Abstract
Cancer derived microarray data sets are routinely produced by various platforms that are either commercially available or manufactured by academic groups. The fundamental difference in their probe selection strategies holds the promise that identical observations produced by more than one platform prove to be more robust when validated by biology. However, cross-platform comparison requires matching corresponding probe sets. We are introducing here sequence-based matching of probes instead of gene identifier-based matching. We analyzed breast cancer cell line derived RNA aliquots using Agilent cDNA and Affymetrix oligonucleotide microarray platforms to assess the advantage of this method. We show, that at different levels of the analysis, including gene expression ratios and difference calls, cross-platform consistency is significantly improved by sequence- based matching. We also present evidence that sequence-based probe matching produces more consistent results when comparing similar biological data sets obtained by different microarray platforms. This strategy allowed a more efficient transfer of classification of breast cancer samples between data sets produced by cDNA microarray and Affymetrix gene-chip platforms.
INTRODUCTION
From its inception, microarray technology for gene expression measurements has developed in several complementary tracks. One of the most widely used approaches, first developed by P. Brown’s group at Stanford and also sold by commercial sources such as Agilent, uses cDNA clones as probes (1). In this method, probes are produced by DNA polymerase using several hundred base-pair long nucleotide chains as templates. This probe selection strategy has several appealing features, including high hybridization stringency and low susceptibility to gene polymorphisms. However, according to various estimates, up to 30% of the probes can be misidentified (2). The most frequently used competing technology, developed by Affymetrix Inc., utilizes short, 25mer DNA oligonucleotides as probes that are chemically synthesized using sequence information stored in various genomic data bases. In this case, probes are only as reliable as the deposited sequence information that is used to design the probes. A recent study has indicated that as much as 50% of Affymetrix probes do not have a matching sequence in the Reference Sequence database (Refseq), casting doubt on the reliability of this subset of probes (B. H. Mecham, D. Z. Wetmore, Z. Szallasi, Y. Sadovsky, I. Kohane and T. J. Mariani, submitted for publication). Combining the uncertainty regarding probe sets in both types of microarray platforms with their well documented experimental noise, such as compression of gene expression ratios (3), necessitates a cautious approach when interpreting and generalizing microarray-based data. For example, the development of massively parallel gene expression measurements holds great promise in cancer diagnostics, but it is less than clear how results derived by one microarray platform can be transferred to data sets produced by another platform. In the case of breast cancer, there are data sets available using three fundamentally different microarray technologies: platforms using cDNA clones as probes (4), platforms using 25mers as probes (Affymetrix) (5) and platforms using 60mer oligonucleotides as probes (6). Attempts at merging the key observations into a set of microarray platform independent results have met with limited success (7). Sorlie et al. (7), for example, found that classification of breast cancer based on gene expression measurements can be used as a prognostic marker. They have extracted a set of about 500 informative genes that produced reproducible and clinically relevant unsupervised classification within their data set. In order to transfer their observations to other platforms they needed to match the corresponding probe sets. This requires a common denominator, which is usually the Unigene ID, as used in several publications and by Sorlie et al. (7). Corresponding probes and probe sets, however, can be matched by sequence information as well. We report here that restricting analysis to sequence-matched probes produces a higher level of consistency between results derived from alternative microarray platforms at all levels of analysis examined.
MATERIALS AND METHODS
Cell culture
The HCC 1954 and MDA-MB-436 human breast tumor cell lines were obtained from American Type Culture Collection (Manassas, VA), and human mammary epithelial cells (HMEC) were obtained from Cambrex Bio Science (Walkersville, MD). Cells were cultured as recommended by the suppliers. All cultures were maintained in 150 mm dishes at 37°C with 5% CO2, and were harvested for RNA isolation when dishes were 60–90% confluent.
Isolation of RNA and Affymetrix hybridization
For each 150 mm dish of cells, media was first removed and the cell monolayer washed briefly in phosphate buffered saline at room temperature. Next, cells were solubilized in 4 ml of TRIzol LS (Invitrogen, Carlsbad, CA), and then, after preparation of a supernatant from the extracts according to the manufacturer’s instructions, total cellular RNA was recovered in the upper phase. To achieve higher purity, this supernatant was then applied to a RNeasy midi column (Qiagen, Valencia, CA) by centrifugation and processed according to the manufacturer’s protocol, beginning with the wash with Buffer RW1. Finally, the volume of the aqueous RNA solution was reduced, when necessary, using a Microcon 30 concentrator (Millipore, Billerica, MA) until a concentration of 0.5–12.0 µg/µl was obtained, as measured by UV spectrophotometry, and RNA was stored at –80°C.
RNA was labeled and hybridized to microarrays from Affymetrix (U95Av.2, U133A and U133B Genechips, 25mer oligonucleotide probe sets) and Agilent (Human 1, cDNA probes) according to the manufacturer’s instructions. For the Affymetrix platform RNA from each cell line was hybridized in duplicates on each of the three different Affymetrix arrays. For the Agilent (double channel) array RNA from the MDA-MB-436 cells were co-hybridized with RNA from the normal HMEC cells on a single array. This experiment was performed in duplicates. Similarly, in a set of duplicate experiments RNA from HCC-1954 was co-hybridized with RNA from HMEC on the Agilent array.
cDNA microarray hybridization and feature extraction
For each cDNA microarray measurement of expression ratios, the combined Cy3- and Cy5-labeled cDNAs were hybridized to an Agilent Human 1 cDNA microarray according to the manufacturer’s protocol, and the arrays scanned using an Agilent Microarray Scanner. Expression ratios were obtained using the feature extraction software that comes with the scanner. For some targets, expression ratios were verified by comparison with ratios determined with the ArraySuite software package, which is described at http://research.nhgri.nih.gov/microarray/main.html.
The microarray data are available at the Gene Expression Omnibus, http://www.ncbi.nlm.nih.gov/geo/ under GEO accession GSE1299.
Mapping of Affymetrix probe sequences to Unigene clusters
All mRNA sequences were retrieved from the NCBI Unigene molecular database build 162 (16 September 2003). Agilent provides probe information, including the GenBank sequence identifier that is most similar to the clone used on the array. The location of all Affymetrix probe sequences was identified in their corresponding mRNAs with the use of map files available at http://lungtranscriptome.bwh.harvard.edu. Microarray data was used only if the Affymetrix probe set and the Agilent clone corresponded to an identical Unigene. These Unigene-matched measurements were further classified as ‘sequence-matched’ if the 25 nt Affymetrix probe was contained within the Agilent clone sequence. Any Unigene-matched measurement for which the Affymetrix probe was not contained within the Agilent clone sequence was defined as ‘non-sequence-matched’.
Since Affymetrix utilizes multiple probe measurements (probe sets) to query a single Unigene, the probe sets and clones were also matched. Affymetrix probe sets and Agilent clones were defined as ‘non-overlapping’ if, for this clone, the probe set contains only non-sequence-matched probes. In this case, the probe set and clone contain Unigene-matched measurements, but measure different segments of the same Unigene. In a similar manner, probe sets that contain at least one single sequence-matched probe were defined as ‘overlapping’ with this sequence-matched Agilent clone. These measurements are Unigene-matched, contain at least one sequence-matched probe and therefore measure identical segments of the same molecule.
Affymetrix microarray data processing and analysis
For each Affymetrix chip, image files were analyzed with Microarray Suite 5.0 (MAS 5.0) software. Bioconductor (8) was used to generate the normalized probe values (using the constant, contrasts, invariant set, loess, qspline, quantiles robust and quantiles normalization methods for both PM and MM intensities) as well as RMA, dChip and MAS 5.0 expression values (9). The default software settings were used for all calculations. Expression measurements from Affymetrix technology are expressed as a single measurement for each gene. As Agilent technology reports expression levels as a ratio between two samples, for comparisons across technologies the Affymetrix data had to be transformed. Here, the expression level for each Affymetrix probe and probe set was transformed into the log base 2 of the ratio between its signal intensity in a cancer sample and its signal intensity in the normal sample. Pearson correlation coefficients for each Affymetrix platform and their corresponding Agilent data were calculated for both the ‘sequence-matched’ and ‘non-sequence-matched’ probes as well as for both ‘overlapping’ and ‘non-overlapping’ probe sets.
Difference calls were obtained from each manufacturer’s standard software package. We limited the data to only those measurements that had an identical call in both sets of replicate comparisons in order to compare only those genes that exhibited consistent changes. 3 × 3 contingency tables were created with difference calls for both the ‘sequence-matched’ and ‘non-sequence-matched’ probe sets with their corresponding Agilent data. A t-statistic was calculated that measured the independence of Affymetrix’s difference calls from Agilent’s. In order to determine whether sequence-matched probe sets provide more consistent change calls with the cDNA platform, the t-statistic (simply a measure of independence, not concordance) needed to be further interpreted using Cramer’s contingency coefficient (10). The coefficient is restricted to the interval between –1 and 1 and is at its maximal value when the counts for each row of the matrix tend to accumulate in one column, but in a different column for each row (indicating a preference for specific call relationships). If the counts for each row do not collect in different columns the value is closer to –1.
Breast cancer clustering
Using the approximately 500 classifiers, or intrinsic genes, specified by Sorlie et al. (4) we identified two sets of informative genes. The first set was prepared as described by Sorlie et al. (7) by matching cDNA clones with their corresponding Affymetrix probe sets using Unigene IDs. The second set was based on those Affymetrix probe sets that contain at least one sequence-matched probe with the Unigene monitored by the intrinsic genes of Sorlie et al. (4). A median centroid was calculated from the cDNA data for each tumor type as described by Sorlie et al. (4,7) and used to classify the resulting Affymetrix sequence-matched and Unigene-matched data set. Median normalized MAS 5.0 expression values for overlapping and Unigene-matched Probe Sets were taken from the data set published by West et al. (5), and clustered using average linkage hierarchical clustering with the Pearson correlation coefficient as the distance metric.
RESULTS
Probe sequence mapping
In order to align measurements between Affymetrix and Agilent technologies, we used the Affymetrix probe mapping files available at http://lungtranscriptome.bwh.harvard.edu. (B. H. Mecham, D. Z. Wetmore, Z. Szallasi, Y. Sadovsky, I. Kohane and T. J. Mariani, submitted for publication). These files contain the location of every Hu95A and Hu133 probe in the Human Unigene database that could be matched to a Unigene sequence. We limited these mapping files to the Unigenes monitored on the Agilent Human 1 cDNA array. If a probe set mapped to multiple Unigene clusters or if its full compliment of probes did not map to the same Unigene ID it was removed from further analysis. This enabled us to distinguish between Affymetrix and Agilent measurements that are derived from identical sequences and those that are associated with the same Unigene cluster without an actual identical sequence. Signal from a given Affymetrix probe was classified as ‘sequence-matched’ if the probe could be mapped to the corresponding Agilent clone and ‘non-sequence-matched’ if there was no sequence overlap but it could be mapped to some other sequence in the Unigene cluster associated with that clone. For example, on the Hu133A platform, the sequence for probe number one of probe set 200011_s_at matches a region in the Agilent clone that corresponds to the mRNA sequence M74491. This probe and clone were classified as sequence-matched. An example of a non-sequence-matched measurement from the Hu133A platform is probe number one of probe set 200598_s_at. It measures Unigene Hs.100058, but does not measure the clone (AB006713) Agilent used to measure this Unigene. Therefore, this Affymetrix probe and Agilent clone were classified as non-sequence-matched measurements. For Affymetrix Hu133A, 36% of the probes were sequence-matched and 16% were non-sequence-matched (see Tables 1 and 2 for the numbers of sequence-matched probes on other Affymetrix chips). There are a large number of probes on both the Affymetrix and Agilent platforms that covered Unigene clones without a corresponding probe set on the other platform (i.e. ∼37% of the Agilent clones do not measure a Unigene monitored on the Hu133A platform and 45% of the Hu133A probe sets do not measure a Unigene monitored by an Agilent clone). These were omitted from further analysis.
Table 1. Statistical significance of the higher correlation for overlapping probe measurements on the various Affymetrix platforms with gene expression measurements produced by the Agilent Human 1 cDNA microarray.
| Platform | Data type 1 Title | Number of probes | Data type 2 Title | Number of probes | Mean Pearson | P-value | ||
|---|---|---|---|---|---|---|---|---|
| Data type 1 | Data type 2 | Paired t-test | Eqvar t-test | |||||
| 133A | MM non-overlapping | 53193 | MM overlapping | 108744 | 0.22 | 0.43 | <0.001 | <0.001 |
| PM non-overlapping | 53193 | MM overlapping | 108744 | 0.35 | 0.43 | 0.01 | 0.03 | |
| PM non-overlapping | 53193 | PM overlapping | 108744 | 0.35 | 0.61 | <0.001 | <0.001 | |
| 133B | MM non-overlapping | 11382 | MM overlapping | 11943 | 0.14 | 0.35 | 0.00 | 0.001 |
| PM non-overlapping | 11382 | MM overlapping | 11943 | 0.29 | 0.35 | 0.10 | 0.28 | |
| PM non-overlapping | 11382 | PM overlapping | 11943 | 0.29 | 0.56 | <0.001 | 0.003 | |
| 95A | MM non-overlapping | 36398 | MM overlapping | 103728 | 0.24 | 0.43 | <0.001 | 0.001 |
| PM non-overlapping | 36398 | MM overlapping | 103728 | 0.38 | 0.43 | 0.08 | 0.28 | |
| PM non-overlapping | 36398 | PM overlapping | 103728 | 0.38 | 0.63 | <0.001 | 0.003 | |
Relative expression ratios were measured and calculated as described in the caption to Figure 1. Correlation coefficients were determined for different subsets of probes including overlapping PM, overlapping MM, non-overlapping PM, as described in the text, and the difference between these probe sets was analyzed by the t-test.
Table 2. Overlapping probe sets produce increased correlation between gene expression measurements produced by the various Affymetrix chips and the Agilent Human 1 cDNA microarray.
| Platform | Metric | Mean Pearson Overlapping | Number of probe sets Overlapping | Mean Pearson Non-Overlapping | Number of probe sets Non-Overlapping | P-value paired t-test | P-value-Eqvar t-test |
|---|---|---|---|---|---|---|---|
| Overlapping | Overlapping | Non-Overlapping | Non-Overlapping | ||||
| 133A | Mas5 | 0.60 | 6134 | 0.40 | 2000 | <0.001 | <0.001 |
| RMA | 0.70 | 6134 | 0.51 | 2000 | <0.001 | <0.001 | |
| dCHIP | 0.69 | 6134 | 0.49 | 2000 | <0.001 | <0.001 | |
| 133B | Mas5 | 0.58 | 751 | 0.36 | 556 | <0.001 | <0.001 |
| RMA | 0.67 | 751 | 0.46 | 556 | <0.001 | <0.001 | |
| dCHIP | 0.67 | 751 | 0.43 | 556 | <0.002 | <0.001 | |
| 95A | Mas5 | 0.62 | 4371 | 0.38 | 730 | <0.001 | <0.001 |
| RMA | 0.69 | 4371 | 0.46 | 730 | <0.001 | <0.001 | |
| dCHIP | 0.70 | 4371 | 0.43 | 730 | <0.001 | <0.001 |
Relative expression ratios were measured and calculated while comparing RNA from MDA-MB-436 and HMEC cells and from the HCC1954 and HMEC cells using three different Affymetrix platforms and the Agilent Human 1 cDNA microarray chip. Gene expression levels based on the Affymetrix chips were calculated using three different algorithms as indicated in the table and the text. The statistical significance was calculated using the t-test as indicated.
Sequence-matched Affymetrix probes show higher correlation with cDNA microarray measurements
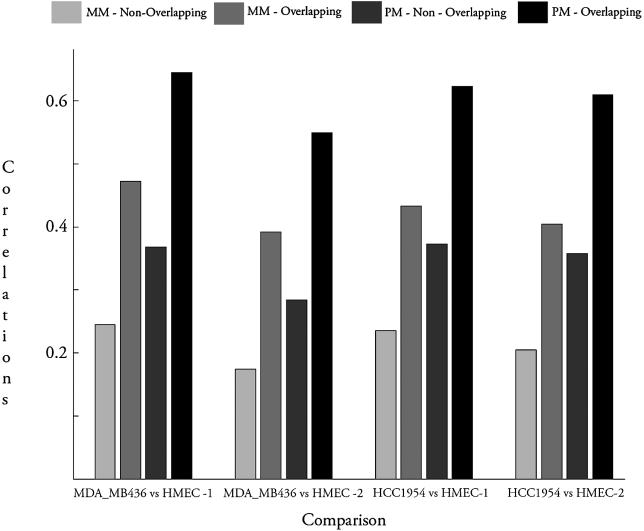
First, we analyzed the relevance of sequence-matching at the level of individual Affymetrix probes in a side-by-side comparison when aliquots of the same RNA were hybridized to both types of platforms. Pearson correlation coefficients were calculated for sequence-matched and non-sequence-matched PM and MM signals with the corresponding Agilent data (Fig. 1 and Table 1) (MM probes were classified as sequence-matched based on the sequence overlap of their perfect match counterpart). An increased correlation was detected in the sequence-matched PM probes versus the non-sequence-matched PM probes (Hu133A, P < 0.001). Interestingly, sequence-matched MM measurements are also more highly correlated with cDNA data than non-sequence-matched PM measurements (Hu133A, P < 0.015). This was not entirely unexpected, since it has been shown that 60–70% of the MM probe signal intensity reflects signals of the PM probes (11).
Figure 1.
Overlapping probes show increased correlation between gene expression measurements produced by the Affymetrix Hu133A chip and the Agilent Human 1 cDNA microarray. Relative expression ratios were measured and calculated while comparing RNA from the cell cultures MDA-MB-436 and HMEC and from the cell cultures HCC1954 and HMEC. Aliquots of the same RNA sample were hybridized to different platforms. Relative expression was calculated at the level of individual probes on the Affymetrix platforms. Pearson correlation coefficients were calculated between ratios determined by individual Affymetrix probe intensities and the ratios obtained from cDNA microarray. Correlation coefficients were determined for different subsets of probes including overlapping PM, overlapping MM, non-overlapping PM and non-overlapping MM.
Recently, several probe normalization techniques have been recommended to remove some aspects of the noise inherent to Affymetrix microarray measurements (9,12,13). In order to test the effects of these probe normalization techniques, the Affymetrix data were normalized using seven different methods (constant, contrasts, invariant set, loess, qspline, quantiles robust and quantiles) and Pearson correlation coefficients with the cDNA microarray data were calculated again. As Supplementary Table 1 indicates, the various probe normalization methods provide no significant improvement in the correlation of non-overlapping probe signals with cDNA microarray measurements. The effect of sequence matching far outweighs the effect of any of the normalization techniques.
Gene expression measurements from sequence-matched Affymetrix probe sets show increased correlation with cDNA microarray measurements in cross-platform comparison of RNA aliquots
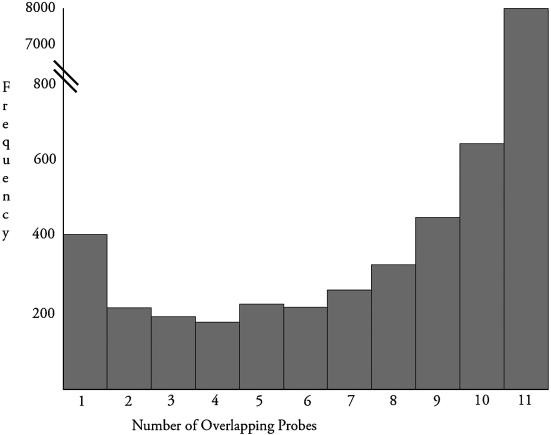
Since Affymetrix technology uses entire probe sets to quantify transcripts, we also classified probe sets and Agilent clones based on their sequence overlap. A probe set was classified as ‘overlapping’ if it contained only sequence-matched probes, or non-overlapping if it contained only ‘non-sequence-matched’ probes. These latter probe sets were, as indicated before, Unigene-matched. However, for each Affymetrix platform, there is a large number of probe sets (e.g. ∼20% for Hu133A chip) that have a partial set of probes overlapping an Agilent clone as shown in Figure 2 and in Supplementary Figures 1A and B. Since the number of probe sets with any given number of sequence-matched probes (i.e. between 1 and 15 for U95Av2 and between 1 and 10 for U133A and U133B) is much lower than the total number of either completely overlapping or non-overlapping probe sets, we decided to pool all ‘partially overlapping’ probes in the first round of analysis. We observed that the partially overlapping probe sets produce a similar correlation with the cDNA microarray data as the completely overlapping probe sets do, which was consistently higher than the correlation between non-overlapping probe sets and cDNA microarray data. (The number of overlapping, non-overlapping and partially overlapping probe sets is listed in Supplementary Table 2.) Therefore, for further analysis all ‘partially overlapping’ probe sets were pooled into the ‘overlapping’ class of probe sets. According to this classification probe set 200042_at _s_at and Agilent clone AI359487 are classified as overlapping since 10 out of 11 probes are overlapping.
Figure 2.
Distribution of the number of overlapping probes between probe sets on the Hu133A Affymetrix chip and the clones serving as probes on the Agilent Human 1 cDNA microarray. The number of overlapping probes for each probe set was calculated as described in the text.
Affymetrix experiments have been traditionally analyzed using information generated by combining multiple probe measurements into a single expression value. MAS 5.0, RMA (13) and dChip (12) are the three most commonly used methods and we tested each of them to determine their relative merit. Pearson correlation coefficients for the expression ratios across all genes between the cDNA microarray and Affymetrix platform were calculated for the overlapping and non-overlapping probe sets (Table 2). The data showed a significantly higher correlation with overlapping than with non-overlapping, Unigene-matched probe sets (e.g. for Hu133A-MAS5, P < 0.0001). Of the three expression calculation metrics, MAS 5.0 was outperformed by both RMA and dChip, the two latter methods producing similar results (Table 2). This is probably due to the lack of advanced non-linear probe normalization in the MAS 5.0 algorithm (13). Moreover, the correlations between data from the Hu133B chip and Agilent microarray are lower than the correlations for either the Hu95A or the Hu133A and their corresponding Agilent data. This is probably due to the fact that the Hu133B platform contains a higher number of unreliable sequences (B. H. Mecham, D. Z. Wetmore, Z. Szallasi, Y. Sadovsky, I. Kohane and T. J. Mariani, submitted for publication). These results indicate that overlapping Affymetrix probe sets produce more correlated results with cDNA microarray data than non-overlapping, Unigene-matched measurements.
Overlapping probe sets produce more consistent difference calls between different microarray platforms
Affymetrix and Agilent technologies provide proprietary algorithms to identify differentially expressed genes. Both methods translate continuous probe intensity values into a discrete ‘difference call’ such as no change (NC), increase (I) or decrease (D) with an associated confidence level. We compared the consistency of difference calls for overlapping and non-overlapping probe sets. Each microarray experiment was performed in duplicates. For any given gene the difference calls were reproducible between replicates in 80–98 % of the cases, depending on the actual experiment and platform (data not shown). We decided to further analyze only those difference calls that were identical in both replicate comparisons in a single technology.
Difference calls for the overlapping and non-overlapping probe sets were used to create 3 × 3 contingency tables (Supplementary Table 3). A t-statistic was calculated to measure the independence of the rows (Affymetrix Decision) and columns (Agilent Decision) of each table (Supplementary Table 3). The t-values for both the overlapping and non-overlapping measurements indicate that the difference calls are not independent of one another (Table 3). While the t-statistic assesses the independence of the two difference calls, it does not provide any measure of their concordance, which was quantified using Cramer’s coefficient (10). The difference in Cramer coefficients (e.g. Hu133A 0.18 overlapping versus 0.04 non-overlapping) indicates that the difference calls between overlapping probe sets and corresponding Agilent data are more similar than those for non-overlapping probe sets.
Table 3. Overlapping probe sets produce more consistent difference calls between Affymetrix and Agilent cDNA microarray platforms.
| Platform | Type | t-value | P-value | Cramer’s coefficient |
|---|---|---|---|---|
| 133A | Overlapping | 3805 | <0.001 | 0.18 |
| Non-overlapping | 511 | <0.001 | 0.07 | |
| 95A | Overlapping | 3702 | <0.001 | 0.24 |
| Non-overlapping | 160 | <0.001 | 0.06 | |
| 133B | Overlapping | 478 | <0.001 | 0.18 |
| Non-overlapping | 80 | <0.001 | 0.04 |
Difference calls were calculated for RNA samples comparing MDA-MB-436 cells to the HMEC cells and comparing the HCC1954 cells to the HMEC cells using the manufacturer’s algorithm for both platform types. Only consistent calls between duplicate measurements were included in the analysis. t-Values and Cramer coefficients were calculated as described in the text for 3 × 3 contingency tables.
Overlapping probe sets provide higher cross-platform consistency in breast cancer associated gene expression data sets
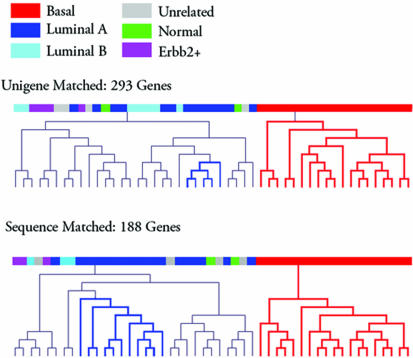
Microarray analysis has improved the molecular classification of cancer subtypes. Since each microarray platform carries a certain amount of technology specific noise, the crucial criterion of reliable classification is whether it could be reproduced in a platform independent manner. A recent paper attempted to reproduce the classification of breast cancer samples into subtypes based on data sets produced by the Affymetrix and cDNA microarray platforms (7). The results indicated that technology-specific noise overwhelmed the underlying shared biology of the two studies. Classification produced by cDNA microarrays could be transferred only to a limited extent to Affymetrix gene chip derived data sets (7) (Fig. 3). We tested the hypothesis whether sequence-matching probes across different microarray technologies removes at least some of the platform-specific noise and helps to better identify similarities and differences between breast cancer samples. Beginning with a set of intrinsic genes that have been shown to identify five distinct breast cancer tumor types (4,7), we determined which of these had a corresponding overlapping probe set on the Affymetrix HuFL chip, which was used in the corresponding studies. Two sets of data were then constructed. The first contains both overlapping and non-overlapping probe sets and produces a group of Unigene-matched measurements. The second contains only overlapping probe sets and produces the set of sequence-matched measurements. As explained in the Materials and Methods, centroids composed of the median expression cDNA values for the intrinsic genes were used to classify each Affymetrix sample as one of the five tumor types (or unrelated if it was not significantly related to any centroid). The Unigene-matched and sequence-matched samples were classified independently and did produce different classifications for identical samples. Figure 3 indicates the clustering diagrams generated by clustering the sequence-matched and Unigene-matched MAS 5.0 values. It shows that sequence-matched probe sets produce a significant improvement without perfectly reproducing the cDNA microarray-based subtype classification. Both clusterings produce a sharp separation between the basal subtype and all other classes. However, the sequence-matched clustering also contains a single node composed of luminal sub-type-A samples. Neither clustering produced a clear separation of the luminal type B, erbB2 or normal tumor types. However, the normal samples are positioned closer in the hierarchical clustering using the sequence-matched probe sets than using only Unigene-matched probe sets. In order to test if we have simply removed too many genes to identify these tumor types we clustered the corresponding cDNA data for each of the Unigene- and sequence-matched measurements. In both clustering results the five distinct tumor types are still readily identifiable indicating a potential role of platform-specific noise on the viability of these intrinsic genes to accurately classify tumors (see Supplementary Figure 2). These results, in combination with the cross-platform comparison on aliquots of breast cancer cell line derived RNA, suggest that sequence matching is a reasonable computational method to improve cross-platform consistency of biological results obtained with different microarray technologies.
Figure 3.
Sequence-matched probe sets provide more consistent classification results derived from breast cancer associated gene expression data sets obtained by different types of microarray platforms. Hierarchical clustering of two different subsets of genes taken from the data set published by West et al. (5). (Top) The result using the 293 genes as suggested by Sorlie et al. (4) after matching genes between the Affymetrix and cDNA microarray platforms using Unigene IDs. (Bottom) The clustering result by reducing the gene set to only those that are also sequence-matched between the two platforms. The color code of the samples was assigned as described in Sorlie et al. (4,7).
DISCUSSION
In this paper, we have shown that overlapping probe sets produce higher consistency between gene expression profiles produced by the Affymetrix and cDNA microarray platforms. With the continuous improvement of microarray technology, it is expected that signals produced by an overlapping Affymetrix probe set and a cDNA clone will be consistent between the two platforms. We were pleased to confirm this expectation in a side-by-side cross-platform comparison. The Pearson correlation coefficient of ∼0.7, and the highly similar difference calls across several thousand genes provides a much improved correlation relative to that seen with earlier versions of these technologies (14). The lower correlation shown by non-sequence-overlapping but Unigene-matched probes is probably due to a number of previously described factors. It may reflect splice variants (15) and the well documented 3′-5′ degradation of microarray signals along genes (16). Unigene clusters assemble putative genes from cDNA clones using a variety of algorithms; however, it has been shown that a subset of these clusters are incorrect (17). A significant fraction of these errors have been eliminated in more recent updates of Unigene and by alternative information sources, such as the human genome. However, the actual Unigene build we used may still contain several cases when two cDNA clones (designated for the moment as A and B), are incorrectly listed as part of the same Unigene cluster. In such a case, cDNA clone A, which is used on the spotted microarray and the Affymetrix probe set, designed against cDNA clone B as a target, will measure the expression of two different transcripts. We conclude that the lower correlation between non-overlapping probe sets are largely due to situations like this. This conclusion is supported by our observation that if there is at least one overlapping probe with a cDNA clone, then the correlation between the two platforms is as high as for completely overlapping probe sets. The overlapping probe(s) seems to ensure the sequence contiguity, required for measuring the same transcript between the two platforms. We have also confirmed, that advanced statistical methods such as RMA (13) provide an advantage for the analysis of Affymetrix chips. This method has previously outperformed both dChip and MAS 5.0 in spike-in studies (13). We assessed the performance of these methods with cross-platform comparison of RNA aliquots, which is a less stringent method than that previously applied by Irizarry et al. (13). This might explain why dCHIP performed almost as well as RMA in our side-by-side comparisons.
In addition to the improvement shown in the cross-platform comparison using RNA aliquots, our analysis produced an important practical result for large-scale, disease-related microarray studies as well. Gene expression profiling of disease states is usually performed on a single microarray platform by any given research group (7). Therefore, it is important to provide practical guidelines for cross-platform, cross-study comparisons. Sequence-matched probe sets provide a relatively easy computational method to ensure the highest possible consistency between data sets produced by different types of microarray platforms.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Atul Butte for helpful suggestions and Travis Burleson for excellent technical assistance. I.S.K. was supported in part by the National Institute of Health through grants HL66805-01 and NS40828-01A1. T.J.M. was supported by the NIH grant HL071885. Z.S. was supported in part by the National Institutes of Health through grants HL066582-01 and 1PO1CA-092644-01.
REFERENCES
- 1.Schena M., Shalon,D., Davis,R.W. and Brown,P.O. (1995) Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science, 270, 467–470. [DOI] [PubMed] [Google Scholar]
- 2.Watson A., Mazumder,A., Stewart,M. and Balasubramanian,S. (1998) Technology for microarray analysis of gene expression. Curr. Opin. Biotechnol., 9, 609–614. [DOI] [PubMed] [Google Scholar]
- 3.Yuen T., Wurmbach,E., Pfeffer,R.L., Ebersole,B.J. and Sealfon,S.C. (2002) Accuracy and calibration of commercial oligonucleotide and custom cDNA microarrays. Nucleic Acids Res., 30, e48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sorlie T., Perou,C.M., Tibshirani,R., Aas,T., Geisler,S., Johnsen,H., Hastie,T., Eisen,M.B., van de Rijn,M., Jeffrey,S.S., Thorsen,T., Quist,H., Matese,J.C., Brown,P.O., Botstein,D., Eystein Lonning,P. and Borresen-Dale,A.L. (2001) Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl Acad. Sci. USA, 98, 10869–10874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.West M., Blanchette,C., Dressman,H., Huang,E., Ishida,S., Spang,R., Zuzan,H., Olson,J.A.,Jr, Marks,J.R. and Nevins,J.R. (2001) Predicting the clinical status of human breast cancer by using gene expression profiles. Proc. Natl Acad. Sci. USA, 98, 11462–11467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van‘t Veer L.J., Dai,H., van de Vijver,M.J., He,Y.D., Hart,A.A., Mao,M., Peterse,H.L., van der Kooy,K., Marton M.J., Witteveen,A.T., Schreiber,G.J., Kerkhoven,R.M., Roberts,C., Linsley,P.S., Bernards,R. and Friend SH. (2002) Gene expression profiling predicts clinical outcome of breast cancer. Nature, 415, 530–536. [DOI] [PubMed] [Google Scholar]
- 7.Sorlie T., Tibshirani,R., Parker,J., Hastie,T., Marron,J.S., Nobel,A., Deng,S., Johnsen,H., Pesich,R., Geisler,S., Demeter,J., Perou,C.M., Lonning,P.E., Brown,P.O., Borresen-Dale,A.L. and Botstein D. (2003) Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc. Natl Acad. Sci. USA, 100, 8418–8423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gautier L., Cope,L., Bolstad,B.M. and Irizarry,R.A. (2004) Affy-analysis of Affymetrix GeneChip data at the probe level. Bioinformatics, 20, 307–315. [DOI] [PubMed] [Google Scholar]
- 9.Bolstad B.M., Irizarry,R.A., Astrand,M. and Speed,T.P. (2003) A comparison of normalization methods for high density oligonucleotide array data based on bias and variance. Bioinformatics, 19, 185–193. [DOI] [PubMed] [Google Scholar]
- 10.Iman R.L. (1994) Analysis of Enumerative Data. A Data-Based Approach To Statistics. Duxbury Press, Belmont, CA, pp. 436–444. [Google Scholar]
- 11.Chudin E., Walker,R., Kosaka,A., Wu,S.X., Rabert,D., Chang,T.K. and Kreder,D.E. (2002) Assessment of the relationship between signal intensities and transcript concentration for Affymetrix GeneChip arrays. Genome Biol., 3, RESEARCH0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li C. and Wong,W.H. (2001) Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc. Natl Acad. Sci. USA, 98, 31–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Irizarry R.A., Bolstad,B.M., Collin,F., Cope,L.M., Hobbs,B. and Speed,T.P. (2003) Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res., 31, e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuo W.P., Jenssen,T.K., Butte,A.J., Ohno-Machado,L. and Kohane,I.S. (2002) Analysis of matched mRNA measurements from two different microarray technologies. Bioinformatics, 18, 405–412. [DOI] [PubMed] [Google Scholar]
- 15.Wang H., Hubbell,E., Hu,J.S., Mei,G., Cline,M., Lu,G., Clark,T., Siani-Rose,M.A., Ares,M., Kulp,D.C. and Haussler,D. (2003) Gene structure-based splice variant deconvolution using a microarray platform. Bioinformatics, 19 (Suppl. 1), I315–I322. [DOI] [PubMed] [Google Scholar]
- 16.Auer H., Lyianarachchi,S., Newsom,D., Klisovic,M.I., Marcucci,G., Kornacker,K. and Marcucci,U. (2003) Chipping away at the chip bias: RNA degradation in microarray analysis. Nature Genet., 35, 292–293. [DOI] [PubMed] [Google Scholar]
- 17.Burke J., Davison,D. and Hide,W. (1999) d2_cluster: a validated method for clustering EST and full-length cDNAsequences. Genome Res., 9, 1135–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.