochemical and mutational analysis of a plant virus polyprotein cleavage site. EMBO J. 1988;7:1281–1287. doi: 10.1002/j.1460-2075.1988.tb02942.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kapust RB, Tozser J, Copeland TD, Waugh DS. The P1′ specificity of tobacco etch virus protease. Biochem Biophys Res Commun. 2002;294:949–955. doi: 10.1016/S0006-291X(02)00574-0. [DOI] [PubMed] [Google Scholar]
- 6.Cordingley MG, Callahan PL, Sardana VV, Garsky VM, Colonno RJ. Substrate requirements of human rhinovirus 3C protease for peptide cleavage in vitro. J Biol Chem. 1990;265:9062–9065. [PubMed] [Google Scholar]
- 7.Kim Y, Babnigg G, Jedrzejczak R, Eschenfeldt WH, Li H, Maltseva N, Hatzos-Skintges C, Gu M, Makowska-Grzyska M, Wu R, An H, Chhor G, Joachimiak A. High-throughput protein purification and quality assessment for crystallization. Methods (Amsterdam, Neth) 2011;55:12–28. doi: 10.1016/j.ymeth.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bryan CM, Bhandari J, Napuli AJ, Leibly DJ, Choi R, Kelley A, Van Voorhis WC, Edwards TE, Stewart LJ. High-throughput protein production and purification at the Seattle Structural Genomics Center for Infectious Disease, Acta Crystallogr. Sect F: Struct Biol Cryst Commun. 2011;67:1010–1014. doi: 10.1107/S1744309111018367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rubio V, Shen Y, Saijo Y, Liu Y, Gusmaroli G, Dinesh-Kumar SP, Deng XW. An alternative tandem affinity purification strategy applied to Arabidopsis protein complex isolation. Plant J. 2005;41:767–778. doi: 10.1111/j.1365-313X.2004.02328.x. [DOI] [PubMed] [Google Scholar]
- 10.Kapust RB, Tozser J, Fox JD, Anderson DE, Cherry S, Copeland TD, Waugh DS. Tobacco etch virus protease: mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency. Protein Eng. 2001;14:993–1000. doi: 10.1093/protein/14.12.993. [DOI] [PubMed] [Google Scholar]
- 11.Walker PA, Leong LE, Ng PW, Tan SH, Waller S, Murphy D, Porter AG. Efficient and rapid affinity purification of proteins using recombinant fusion proteases. Bio/Technology. 1994;12:601–605. doi: 10.1038/nbt0694-601. [DOI] [PubMed] [Google Scholar]
- 12.Boross P, Bagossi P, Copeland TD, Oroszlan S, Louis JM, Tozser J. Effect of substrate residues on the P2′ preference of retroviral proteinases. Eur J Biochem. 1999;264:921–929. doi: 10.1046/j.1432-1327.1999.00687.x. [DOI] [PubMed] [Google Scholar]