Abstract
Angiogenin (ANG) undergoes nuclear translocation and promotes ribosomal RNA (rRNA) transcription thereby enhancing cell growth and proliferation. However, the mode of action of ANG in stimulating rRNA transcription is unclear. Here, we show that ANG enhances the formation of RNA polymerase I (Pol I) pre-initiation complex at the ribosomal DNA (rDNA) promoter. ANG binds at the upstream control element (UCE) of the promoter and enhances promoter occupancy of RNA Pol I as well as the selectivity factor SL1 components TAFI48 and TAFI110. We also show that ANG increases the number of actively transcribing rDNA by epigenetic activation through promoter methylation and histone modification. ANG binds to histone H3, inhibits H3K9 methylation, and activates H3K4 methylation as well as H4 acetylation at the rDNA promoter. These data suggest that one of the mechanisms by which ANG stimulates rRNA transcription is through an epigenetic activation of rDNA promoter.
Keywords: Angiogenin, rRNA transcription, RNA Pol I pre-initiation complex, DNA methylation, histone modification
Introduction
Angiogenin (ANG), also known as ribonuclease 5 (RNASE5), is a member of the vertebrate-specific, secreted ribonuclease superfamily. ANG was originally isolated from the conditioned medium of a tumor cell line as a tumor angiogenic factor (Fett et al., 1985), but since then has been shown to have a more widespread function and plays an important role in a number of physiological and pathological conditions (Li and Hu, 2010). It promotes cancer progression by stimulating both tumor angiogenesis and cancer cell proliferation (Yoshioka et al., 2006). It also possesses neurogenic and neuroprotective activity (Li et al., 2013b) and its deficiency has been related to pathogenesis underlying neurodegenerateive diseases including amyotrophic lateral sclerosis (ALS) (Greenway et al., 2006) and Parkinson’s disease (PD) (van Es et al., 2011). Studies in various animal models have shown that ANG inhibitors and activators are effective therapeutics for cancer (Ibaragi et al., 2009a; Ibaragi et al., 2009b) and for ALS (Kieran et al., 2008), respectively. Mechanistic studies indicate that nuclear localization is necessary for ANG to exert its biological activities (Moroianu and Riordan, 1994b; Tsuji et al., 2005; Wu et al., 2007). ANG undergoes nuclear translocation, accumulates in nucleolus (Moroianu and Riordan, 1994b), and stimulates ribosomal RNA (rRNA) transcription (Xu et al., 2002). Inhibition of nuclear translocation abolishes ANG-mediated angiogenesis (Hu, 1998; Moroianu and Riordan, 1994a). In endothelial cells, ANG-mediated rRNA transcription is essential for angiogenesis induced by other angiogenic factors including acidic fibroblast growth factor (aFGF), basic fibroblast growth factor (bFGF), and vascular endothelial growth factor (VEGF) (Kishimoto et al., 2005). In cancer cells, ANG undergoes constitutive nuclear translocation resulting in a surplus of rRNA, which contributes to the uncontrolled growth and proliferation of cancer cells (Hu et al., 2000; Tsuji et al., 2005). In motor neurons, ANG variants deficient in nuclear translocation fail to protect stress-induced degeneration (Subramanian et al., 2008).
Nuclear ANG has been show to bind to the CT-rich repeats at the promoter and intergenic sequence (IGS) region of ribosomal DNA (rDNA) (Xu et al., 2003). An ANG-binding element (ABE) has been identified from the IGS of rDNA and shown to have ANG-dependent promoter activity in a luciferase reporter assay. However, it is unclear where exactly ANG binds on rDNA in vivo and how ANG promotes rRNA transcription.
rRNA synthesis is complex process and is regulated at multiple levels, including alteration of the epigenetic status of rDNA, assembly of transcription initiation and elongation complex, and pre-rRNA processing (McStay and Grummt, 2008). Each human cell contains ~400 copies of the rRNA gene arranged in head-to-tail tandem repeats on 5 chromosomes (Long and Dawid, 1980). These rDNA repeats exist in either an active or a silent state, depending on its epigenetic influences. The ratio between the active and silent rDNA copies affects the overall rRNA transcription level (Birch and Zomerdijk, 2008). Active rDNA is associated with RNA Pol I and a number of rRNA-specific trans-acting regulatory proteins such as TATA-binding protein-TAFI complex SL1 and upstream control element (UCE) binding factor (UBF), and is characterized by DNA hypomethylation, histone H4 acetylation, and H3K4 methylation. By contrast, silent rDNA correlates with DNA hypermethylation, histone H4 hypoacetylation, and H3K9 and H4K20 methylation (Birch and Zomerdijk, 2008; Grummt, 2007; Preuss and Pikaard, 2007). Several proteins have been identified as rDNA epigenetic regulators. For example, the NoRC complex, which consists of SWI/SNF proteins, histone deacetylases, and TTF-1-interacting protein 5, induces histone H4 deacetylation and histone H3K9 methylation, thereby silences rRNA transcription (Santoro and Grummt, 2005).
In order to know how ANG regulates rRNA transcription, we examined the interaction between ANG and rDNA and the effect of ANG on epigenetic status of rDNA. We found that ANG binds at both ABE and the UCE region of rDNA promoter. We also found that knockdown of ANG enhances rDNA promoter methylation and decreases the number of active rDNA copies. ANG knockdown also resulted in a decrease in histone H4 acethylation and H3K4 methylation, and an increase in H3K9 methylation. Moreover, we found that ANG interacts with H3 and is able to inhibit H3 methylation directly. These results suggest that ANG promotes rRNA transcription through epigenetic modification of the rDNA promoter.
Materials and methods
Preparation of recombinant proteins and iodination of ANG
Recombinant histone H1, H2a, H2b, H3, and H4 was prepared as GST fusion proteins in the pGEX mammalian expression system and purified with GSH-Sephadex (GE Healthcare Life Sciences) affinity chromatography. Thrombin was used to generate histone proteins without the GST tag. The N-terminal peptide (1-40) of H3 was expressed in the pGEX system and purified as GST-fusion protein. K9A mutation of the H3 N-terminal peptide was prepared by site-directed mutagenesis. Recombinant ANG was prepared from an E. coli expression system and purified by HPLC (Shapiro et al., 1988). 125I-ANG was prepared using Iodo-beads (Pierce). Na125I (0.5 mCi, 17.4 Ci/g, PerkinElmer NEN) was mixed with 1 Iodo-bead and incubated at room temperature (RT) in PBS for 15 min. ANG (50 μg) was then added and incubated for another 15 min at RT. Iodinated ANG was separated from free iodine by a PD-10 desalting column. Radioactivity was determined by a gamma counter. The specificity of 125I-ANG was 2.1 μCi/μg.
Cell culture and shRNA induction
HeLa cells were cultured in DMEM supplemented with 10% FBS, 100 U/ml penicillin, and 100 μg/ml streptomycin. GIPZ shRNAmirs specific to human ANG (ANGi-1 and ANGi-2) and the non-silencing GIPZ shRNAmir control were purchased from Thermo Scientific Open Biosystems. The sequences of ANGi-1 and ANGi-2 are 5′-CGCATCAAGGCCATCTGTGAAA-3′ and 5′-CAACGTTGTTGTTGCTTGTGAA-3′, respectively. Lentiviral particles were packaged in 293T/17 cells with the generation II packaging plasmids. HeLa cells were infected with lentiviral particles for 24 h in the presence of 8 μg/ml polybrene (Millipore). The medium was replaced with the complete growth medium and incubated for 24 h and then cells were selected for 4 days in the presence of 1μg/ml puromycin. Stable ANG knockdown cells were established and used for all experiments.
ANG affinity chromatography and amino acid sequence analyses
The nuclear fraction of HeLa cells were prepared as described previously (Xu et al., 2003). Nuclear proteins were extracted by homogenizing in 100 mM Tris, pH 7.4, containing 2% Triton X-100, 0.1% SDS, 0.5% sodium deoxycholate, and protease inhibitor cocktail. The nuclear extracts were diluted in 4 volume of 25 mM Tris, pH 7.4, and passed through an RNaseA-Sepharose and a DNase I-Sepharose column (1 ml Sepharose coupled with 2 mg) successively. The flow-through fraction was applied to an ANG-Sepharose column (1 ml Sepharose coupled with 2 mg ANG), washed with 40 mM Tris, pH 7.5, and eluted with a 0 – 1 M NaCl gradient in 40 mM Tris, pH 7.5. Fractions of 0.5 ml were collected and analyzed by SDS-PAGE. Commasie blue-stained bands were excised, homogenized, and eluted in water by ultrafiltration through a XM300 membrane (Millipore). Protein samples were dried onto isothiocyanato sequencing membranes (Millipore) according to the manufacturer’s protocol, rinsed briefly with water (2x) and methanol (2x), and installed in a sequencer cartridge (Millipore). A Millipore ProSequencer, using the Cov-100 program, was used for sequencing. All the materials from each cycle was injected for HPLC identification of the phenylthiohydantoin amino acids (Strydom and Kandror, 1994).
In vitro methylation of Histone H3
The S100 fraction from HeLa cells was prepared according to the published method (Graveley et al., 1996). Recombinant histone proteins (1 μg) was mixed with 100 μl of S100 fraction and 0.125 μCi 14C-labeled S-Adenosyl-L-methionine (methyl-14C-AdoMet) (50 μCi/μmol, PerkinElmer NEN) at RT for 30 min. The reaction was stopped by adding an equal volume of 2X SDS-PAGE sample buffer. Methylated histone was separated from free methyl-14C-AdoMet by SDS-PAGE and visualized by autoradiography.
Antibodies
Rabbit polyclonal antibodies against ANG were prepared according to a standard protocol and affinity purified. Antibodies to RNA polymerase I (RPA194), TAFI110, TAFI48, UBF, β-actin and α-tubulin were purchased from Santa Cruz. Antibodies against histone H1, H3, H3K4 di-methylation (H3K4me2), H3K9 di-methylation (H3K9me2), H4 acetylation (H4ac), and H3K9/K18 acetylation (H3K9/18ac) were purchased from Millipore.
RNA isolation, qPCR and Northern blot analysis
Total RNA was extracted using the TRIZOL reagent (Invitrogen) and reverse transcribed (0.5 μg) using the high capacity cDNA reverse transcription kit (Applied Biosystems). qPCR analysis was performed in 10 μL reactions using the ABI7900HT and SYBR green PCR master mix (Applied Biosystems). The sequences of primers for ANG are: forward, 5′-AGAAGCGGGTGAGAAACAA-3′; reverse, 5′-CTTCCAACACAGGCTCCTCG-3′. The sequences of the primers for β-actin are: forward, 5′-AGCGAGCATCCCCCAAAGTT-3′; reverse, 5′-GGGCACGAAGGCTCATCATT-3′. The relative ANG mRNA level was determined using the 2−ΔΔCt method with β-actin as the internal control (Livak and Schmittgen, 2001). For Northern blot analysis, samples of RNA (10 μg) were separated by electrophoresis through a 1% agarose/2% formaldehyde gel, and transferred overnight in 20X SSC to a positively charged nylon membrane (Ambion). Digoxigenin (DIG)-labeled DNA probes were used for detection. The probe for the 47S rRNA precursor is 5′-GGTCGCCAGAGGACAGCGTGTCAG-3′ that hybridizes to the first 25 nucleotides of the 47S pre-rRNA (Kishimoto et al., 2005). The probe for β-actin is 5′-AGGGATAGCACAGCCTGGATAGCAAC-3′ that hybridizes to nucleotides 484-509 of β-actin mRNA. DIG Northern Starter Kit from Roche Applied Science was used to visualize the signals.
Chromatin immunoprecipitation (ChIP)
Cells were cross-linked with 1% formaldehyde for 10 min at 37 °C. Cross-linking reaction was quenched with 0.125 M glycine. The cells were collected and re-suspended in 50 mM Tris, pH 8.1, containing 1% SDS, 10 mM EDTA, and protease inhibitor cocktail. The lysates were sonicated to yield DNA fragments of 200 to 1,000 bp (examined by electrophoresis), cleared by centrifugation, and diluted with 10 volume of ChIP dilution buffer containing 16.7 mM Tris, pH 8.1, 0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM NaCl, and protease inhibitor cocktail. The diluted lysates were mixed with salmon sperm DNA/protein G/A agarose at 4 °C for 1 h. For each IP reaction, the lysates were incubated with 5 μg of antibody overnight at 4°C with rotation. The immuno-complexes were collected with protein G agarose, eluted and de-crosslinked at 65°C. After RNase A and proteinase digestion, immunoprecipitated DNA was extracted and used for PCR or qPCR analyses with the primers designed using online Primer-Blast software (http://www.ncbi.nlm.nih.gov) or as described (Grandori et al., 2005). The sequences of the ChIP-qPCR primers are as follows. UCE: forward, 5′-TTGACCAGAGGGACCCCGGG-3′ corresponding to position 42,818 to 42,837 of rDNA sequence; reverse, 5′-GTCACCGACCACGCCGCC-3′ corresponding to position 42,912 to 42,929. CORE: forward, 5′-CGGGGGAGGTATATCTTTCG-3′ (position 42,945 to 42,964); reverse, 5′-GTCACCGTGAGGCCAGAG-3′ (position 64-81). Promoter region (including both UCE and CORE): forward, 5′-TTGACCAGAGGGACCCCGGG-3′ (position 42,818 to 42,837); reverse, 5′-CCAACCTCTCCGACGACA-3′ (position 26-43). Coding region (within 5.8S rRNA): forward, 75′-AGTCGGGTTGCTTGGGAATGC-3′ (position 8,204 to 8,224); reverse, 5′-CCCTTACGGTACTTGTTGACT-3′ (position 8,280-8,300). IGS: forward 5′-GTTGACGTACAGGGTGGACTG-3′ (position 18,155-18,175); reverse, 5′-GGAAGTTGTCTTCACGCCTGA-3′ (position 18,260-18,280). ABE: forward, 5′-CTACTGGGCTAGGGCCTTCT-3′ (position 22,058-22,077); reverse, 5′-CAGACAGGGAGGGAGAGAGA-3′ (position 22,282-22,301).
DNA methylation analysis
DNA samples were prepared as described above in the ChIP assay and were digested with HpaII (New England Biolabs). qPCR was carried out using the rDNA promoter region primers described above. The relative resistance to HpaII digestion was calculated after normalization to mock-digested DNA (total rDNA) (Yuan et al., 2007).
Statistical analysis
Data are presented as means ± SD. The significance between experimental groups was determined by two-tailed Student’s t-test and expressed as * and ** representing a P value of <0.05 and <0.01, respectively.
Results
ANG binds to the promoter and ABE of rDNA in vivo
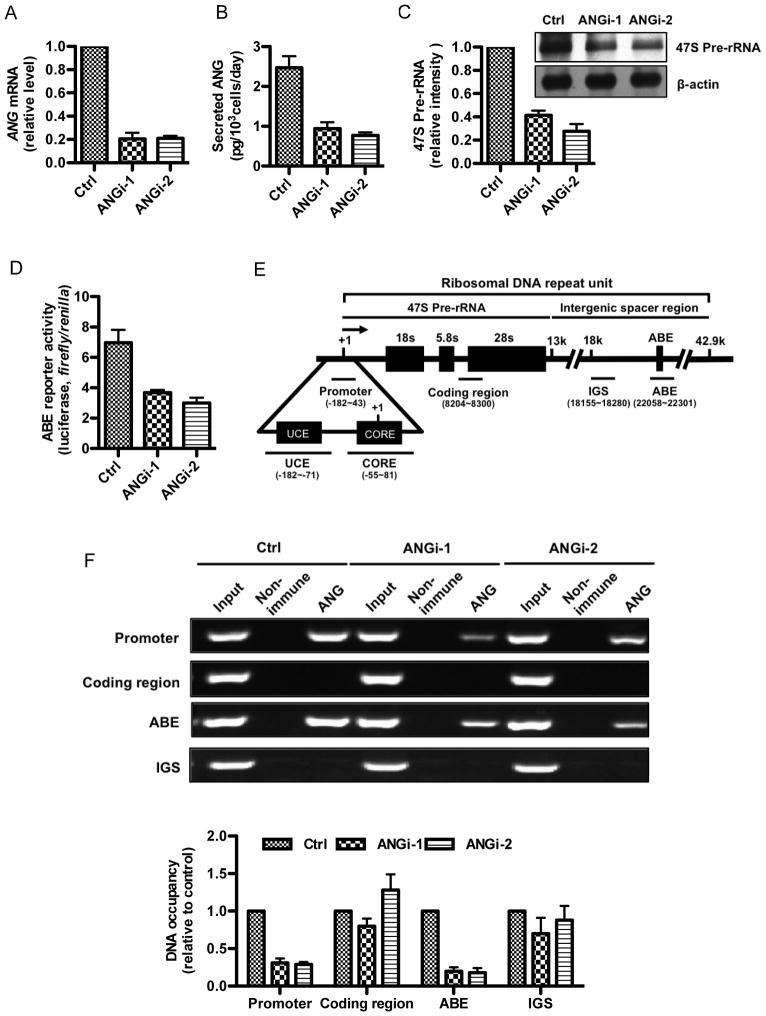
ChIP experiments were performed to characterize the interaction between ANG and rDNA in control and in ANG knockdown cells. Two ANG-specific shRNA constructs were used, which knocked down ANG expression by 80 and 79% at the mRNA level (Fig. 1A), and 62 and 69% at the protein level (Fig. 1B), respectively. Knockdown of ANG decreased the steady-state level of 47S rRNA precursor (Fig. 1C) by 59 and 73%, respectively, confirming the role of ANG in rRNA transcription. Consistently, the reporter activity of ABE was also decreased by 47 and 57%, respectively, in the two ANG knockdown cell lines (Fig. 1D). These data confirmed that down-regulation of ANG expression in HeLa cells is accompanied by a decrease in ANG-dependent transcription activities. These two ANG knockdown cell lines were therefore used for characterization of the effect of ANG on rRNA transcription.
Fig. 1.
ANG binds at the promoter and ABE region of rDNA. A: ANG mRNA levels in control and shRNA knockdown HeLa cells determined by qRT-PCR. B: Secreted ANG protein levels in control and shRNA knockdown HeLa cells determined by ELISA. C: 47S pre-rRNA level is decreased in ANG knockdown cells. Top panel, Northern blots gels detected with DIG-labeled probes specific to the 47S pre-rRNA and β-actin mRNA. Bottom panel, quantification of the 47S pre-rRNA relative to β-actin mRNA by ImageJ analysis. D: ABE reporter activity is decreased in ANG knockdown cells. pGL3E-ABE firefly-luciferase and pRL-TK Renilla luciferase constructs were co-transfected and cultured for 24 hours. Firefly and Renilla luciferase activities were determined by the Dual-Luciferase Reporter Assay System (Promega). E. Schematic representation of a single human rDNA repeat. Within each of the rDNA repeats (44 kb) there is a transcription unit (13.3 kb) for 47S rRNA transcript and a non-transcribed spacer (30.7 kb) where all the known transcription regulatory elements such as UCE, CORE, the enhancers, the spacer promoters and the proximal transcription termination sequence, are located (Bell et al., 1988). Because rDNAs are arranged in tandem repeats, the downstream of one coding region is an upstream of another coding region. The positions of the primers used in ChIP-qPCR were shown in parentheses. The black lines above the primer sets indicate the positions of the PCR amplicons. F: ChIP analysis of ANG binding to rDNA. Crosslinked chromatin was prepared and precipitated with a non-immune or an ANG-specific IgG, and analyzed by conventional PCR (top panels) or qPCR (bottom panel) with primer sets shown in (E). Data shown are representative ChIP gels (top panels) or means ± SD of three independent experiments in ChIP-qPCR (bottom panel).
ChIP was carried out in control and in ANG knockdown cells to examine the binding of ANG to rRNA gene. Four sites were examined (Fig. 1E). Besides the ABE site where ANG is anticipated to bind based on the previous in vitro results (Xu et al., 2003), we also examined the promoter region consisting of the UCE and the core promoter (CORE) where RNA Pol I pre-initiation complex is assembled, the coding region, and the IGS (Fig. 1E). Fig. 1F shows that binding of ANG to the promoter and to ABE was detected (Fig. 1F, panels 1 and 3 from the top, lane 3). As expected, no binding of ANG was detected in the coding region and in the IGS (Fig. 1F, panels 2 and 4 from the top, lane 3). Significantly, ChIP-qPCR analysis showed that ANG occupancy at the promoter decreased by 69 and 71% by, respectively, in the two ANG knockdown cell lines (Fig. 1F, bottom panel). Similarly, ANG knockdown resulted in an 80 and 71% decrease, respectively, in the binding of ANG to ABE (Fig. 1F, bottom panel). These results indicate that the binding of ANG to the promoter and to ABE of rDNA is specific.
ANG level is correlated with the formation of RNA Pol I pre-initiation complex
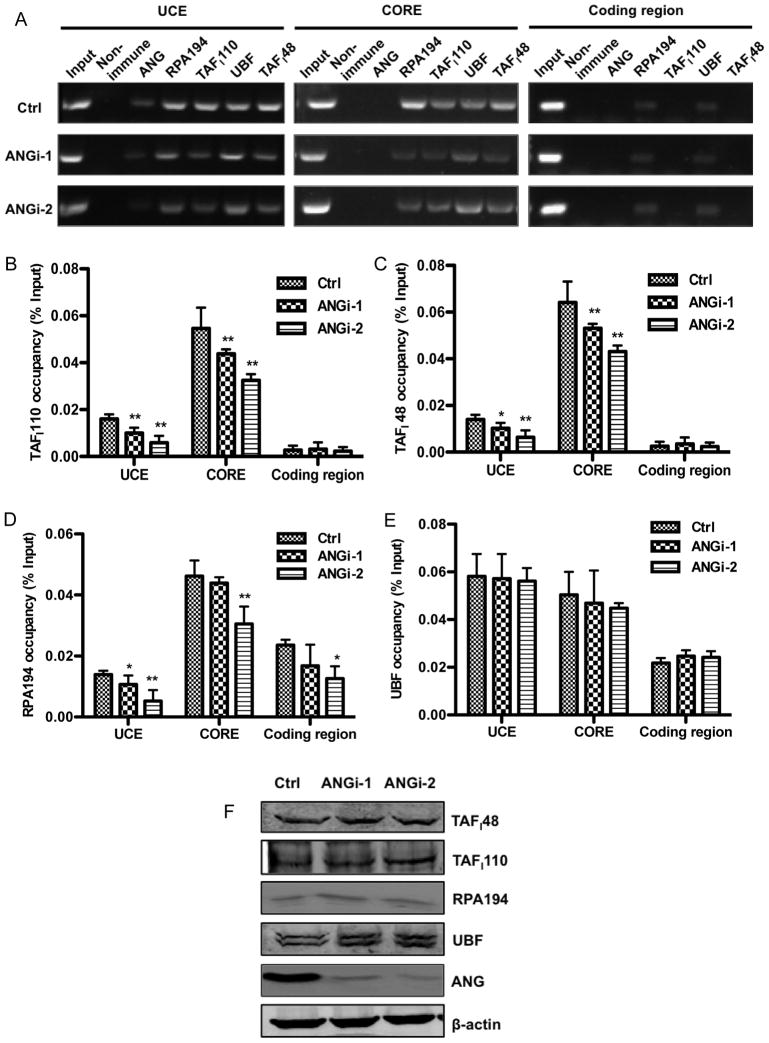
Human RNA Pol I initiates transcription at the rDNA promoter (−182 to 43), which contains the CORE (−45 to +18) and UCE (−156 to −107), and requires at least two auxiliary factors, the UBF and the promoter selectivity factor SL1 (Drygin et al., 2010). The rate of rRNA transcription is primarily determined by the rate of RNA Pol I pre-initiation complexes formation (Panov et al., 2001), which is reflected by the abundance of SL1 at the promoter region. SL1 is the complex of TBP-TAFs, an essential core promoter-binding factor that nucleates productive Pol I preinitiation complex formation. Therefore, promoter occupancy by SL1 directly correlates with rRNA transcription rate. To examine whether ANG stimulates rRNA transcription by enhancing pre-initiation complex formation, we used ChIP-qPCR to examine the effect of ANG on the binding of Pol I subunit RPA194, SL1 subunit TAFI48 and TAFI110, and UBF to the promoter and coding region of rDNA (Fig. 2). Conventional ChIP confirmed that RPA194 and UBF bind to UCE and CORE, as well as the coding region (Fig. 2A). However, TAFI110 and TAFI48 bind only to the UCE and to the CORE (Fig. 2A). These results are consistent with previous findings that TAFI110 and TAFI48, as the essential components of the pre-initiation complex, are located only in the promoter region, and that UBF and RPA194 are also in the elongation complex and are located across the entire rDNA gene repeat unit (O’Sullivan et al., 2002). Interestingly, ANG was only detected in the UCE part but not in the CORE part of the promoter, suggesting that ANG may play a more prominent role in the formation of pre-initiation complex than in transcription elongation.
Fig. 2.
ANG Knockdown decreases promoter occupancy of RNA Pol I pre-initiation transcription machinery on rDNA promoter. A: ChIP analyses of RNA Pol I pre-initiation complex formation. The abundance of RNA Pol I pre-initiation complex components (RPA194, TAFI110, TAFI48 and UBF) at the UCE, CORE and coding region of rDNA was determined by conventional ChIP experiments. B-E: Quantitative analyses of TAFI110 (B), TAFI48 (C), RPA194 (D), and UBF (E) at the UCE, CORE and coding region of rDNA by ChIP-qPCR. Data shown are means ± SD of three independent experiments, each normalized to the value of its own input control. * P < 0.05; ** P < 0.01). F: Cellular protein levels of RNA Pol I pre-initiation complex components (TAFI48, TAFI110, RPA194, and UBF). β-Actin was used as a loading control. ANG was included to ensure that its expression was successfully knocked down.
Significantly, binding of TAFI110 and TAFI48 to UCE and CORE was decreased in ANG knockdown cells. Occupancy of TAFI110 at UCE was 0.01 ± 0.002 and 0.006 ± 0.003 % of the input control in the two ANG knockdown cell lines, respectively, representing a 38 and 63% decrease from that in the control cells (0.016 ± 0.002) (Fig. 2B). Similarly, binding of TAFI110 to the CORE was also decreased by 20 and 40%, respectively, in the two knockdown cell lines (Fig. 2B). Consistently, binding of TAFI48 to UCE and to the CORE was also decreased to a similar extent in ANG knockdown cells (Fig. 2C). Binding to the coding region, which was not a significantly amount, was not altered in ANG knockdown cells (Fig. 2B and 2C). Binding of RPA194 to the UCE was significantly decreased in both ANG knockdown cells. However, the decrease in RPA194 binding to the CORE and to the coding region was significant only in ANGi-2 but not in ANGi-1 knockdown cell line (Fig. 2D). This is probably due to a more robust knockdown efficiency in ANGi-2 cells (Fig. 1B). Consistently, suppression of rRNA transcription was also more significant in ANGi-2 cells (Fig. 1C) where both initiation and elongation are affected. As expected, UBF binding was not affected by ANG level in all three sites (Fig. 2E). The protein levels of RPA194, TAFI48, TAFI110 and UBF were not changed in ANG knockdown cells (Fig. 2F), indicating that the reduced formation of pre-initiation complex in ANG knockdown cells is not a result of decreased protein level of the components.
ANG knockdown decreases the number of active rDNA repeats
A unique feature of the RNA Pol I system is that only a subset of the ~400 rDNA copies in eukaryotic cells are actively transcribing. This results in the coexistence of transcriptionally active and silent rDAN copies. We therefore examined the effect of ANG on the ratio of active verses silent rDNA by examining the methylation status of the rDNA in control and ANG knockdown cells. Genomic DNA was digested with methylation-sensitive restriction enzymes HpaII followed by PCR amplification at the UCE, CORE and coding regions of rDNA. Methylated chromatin are resistant to HpaII digestion and is a marker for silent gene (Santoro and Grummt, 2001). Therefore, HpaII-sensitive and -resistant DNA represents active and silent genes, respectively. Fig. 3 shows that HpaII sensitivity decreased significantly in ANG knockdown cells, indicating that ANG level is positively correlated with the copy numbers of active rDNA. In the control cells, 58 ± 4% of rDNA at the UCE region were active, which was decreased to 49 ± 8% and 45 ± 4%, respectively, in the two ANG knockdown cell lines (Fig. 3). A similar decrease in active chromatin was observed at the CORE but not at the coding region. These results suggest that one of the mechanisms by which ANG enhances RNA Pol I pre-initiation complex formation is via modulation of DNA methylation at rDNA promoter.
Fig. 3.
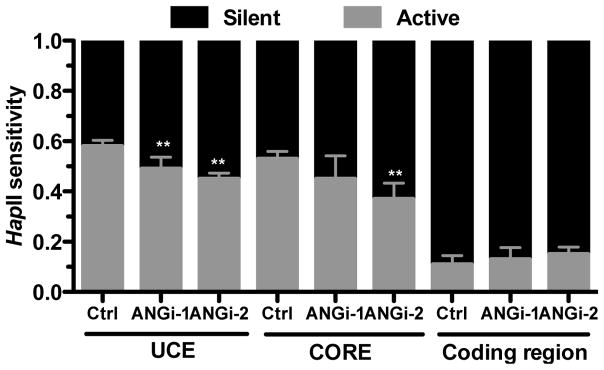
ANG knockdown decreases the ratio of active to silent rDNA genes. ChIP input DNA from the control and ANG knockdown cells were digested with HpaII. qPCR reactions were carried out on both mock- and HpaII-digested DNA. The value from HpaII-digested DNA represents the level of silent chromatin (HpaII resistant, black bar). The value from mock-digested DNA represents the total input DNA. The difference between mock- and HpaII-digested DNA represents the level of active chromatinn (HpaII sensitive, light bar).
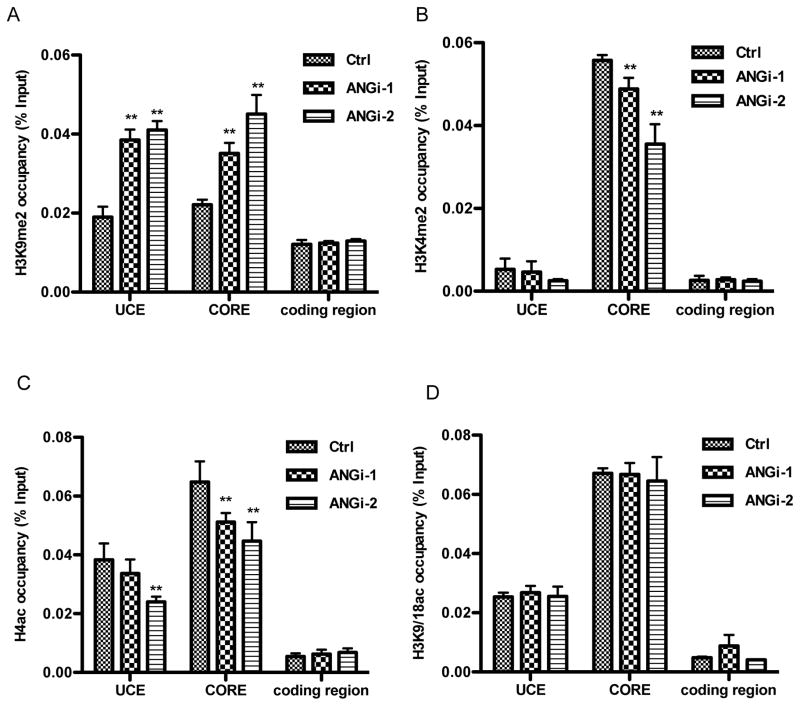
ANG alters histone modification at rDNA promoter
Transcriptional regulation of rRNA has also been reported to be regulated by epigenetic modification of rDNA through histone modification (Grummt and Langst, 2013). We therefore examined the effect of ANG on histone modification at the UCE, CORE, and coding region of rDNA. ChIP-qPCR analyses were performed to determine the status of histone modifications related to gene activation including H4ac, H3K4me2, and (H3K9/18ac) (Gao et al., 2010; Koch et al., 2007). ANG knockdown resulted in a significant increase in H3K9me2, a marker of transcription repression (Rosenfeld et al., 2009), at both UCE and CORE in ANG knockdown cells (Fig. 4A), indicating that ANG deficiency resulted in a more silent rDNA. Consistently, we observed a significant decrease in both H3K4me2 (Fig. 4B) and H4ac (Fig. 4C), markers for transcription activation, at the CORE and UCE (ANGi-2 cells only) in ANG knockdown cells. The status of H3K9/18ac, another active chromatin marker, was not altered in ANG knockdown cells (Fig. 3D). No difference in histone modification was observed in the coding region of rDNA in ANG knockdown cells, consistent with the finding that ANG promotes rRNA transcription mainly by enhancing the formation of pre-initiation complex rather than by affecting elongation (Fig. 2).
Fig. 4.
ANG alters histone modification in rDNA promoter. Control and ANG knockdown cells were subjected to ChIP-qPCR analyses for the abundance of H3K9me2 (A), H3K4me2 (B), H4ac (C) and H3K9/K18ac (D) at UCE, CORE and coding region of rDNA. Data shown are means ± SD of three independent experiments, each normalized to its own input control. * P < 0.05; ** P < 0.01.
ANG interacts with histone H3 and inhibits H3 methylation
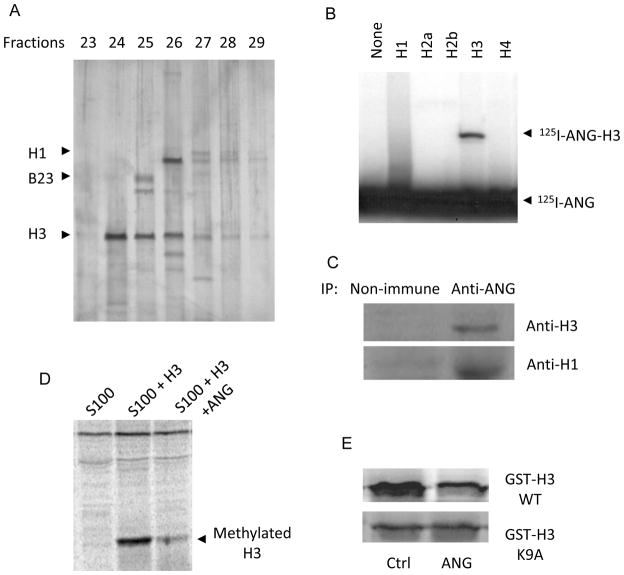
To gain more mechanistic insight of epigenetic modification of rDNA by ANG, we attempted to identify ANG-binding proteins in the nucleus. We isolated HeLa cell nuclei by subcellular fractionation, extracted nuclear proteins and subjected them to ANG affinity chromatography. Our previous experience with ANG affinity chromatography on plasma membrane fractions (Hu et al., 1991; Hu et al., 1997) indicated that non-specific charge interactions with ANG (pI>9.5) will result in too many proteins binding to the ANG column, which may mask specific interactions. Therefore, we first passed the nuclear extract through a ribonuclease A (RNase A)-Sepharose column to remove nonspecific acidic proteins. RNase A (pI>9.5) is an ANG analogue with 56% sequence similarity but does not bind to ABE, is not angiogenic and unable to stimulate rRNA synthesis. ANG is also known to bind to actin (Hu et al., 1993). Therefore, we used a DNase I-Sepharose column to remove any actins and actin-like molecules because DNase I binds to actin tightly and specifically (Lazarides and Lindberg, 1974). The post-RNase A, post-DNase I material was then applied to the ANG-Sepharose affinity column. Bound material was eluted with a linear gradient of 0–1 M NaCl and the fractions were subjected to SDS-PAGE. Fig. 5A shows that approximately 7 bands were visible. The identities of the three most prominent bands were determined by amino acid sequencing analyses to be histone H3 (fraction 24), B23 (fraction 25, top band), and histone H1 (fraction 26, top band).
Fig. 5.
ANG binds to H3 and inhibits H3K9 methylation. A: Nuclear proteins were subject to ANG-Sepharose affinity chromatography and the fractions were analyzed by SDS-PAGE. Identities of 3 bands were determined by N-terminal amino acid sequencing. B: In vitro binding of ANG to Histones. 125I-ANG (0.5 μci) was incubated with histones (1 μg) in 50 μl of PBS and crosslinked with 0.5 mM EDC. The crosslinked products were separated by SDS-PAGE and visualized by autoradiography. C: In vivo binding of ANG to histone H3 examined by Co-IP. Nuclear proteins were precipitated by ANG mAb 26-2F or an isotype-matched non-immune IgG, followed by immunoblotting detection with H3 and H1 antibodies. D: Inhibition of histone H3 methylation by ANG. Recombinant H3 (1 μg) was mixed with 14C-AdoMet (0.5 μci) and incubated with HeLa S100 fraction (100 μl) in the absence or presence of ANG (1 μg) for 1 hour. The product was separated on SDS-PAGE and visualized by fluorography. E: Inhibition of GSH-H3 tag by ANG. The fusion proteins of GST and the N-terminal 40 residues of H3 with or without K9→A mutation were incubated with 14C-AdoMet and S100 fraction in the absence or presence of ANG. The products were separated by SDS-PAGE and visualized by autoradiography.
Because histone H3 modification is altered by ANG in rDNA promoter (Fig. 4), we examined whether ANG is able to bind to histones directly by chemical crosslinking experiments. 125I-ANG was mixed with recombinant histone proteins and the complexes were covalently crosslinked by 1-Ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC). The crosslinked complexes were separated from free proteins by SDS-PAGE and visualized by autoradiography. Fig. 5B shows that ANG forms a complex with H3 but not with H2a, H2b or H4. A smeared area was visible between ANG and H1 but no clear bands could be identified.
We therefore performed a co-IP experiment to examine in vivo binding of ANG to H1 and H3. Nuclear proteins from HeLa cells were precipitated by ANG monoclonal antibody 26-2F and blotted with anti-H1 and anti-H3 IgG. Fig. 5C shows that both H1 and H3 could be precipitated by ANG antibody, indicating that ANG is indeed bond to H1 and H3 in vivo. We next examined the effect of ANG on methylation of H3 catalyzed by HeLa cell extracts (S100 fraction). Fig. 5D shows that ANG inhibits H3 methylation. Therefore, one possible mechanism by which ANG modulates the chromatin status of rDNA is through direct binding to H3 and inhibits its methylation.
Results presented in Fig. 4 have shown that ANG inhibits H3K9 methylation at the rDNA promoter in vivo (Fig. 4A). To confirm that these in vivo findings can be recapitulated by a more defined in vitro system, we prepared GST-H3 tags containing the N-terminal 40 residues with and without K9A mutation. In vitro methylation experiment shows that ANG no longer inhibited the methylation of GST-H3 with K to A mutation at position 9, indicating that K9 is the residue whose methylation is inhibited directly by ANG binding.
Discussion
ChIP analyses demonstrated that ANG binds at the ABE and the UCE region of rDNA promoter in vivo, confirming previous findings (Li et al., 2013a; Xu et al., 2003). Binding of ANG to ABE is probably a direct protein-DNA interaction as ANG is known to be able to bind to ABE with an affinity of 0.15 μM in an in vitro equilibrium dialysis assay (Xu et al., 2003). However, binding of ANG to the UCE region is probably not a direct interaction to the DNA sequence but rather mediated by some co-factors. UCE has not been shown to be able to bind to ANG either by ANG affinity chromatography (Xu et al., 2003) and or by electrophoretic mobility shift assay (data not shown). At this time, the co-factors that mediate the interaction of ANG to the UCE region on rDNA promoter are unknown. But it is clear that a reduction in binding of ANG to the UCE is correlated with a decrease in promoter occupancy of SL1 (TAFI110 and TAFI48) and RNA Pol I at the rDNA promoter, which slows down the formation of RNA Pol I pre-initiation complex and suppresses rRNA transcription. These results suggest that stimulation of RNA Pol I pre-initiation complex formation could be one of the mechanisms by which ANG promotes rRNA transcription.
It is interesting that ANG enhances rDNA promoter occupancy of RNA Pol I, TAFI110 and TAFI48 but not UBF. UBF is a multifunctional transcription factor with one of the functions to be maintenance of the euchromatic state of rDNA (Sanij and Hannan, 2009). The finding that UBF binding to rDNA is not influenced by ANG level suggests that ANG enhances pre-initiation complex formation in an UBF-independent manner. One possible mechanism could be through alteration of chromatin activation status of the rDNA loci. ANG knockdown reduced the numbers of active rDNA copies, suggesting a chromatin remodeling activity of ANG. Consistently, we found that ANG level is positively correlated with euchromatin histone markers such H4 acetylation and H3K4 methylation, and inversely correlated with heterochromatin histone markers such as H3K9 methylation. Based on the findings that ANG is able to bind H3 and inhibit H3 methylation at K9, it is probable that a direct binding of ANG to H3 is one of the causes for inhibition of H3K9 methylation thereby activating rDNA, enhancing pre-initiation complex formation, and promoting rRNA transcription.
Interplay between histone methylation and acetylation has been established (Schubeler et al., 2004). It is known that deacetylation of H3K9 is a prerequisite for H3K9 methylation and subsequent recruitment of heterochromatin protein HP1. It is interesting to note that ANG did not affect acetylation of H3K9 and H3K18 but did affect H4 acetylation. Although it is unclear at present whether increased H4 acetylation is dependent on decreased H3K9 methylation in response to ANG, the fact that both are affected by ANG strongly indicate a profound effect of ANG on chromatin structure of the rDNA loci.
The histone methytransferase involved in ANG-mediated inhibition of H3K9 methylation is unknown. Four human enzymes, SUV39H1 (Rea et al., 2000), G9a (Tachibana et al., 2002), Eu-HMT1 (Ogawa et al., 2002), and SETDB1 (Schultz et al., 2002), have been characterized to catalyze H3K9 methylation. It has been shown that SUV39H1-catalyzed H3K9 methylation plays a dominant role in pericentric heterochromatin formation and transcription silencing, whereas the other three enzymes all target the euchromatin to repress transcription. ChIP experiments are underway to determine the abundance of these enzymes at the promoter region of rDNA in ANG sufficient and deficient cells. The amount of methyltransferase located at the rDNA promoter will reveal which enzyme is inhibited by ANG and which type of the chromatin structures (heterochromatin verse euchromatin) of rDNA is likely affected by ANG through modification of H3K9 methylation. However, no matter which methytransferase is involved ANG-mediated inhibition of H3K9 methylation, the current results presents a plausible mechanism that ANG-induced inhibition of H3K9 methylation remodels the chromatin structure of rDNA, results in an increase in the number of active copies of rDNA repeats, enhances the formation of RNA Pol I pre-initiation complex, and thereby promotes rRNA transcription.
Acknowledgments
Contract grant sponsor: National Institute of Health
Contract grant numbers: R01NS065237, R01CA105241
Contract grant sponsor: National Natural Science Foundation of China
Contract grant numbers: 30400227, 31170721
Footnotes
Conflicts of interest: Nothing to declare
References
- Bell SP, Learned RM, Jantzen HM, Tjian R. Functional cooperativity between transcription factors UBF1 and SL1 mediates human ribosomal RNA synthesis. Science. 1988;241:1192–1197. doi: 10.1126/science.3413483. [DOI] [PubMed] [Google Scholar]
- Birch JL, Zomerdijk JC. Structure and function of ribosomal RNA gene chromatin. Biochem Soc Trans. 2008;36:619–624. doi: 10.1042/BST0360619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drygin D, Rice WG, Grummt I. The RNA polymerase I transcription machinery: an emerging target for the treatment of cancer. Annual review of pharmacology and toxicology. 2010;50:131–156. doi: 10.1146/annurev.pharmtox.010909.105844. [DOI] [PubMed] [Google Scholar]
- Fett JW, Strydom DJ, Lobb RR, Alderman EM, Bethune JL, Riordan JF, Vallee BL. Isolation and characterization of angiogenin, an angiogenic protein from human carcinoma cells. Biochemistry. 1985;24:5480–5486. doi: 10.1021/bi00341a030. [DOI] [PubMed] [Google Scholar]
- Gao X, Wei S, Lai K, Sheng J, Su J, Zhu J, Dong H, Hu H, Xu Z. Nucleolar follistatin promotes cancer cell survival under glucose-deprived condition through inhibiting cellular rRNA synthesis. J Biol Chem. 2010;285:36857–36864. doi: 10.1074/jbc.M110.144477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandori C, Gomez-Roman N, Felton-Edkins ZA, Ngouenet C, Galloway DA, Eisenman RN, White RJ. c-Myc binds to human ribosomal DNA and stimulates transcription of rRNA genes by RNA polymerase I. Nat Cell Biol. 2005;7:311–318. doi: 10.1038/ncb1224. [DOI] [PubMed] [Google Scholar]
- Graveley BR, Fleming ES, Gilmartin GM. Restoration of both structure and function to a defective poly(A) site by in vitro selection. J Biol Chem. 1996;271:33654–33663. doi: 10.1074/jbc.271.52.33654. [DOI] [PubMed] [Google Scholar]
- Greenway MJ, Andersen PM, Russ C, Ennis S, Cashman S, Donaghy C, Patterson V, Swingler R, Kieran D, Prehn J, Morrison KE, Green A, Acharya KR, Brown RH, Jr, Hardiman O. ANG mutations segregate with familial and ‘sporadic’ amyotrophic lateral sclerosis. Nature genetics. 2006;38:411–413. doi: 10.1038/ng1742. [DOI] [PubMed] [Google Scholar]
- Grummt I. Different epigenetic layers engage in complex crosstalk to define the epigenetic state of mammalian rRNA genes. Hum Mol Genet. 2007;16(Spec No 1):R21–27. doi: 10.1093/hmg/ddm020. [DOI] [PubMed] [Google Scholar]
- Grummt I, Langst G. Epigenetic control of RNA polymerase I transcription in mammalian cells. Biochim Biophys Acta. 2013;1829:393–404. doi: 10.1016/j.bbagrm.2012.10.004. [DOI] [PubMed] [Google Scholar]
- Hu G, Xu C, Riordan JF. Human angiogenin is rapidly translocated to the nucleus of human umbilical vein endothelial cells and binds to DNA. J Cell Biochem. 2000;76:452–462. doi: 10.1002/(sici)1097-4644(20000301)76:3<452::aid-jcb12>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- Hu GF. Neomycin inhibits angiogenin-induced angiogenesis. Proc Natl Acad Sci U S A. 1998;95:9791–9795. doi: 10.1073/pnas.95.17.9791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu GF, Chang SI, Riordan JF, Vallee BL. An angiogenin-binding protein from endothelial cells. Proc Natl Acad Sci U S A. 1991;88:2227–2231. doi: 10.1073/pnas.88.6.2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu GF, Riordan JF, Vallee BL. A putative angiogenin receptor in angiogenin-responsive human endothelial cells. Proc Natl Acad Sci U S A. 1997;94:2204–2209. doi: 10.1073/pnas.94.6.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu GF, Strydom DJ, Fett JW, Riordan JF, Vallee BL. Actin is a binding protein for angiogenin. Proc Natl Acad Sci U S A. 1993;90:1217–1221. doi: 10.1073/pnas.90.4.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibaragi S, Yoshioka N, Kishikawa H, Hu JK, Sadow PM, Li M, Hu G-f. Angiogenin-stimulated Ribosomal RNA Transcription Is Essential for Initiation and Survival of AKT-induced Prostate Intraepithelial Neoplasia. Mol Cancer Res. 2009a;7:415–424. doi: 10.1158/1541-7786.MCR-08-0137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibaragi S, Yoshioka N, Li S, Hu MG, Hirukawa S, Sadow PM, Hu G-f. Neamine inhibits prostate cancer growth by suppressing angiogenin-mediated ribosomal RNA transcription. Clin Cancer Res. 2009b;15:1981–1988. doi: 10.1158/1078-0432.CCR-08-2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieran D, Sebastia J, Greenway MJ, King MA, Connaughton D, Concannon CG, Fenner B, Hardiman O, Prehn JH. Control of motoneuron survival by angiogenin. J Neurosci. 2008;28:14056–14061. doi: 10.1523/JNEUROSCI.3399-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishimoto K, Liu S, Tsuji T, Olson KA, Hu GF. Endogenous angiogenin in endothelial cells is a general requirement for cell proliferation and angiogenesis. Oncogene. 2005;24:445–456. doi: 10.1038/sj.onc.1208223. [DOI] [PubMed] [Google Scholar]
- Koch CM, Andrews RM, Flicek P, Dillon SC, Karaoz U, Clelland GK, Wilcox S, Beare DM, Fowler JC, Couttet P, James KD, Lefebvre GC, Bruce AW, Dovey OM, Ellis PD, Dhami P, Langford CF, Weng Z, Birney E, Carter NP, Vetrie D, Dunham I. The landscape of histone modifications across 1% of the human genome in five human cell lines. Genome Res. 2007;17:691–707. doi: 10.1101/gr.5704207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarides E, Lindberg U. Actin is the naturally occurring inhibitor of deoxyribonuclease I. Proc Natl Acad Sci U S A. 1974;71:4742–4746. doi: 10.1073/pnas.71.12.4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Hu GF. Angiogenin-mediated rRNA transcription in cancer and neurodegeneration. Int J Biochem Mol Biol. 2010;1:26–35. [PMC free article] [PubMed] [Google Scholar]
- Li S, Hu MG, Sun Y, Yoshioka N, Ibaragi S, Sheng J, Sun G, Kishimoto K, Hu GF. Angiogenin mediates androgen-stimulated growth of prostate cancer cells and correlates with castration resistance. Mol Cancer Res. 2013a Jul 12; doi: 10.1158/1541-7786.MCR-13-0072. Epub aheas of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Sheng J, Hu JK, Yu W, Kishikawa H, Hu MG, Shima K, Wu D, Xu Z, Xin W, Sims KB, Landers JE, Brown RH, Jr, Hu GF. Ribonuclease 4 protects neuron degeneration by promoting angiogenesis, neurogenesis, and neuronal survival under stress. Angiogenesis. 2013b;16:387–404. doi: 10.1007/s10456-012-9322-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Long EO, Dawid IB. Repeated genes in eukaryotes. Annu Rev Biochem. 1980;49:727–764. doi: 10.1146/annurev.bi.49.070180.003455. [DOI] [PubMed] [Google Scholar]
- McStay B, Grummt I. The epigenetics of rRNA genes: from molecular to chromosome biology. Annu Rev Cell Dev Biol. 2008;24:131–157. doi: 10.1146/annurev.cellbio.24.110707.175259. [DOI] [PubMed] [Google Scholar]
- Moroianu J, Riordan JF. Identification of the nucleolar targeting signal of human angiogenin. Biochem Biophys Res Commun. 1994a;203:1765–1772. doi: 10.1006/bbrc.1994.2391. [DOI] [PubMed] [Google Scholar]
- Moroianu J, Riordan JF. Nuclear translocation of angiogenin in proliferating endothelial cells is essential to its angiogenic activity. Proc Natl Acad Sci U S A. 1994b;91:1677–1681. doi: 10.1073/pnas.91.5.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Sullivan AC, Sullivan GJ, McStay B. UBF binding in vivo is not restricted to regulatory sequences within the vertebrate ribosomal DNA repeat. Molecular and cellular biology. 2002;22:657–668. doi: 10.1128/MCB.22.2.657-668.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa H, Ishiguro K, Gaubatz S, Livingston DM, Nakatani Y. A complex with chromatin modifiers that occupies E2F- and Myc-responsive genes in G0 cells. Science. 2002;296:1132–1136. doi: 10.1126/science.1069861. [DOI] [PubMed] [Google Scholar]
- Panov KI, Friedrich JK, Zomerdijk JC. A step subsequent to preinitiation complex assembly at the ribosomal RNA gene promoter is rate limiting for human RNA polymerase I-dependent transcription. Molecular and cellular biology. 2001;21:2641–2649. doi: 10.1128/MCB.21.8.2641-2649.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss S, Pikaard CS. rRNA gene silencing and nucleolar dominance: insights into a chromosome-scale epigenetic on/off switch. Biochim Biophys Acta. 2007;1769:383–392. doi: 10.1016/j.bbaexp.2007.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea S, Eisenhaber F, O’Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, Jenuwein T. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- Rosenfeld JA, Wang Z, Schones DE, Zhao K, DeSalle R, Zhang MQ. Determination of enriched histone modifications in non-genic portions of the human genome. BMC Genomics. 2009;10:143. doi: 10.1186/1471-2164-10-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanij E, Hannan RD. The role of UBF in regulating the structure and dynamics of transcriptionally active rDNA chromatin. Epigenetics. 2009;4:374–382. doi: 10.4161/epi.4.6.9449. [DOI] [PubMed] [Google Scholar]
- Santoro R, Grummt I. Molecular mechanisms mediating methylation-dependent silencing of ribosomal gene transcription. Mol Cell. 2001;8:719–725. doi: 10.1016/s1097-2765(01)00317-3. [DOI] [PubMed] [Google Scholar]
- Santoro R, Grummt I. Epigenetic mechanism of rRNA gene silencing: temporal order of NoRC-mediated histone modification, chromatin remodeling, and DNA methylation. Molecular and cellular biology. 2005;25:2539–2546. doi: 10.1128/MCB.25.7.2539-2546.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubeler D, MacAlpine DM, Scalzo D, Wirbelauer C, Kooperberg C, van Leeuwen F, Gottschling DE, O’Neill LP, Turner BM, Delrow J, Bell SP, Groudine M. The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes & development. 2004;18:1263–1271. doi: 10.1101/gad.1198204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ., 3rd SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes & development. 2002;16:919–932. doi: 10.1101/gad.973302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro R, Harper JW, Fox EA, Jansen HW, Hein F, Uhlmann E. Expression of Met-(-1) angiogenin in Escherichia coli: conversion to the authentic less than Glu-1 protein. Analytical Biochemistry. 1988;175:450–461. doi: 10.1016/0003-2697(88)90569-6. [DOI] [PubMed] [Google Scholar]
- Strydom DJ, Kandror K. Identification of actin isoforms after in situ hydroxylamine cleavage on sequencer membranes: serum actin is a cytoplasmic isoform. Biochem Biophys Res Commun. 1994;205:195–201. doi: 10.1006/bbrc.1994.2649. [DOI] [PubMed] [Google Scholar]
- Subramanian V, Crabtree B, Acharya KR. Human angiogenin is a neuroprotective factor and amyotrophic lateral sclerosis associated angiogenin variants affect neurite extension/pathfinding and survival of motor neurons. Hum Mol Genet. 2008;17:130–149. doi: 10.1093/hmg/ddm290. [DOI] [PubMed] [Google Scholar]
- Tachibana M, Sugimoto K, Nozaki M, Ueda J, Ohta T, Ohki M, Fukuda M, Takeda N, Niida H, Kato H, Shinkai Y. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes & development. 2002;16:1779–1791. doi: 10.1101/gad.989402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji T, Sun Y, Kishimoto K, Olson KA, Liu S, Hirukawa S, Hu GF. Angiogenin is translocated to the nucleus of HeLa cells and is involved in ribosomal RNA transcription and cell proliferation. Cancer Res. 2005;65:1352–1360. doi: 10.1158/0008-5472.CAN-04-2058. [DOI] [PubMed] [Google Scholar]
- van Es MA, Schelhaas HJ, van Vught PW, Ticozzi N, Andersen PM, Groen EJ, Schulte C, Blauw HM, Koppers M, Diekstra FP, Fumoto K, LeClerc AL, Keagle P, Bloem BR, Scheffer H, van Nuenen BF, van Blitterswijk M, van Rheenen W, Wills AM, Lowe PP, Hu GF, Yu W, Kishikawa H, Wu D, Folkerth RD, Mariani C, Goldwurm S, Pezzoli G, Van Damme P, Lemmens R, Dahlberg C, Birve A, Fernandez-Santiago R, Waibel S, Klein C, Weber M, van der Kooi AJ, de Visser M, Verbaan D, van Hilten JJ, Heutink P, Hennekam EA, Cuppen E, Berg D, Brown RH, Jr, Silani V, Gasser T, Ludolph AC, Robberecht W, Ophoff RA, Veldink JH, Pasterkamp RJ, de Bakker PI, Landers JE, van de Warrenburg BP, van den Berg LH. Angiogenin variants in Parkinson disease and amyotrophic lateral sclerosis. Ann Neurol. 2011;70:964–973. doi: 10.1002/ana.22611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Yu W, Kishikawa H, Folkerth RD, Iafrate AJ, Shen Y, Xin W, Sims K, Hu GF. Angiogenin loss-of-function mutations in amyotrophic lateral sclerosis. Ann Neurol. 2007;62:609–617. doi: 10.1002/ana.21221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu ZP, Tsuji T, Riordan JF, Hu GF. The nuclear function of angiogenin in endothelial cells is related to rRNA production. Biochem Biophys Res Commun. 2002;294:287–292. doi: 10.1016/S0006-291X(02)00479-5. [DOI] [PubMed] [Google Scholar]
- Xu ZP, Tsuji T, Riordan JF, Hu GF. Identification and characterization of an angiogenin-binding DNA sequence that stimulates luciferase reporter gene expression. Biochemistry. 2003;42:121–128. doi: 10.1021/bi020465x. [DOI] [PubMed] [Google Scholar]
- Yoshioka N, Wang L, Kishimoto K, Tsuji T, Hu GF. A therapeutic target for prostate cancer based on angiogenin-stimulated angiogenesis and cancer cell proliferation. Proc Natl Acad Sci USA. 2006;103:14519–14524. doi: 10.1073/pnas.0606708103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X, Feng W, Imhof A, Grummt I, Zhou Y. Activation of RNA polymerase I transcription by cockayne syndrome group B protein and histone methyltransferase G9a. Mol Cell. 2007;27:585–595. doi: 10.1016/j.molcel.2007.06.021. [DOI] [PubMed] [Google Scholar]