Abstract
A subset of mammalian genes differ functionally between two alleles due to genomic imprinting, and seven such genes (Peg3, Usp29, APeg3, Zfp264, Zim1, Zim2, Zim3) are localized within the 500-kb genomic interval of the human and mouse genomes, constituting the Peg3 imprinted domain. This Peg3 domain shares several features with the other imprinted domains, including an evolutionarily conserved domain structure, along with transcriptional co-regulation through shared cis regulatory elements, as well as functional roles in controlling fetal growth rates and maternal-caring behaviors. The Peg3 domain also displays some unique features, including YY1-mediated regulation of transcription and imprinting; conversion and adaptation of several protein-coding members as ncRNA genes during evolution; and its close connection to human cancers through the potential tumor suppressor functions of Peg3 and Usp29. In this review, we summarize and discuss these features of the Peg3 domain.
Keywords: genomic imprinting, ncRNA, Peg3, tumor suppressor genes, YY1 transcription factor
Introduction
In mammals, a small subset of autosomal genes are functionally different between two alleles due to an epigenetic mechanism, termed genomic imprinting [1, 2]. The number of imprinted genes is estimated to be 100-200 per mammalian genome. These genes are usually clustered in specific chromosomal regions; thus, several genes from a given genomic interval are imprinted together as a group. Each imprinted domain, harboring 5 to 10 members, spans a relatively large genomic interval, ranging from a few hundred kilobases to megabases in length [3]. The genomic structure of a given domain, such as gene content, order, and distance, is well conserved between different mammalian lineages. The evolutionary constraint maintaining this domain structure is likely related to the fact that the imprinted genes within one domain are usually co-regulated through shared cis regulatory elements. These regulatory regions are termed imprinting control regions (ICRs) [4]. Several ICRs have been identified from individual domains by a series of human and mouse genetic studies in the past 20 years. Genetic and molecular studies investigating these ICRs, particularly the ICR of the H19/Igf2 domain, have been very insightful with regard to our understanding of epigenetics, as well as long-distance transcriptional control mechanisms in humans and mice [2].
Paternally expressed gene 3 (Peg3) is one of the imprinted genes that were discovered 20 years ago through subtractive pooling schemes of mRNA between paternal and maternal disomic embryos [5]. An independent study also discovered Peg3 as an important gene for muscle development; it was thus named Pw1 [6]. Shortly after these simultaneous discoveries of mouse Peg3, the human homolog was also identified as one of the zinc finger genes located in human chromosome 19q13.4 [7]. Since then, six additional imprinted genes have been identified from the 500-kb genomic interval surrounding the Peg3 locus, confirming the presence of an imprinted domain in the human and mouse genomes; it was thus named the Peg3 imprinted domain. A series of follow-up investigations have been conducted to characterize the genomic structures and regulatory mechanisms, as well as the functions of individual genes located within the Peg3 imprinted domain. In this short review, we intend to 1) summarize the information that we have learned so far regarding the Peg3 domain and 2) discuss the similarities and differences between the Peg3 domain and other imprinted domains.
Evolutionarily Conserved Genomic Structure
The 500-kb genomic interval of the Peg3 domain harbors seven imprinted genes: paternally expressed Peg3; Usp29 (ubiquitin-specific hydrolase 29); APeg3 (antisense Peg3); Zfp264 (zinc finger protein 264); and maternally expressed Zim1 (zinc finger gene, imprinted 1), Zim2, and Zim3 (Fig. 1) [8,9,10,11,12]. All of these imprinted genes, except Zim1, are conserved among individual mammalian species. Zim1, however, is found only in mammals with large litter sizes, such as rodents and canines [13]. The order, spacing, and orientation of these imprinted genes are also well conserved between different mammalian species. This is quite unusual, since the Peg3 domain itself is part of a large zinc finger gene family cluster, which is known to go through dynamic genomic changes during evolution. This is even more evident in the genomic regions immediately surrounding the Peg3 domain, which are occupied by two gene families: olfactory receptor and vomeronasal organ receptor. These two gene families show many more frequent lineage-specific expansions and deletions during mammalian evolution, resulting in quite different structures between different species [14]. The changes observed from the surrounding genomic regions are in stark contrast to the structural conservation observed in the Peg3 domain.
Fig. 1.
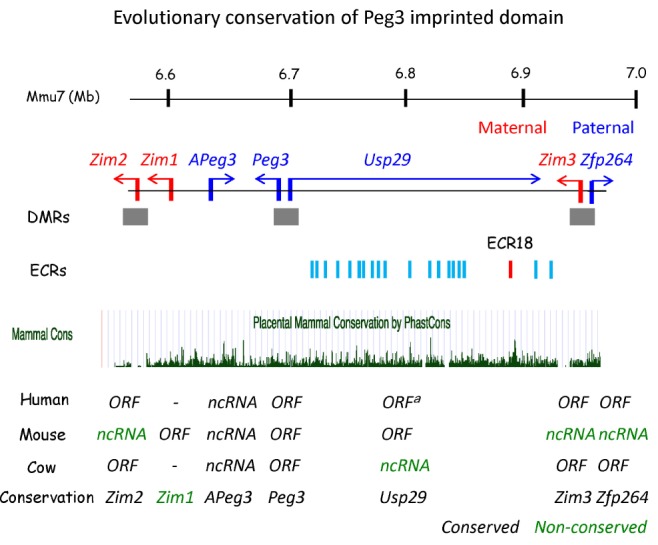
Evolutionary conservation of the Peg3 imprinted domain. The 500-kb genomic interval of the Peg3 domain is shown using the mouse genomic region as a representative locus, along with its relative position in mouse chromosome 7 (Mmu7). The paternally expressed and maternally expressed genes are indicated with blue and red arrows, respectively. The three differentially methylated regions (DMRs) are indicated with grey boxes. The 20 evolutionarily conserved regions (ECRs) are shown with vertical blue lines, except ECR18 in red. The graph in the middle represents the sequence conservation levels of the Peg3 domain among placental mammals. The table below summarizes the evolutionary conservation of gene content and their open reading frames (ORFs) with different colors: black for conserved and green for non-conserved. The first two exons of mouse Usp29 were previously named ossification center-associated transcript (Ocat), while those in humans and cow are named MER1 repeat-containing imprinted transcript 1 (MIMT1). aThe ORF indicates the loss of the USP29 ORF in one human population.
Within the 500-kb region of the Peg3 domain, the seven imprinted genes are unevenly distributed: three genes are clustered together on one side, and the remaining four genes are on the other side (Fig. 1). By contrast, the middle 250-kb region is more or less empty, lacking any obvious open reading frame (ORF). Yet, this overall genomic layout is well conserved among all mammals [8]. According to the results from a sequence comparison of several mammalian species, this middle 250-kb region is filled with 20 small genomic fragments, ranging from 100-300 bp in length, that show high levels of sequence conservation between species. These regions, termed evolutionarily conserved regions (ECRs), are likely cis regulatory elements that may be involved in controlling the transcription and imprinting of the Peg3 domain [15]. This prediction has been recently confirmed through the epigenomic results in which the majority of these ECRs is associated with two histone modifications, H3K4me1 and H3K27ac (monomethylation of lysine 4 and acetylation of lysine 27, respectively, of histone 3) [16], which are closely associated with enhancers [17]. The relative positions, spacing, and orientations of these potential cis regulatory elements are also well conserved among all mammals. This large number of potential cis regulatory elements within the 250-kb region hints at one possibility that unknown, but complex, regulatory mechanisms exist to control transcription and imprinting of the Peg3 domain.
Although the 500-kb domain has been structurally well preserved during mammalian evolution, the protein-coding capacity of each imprinted gene has been quite often lost independently in the different lineages of mammals (Fig. 1). For example, three zinc finger genes, Zim2, Zim3, and Zfp264, all have lost their ORFs in the rodent lineage [9, 10]. The current sequences of these genes in rodents have diverged substantially from their original coding sequences, so that they are barely recognizable as zinc finger genes. Thus, these genes may have lost their ORFs quite a long time ago, at the early stages of rodent evolution. Nevertheless, they are still transcriptionally active, indicating that these genes might have functionally adapted as non-coding RNA genes. In fact, one of these genes, Zim3, has become an antisense gene to Usp29: the transcribed regions overlap quite extensively between maternally expressed Zim3 and paternally expressed Usp29. Usp29 also has taken a similar evolutionary path as seen in the three zinc finger genes. In the bovine lineage, the overall sequence structure of Usp29 is still intact, without any major deletions and insertions, but it contains several base substitutions causing STOP codons, indicating that these substitutions likely occurred in recent evolutionary times in the bovine lineage [13]. Interestingly, a similar situation has also been observed in one human population: one substitution causing a STOP codon has been found within the coding region of USP29 (http://browser.1000genomes.org/index.html). So far, Peg3 is the only gene that has maintained its ORF in all lineages of mammals, while the other imprinted genes have lost ORFs in the different lineages of mammals. This suggests that the functions of Peg3 might be the most critical for the survival of mammals as a protein-coding gene. On the other hand, the other members of the Peg3 domain might be less critical for the survival of mammals, which subsequently allows them to have adapted as ncRNA genes during mammalian evolution.
Co-regulation of Transcription and Imprinting
The transcription of the Peg3 domain is regulated through at least five promoters: two bidirectional promoters for Peg3/Usp29 and Zim3/Zfp264 and three individual promoters for APeg3, Zim1, and Zim2. Among these promoter regions, three regions are differentially methylated between two alleles; thus they are named differentially methylated regions (DMRs). These include the DMRs of Peg3, Zim2, and Zim3 (Fig. 1). According to previous studies, the PEG3-DMR is a gametic DMR, such that its allele-specific methylation is established during gametogenesis in parents-in this case, the maternal-specific methylation during oogenesis [18]. In contrast, the DMRs of Zim2 and Zim3 are established after fertilization and are thus categorized as somatic DMRs [19]. The promoter of Zim1, on the other hand, is not methylated at all on either allele, whereas the methylation status of the APeg3 promoter is currently unknown. Among the three DMRs, the Peg3-DMR is the most critical for transcription and imprinting of the Peg3 domain. Deletion of part of this DMR causes changes in the transcriptional levels of all genes in this domain [20]. The same mutation also affects the monoallelic expression (imprinting) pattern of two genes, Zfp264 and Zim2: a switch from paternal- to maternal-specific expression for Zfp264 and a change from maternal to biallelic expression for Zim2. These results demonstrate that the Peg3-DMR is an ICR for this domain. It is, however, currently unknown how the Peg3-DMR obtains DNA methylation during oogenesis and how the Peg3-DMR serves as a major cis regulatory region governing the overall transcription of the Peg3 domain.
The 4.0-kb Peg3-DMR can be divided into two small regions: the first 1.5-kb region harbors the bidirectional promoter and the first exon for both Peg3 and Usp29, and the second 2.5-kb region covers the first intron of Peg3. The 2.5-kb intron region displays an unusual tandem repeat sequence structure, and one small motif (GGCGCCATCTT) within these repeats turns out to be a DNA-binding site for the transcription factor YY1 [21, 22]. Since this YY1-binding motif is part of the tandem repeats, multiple YY1 binding sites, 10 sites on average, are easily found within the 2.5-kb region of the Peg3-DMR for any given mammal [23]. A series of studies have been conducted to understand the potential roles for YY1 in the function of the Peg3-DMR. First, according to the results, conditional knockdown of YY1 during oogenesis results in hypomethylation of the Peg3-DMR, suggesting that the protein level of YY1 is very critical for the establishment of DNA methylation in the Peg3-DMR [24]. A similar outcome has also been observed in somatic cells using in vivo and in vitro systems [25, 26]. Thus, YY1 is likely to be involved in setting DNA methylation and also in the maintenance of the Peg3-DMR. Second, the expression of Peg3 is usually up-regulated in response to the reduced levels of YY1 protein. This suggests that YY1 may function as a repressor of the transcription of Peg3 in somatic cells. This outcome is usually accompanied by global changes in the expression levels of other imprinted genes in the Peg3 domain, confirming a major role played by YY1 in the entire Peg3 domain [26]. Collectively, the results obtained so far confirm that YY1 is a major regulator of the Peg3 domain, possibly through the Peg3-DMR.
Along with the five promoters, the 20 ECRs located in the middle 250-kb region are also predicted to be involved in the transcriptional control of the Peg3 domain. To further follow up on this prediction, a series of 3C (chromosome conformation capture) experiments were conducted using various adult tissues harvested from mouse [15]. The results revealed that one of these ECRs interacts with the bidirectional promoter of Peg3/Usp29. This particular one, ECR18, is located 200 kb upstream of the bidirectional promoter, which is actually positioned closer to the bidirectional promoter of Zim3/Zfp264. The interaction is the most prominent in the brain but is also detected in other tissues, such as testis and liver. As described earlier, ECR18 is associated with two histone modifications, H3K4me1 and H3K27ac, which mark active enhancers. Subsequent in vitro promoter assays indeed demonstrated that ECR18 functions as an enhancer for the promoter of Peg3/Usp29 in several cell lines tested. The 3C experiments further indicated that ECR18 also interacts with the other promoters of the Peg3 domain. In particular, the bidirectional promoter of Zim3/Zfp264 and the promoter of Zim2 were shown to interact with ECR18 in the testis. Thus, ECR18 might be a shared enhancer for several genes in the Peg3 domain, not just for Peg3 and Usp29.
The transcription and imprinting of the Peg3 domain is likely to be co-regulated through the Peg3-DMR, given the observations described above, such as global changes in the expression levels of the Peg3 domain either by targeting of the Peg3-DMR or by targeting of the transcription factor YY1. This predicted mechanism might be explained in the following manner, which involves ECR18 as a shared enhancer for the Peg3 domain (Fig. 2). On the paternal allele, the Peg3-DMR is unmethylated, and functions as a dominant promoter for both Peg3 and Usp29, thus outcompeting the promoters of the remaining genes, Zim1 and Zim2, for usage of the shared enhancer ECR18. On the maternal allele, the Peg3-DMR is methylated and silent, so that the promoters of Zim1 and Zim2 can access ECR18 for their transcription. This results in the paternal expression of Peg3 and Usp29 and the maternal expression of Zim1 and Zim2. This is particularly true in the brain, as strict imprinting and transcription of these four genes are usually observed in the neonatal brain. On the other hand, the imprinting and transcription of the remaining genes are either bi-allelic or not detectable in the brain; thus, the proposed model is more applicable for explaining the imprinting of the four genes in the brain. This model could explain some of the effects observed from the deletion mutant of the Peg3-DMR, showing bi-allelic expression of Zim2 in carriers inheriting the mutation paternally [20]. Deletion of part of the Peg3-DMR, the 2.5-kb YY1-binding sites, is likely to be responsible for the down-regulation of Peg3 expression and subsequently allows the access of Zim2 to the shared enhancer ECR18 on the paternal allele, resulting in the bi-allelic expression of Zim2. Although reasonable, this model needs to be revised with additional data in the near future in order to explain many aspects of the imprinting and transcription of the Peg3 domain that cannot be explained at the moment.
Fig. 2.
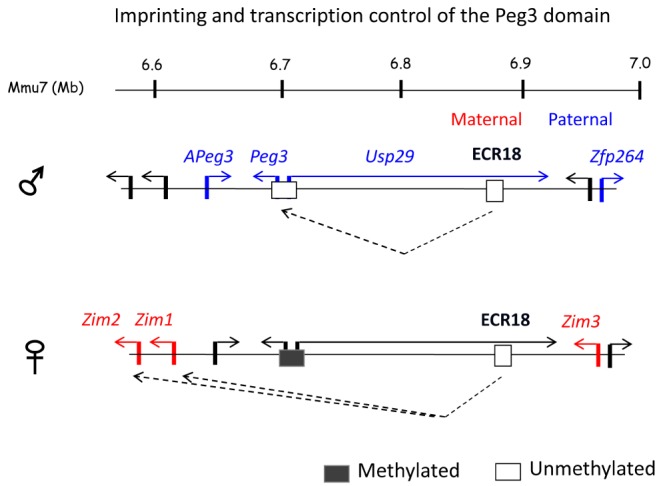
Transcription and imprinting control of the Peg3 domain. The schematic representation illustrates a potential mode of the Peg3-DMR and ECR18 in the co-regulation and mono-allelic expression (imprinting) of four genes in the mouse brain: the paternally expressed Peg3 and Usp29 and the maternally expressed Zim1 and Zim2. On the paternal allele, the Peg3-DMR becomes a dominant user for the shared enhancer ECR18 and out-competes the remaining two genes, Zim1 and Zim2. Thus, Peg3 and Usp29 are expressed from the paternal allele, while Zim1 and Zim2 become silent on the paternal allele. On the maternal allele, the Peg3-DMR is methylated, thus causing the repression of both Peg3 and Usp29. As a consequence, the two genes Zim1 and Zim2 can access ECR18 and be expressed from the maternal allele. However, the current model cannot explain the imprinting of the three remaining genes, APeg3, Zim3 and Zfp264, since some of these genes are expressed mainly in testis but not in brain.
Functions of Imprinted Genes
At the organismal level, mammalian imprinted genes usually have similar functions, controlling fetal growth rates and nurturing behaviors [27, 28]. This is likely the case for the imprinted genes located within the Peg3 domain, based on the following lines of evidence. First, most genes in the Peg3 domain are expressed in early embryogenesis and the brain. The expression levels of Peg3 and Zim1 are very high in embryos and placenta [5, 11]. In brain, Peg3, Zim1, Zim2, and APeg3 are all expressed in the hypothalamus [29, 30], which is known to be involved in controlling maternal-caring behaviors and milk provision [31]. Second, mutations in the Peg3 locus tend to have two consistent phenotypic effects [29, 32]. The pups with mutant alleles have defects in milk-suckling behaviors. As a consequence, the carriers become progressively weaker than their littermates due to the insufficient uptake of milk, which quite often results in perinatal lethality. The adult mice of both genders also have various reproductive and behavioral problems [32,33,34]. Mutant females produce a relatively small number of mature eggs, resulting in reduced litter sizes. Also, pregnant females have problems in milk letdown and in maternal-caring behaviors, thus causing the perinatal lethality observed among wild-type pups. It is interesting to point out that Peg3 mutations have effects on a set of paired behaviors between the mothers and their pups for milk provision, milk suckling, and letdown. This has been a basis for the 'co-adaptation' theory of mammalian genomic imprinting [35]. In the case of bovine Usp29, a mutation deleting a 100-kb interval within the middle 250-kb region has been shown to be responsible for the stillbirth of calves [36]. The exact mechanism by which this deletion causes stillbirth is currently unknown, but this mutational effect is overall consistent with the phenotypes that have been observed from the Peg3 domain, controlling the growth and development of the fetus during the gestational period.
At the cellular level, the predicted functions of Peg3 are worthwhile to be discussed. Peg3 is a potential tumor suppressor, given the observation that many human patients with ovarian and breast cancers lose the expression of PEG3 due to DNA hypermethylation on the PEG3-DMR [37,38,39]. This was further demonstrated by in vitro studies in which reintroduction of human PEG3 into primary cell lines derived from ovarian cancer patients resulted in the inhibition of cell growth [39]. According to recent data from in vitro studies, Peg3 also appears to be involved in autophagy in endothelial cells [40]. Soluble forms of several structural proteins, including decorin, are shown to trigger autophagy in endothelial cells as part of the apoptotic pathways that are designed to remove stressed and damaged cells. Interestingly, PEG3 has been found to play a major role in conveying this external signal through vesicular endothelial growth factor receptor 2 (VEGFR2) to induce the transcription of two core components, BECLIN1 and LC3, for autophagosomes. The potential roles of Peg3 in autophagy are particularly intriguing, since Peg3 is predicted to play important roles in the placenta, where autophagy is critical for the constant turnover of protein and other nutritional materials [41]. It is also relevant to note that nutritional starvation of pregnant females causes changes in the expression levels of a subset of mammalian genes. Interestingly, the majority of these genes is also affected by mutations on Peg3, suggesting that these genes are likely either direct or indirect downstream genes of Peg3 [41]. Thus, Peg3 may be a major player in controlling in nutritional supply between females and pups during pregnancy. Since autophagy is regarded as a part of apoptotic pathways, the potential roles for Peg3 in autophagy are also consistent with previous observations hinting at the functions of Peg3 in apoptosis. These include up-regulation of Peg3 during p53-mediated apoptosis, muscle cachexia (or wasting) in response to tumor load, and induction of Peg3 in response to hypoxic conditions [42,43,44]. Taken together, these results support the idea that Peg3 is a tumor suppressor with a potential connection to autophagy.
At the molecular level, the imprinted genes in the Peg3 domain code for two different types of proteins: a ubiquitin-specific protease for Usp29 and zinc finger proteins for Peg3, Zim1, Zim2, Zim3, and Zfp264. According to recent studies, USP29 might also be a tumor suppressor, given the observation that USP29 is required for the stability of Claspin, a key protein for the DNA damage checkpoint [45]. This is quite intriguing, since DNA hypermethylation of the PEG3-DMR has always been regarded as an epigenetic abnormality affecting the expression of PEG3 but not USP29, although the PEG3-DMR controls both genes. Thus, it would be interesting to test whether the outcome of this DNA hypermethylation manifests as down-regulation of PEG3 only or PEG3 and USP29 together. Among the four zinc finger proteins within the Peg3 domain, Peg3 is the only gene that has been studied so far at the protein level. According to recent studies, PEG3 binds to many genomic loci as a DNA-binding protein with transcriptional repression activity [46]. It is well known that many zinc finger proteins, including those in the Peg3 domain, repress their downstream genes through interaction with KRAB-associated protein 1 (KAP-1) and SET domain-containing histone methylase at H3K9 (SETDB1) [47, 48]. Thus, we predict that PEG3 may also repress its downstream genes through SETDB1/KAP-1-driven H3K9me3. According to recent studies, several placenta-specific gene families are de-repressed in mouse embryos with the Peg3-null mutant allele; yet, these gene families are usually repressed through H3K9me3-mediated mechanisms [49, 50]. Thus, this result again supports the idea that PEG3 controls its downstream genes through H3K9me3-mediated mechanisms.
At the molecular level, several genes within the Peg3 domain are also predicted to function as ncRNA genes (Fig. 1). Among these genes, APeg3 is quite unique as an RNA gene due to the following reasons. First, it is well conserved among all mammals [51]. Second, it is an antisense transcript gene localized in the 3'-UTR of another proteincoding gene-in this case, Peg3 [12]. Third, APeg3 is expressed only in the hypothalamus but highly up-regulated in response to osmotic challenge [30]. Given these special circumstances, it has been predicted that APeg3 may be designed to control the expression of Peg3. Consistent with this prediction, in vitro studies demonstrated that APeg3 has the potential to down-regulate the mRNA and protein levels of Peg3 [51]. Although the mechanistic basis for the observed down-regulation needs to be investigated further, the in vivo functions of APeg3 are most likely connected to those of Peg3.
Shared Versus Unique Features of the Peg3 Domain
The Peg3 domain displays unique features and also shares several features with the other imprinted domains. First, the domain structure has been well preserved during mammalian evolution, as seen in the other domains. This makes sense with the prediction that the imprinting and transcription of the Peg3 domain are likely controlled, as a group, through a shared cis regulatory element-in this case, the Peg3-DMR. As part of the conserved genomic interval, many ECRs are found to be localized within the middle 250-kb region and are also predicted to be enhancers for individual imprinted genes within the Peg3 domain (Fig. 1). Coordinating these potential enhancers with individual promoters may be needed, since some of these ECRs are likely to be shared among individual imprinted genes. As an ICR, the Peg3-DMR might be responsible for this coordinating task. According to a situation observed in the H19/Igf2-imprinted domain, its ICR functions as an enhancer blocker through another transcription factor, CTCF, controlling the accessibility of a shared enhancer between H19 and Igf2 [1, 2]. Interestingly, the ICR of the H19/Igf2 domain also has an unusual cluster of CTCF binding sites, which is very similar to the localization of a tandem array of multiple YY1 binding sites within the Peg3-DMR [52]. Although both ICRs are similar in many ways, their roles as ICRs are likely different from each other. This prediction is based on the result that YY1 and YY1 binding sites do not display enhancer-blocking activity [53]. One plausible role for YY1 might be 'linking' or 'tethering' long-distance enhancers to the corresponding target promoters [54]. As described earlier, ECR18 interacts with the bidirectional promoter of Peg3/Usp29. This long-distance interaction, or linking, between ECR18 and the bidirectional promoter may be mediated through the YY1/YY1-binding sites within the Peg3-DMR (Fig. 2). A similar scenario is also possible for the other ECRs found within the Peg3 domain, which warrants a series of systematic and careful investigations in the near future. Overall, as seen in other domains, the conserved genomic structure of the Peg3 domain makes sense with the presence of co-regulation through the Peg3-DMR, but the mechanisms governing the Peg3 domain by this ICR are likely to be unique, based on the involvement of the transcription factor YY1.
Second, the overall functional outcomes of the Peg3 domain are similar to those observed from other domains, such as controlling fetal growth rates and maternal-caring behaviors [27, 28, 35]. In particular, this has been consistently demonstrated through functional studies of the Peg3 locus [29, 32]. Despite these similar outcomes at the organismal level, the potential function of each imprinted gene is likely to be unique at the molecular and cellular levels. For instance, Peg3 codes for a DNA-binding protein with repression activity at the molecular level and inhibits cell growth as a tumor suppressor at the cellular level. Yet, Peg3 is regarded as a growth stimulator at the organismal level, since the down-regulation of Peg3 usually results in reduction in body size. The functional outcomes of Peg3 seem to be somewhat contradictory between the cellular and organismal levels. This is quite unique compared to other imprinted genes encoding proteins with different biochemical properties, such as paternally expressed insulin-like growth factor 2 (Igf2) and maternally expressed insulin-like growth factor 2 receptor (Igf2r). The phenotypic outcomes of this pair of ligand and receptor are overall similar at the cellular and organismal levels [27, 28]. Igf2 stimulates cell growth, and thus, higher levels of Igf2 result in bigger body size. Igf2r derives exactly the opposite phenotypes to those of Igf2, inhibiting cell growth, and thus, more Igf2r results in smaller body size. As proposed earlier, the conflicting phenotypes of Peg3 at the cellular and organismal levels could be explained in the following manner [55]. Peg3 might be involved in programmed cell death during the development of neuron cells-specifically, a set of neuron cells producing growth suppressors. The absence of Peg3 could result in the overgrowth of these neuron cells with an excessive amount of growth suppressors at the cellular level, causing smaller body size at the organismal level. Although this remains to be tested, it is feasible, since many imprinted genes are highly expressed in the neuroendocrine system, such as the hypothalamus, which is responsible for controlling body size through growth hormones [31]. In that regard, it is interesting to note that another paternally expressed DNA-binding gene, Zac1 (zinc finger protein regulating apoptosis and cell cycle arrest), also shows similar conflicting outcomes-it is a growth suppressor at the cellular level but a growth stimulator at the organismal level [56, 57]. Since both Peg3 and Zac1 are growth suppressors at the cellular level, these genes have been quite often identified as tumor suppressors. Although the overall functional contributions by the Peg3 domain are similar to those of other domains at the organismal level, the detailed function of each imprinted gene is predicted to be unique at the cellular and molecular levels.
Third, the Peg3 domain also contains several ncRNA genes, as seen in other imprinted domains. These ncRNA genes include Zim2, Zim3, and Zfp264 in the rodent lineage (Fig. 1). These ncRNA genes are unique due to the following reasons. First, these ncRNA genes are of relatively recent evolutionary origin, so that their ancestral genes can be identified easily through sequence similarity to their corresponding protein-coding sequences. It is currently unknown whether these ncRNA genes are in the process of becoming pseudogenes or have already adapted as ncRNA genes with some functions. Regardless, they have maintained their transcriptional activity for quite a long time, given the accumulation of mutations disrupting their ORFs, suggesting that their gene products, ncRNA, or transcription itself is now part of the Peg3 domain. If some of them have already adapted as ncRNA genes, examining their sequences should provide some clues regarding how protein-coding sequences become ncRNA genes during evolution. Thus, the ncRNA genes of the Peg3 domain might provide a very unique opportunity to understand the evolutionary paths of mammalian genes. In that regard, it is relevant to note that mammalian X chromosome inactivation-specific transcript (Xist) represents another well-known case where a proteincoding gene has become an ncRNA gene [58]. Second, some functions of these ncRNA genes should be lineage-specific to rodents, since the original genes of these ncRNA genes are still protein-coding genes in all other mammals. This level of lineage-specific change has not been observed from other imprinted domains. This might be related to the fact that the Peg3 domain has been part of a large zinc finger gene family, which is known to be vulnerable to genomic rearrangements, including deletion and duplication [14]. Also, the functional redundancy afforded by the zinc finger gene family might have allowed its members to drift away from their proteincoding sequences without severe phenotypic consequences. Overall, the structurally conserved genomic interval of the Peg3 domain might have allowed several protein-coding members to adapt into ncRNA genes, which seems to be quite different from the evolutionary patterns observed from other imprinted domains.
Future Directions
The Peg3 domain shows many similar features as seen in other imprinted domains, which include its conserved genomic structure, allowing long-distance transcriptional regulation through ICRs, as well as allele-specific DNA methylation and subsequent monoallelic expression. In the past two decades, these unusual features have been studied extensively using other imprinted domains as model loci. At the same time, the Peg3 domain displays several unique features that have not been studied in other imprinted domains. These unique features are thus worthwhile to study in the future in increase our understanding of genomic imprinting and imprinting-related human disorders. First, YY1/YY1-binding sites appear to be a major player in the Peg3 domain. In particular, a tandem array of YY1-binding sites has been selected for the function of the Peg3-DMR; yet, the molecular basis for this function is still unknown. One possibility would be that these multiple YY1-binding sites might be involved in setting DNA methylation during oogenesis. The other possibility would be that these YY1-binding sites might be required for the transcriptional control of Peg3 and adjacent imprinted genes. Targeting these YY1-binding sites through mutagenesis experiments should provide a clear and direct answer regarding the in vivo role of multiple YY1-binding sites. Second, Peg3 has been recognized as a potential tumor suppressor for some time, without any progress in our understanding. According to recent studies, Peg3 may play a major role in regulating autophagy of endothelial cells, which is very intriguing, due to the following reasons: 1) Peg3 is highly expressed in the organs that are regarded as endothelial cell-rich tissues, such as placenta, and 2) autophagy is also regarded as part of apoptotic pathways. Thus, characterizing the potential roles of Peg3 in autophagy should be a worthy project in the immediate future. Third, the Peg3 domain appears to have evolved very rapidly during mammalian evolution. As a consequence, several members of this 500-kb domain have changed from protein-coding to ncRNA genes. It is well known that the ancestral state of a given genomic interval cannot be easily reconstructed, due to the many changes that have accumulated during long evolutionary times. In the case of the Peg3 domain, its 500-kb genomic structure has been so well preserved that its ancestral structure can be readily reconstructed by comparing the Peg3 domains of several mammals. Thus, careful sequence analyses should provide immediate insights regarding the evolutionary paths of mammalian genes, especially those that have undergone the transformation from protein-coding to non-coding RNA genes. Overall, these directions are worthwhile to pursue in the near future, which should most likely provide additional exciting information regarding how the Peg3 domain is regulated and what functional roles the Peg3 domain plays in mammalian biology.
Acknowledgments
We would like to thank the members of the JooKim Lab for their thoughtful feedback and discussion on the manuscript. This research was supported by the National Institute of Health (J.K. R01-GM066225 and R01-GM097074).
References
- 1.Bartolomei MS, Ferguson-Smith AC. Mammalian genomic imprinting. Cold Spring Harb Perspect Biol. 2011;3:pii: a002592. doi: 10.1101/cshperspect.a002592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barlow DP, Bartolomei MS. Genomic imprinting in mammals. Cold Spring Harb Perspect Biol. 2014;6:pii: a018382. doi: 10.1101/cshperspect.a018382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Edwards CA, Ferguson-Smith AC. Mechanisms regulating imprinted genes in clusters. Curr Opin Cell Biol. 2007;19:281–289. doi: 10.1016/j.ceb.2007.04.013. [DOI] [PubMed] [Google Scholar]
- 4.Spahn L, Barlow DP. An ICE pattern crystallizes. Nat Genet. 2003;35:11–12. doi: 10.1038/ng0903-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuroiwa Y, Kaneko-Ishino T, Kagitani F, Kohda T, Li LL, Tada M, et al. Peg3 imprinted gene on proximal chromosome 7 encodes for a zinc finger protein. Nat Genet. 1996;12:186–190. doi: 10.1038/ng0296-186. [DOI] [PubMed] [Google Scholar]
- 6.Relaix F, Weng X, Marazzi G, Yang E, Copeland N, Jenkins N, et al. Pw1, a novel zinc finger gene implicated in the myogenic and neuronal lineages. Dev Biol. 1996;177:383–396. doi: 10.1006/dbio.1996.0172. [DOI] [PubMed] [Google Scholar]
- 7.Kim J, Ashworth L, Branscomb E, Stubbs L. The human homolog of a mouse-imprinted gene, Peg3, maps to a zinc finger gene-rich region of human chromosome 19q13.4. Genome Res. 1997;7:532–540. doi: 10.1101/gr.7.5.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim J, Noskov VN, Lu X, Bergmann A, Ren X, Warth T, et al. Discovery of a novel, paternally expressed ubiquitin-specific processing protease gene through comparative analysis of an imprinted region of mouse chromosome 7 and human chromosome 19q13.4. Genome Res. 2000;10:1138–1147. doi: 10.1101/gr.10.8.1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim J, Bergmann A, Wehri E, Lu X, Stubbs L. Imprinting and evolution of two Kruppel-type zinc-finger genes, ZIM3 and ZNF264, located in the PEG3/USP29 imprinted domain. Genomics. 2001;77:91–98. doi: 10.1006/geno.2001.6621. [DOI] [PubMed] [Google Scholar]
- 10.Kim J, Bergmann A, Lucas S, Stone R, Stubbs L. Lineage-specific imprinting and evolution of the zinc-finger gene ZIM2. Genomics. 2004;84:47–58. doi: 10.1016/j.ygeno.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 11.Kim J, Lu X, Stubbs L. Zim1, a maternally expressed mouse Kruppel-type zinc-finger gene located in proximal chromosome 7. Hum Mol Genet. 1999;8:847–854. doi: 10.1093/hmg/8.5.847. [DOI] [PubMed] [Google Scholar]
- 12.Choo JH, Kim JD, Kim J. Imprinting of an evolutionarily conserved antisense transcript gene APeg3. Gene. 2008;409:28–33. doi: 10.1016/j.gene.2007.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim J, Bergmann A, Choo JH, Stubbs L. Genomic organization and imprinting of the Peg3 domain in bovine. Genomics. 2007;90:85–92. doi: 10.1016/j.ygeno.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 14.Kim J, Gordon L, Dehal P, Badri H, Christensen M, Groza M, et al. Homology-driven assembly of a sequence-ready mouse BAC contig map spanning regions related to the 46-Mb gene-rich euchromatic segments of human chromosome 19. Genomics. 2001;74:129–141. doi: 10.1006/geno.2001.6521. [DOI] [PubMed] [Google Scholar]
- 15.Thiaville MM, Kim H, Frey WD, Kim J. Identification of an evolutionarily conserved cis-regulatory element controlling the Peg3 imprinted domain. PLoS One. 2013;8:e75417. doi: 10.1371/journal.pone.0075417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shen Y, Yue F, McCleary DF, Ye Z, Edsall L, Kuan S, et al. A map of the cis-regulatory sequences in the mouse genome. Nature. 2012;488:116–120. doi: 10.1038/nature11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lucifero D, Mann MR, Bartolomei MS, Trasler JM. Gene-specific timing and epigenetic memory in oocyte imprinting. Hum Mol Genet. 2004;13:839–849. doi: 10.1093/hmg/ddh104. [DOI] [PubMed] [Google Scholar]
- 19.Huang JM, Kim J. DNA methylation analysis of the mammalian PEG3 imprinted domain. Gene. 2009;442:18–25. doi: 10.1016/j.gene.2009.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim J, Ekram MB, Kim H, Faisal M, Frey WD, Huang JM, et al. Imprinting control region (ICR) of the Peg3 domain. Hum Mol Genet. 2012;21:2677–2687. doi: 10.1093/hmg/dds092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim J, Kollhoff A, Bergmann A, Stubbs L. Methylation-sensitive binding of transcription factor YY1 to an insulator sequence within the paternally expressed imprinted gene, Peg3. Hum Mol Genet. 2003;12:233–245. doi: 10.1093/hmg/ddg028. [DOI] [PubMed] [Google Scholar]
- 22.Kim JD, Hinz AK, Bergmann A, Huang JM, Ovcharenko I, Stubbs L, et al. Identification of clustered YY1 binding sites in imprinting control regions. Genome Res. 2006;16:901–911. doi: 10.1101/gr.5091406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim JD, Kim J. YY1's longer DNA-binding motifs. Genomics. 2009;93:152–158. doi: 10.1016/j.ygeno.2008.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim JD, Kang K, Kim J. YY1s role in DNA methylation of Peg3 and Xist. Nucleic Acids Res. 2009;37:5656–5664. doi: 10.1093/nar/gkp613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim J, Kim JD. In vivo YY1 knockdown effects on genomic imprinting. Hum Mol Genet. 2008;17:391–401. doi: 10.1093/hmg/ddm316. [DOI] [PubMed] [Google Scholar]
- 26.Kim JD, Hinz AK, Choo JH, Stubbs L, Kim J. YY1 as a controlling factor for the Peg3 and Gnas imprinted domains. Genomics. 2007;89:262–269. doi: 10.1016/j.ygeno.2006.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moore T, Haig D. Genomic imprinting in mammalian development: a parental tug-of-war. Trends Genet. 1991;7:45–49. doi: 10.1016/0168-9525(91)90230-N. [DOI] [PubMed] [Google Scholar]
- 28.Tilghman SM. The sins of the fathers and mothers: genomic imprinting in mammalian development. Cell. 1999;96:185–193. doi: 10.1016/s0092-8674(00)80559-0. [DOI] [PubMed] [Google Scholar]
- 29.Li L, Keverne EB, Aparicio SA, Ishino F, Barton SC, Surani MA. Regulation of maternal behavior and offspring growth by paternally expressed Peg3. Science. 1999;284:330–333. doi: 10.1126/science.284.5412.330. [DOI] [PubMed] [Google Scholar]
- 30.Glasgow E, Ryu SL, Yamashita M, Zhang BJ, Mutsuga N, Gainer H. APeg3, a novel paternally expressed gene 3 antisense RNA transcript specifically expressed in vasopressinergic magnocellular neurons in the rat supraoptic nucleus. Brain Res Mol Brain Res. 2005;137:143–151. doi: 10.1016/j.molbrainres.2005.02.030. [DOI] [PubMed] [Google Scholar]
- 31.Ivanova E, Kelsey G. Imprinted genes and hypothalamic function. J Mol Endocrinol. 2011;47:R67–R74. doi: 10.1530/JME-11-0065. [DOI] [PubMed] [Google Scholar]
- 32.Kim J, Frey WD, He H, Kim H, Ekram MB, Bakshi A, et al. Peg3 mutational effects on reproduction and placenta-specific gene families. PLoS One. 2013;8:e83359. doi: 10.1371/journal.pone.0083359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Swaney WT, Curley JP, Champagne FA, Keverne EB. The paternally expressed gene Peg3 regulates sexual experience-dependent preferences for estrous odors. Behav Neurosci. 2008;122:963–973. doi: 10.1037/a0012706. [DOI] [PubMed] [Google Scholar]
- 34.Swaney WT, Curley JP, Champagne FA, Keverne EB. Genomic imprinting mediates sexual experience-dependent olfactory learning in male mice. Proc Natl Acad Sci U S A. 2007;104:6084–6089. doi: 10.1073/pnas.0609471104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Curley JP, Barton S, Surani A, Keverne EB. Coadaptation in mother and infant regulated by a paternally expressed imprinted gene. Proc Biol Sci. 2004;271:1303–1309. doi: 10.1098/rspb.2004.2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Flisikowski K, Venhoranta H, Nowacka-Woszuk J, McKay SD, Flyckt A, Taponen J, et al. A novel mutation in the maternally imprinted PEG3 domain results in a loss of MIMT1 expression and causes abortions and stillbirths in cattle (Bos taurus) PLoS One. 2010;5:e15116. doi: 10.1371/journal.pone.0015116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dowdy SC, Gostout BS, Shridhar V, Wu X, Smith DI, Podratz KC, et al. Biallelic methylation and silencing of paternally expressed gene 3 (PEG3) in gynecologic cancer cell lines. Gynecol Oncol. 2005;99:126–134. doi: 10.1016/j.ygyno.2005.05.036. [DOI] [PubMed] [Google Scholar]
- 38.Feng W, Marquez RT, Lu Z, Liu J, Lu KH, Issa JP, et al. Imprinted tumor suppressor genes ARHI and PEG3 are the most frequently down-regulated in human ovarian cancers by loss of heterozygosity and promoter methylation. Cancer. 2008;112:1489–1502. doi: 10.1002/cncr.23323. [DOI] [PubMed] [Google Scholar]
- 39.Chen MY, Liao WS, Lu Z, Bornmann WG, Hennessey V, Washington MN, et al. Decitabine and suberoylanilide hydroxamic acid (SAHA) inhibit growth of ovarian cancer cell lines and xenografts while inducing expression of imprinted tumor suppressor genes, apoptosis, G2/M arrest, and autophagy. Cancer. 2011;117:4424–4438. doi: 10.1002/cncr.26073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Buraschi S, Neill T, Goyal A, Poluzzi C, Smythies J, Owens RT, et al. Decorin causes autophagy in endothelial cells via Peg3. Proc Natl Acad Sci U S A. 2013;110:E2582–E2591. doi: 10.1073/pnas.1305732110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Broad KD, Keverne EB. Placental protection of the fetal brain during short-term food deprivation. Proc Natl Acad Sci U S A. 2011;108:15237–15241. doi: 10.1073/pnas.1106022108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Relaix F, Wei X, Li W, Pan J, Lin Y, Bowtell DD, et al. Pw1/Peg3 is a potential cell death mediator and cooperates with Siah1a in p53-mediated apoptosis. Proc Natl Acad Sci U S A. 2000;97:2105–2110. doi: 10.1073/pnas.040378897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schwarzkopf M, Coletti D, Sassoon D, Marazzi G. Muscle cachexia is regulated by a p53-PW1/Peg3-dependent pathway. Genes Dev. 2006;20:3440–3452. doi: 10.1101/gad.412606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yamaguchi A, Taniguchi M, Hori O, Ogawa S, Tojo N, Matsuoka N, et al. Peg3/Pw1 is involved in p53-mediated cell death pathway in brain ischemia/hypoxia. J Biol Chem. 2002;277:623–629. doi: 10.1074/jbc.M107435200. [DOI] [PubMed] [Google Scholar]
- 45.KMartin Y, Cabrera E, Amoedo H, Hernández-Pérez S, Domínguez-Kelly R, Freire R. Oncogene. 2014. Mar 17, USP29 controls the stability of checkpoint adaptor Claspin by deubiquitination. [Epub]. http://dx.doi.org/10.1038/onc.2014.38. [DOI] [PubMed] [Google Scholar]
- 46.Thiaville MM, Huang JM, Kim H, Ekram MB, Roh TY, Kim J. DNA-binding motif and target genes of the imprinted transcription factor PEG3. Gene. 2013;512:314–320. doi: 10.1016/j.gene.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ., 3rd SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 2002;16:919–932. doi: 10.1101/gad.973302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Frietze S, O'Geen H, Blahnik KR, Jin VX, Farnham PJ. ZNF274 recruits the histone methyltransferase SETDB1 to the 3' ends of ZNF genes. PLoS One. 2010;5:e15082. doi: 10.1371/journal.pone.0015082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.ENCODE Project Consortium. Birney E, Stamatoyannopoulos JA, Dutta A, Guigó R, Gingeras TR, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 51.Frey WD, Kim J. APeg3: regulation of Peg3 through an evolutionarily conserved ncRNA. Gene. 2014;540:251–257. doi: 10.1016/j.gene.2014.02.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim J. Multiple YY1 and CTCF binding sites in imprinting control regions. Epigenetics. 2008;3:115–118. doi: 10.4161/epi.3.3.6176. [DOI] [PubMed] [Google Scholar]
- 53.Kim JD, Yu S, Choo JH, Kim J. Two evolutionarily conserved sequence elements for Peg3/Usp29 transcription. BMC Mol Biol. 2008;9:108. doi: 10.1186/1471-2199-9-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Calhoun VC, Levine M. Long-range enhancer-promoter interactions in the Scr-Antp interval of the Drosophila Antennapedia complex. Proc Natl Acad Sci U S A. 2003;100:9878–9883. doi: 10.1073/pnas.1233791100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Broad KD, Curley JP, Keverne EB. Increased apoptosis during neonatal brain development underlies the adult behavioral deficits seen in mice lacking a functional paternally expressed gene 3 (Peg3) Dev Neurobiol. 2009;69:314–325. doi: 10.1002/dneu.20702. [DOI] [PubMed] [Google Scholar]
- 56.Spengler D, Villalba M, Hoffmann A, Pantaloni C, Houssami S, Bockaert J, et al. Regulation of apoptosis and cell cycle arrest by Zac1, a novel zinc finger protein expressed in the pituitary gland and the brain. EMBO J. 1997;16:2814–2825. doi: 10.1093/emboj/16.10.2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Varrault A, Gueydan C, Delalbre A, Bellmann A, Houssami S, Aknin C, et al. Zac1 regulates an imprinted gene network critically involved in the control of embryonic growth. Dev Cell. 2006;11:711–722. doi: 10.1016/j.devcel.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 58.Duret L, Chureau C, Samain S, Weissenbach J, Avner P. The Xist RNA gene evolved in eutherians by pseudogenization of a protein-coding gene. Science. 2006;312:1653–1655. doi: 10.1126/science.1126316. [DOI] [PubMed] [Google Scholar]


