Abstract
The prevalence of oral squamous cell carcinoma (OSCC) has significantly increased over decades in several countries and human papilloma virus (HPV) has been indicated as one of the underlying causes. This suggests that HPV plays a role in the early stages of carcinogenesis but is not a requisite for the maintenance and progression of malignant state. p53 is a tumor suppressor gene that checks the cell and promotes apoptosis and cell repair that can be deactivated by mutations and a viral interaction leading to cancer and individuals with particular polymorphic variant of p53 is more susceptible to HPV-induced carcinogenesis. The present study has been carried out to detect and correlate p53 polymorphism/mutation, HPV DNA in the biopsy samples of oral cancer patients who had tobacco habits.
Keywords: Human papilloma virus, oral squamous cell carcinoma, p53 polymorphism/mutation
INTRODUCTION
Oral cancer is one of the ten most common cancers in the world and is the most common cancer in the South East Asia (India, Bangladesh, Pakistan and Sri Lanka) where it accounts for one- third of all body cancers.[1] In the oral cavity, most common malignancy is squamous cell carcinoma (OSCC) representing 90-95% of all oral malignancies.[2]
Mass ignorance and poverty coupled with long standing deleterious oral habits resulted in an alarming increase in the number of OSCC patients in India.[3] Annually about 2,70,000 cases of oral cancers are reported worldwide and about 82,000 of them are diagnosed in India alone.[4]
In recent years, reports have emphasized the correlation between the immune status of a person and its concomitant increase in the incidence of neoplasia.[5] So far, 120 types of HPV have been identified and they are classified into high risk and low risk HPV viruses. Among the high risk, HPV-16 and HPV-18 are known to be involved in cancer cervix.[6] It has been established that HPV-16 and HPV-18 are associated with oral carcinogenesis.[7] Their genomes are integrated into the host DNA and are transcriptionally active in both tumor and tumor derived cell lines.[8,9]
The most intensely involved gene in carcinogenesis is tumor suppressor gene called p53. The p53 locus is the most commonly mutated locus in human cancers. Located on the short arm of chromosome 17, p53 is a 393 amino acid protein that is expressed in all the tissues of the body.[10] Its biological role is to protect cells from DNA damage caused by radiation, chemical carcinogens or other mechanisms. p53 does this either by arresting the cell cycle so that DNA repair can occur or by inducing programmed cell death.[11] There has been considerable debate regarding the immortalization of kertainocytes, especially intermediate cytokeratin filaments 7 and 19, by the HPV type E7 which associates it with squamous cell carcinoma.[12,13] It also seems clear that mutation of the p53 gene is one of the most common abnormalities in Squamous Cell Carcinoma of the Head and Neck (SCCHN), with approximately 50% of cases associated with this mutation.[14]
The aim of the present study was:
Detection of HPV DNA in the biopsy samples of oral cancer patients who had tobacco habits
Detection of HPV DNA in intraoral smears from tobacco using healthy controls
Study of p53 polymorphism in the genomic DNA from cancer patients and healthy controls
Study of p53 gene mutation in DNA extracted from the biopsy samples of cancer patients and intraoral smears from healthy controls
To find out the risk of cancer due to HPV infection, p53 polymorphism and mutation at p53.
MATERIALS AND METHODS
This study was carried out in the Department of Oral and Maxillofacial Pathology.
Selection of subjects: Cases (cancer patients): The patients attending the Out-patient Department, during 2006-08 were examined for the presence of tobacco related oral cancer.
Controls (Healthy individuals): The ‘control’ individuals, having tobacco chewing, smoking habits and corroborative age but without any oral lesions, were selected from the same hospital.
Examination of patients and controls: Of all, 83 were selected as patients and 100 as controls as their habit data were available. Proper informed consents were obtained from the 83 patients and 100 controls individuals. Tobacco Habit: All patients and controls had tobacco habit.
Tobacco habits were classified into three groups –
-
(i)
Smoking tobacco (e.g. cigarettes, Bidi, Chutta etc.)
-
(ii)
Chewable tobacco (e.g. Catechu, Jarda, Gurakhu, Khaini and Snuff etc.)
-
(iii)
Mixed tobacco habits (i.e. both tobacco smoking and chewing).
Unit used to express the amount of smoking tobacco is pack year (PY) (pack year = duration of habit in year X number of pack daily smoked). One pack = 20 Bidi or 10 Cigarettes.
Unit used to express the amount of chewable tobacco consumed by an individual is chewing year (CY). Chewing year = Duration of habit in year X Frequency of daily intake.
Blood collection: Approximately 3 ml venous blood was collected from each of the aforesaid patients and controls. Blood was mixed slowly with ethylenediaminetetraacetic acid (EDTA) for 1 min and stored at −20ºC for DNA isolation and subsequent procedure.
Biopsy procedure: Biopsy done at the representative site of the lesion from all the aforesaid patients. The tissue was immediately fixed and used to confirm the histopathological diagnosis of OSCC. Genomic DNA was isolated using QIAamp DNA isolation kit (Qiagen GmbH, Germany) from blood, exfoliated cells from oral cavity of controls and processed tissues of cancer patients.
Genotyping at codon 72 (Arg/Pro) polymorphism of p53:
(1) Polymerase chain reaction (PCR):
The double-stranded DNA was dissociated into single-stranded DNA at a denaturing temperature of 94ºC for 5 minutes and then for 30 seconds. The cycle was repeated 30 times, which increased the amount of target DNA exponentially.
Primer sequences (p53 codon 72) at exon 4
Forward Primer:
5′-TTTTCACCCATCTACAGTCCCCCTTG-3′
Reverse Primer:
5′- TAGGAGCTGCTGGTGCAGGGGCCCCG-3′
(3) Mutation detection at p53 (exon4-exon9): Mutation at p53 gene was checked by DNA sequencing of the PCR products using ABI - 3100 automated DNA sequencer. Primer sequences for p53 sequencing: P53 exon 5-6 F: 5′ -TGTTCACTTGTGCCCTGACT-3′ [Figure 1].
Figure 1.
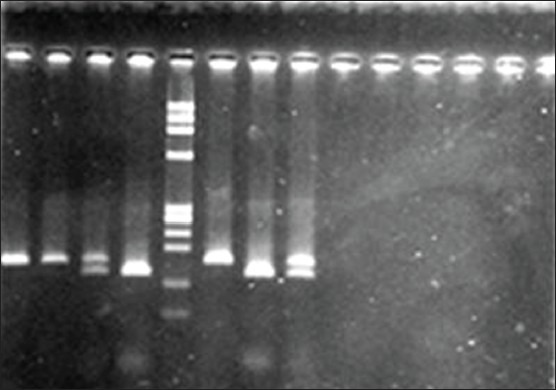
DNA bands after digestion: 135 and 27 base pairs of pro and uncut 162 base pairs for Arg
Screening of HPV
Detection of HPV-16 and HPV-18: Initially, part of the β-globin gene was amplified to check the presence and integrity of the DNA in the samples (Tachezy et al., 1999).[15] The presence of HPV in the cancer tissues was detected by PCR using primers (MY09 and MY11) from the consensus L1 region. Detection of HPV16 and 18 in the L1 positive samples were carried out in separate tubes using type specific primers (HPV16: TCAAAAGCCACTGTGTCCTG and CGTGTTCTTGATGATCTGCA; HPV18: ACCTTAATGAA AAACCACGA and CGTCGTTGGAGTCGTTCCTG) [Figure 2] homologous to a region of the E6 gene (Haraf et al., 1996).[16] For final confirmation of the HPV types, the PCR products were transferred from agarose gel to Gene Screen nylon membrane for southern hybridization with [32P] labeled HPV type specific probes.
Figure 2.
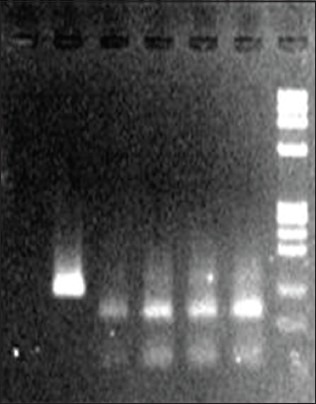
DNA bands after digestion: 81and 39bp for HPV 16; 51 and 49 bp for HPV 18
Statistical tests
Chi square test, Odds Ratio (OR) and its confidence interval (CI).
RESULTS AND OBSERVATIONS
All the patients and controls were living in and around the city located in the eastern region of India. None of the patients and controls were exposed to specific occupational or environmental carcinogens and so environmental effects, other than tobacco use were similar. A total of 83 cancer patients and 100 controls were included in our study. Distribution of age, sex and tobacco habits in patients and controls were shown in Table 1. Numbers of smokers were significantly less (P < 0.0001) in cancer patients than control groups. Numbers of smokeless tobacco users were significantly more in cancer patients (P = 0.005). But, the numbers of mixed habitués were similar in two groups. Mean smoking and smokeless tobacco doses were significantly different in patients and controls.
Table 1.
Demography and tobacco exposure of cancer patients and controls

Restriction length polymorphism was observed after digestion with Sma I enzyme using Agarose gel electrophoresis. The length of the PCR product was 162 base pairs. In the case of pro/pro genotype, the PCR product DNA was cut at 135 and 27 base pairs. In the case of pro/arg genotype, pro allele containing DNA was cut at 135 and 27 base pairs and the other allele remained uncut (i.e. 175 bp). In the case of arg/arg genotype, the PCR product DNA remained uncut (i.e. 175 bp). Genotyping at codon 72 at p53 was done by PCR and restriction fragment length polymorphism (RFLP). But, 10% of the PCR products were re-sequenced to confirm the genotypes at this locus. The re-sequencing results matched exactly with the genotype data from PCR RFLP analysis.
No significant association between codon 72 polymorphism and cancer was observed [Table 2]. It is observed that HPV infection was associated with increased risk of cancer (OR = 5.5, 95%, CI = 1.6-1.9) [Table 3].
Table 2.
Distribution of genotypes at p53 exon 4 among cancer patients and controls
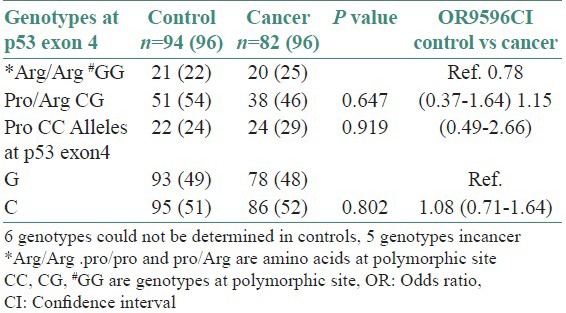
Table 3.
HPV infection status in cancer patients and control

Histopathologically, all malignancies were diagnosed as Squamous Cell Carcinoma (SCC) and morphologically they were well (79% and 67%), moderately (18% and 29%) and poorly differentiated (3% and 4%) carcinoma in HPV infected and non-infected tumors, respectively [Table 4]. Table 5 shows the tobacco habits of the HPV infected samples in cancer and controls, 26% of the 38 cancer were found to be smokers, 42% were smokeless tobacco chewers and 32% had mixed habits. Whereas 35% of the 23 controls were smokers, 39% were smokeless tobacco chewers and 26% had mixed habits. The presence of p53 mutation in cancer and control patients is highlighted in Table 6. Five mutations in p53 gene were detected in cancer tissue DNA samples and the changes are: C > T (Arg > Cys), C > T (Arg > Trp) and C > T (Arg > stop) at exon 8, G > A (Arg > His) at exon 5 and C > T at intron 9.
Table 4.
Histopathological findings in HPV infected cancer samples N=38%
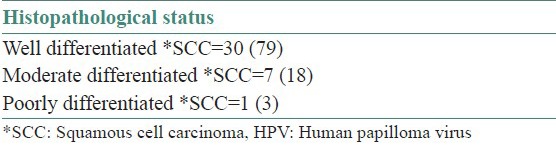
Table 5.
Tobacco habits in HPV infected samples
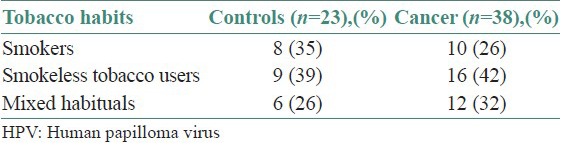
Table 6.
Presence of Mutation in P53 Gene among cancer and controls

DISCUSSION
Recently developed molecular biology techniques have allowed investigators to identify human papilloma virus oral cancer precisely (Greer et al., 1989, Kashima et al., 1990).[17,18] It is, nevertheless, very significant that HPV type 16, a type demonstrated in oral carcinomas, has been shown to induce dysplasia in squamous epithelium in an otherwise sterile in vitro environment (McCance et al., 1988).[19] Enormous ranges of detection rates have been reported, ranging from 0% to 81% (Scully et al., 2002, Bouda et al., 2000, Sugiyama et al., 2003).[20,21,22] Terai et al. pointed out that the rate depends on a number of factors including the source of the specimen, the detection method and the number of subjects and their ages. They found HPV in 30 out of their 37 specimens (Terai et al., 2002, Sugiyama et al., 2003).[22,23] Review of literature exhibits a wide range of p53 alterations in head and neck tumors from 33-76% (Kozomora et al., 2005, Wilson et al., 1995, Chomchai et al., 1999).[24,25,26] As in other cancers, the p53 mutations show overexpression in most of the cases and it is the mutated form rather than the wild type gene, which is overexpressed. However, in some cases it is possible to observe overexpression but not to find mutation in the gene (Taylor et al. 1990, Patridge et al., 1999, Donna et al., 2002).[27,28,29] The mutation is often observed as an abnormal expression of the gene since immuno-cytochemical staining can detect p53 readily in many oral cancers but not in normal tissues (Ravi et al., 1996, Ogden et al., 1992, Donna et al., 2002).[29,30,31] In an International Agency for Research on Cancer (IARC), multicentric study conducted by Min Dai et al., the results showed an inverse association between the presence of p53 mutations and HPV 16 infections in tissue specimens from cancers of the oral cavity and oropharynx (Min Dai et al., 2004).[32] The biological connection between HPV and p53 proteins has already been established by the fact that viral E6 protein can inactivate wild type p53 protein (Munger et al., 2002, Kozomora et al., 2005).[33,34] Therefore, it is questionable whether a mutated p53 gene can significantly influence the prognosis of patients already affected with HPV. Patients with HPV infection plus p53 mutation had the worst prognosis. Other authors have also studied the relationship between HPV infection and mutation of the p53 gene concerning prognosis of tumor patients.
The biochemical and biological activities of p53 that are affected by codon 72 polymorphism have been evaluated in a series of studies. It was first shown that this polymorphism had a profound effect on the primary structure of p53 (Mitra et al., 2005). More recently, it has been shown that the ability of p53 to interact with the TFID associated factors is much stronger for Pro-72 associated form than for Arg-72 form whereas Pro-72 form has increased transcription transactivation capacity (Mitra et al., 2005, Marin et al., 2000).[35,36] In contrast, analysis of the ability of the two polymorphic varieties to induce apoptosis suggests that the two polymorphic varieties though differing in kinetics are capable of inducing equal level of apoptosis (Mitra et al., 2005, Marin et al., 2000).[19,33] Recent studies using temperature sensitive form of p53 protein further supports these observations and shows that the Arg-72 form has a much more stronger capacity to induce apoptosis than the Pro-72 form of p53 in tumor cells but not in normal cells (Mitra et al., 2005, Dumont et al., 2003);[32,37] however, in primary head and neck tumors, retention of the arginine allele in codon 72 of p53, correlates with poor apoptosis (Mitra et al., 2005, Schneider et al., 1986).[20,33] Finally, the Pro-72 form appears to induce a higher level of G1 arrest as compared to the Arg-72 form (Mitra et al., 2005, Pim et al., 2004).[33,38] In summary, it may be stated that Arg-72 form induces a lower level of G1 arrest, has reduced transcriptional transactivational capacity of downstream target genes and has a lower apoptotic potential in primary tumors as compared to the Pro-72 form. Thus, we argue that individuals with the Arg-72 form of p53 are less efficient in processing tobacco associated DNA damage signals and hence are more susceptible to develop leukoplakia (Mitra et al., 2005).[33] Tao et al. (2002)[27] conducted a study on polymorphism of p53 gene and observed that polymorphism at codon 72 of exon 4 which results in either an arginine residue (CGC) or a proline residue (CCC), modulate risk of cancer differentially. Prevalence of mutation was observed to be more in patients having increased tobacco smoking habit. The p53 gene is mutated in over one-half of oral cancers. The mutation is often observed as an abnormal expression of the gene since immuno-cytochemical staining can detect p53 readily in many oral cancers but not in normal tissues (Donna A et al., 2002).[11] The majority of the mutation was observed in the region of exons 5–8 of p53 gene. Besides this, a small percentage of p53 mutations can be found outside this region.
SUMMARY AND CONCLUSION
The present study was undertaken with a view to assess the association among HPV infection, p53 polymorphisms, p53 mutation and the risk of development of oral cancer in patients from Kolkata. The findings thereafter were recorded carefully, analyzed and corroborated in reference to the aims and objectives of this study and the following observations and conclusions were drawn.
HPV infection increases the risk of oral cancer
Genotype of p53 at codon 72 did not modulate the risk of cancer
Mutations at p53 gene were found in 5 of the 83 cancer and none in 100 control subjects. To detect mutations, DNA sequence from blood and tissue samples from same individuals were compared. Five mutations in TP53 gene were detected in tissue samples and the changes are: C >T (Arg > Cys), C > T (Arg > Trp) and C > T (Arg > stop) at exon 8, G > A (Arg > His) at exon 5 and C > T at intron 9.
ACKNOWLEDGMENT
Dr. Bidyut Roy. Head of the Department, Department of Genetics, ISI, Kolkata
Footnotes
Source of Support: Nil.
Conflict of Interest: None declared.
REFERENCES
- 1.Atkinson L, Chester IC, Smyth FG, Ten S. Oral cancer in New Guinea. A study in demography and etiology. Cancer. 1964;17:1289–98. doi: 10.1002/1097-0142(196410)17:10<1289::aid-cncr2820171011>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 2.Kurokawa H, Matsumoto S, Murata T, Yamashita Y, Tomoyose T, Zhang M, et al. Immunohistochemical study of syndecan -1 down regulation and the expression of p53 protein or Ki-67 antigen in oral leukoplakia with or without epithelial dysplasia. J Oral Pathol Med. 2003;32:513–21. doi: 10.1034/j.1600-0714.2003.00117.x. [DOI] [PubMed] [Google Scholar]
- 3.Bouda M, Gorgoulis VG, Kastrinakis NG, Giannoudis A, Tsoli E, Danassi-Afentaki D, et al. “High risk” HPV types are frequently detected in potentially malignant and malignant oral lesions, but not in normal oral mucosa. Mod Pathol. 2000;13:644–53. doi: 10.1038/modpathol.3880113. [DOI] [PubMed] [Google Scholar]
- 4.Boyd NM, Reade PC. Mechanisms of carcinogenesois with particular reference to the oral mucosa. J Oral Pathol. 1988;17:193–201. doi: 10.1111/j.1600-0714.1988.tb01524.x. [DOI] [PubMed] [Google Scholar]
- 5.Smith EM, Ritchie JM, Summersgill KF, Hoffman HT, Wang DH, Haugen TH, et al. Human papillomavirus in oral exfoliated cells and risk of head and neck cancer. J Natl Cancer Inst. 2004;96:449–55. doi: 10.1093/jnci/djh074. [DOI] [PubMed] [Google Scholar]
- 6.Elledge RM, Lee WH. Life and death by p53. Bioassays. 1995;17:923–30. doi: 10.1002/bies.950171105. [DOI] [PubMed] [Google Scholar]
- 7.Eric J. Pathogenesis and progression of squamous cell carcinoma of the Head and Neck. In: Jeffery N, editor. Cancer of the Head and Neck. Amsterdam: Elsevier Publishers; 2003. pp. 5–27. [Google Scholar]
- 8.Evans SJ, Langdon JD, Rapidis AD, Johnson NW. Prognostic significance of STNMP and velocity of tumor growth in oral cancer. Cancer. 1982;49:773–6. doi: 10.1002/1097-0142(19820215)49:4<773::aid-cncr2820490428>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 9.Fahmy MS, Sadeghi A, Behmard S. Epidemiologic Study of oral cancer in Fars Province, Iran. Community Dent Oral Epidemiol. 1983;11:50–8. doi: 10.1111/j.1600-0528.1983.tb01354.x. [DOI] [PubMed] [Google Scholar]
- 10.Ibrahim SO, Warnakulasuriya KA, Idris AM, Hirsch JM, Johnson NW, Johannessen AC. Expression of keratin 13, 14 and 19 in oral hyperplastic and dysplastic lesions from Sudanese and Swedish snuff dippers: Association with Human papillomavirus infection. Anticancer Res. 1998;18:635–45. [PubMed] [Google Scholar]
- 11.Whyte DA, Broton CE, Shillitoe EJ. The unexplained survival of cells in oral cancer. What is the role of P53? J Oral Pathol Med. 2002;31:125–33. doi: 10.1034/j.1600-0714.2002._310301.x. [DOI] [PubMed] [Google Scholar]
- 12.Favia G, Kanduc D, Lo Muzio L, Lucchese A, Serpico R. Possible association between HPV16 E7 protein level and cytokeratin 19. Int J Cancer. 2004;111:795–7. doi: 10.1002/ijc.20343. [DOI] [PubMed] [Google Scholar]
- 13.Combes JD, Franceschi S. Role of human papillomavirus in non-oropharyngeal head and neck cancers. Oral Oncol. 2014;50:370–9. doi: 10.1016/j.oraloncology.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 14.Kashima HK, Kutcher M, Kessis T, Levin LS, de Villiers EM, Shah K. Human papillomavirus in squamous cell carcinoma, leukoplakia, lichen planus, and clinically normal epithelium of the oral cavity. Ann Otol Rhinol Laryngol. 1990;99:55–61. doi: 10.1177/000348949009900110. [DOI] [PubMed] [Google Scholar]
- 15.Tachezy R, Mikysková I, Saláková M, Van Ranst M. Correlation between human papillomavirus-associated cervical cancer and p53 codon 72 arginine/proline polymorphism. Hum Genet. 1999;105:564–6. doi: 10.1007/s004399900138. [DOI] [PubMed] [Google Scholar]
- 16.Haraf DJ, Nodzenski E, Brachman D, Mick R, Montag A, Graves D, et al. Human papilloma virus and p53 in head and neck cancer: Clinical correlates and survival. Clin Cancer Res. 1996;2:755–62. [PubMed] [Google Scholar]
- 17.Greer RO, Eversole LR. Leukoplakia, verruciform hyperkeratosis, verrucous carcinoma, and squamous carcinoma associated with human papillomavirus. Oral Surg. 1989;68:598. [Google Scholar]
- 18.Johnson NW. Histological and histochemical studies of oral cancer. Int Dent J. 1977;27:25–34. [PubMed] [Google Scholar]
- 19.Marin MC, Jost CA, Brooks LA, Irwin MS, O’Nions J, Tidy JA, et al. A common polymorphism acts as an intragenic modifier of mutant p53 behavior. Nat Genet. 2000;25:47–54. doi: 10.1038/75586. [DOI] [PubMed] [Google Scholar]
- 20.Schneider A, Oltersdorf T, Schneider V, Gissmann L. Distribution pattern of human papillomavirus 16 genome in cervical neoplasia by molecular in situ hybridization of tissue sections. Int J Cancer. 1986;39:717–21. doi: 10.1002/ijc.2910390611. [DOI] [PubMed] [Google Scholar]
- 21.Boyle JO, Hakim J, Koch W, van der Riet P, Hruban RH, Roa RA, et al. The incidence of p53 mutation increases with progression of head and neck cancer. Cancer Res. 1993;53:4477–80. [PubMed] [Google Scholar]
- 22.Sugerman PB, Shillitoe EJ. The high risk human papilloma virus and oral cancer: Evidence for and against a causal relationship. Oral Dis. 1997;3:130–47. doi: 10.1111/j.1601-0825.1997.tb00025.x. [DOI] [PubMed] [Google Scholar]
- 23.Taylor D, Koch WM, Zahurak M, Shah K, Sidransky D, Westra WH. Immunohistochemical detection of p53 protein accumulation in head and neck cancer: Correlation with p53 gene alterations. Hum Pathol. 1990;30:1221–5. doi: 10.1016/s0046-8177(99)90041-2. [DOI] [PubMed] [Google Scholar]
- 24.Koppikar P, deVilliers EM, Mulherkar R. Identification of human papillomavirus in tumors of the oral cavity in an Indian community. Int J Cancer. 2005;113:946–50. doi: 10.1002/ijc.20664. [DOI] [PubMed] [Google Scholar]
- 25.Waterhouse S, Muir C, Shanmugaratnam K, Powell J. International Agency for Research on Cancer France. IV. France; Lyon: 1982. Cancer incidence of five continents. Basis of adjusted incidence. [Google Scholar]
- 26.Broders AC. The microscopic grading of cancer. Surg Clin North Am. 1941;21:947–62. [Google Scholar]
- 27.Li T, Lu ZM, Guo M, Wu QJ, Chen KN, Xing HP, et al. p53 codon polymorphism and the risk of human papillomavirus associated carcinomas in China. Cancer. 2002;95:2571–6. doi: 10.1002/cncr.11008. [DOI] [PubMed] [Google Scholar]
- 28.Ogden GR, Kiddie RA, Lunny DP, Lane DP. Assessment of p53 protein expression in normal, benign and malignant oral mucosa. J Pathol. 1992;166:389–94. doi: 10.1002/path.1711660411. [DOI] [PubMed] [Google Scholar]
- 29.Chomchai JS, Du W, Sarkar FH, Li YW, Jacobs JR, Ensley JF, et al. Prognostic significance of p53 mutations in laryngeal cancer. Laryngoscope. 1999;109:455–9. doi: 10.1097/00005537-199903000-00021. [DOI] [PubMed] [Google Scholar]
- 30.Rapidis AD, Langdon JD, Patel MF, Harvey PW. STNMP: A new system for the Clinico-pathological classification and identification of intra-oral carcinomata. Cancer. 1977;39:204–9. doi: 10.1002/1097-0142(197701)39:1<204::aid-cncr2820390132>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 31.Nylander K, Dabelsteen E, Hall PA. The p53 molecule and its prognostic role in squamous cell carcinomas of the head and neck. J Oral Pathol Med. 2000;29:413–25. doi: 10.1034/j.1600-0714.2000.290901.x. [DOI] [PubMed] [Google Scholar]
- 32.Miller CS, White DK. Human papillomavirus expression in oral mucosa, premalignant conditions and squamous cell carcinomas: A retrospective review of the literature. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 1996;82:57–68. doi: 10.1016/s1079-2104(96)80378-7. [DOI] [PubMed] [Google Scholar]
- 33.Mitra S, Sikdar N, Misra C, Gupta S, Paul RR, Roy B, et al. Risk assessment of p53 genotypes and haplotypes in tobacco–associated leukoplakia and oral cancer patients from eastern India. Int J Cancer. 2005;117:786–93. doi: 10.1002/ijc.21263. [DOI] [PubMed] [Google Scholar]
- 34.Kozomara R, Jović N, Magić Z, Branković-Magić M, Minić V. p53 mutation and human papillomavirus in oral squamous cell carcinomas: Correlation with overall survival. J Craniomaxillofacial Surg. 2005;33:342–8. doi: 10.1016/j.jcms.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 35.Dai M, Clifford GM, le Calvez F, Castellsagué X, Snijders PJ, Pawlita M, et al. IARC Multicenter Oral Cancer Study Group. Human papillomavirus type 16 and TP53 mutation in oral cancer: Matched analysis of IARC multicenter study. Cancer Res. 2004;64:468–71. doi: 10.1158/0008-5472.can-03-3284. [DOI] [PubMed] [Google Scholar]
- 36.Lynch GA. Cancer of the lip. Ulster Med J. 1967;36:44–50. [PMC free article] [PubMed] [Google Scholar]
- 37.Dumont P, Leu JI, Della Pietra AC, 3rd, George DL, Murphy M. The codon 72 polymorphic variants of p53 have markedly different apoptotic potential. Nat Genet. 2003;33:357–65. doi: 10.1038/ng1093. [DOI] [PubMed] [Google Scholar]
- 38.Pim D, Banks L. p53 polymorphic variants at codon 72 exert different effects on cell cycle progression. Int J Cancer. 2004;108:196–9. doi: 10.1002/ijc.11548. [DOI] [PubMed] [Google Scholar]


