Abstract
The product of the Escherichia coli yadB gene is homologous to the N-terminal part of bacterial glutamyl-tRNA synthetases (GluRSs), including the Rossmann fold with the acceptor-binding domain and the stem-contact fold. This GluRS-like protein, which lacks the anticodon-binding domain, does not use tRNAGlu as substrate in vitro nor in vivo, but aminoacylates tRNAAsp with glutamate. The yadB gene is expressed in wild-type E. coli as an operon with the dksA gene, which encodes a protein involved in the general stress response by means of its action at the translational level. The fate of the glutamylated tRNAAsp is not known, but its incapacity to bind elongation factor Tu suggests that it is not involved in ribosomal protein synthesis. Genes homologous to yadB are present only in bacteria, mostly in Proteobacteria. Sequence alignments and phylogenetic analyses show that the YadB proteins form a distinct monophyletic group related to the bacterial and organellar GluRSs (α-type GlxRSs superfamily) with ubiquitous function as suggested by the similar functional properties of the YadB homologue from Neisseria meningitidis.
Keywords: glutamyl-tRNA synthetase, misacylation, evolution
The aminoacyl-tRNA synthetases (aaRSs) are among the oldest known proteins and were used in phylogenetic studies to confirm the organismal tree of life proposed on the basis of rRNA analyses (1). The central role of this family of enzymes is catalysis of amino acid activation and transfer to tRNA in view of protein synthesis on the ribosomes. On the other hand, some atypical aaRSs and aaRS-like proteins that contain fewer domains and are smaller than the corresponding conventional aaRSs, participate in other functions, such as amino acid biosynthesis and RNA binding (2, 3).
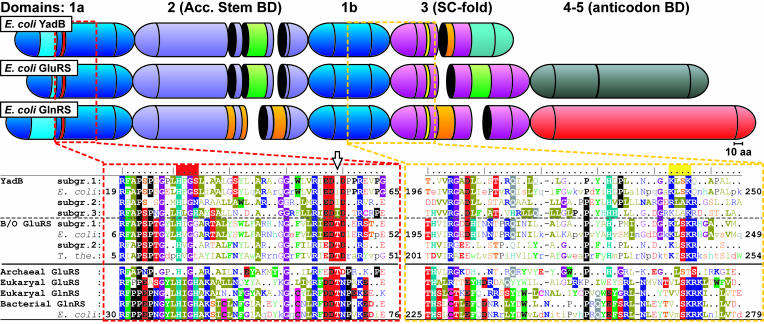
The presence in the Escherichia coli genome of an ORF of 256 codons (ORF256) potentially encoding a protein similar (35% identity) to E. coli glutamyl-tRNA synthetase (GluRS) was reported by Fujita et al. (4). This ORF was later named yadB. It contains 308 codons (5), encoding a protein similar to the N-terminal part of bacterial GluRSs (6). Its structure is modular and includes a Rossmann fold (domains 1a and 1b), a binding domain of the tRNA acceptor stem (domain 2), and a stem-contact (SC) fold (domain 3), but it lacks the region corresponding to the two moieties of the anticodon-binding domain (domains 4–5) (Fig. 1 Upper). A phylogenetic study of GluRSs and glutaminyl-tRNA synthetases (GlnRSs), aaRSs from class Ib (7), including E. coli ORF256, suggests that this ORF is related to bacterial nondiscriminating GluRSs (ND-GluRSs), such as those of Rhizobium meliloti (8). The ND-GluRSs present in organisms that lack the GlnRS gene, like archaea and most bacteria, glutamylate both tRNAGlu and tRNAGln (8–11). The Glu-tRNAGln product is then converted into Gln-tRNAGln by a tRNA-dependent amidotransferase (7, 12). This transamidation pathway is believed to be the ancient way to produce Gln-tRNAGln in bacteria before the recent acquisition of a eukaryotic GlnRS by horizontal gene transfer in bacteria, mostly in the γ- and ε-subdivisions of Proteobacteria (7, 8, 13–16). ORF256 was considered as a possible relic of an ancient ND-GluRS, which was replaced by its modern discriminating relative (8). It is now assumed that GlnRSs arose from the duplication of an ancestral eukaryotic GluRS, which then became specific for Gln and tRNAGln (13). On this basis, a unifying classification scheme was proposed for the GluRS and GlnRS superfamily (GlxRS), with the α- and β-types consisting of the “bacterial/organellar GluRS” group exemplified by Thermus thermophilus GluRS (6) and the “archaeal/eukaryal GluRSs plus all GlnRSs” group exemplified by E. coli GlnRS (17, 18) (Fig. 1 Lower).
Fig. 1.
Comparison of the sequences of YadB proteins with GluRSs and GlnRSs showing the modular arrangements and the structural relationships of the three families of proteins. The catalytic domain (1a and 1b), the acceptor stem-binding domain [2 (Acc. Stem BD)], and the stem-contact fold domain [3 (SC-fold)] are blue, pastel blue, and purple, respectively. The anticodon-binding domain(s) of GluRSs and GlnRSs are gray (α-type, 4–5) and light red (β-type, 4), respectively. Insertions specificto α- and β-type GlxRSs are light green and orange, respectively. The YadB-specific C-terminal sequence is turquoise. The red and yellow strips above the sequences indicate the location of the class I aaRS sequence motifs “HIGH” and “KMSK”, respectively. A structure-based alignment of amino acid consensus sequences of the regions containing these two motifs are presented in red and yellow boxes. The consensus sequences represent the three YadB subgroups, subgroups 1 and 2 of bacterial and organellar (B/O) α-type GluRSs, and the β-type GlxRSs (archaeal and eukaryal GluRSs, and bacterial and eukaryal GlnRSs). Aligned individual sequences of E. coli YadB, GluRS and GlnRS and T. thermophilus GluRS are also presented. Residues are presented by colored characters: highlighted for 100% conservation, bold for 75–99%, normal for 50–74%, and a dot (or lowercase character in the individual sequences) for <50% conservation. The black arrow points at a conserved residue distinct in YadBs and GlxRSs (see the text).
We report here that E. coli YadB is a miniGluRS resembling structurally the catalytic domain of standard GluRSs, but it differs functionally from these enzymes by its capacity to activate Glu in the absence of tRNA and to transfer the activated Glu on tRNAAsp and not tRNAGlu or tRNAGln, the natural substrates of ND-GluRSs.
Materials and Methods
Production of YadB. For expression of the N-terminally His-tagged protein, the ORF was amplified by PCR with E. coli K12 genomic DNA, Pfu DNA polymerase (Stratagene), a forward primer extended by six His codons and the NcoI site, and a reverse primer extended by the EcoRI site. The amplicon was purified with the QIAquick PCR clean-up kit (Qiagen, Valencia, CA) and cloned into pET-28c. The gene extended by six His codons was also cloned in pDest17 vector by using the Gateway recombination system (Invitrogen). For expression of the native protein the ORF was amplified with a forward primer introducing the attBI site, a Shine–Dalgarno sequence, a Kozak site and a start codon, and a reverse primer introducing an attBII site and a stop codon, and cloned into the pDest14 expression vector. The constructs, pET-(His-6)-YadB, pDest17-(His-6)-YadB, and pDest14-YadB, were checked by sequencing. The (His-6)-YadB constructs were used to transform, respectively, E. coli JP1449(DE3)pLysS encoding a thermosensitive GluRS (19, 20) and BL21(DE3), and the His-tagged YadB constructs were purified by affinity chromatography on Ni-NTA agarose (Qiagen). Native YadB was overproduced in the Rosetta (DE3)pLysS strain (Novagen) expressing minor tRNAs and purified by chromatographies on DEAE-cellulose, phosphocellulose, and hydroxyapatite. PAGE of the purified proteins showed the presence of two polypeptide chains, one of full-length and another one of intensity varying with the preparation, starting with Pro-49. The His-tagged version from pDest17-YadB construct was used for the crystallographic analyses and both His-tagged and non-His-tagged versions for the biochemical assays. Because the two proteins exhibit the same catalytic activities, only data obtained with wild-type YadB are reported here.
aaRS Assays. Mixtures for tRNA aminoacylation (200 μl) contained 100 mM Na-Hepes (pH 7.2), 2 mM ATP, 10 mM MgCl2, 30 mM KCl, and 25 μM l-[14C]Glu (330 cpm·pmol-1) or l-[14C]Asp (280 cpm·pmol-1), except for Km measurements with pure tRNAAsp, where 10 μM [3H]Glu or [3H]Asp (2000 cpm·pmol-1) were used, and either 0.5 mg·ml-1 unfractionated E. coli tRNA or 2 μM E. coli tRNAGlu or tRNAAsp and appropriate concentrations of YadB, GluRS, or aspartyl-tRNA synthetase (AspRS) for initial rate or plateau measurements. Km values were determined according to the Lineweaver–Burk representation by using appropriate ranges of substrates concentrations. Unfractionated tRNA was from Roche Diagnostics (Meylan, France) and pure E. coli tRNAGlu (36 nmol·mg-1) from Subriden RNA (Rolling Bay, WA). Pure E. coli tRNAAsp (27 nmol·mg-1) was purified from an overexpressing strain, and enriched tRNAGlu was prepared from E. coli strain XL-1-Blue (pKR15) (21). ATP-[32P]PPi exchange reactions were conducted as described (22).
RNA Purification and Sequencing. The RNA glutamylated by YadB was isolated from nonaminoacylated tRNA after derivatization of the charged amino acid as described (23). Its sequence was determined by a combination of 2D electrophoresis, homochromatography, and enzymatic sequencing (24, 25). The level of the modified nucleotides was determined by a postlabeling procedure after total RNase T2 digestion (26).
Aminoacyl-tRNA Hydrolysis Assays. The protection of aminoacylated tRNAs by elongation factor Tu (EF-Tu) against hydrolysis was measured according to an established procedure (27).
In Vivo mRNA and YadB Detections. mRNA was isolated from E. coli K12 cells grown in a LB medium and harvested in the exponential phase. Transcripts were detected after DNase treatment with the AccessQuick (Promega) RT-PCR system (RNeasy Midi Kit, Qiagen) by using appropriate 20- to 23-nt-long primers for amplification of the proximal portion of dksA, the distal portion of yadB, the full length yadB, and the dksA-yadB operon. Transcripts were analyzed by agarose-gel electrophoresis. YadB was detected in E. coli crude and S100 extracts by Western blot analysis with anti-YadB antibodies obtained from immunized rabbits.
Crystallographic and Bioinformatic Methods. Details on crystallization of the His-tagged YadB, structure determination, and refinement are published elsewhere (22).
ORFs encoding sequences homologous to E. coli YadB, GluRS, and GlnRS were searched in databases by using blastp and tblastn (28) through the ENTREZ portal at the National Institutes of Health (www3.ncbi.nlm.nih.gov/Entrez/index.html). Protein sequences were aligned with clustal x (29) and refined manually in bioedit (30) with the 3D structures of E. coli GlnRS (17) and T. thermophilus GluRS (6, 31). The neighbor-joining phylogeny was based on pairwise distances between amino acid sequences by using neighbor and protdist of the phylip 3.6a2.1 package (32) and the dayhoff 120 matrix (33). The protpars program of the same package was used for the maximum-parsimony analysis. The confidence limits of branching points from 1,000 bootstrap replications were estimated with seqboot and consense, and the branch lengths were recalibrated with fitch. Maximum-likelihood analysis was done with tree-puzzle 5.0 (34). The parameters used were 5,000 puzzling quartets, the JTT matrix (35), gamma-distributed rates over eight categories, and the α-parameter estimated from the data. The trees were reconstructed with treeview (36).
Results and Discussion
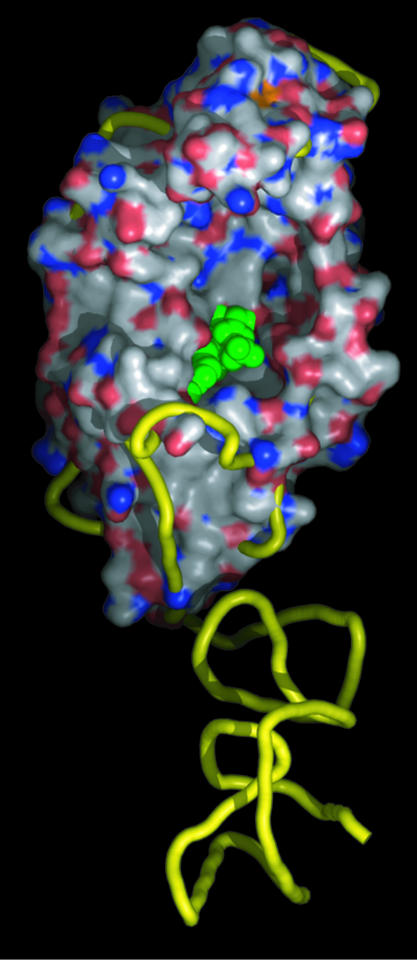
The Crystallographic Structure of YadB Reveals a MiniGluRS Fold with Unexpected Functional Properties. The 3D structure of YadB at 1.5 Å (Fig. 2) shows striking structural similarities to GluRSs and, to a lesser extent, to GlnRSs, with a Rossmann fold, a connective peptide domain, and part of the SC fold. When compared with these standard aaRSs, however, YadB lacks the C-terminal anti-codon-binding domain but contains a potential aminoacyladenylate-binding pocket within its Rossmann-fold domain (22). Thus, it can be anticipated that this truncated GluRS will be unable to aminoacylate a tRNA but might still activate an amino acid. Preliminary functional assays showed indeed activation of Glu by YadB and inability of this protein to aminoacylate tRNAGlu (22).
Fig. 2.
Three-dimensional structure of E. coli YadB and comparison with homologous GluRS. The structures of YadB has been superimposed on that of the T. thermophilus GluRS (1GLN, 6). The figure displays YadB surface (C, white; O, red; N, blue) and GluRS ribbon trace (yellow). The modeled aminoacyl-adenylate is emphasized in green. The coordinates of YadB are deposited in the Protein Data Bank (www.pdb.org; PDB ID code 1NZJ). Figure 2 was made with pymol.
These first assays, however, revealed the unexpected property of YadB to activate Glu in the absence of tRNA, in contrast to canonical GluRSs that require tRNAGlu for this step (37). The comparative kinetic data obtained with wild-type YadB and GluRS (Table 1) fully confirm those obtained with His-tagged YadB (22). They show, in particular, the tRNA-independent activation of Glu by YadB with a strongly decreased affinity for the amino acid but an unaffected reaction rate as compared with the GluRS-catalyzed reaction. Note, the presence of tRNA does not affect the reaction kinetics. Furthermore, both proteins exhibit the same inhibition by glutamol-AMP (GoA), a nonhydrolyzable analog of Glu-AMP (38). We recall that the YadB-catalyzed activation is specific, because no other l-amino acid or d-Glu is activated (22).
Table 1. Kinetic constants of ATP-PPi exchange catalyzed by E. coli YadB and GluRS and inhibition by GoA.
|
Km, mM
|
Kcat, s-1
|
|||||
|---|---|---|---|---|---|---|
| Glutamate
|
ATP
|
|||||
| Enzyme | -tRNA | +tRNA | -tRNA | +tRNA | -tRNA | +tRNA |
| YadB | 5.3 | 5.0 | 0.40 | 0.43 | 18 | 12 |
| GluRS | NR | 0.03 | NR | 0.06 | NR | 10 |
| Ki, μM, for GdA | Kcat, s-1 | |||||
| YadB | 0.44 | ND | 0.20 | ND | 18 | ND |
| GluRS | NR | 2.0 | NR | 1.0 | NR | 10 |
The conditions are described in Materials and Methods. When used, tRNA was an unfractionated bulk from E. coli. ND, not determined; NR, no reaction.
E. coli YadB Glutamylates an RNA Distinct from tRNAGlu and tRNAGln. The former experiments (22) have shown that YadB is neither able to glutamylate cognate E. coli tRNAGlu nor the related tRNAGln, which can be a substrate of ND-GluRSs (8–11). Unexpectedly, control experiments with crude E. coli tRNA indicated charging of 4% of tRNA, suggesting that one or several other tRNA species or a contaminating unknown RNA molecule can be aminoacylated by YadB.
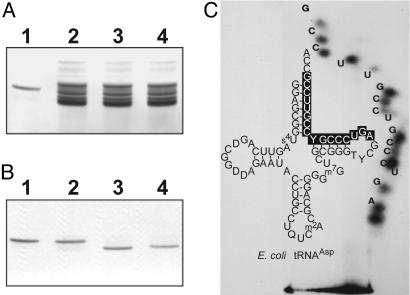
Isolation and Identification of the RNA Species Charged with Glutamate by YadB. To isolate the RNA substrate of YadB from the other components of crude tRNA, the α-NH2 group of the Glu residue of this Glu-RNA was reacted with sulfo-NHS-LC-LC-biotin (23). The biotinylated Glu-RNA was then adsorbed specifically on a streptavidin-agarose column. After deacylation at pH 8.8 on the column, the eluted RNA migrates as a single band in denaturing PAGE (Fig. 3A, lane 1). This RNA is fully glutamylable (1,700 pmol/A260 nm) by YadB, but it is not charged by pure E. coli GluRS or GlnRS. To confirm that this RNA is distinct from the tRNA substrates of GluRS and GlnRS, the electrophoretic mobility at pH 6.5 of its glutamylated form was compared with that of Glu-tRNAGlu and Gln-tRNAGln obtained by charging crude E. coli tRNA with either GluRS or GlnRS from E. coli. Detection of the 14C-labeled aa-tRNAs by phosphorimaging confirmed that aminoacylation of crude tRNA by YadB produces a homogeneous product that migrates as a single band and at the same speed as the purified RNA glutamylated by YadB and that the mobility of this Glu-RNA is slower than those of Glu-tRNAGlu and Gln-tRNAGln (Fig. 3B).
Fig. 3.
Identification and characterization of the tRNA charged by YadB. (A) PAGE in 40 mM Mops (pH 6.5)/10 mM sodium acetate/1 mM EDTA. Lane 1, 2 μg of purified RNA charged with l-[14C]Glu by E. coli YadB; lanes 2–4, 40 μgof unfractionated E. coli tRNA charged with l-[14C]Glu by E. coli YadB or GluRS or with l-[14C]Gln by GlnRS, respectively. The RNAs were stained with methylene blue. (B) PhosphorImager exposure of the gel shown in A. (C) Homochromatogram of RNA fragment specific of E. coli tRNAAsp and cloverleaf fold of this tRNA. The oligonucleotide identifying tRNAAsp is displayed on a black background.
The RNA substrate of YadB was identified by sequencing. We determined that the sequence of the 16 nucleotides of its 3′ end was used as bait for a blast search of E. coli genome. This sequence is found only in the three genes encoding an identical tRNAAsp, revealing that the unknown RNA is tRNAAsp (Fig. 3C). This conclusion was strengthened after identification in this RNA of the seven modified nucleotides (s4U, D, Q, m2A, m7G, T, and Ψ) characterizing E. coli tRNAAsp (39) and by the finding that they are present in the same molar ratios in the RNA substrate of YadB and in tRNAAsp.
Kinetics of tRNAAsp Glutamylation. At first, it appeared that pure tRNAAsp can be fully charged by YadB and by AspRS. The catalytic constants of glutamylation of this tRNA by YadB and those of its aspartylation by E. coli AspRS are compared in Table 2. Both enzymes display similar Km values for tRNAAsp (in the lower μM range) in contrast to the Km values for the amino acid that differ strongly (3 and 0.022 mM for YadB and AspRS). This finding indicates a low affinity of YadB for Glu, in agreement with the kinetic analysis of the ATP-PPi exchange (see Table 1). Despite this difference, the two enzymes charge tRNAAsp with comparable rates (kcat) and catalytic efficiencies (kcat/Km) with respect to tRNA. As to the amino acid, however, the catalytic efficiency of YadB is about three orders of magnitude lower than that of AspRS. Remarkably, however, GoA, which inhibits YadB competitively with respect to Glu and ATP (Table 1), exhibits a strong affinity for this enzyme (in the μM range).
Table 2. Kinetic constants of glutamylation and aspartylation of E. coli tRNAAsp by YadB and AspRS.
|
E. coli tRNAAsp
|
||||||
|---|---|---|---|---|---|---|
|
Km, μM
|
kcat/Km, μM·s-1
|
|||||
| Enzyme | tRNA | ATP | Amino acid | Kcat, s-1 | tRNA | Amino acid |
| YadB | 0.15 | 170 | 3,000 | 2.2 | 14.7 | 0.73 × 10-3 |
| AspRS | 0.2 | ND | 22 | 6.8 | 34 | 0.31 |
| kcat/Km[AspRS]/kcat/Km[YadB] (tRNA) | 2.31 | |||||
| kcat/Km[AspRS]/kcat/Km[YadB] (amino acid) | 424 | |||||
The conditions are described in Materials and Methods.
These results suggest that in vivo the only tRNA significantly glutamylated by YadB is the single tRNAAsp species. Taking into account the high intracellular Glu concentration in E. coli and in several other prokaryotes (40), the low affinity of YadB for Glu in the tRNA-independent activation reaction could allow it to be a good sensor of intracellular Glu concentration, which, for instance, increases significantly in E. coli during osmotic stress (41).
Because the ATP-PPi exchange is promoted only by l-Glu and only tRNAAsp is aminoacylated (22), it can be concluded that YadB is a Glu-activating enzyme that mischarges tRNAAsp.
Glutamylated tRNAAsp Does Not Interact with EF-Tu. Considering the significant kcat value of YadB for tRNAAsp glutamylation (Table 2), and the presence of YadB in E. coli (see below), glutamylated tRNAAsp is likely to be present in these cells. Its participation to protein synthesis would imply interaction with EF-Tu. This possibility was checked by the protection induced by EF-Tu against hydrolysis of the Glu-tRNAAsp ester bond. Although the half-lives of Glu-tRNAGlu and Asp-tRNAAsp are substantially increased in the presence of EF-Tu (from 25 to >100 min at pH 7.4), the half-life of Glu-tRNAAsp is unchanged (7.5 min). This lack of protection suggests that Glu-tRNAAsp binds poorly to EF-Tu and, therefore, is probably not used for ribosome-mediated protein synthesis, a conclusion consistent with the lack of toxicity of YadB overexpressed in E. coli (see below). The behavior of Glu-tRNAAsp resembles that of naturally mischarged tRNAs, such as Asp-tRNAAsn, that are unable to bind to EF-Tu before conversion of the noncognate amino acid to that homologous to the tRNA (42). This result is puzzling, however, because, according to data from Uhlenbeck's group (43), Glu-tRNAAsp may have appreciable affinity for the elongation factor.
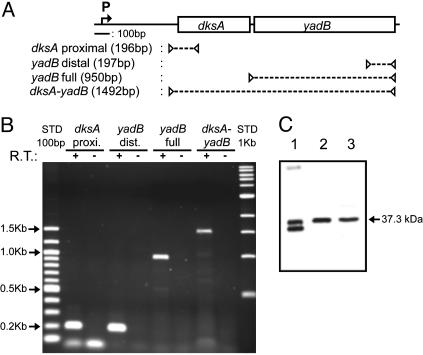
The YadB mRNA and Its Translation Product are Expressed in Vivo. The mRNA. The putative presence of a YadB mRNA in E. coli cells was investigated by RT-PCR with the products corresponding to the 3′ portion of yadB or the entire gene (Fig. 4A). Amplification reactions were positive for both fragments (Fig. 4B). Because no putative promoter could be identified close to the translational initiation site of the yadB ORF, its coexpression with the upstream gene dksA was also investigated with a PCR probe covering both genes (Fig. 4 A and B). The presence of a 1.5-kbp RT-PCR amplification product indicates that the yadB gene is expressed as an operon with dksA in E. coli grown on LB medium (Fig. 4A). Expression was identical in early-log and stationary phases. The conservation of a synteny of the two genes in other bacteria, such as Pseudomonas aeruginosa (44), Yersinia pestis (45), and Vibrio cholerae (46), suggests that they participate to the same metabolic pathway. The dksA gene, nonessential in E. coli, was first identified as a dosage-dependent suppressor of dnaK, which encodes the hsp70-like protein (47). In E. coli, DksA is thought to regulate at a translational level the induction of RpoS, an alternative σ-subunit of RNA polymerase, and referred to as the major regulator of the general stress response (48, 49). Mutations in dksA decrease the virulence of Salmonella typhimurium in mice, consistent with the role for RpoS in pathogenesis (48). In P. aeruginosa, DksA regulates at the posttranscriptional level the production of the extracellular virulence factors encoded by lasB and rhlAB (50). The coexpression of dksA and yadB, together with the properties of YadB reported here, strongly suggests this protein could be involved in this pathway by a yet unknown translational mechanism.
Fig. 4.
In vivo expression of E. coli YadB gene. (A) Physical map of the E. coli dksA-yadB region showing the localization of the RT-PCR amplification targets. The promoter for dksA identified by Kang and Craig (47) is indicated by the letter P. (B) Expression of the YadB mRNA in E. coli K12 grown in LB medium/agarose gel electrophoresis of the products of the RT-PCR reactions and of controls without the reverse polymerase (R.T.). (C) Expression of the YadB protein. Lanes 1–3, analysis by Western blot of, respectively, 10 ng of overexpressed pure YadB and 9 μg of crude and S100 extracts from wild-type E. coli.
The protein. Presence of YadB in E. coli was investigated by immunodetection on Western blots. Analysis of crude protein or S100 extracts identified a polypeptide chain of 37.3 kDa able to react with anti-YadB antibodies. This band is also present in samples containing YadB purified from the overexpressing E. coli strain (Fig. 4C).
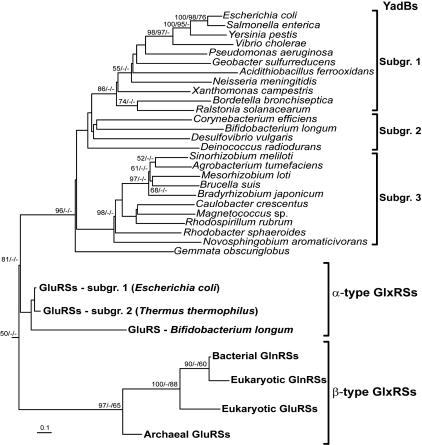
YadB Is a Bacterial-Restricted Protein. Analysis of all available bacterial, archaeal, and eukaryotic genomes revealed sequences similar to that of E. coli YadB only in the bacterial domain, more specifically in some Actinobacteria, some Cyanobacteria, most Proteobacteria, and Deinococcus radiodurans from the Thermus/Deinococcus group (Fig. 5).
Fig. 5.
Rooted phylogenetic tree of YadB proteins and GluRS/GlnRS representatives. Bootstrap values (>50%) calculated from 400 replicates in the maximum-parsimony analysis, 1,000 replicates in the neighbor-joining analysis, and 5,000 puzzling steps in the maximum-likelihood analysis are indicated at their corresponding nodes separated by ′/′ in that specific order. The tree is rooted with P. horikoshi class I LysRS (not shown).
To verify whether the proteins encoded by these genes display the functional characteristics of E. coli YadB, we expressed the protein from Neisseria meningitidis in E. coli. As expected on the basis of the sequence similarities, the purified Neisseria protein showed functional properties similar to those of E. coli YadB (results not shown). The evidence that the identified bacterial genes encoding YadB proteins should have the ubiquitous function of glutamylating tRNAAsp is strong.
Evolutionary Linkage Among YadBs, GluRSs, and GlnRSs. Sequences corresponding to GluRSs (α- and β-type) and GlnRSs were retained by the search for YadB proteins, confirming their similarity with that of E. coli YadB. In nearly all bacteria possessing an ORF encoding a GlnRS, a homologue of E. coli YadB is present, with the exception of the Pasteurellaceae (a subgroup among Proteobacteria) and the Firmicute Clostridium difficile. Also, in a majority of bacteria bearing a YadB homologue but no GlnRS, two GluRS sequences were found and they were easily distinguished from YadB by their larger size and a distinctive sequence feature (see below). In the eukaryota, the α-type organellar GluRS was the closest hit, followed by the cytoplasmic β-type GluRSs and GlnRSs. In the archaea, no YadB homologue was found.
A neighbor-joining phylogenetic tree obtained from aligned amino acid sequences of E. coli class I aaRSs, including YadB sequences (data not shown), indicates that YadB is more closely related to GluRS and GlnRS than to any other aaRS. The structure-based alignment of the N-terminal amino acid sequences of 26 putative YadB proteins and of GluRSs and GlnRSs representatives, reveals the existence of three different subgroups of YadBs (see Fig. 1). They are characterized by the conserved specific HxGS, HxGN, and HxGH sequences aligned with the HIGH consensus of the class I aaRS sequence signature (51) and by most of the α-type-specific insertions and deletions (shown in the Fig. 1 Upper as green segments and gaps, respectively) found in the N-terminal catalytic module (domains 1–3) with the exception of those located in the second half of domain 3, the so-called SC fold (6, 52). This last region, which corresponds to the C terminus of YadBs, is poorly conserved and displays little similarity with either α- or β-type GlxRSs (Fig. 1).
One feature specific to all YadBs is the presence of Ile, Leu, or Val, (position 57 in the E. coli enzyme, shown by the arrow in Fig. 1), substituting a Thr residue in GlxRSs (positions 43, 44, and 68 in T. thermophilus GluRS, E. coli GluRS, and E. coli GlnRS, respectively), in the highly conserved motif “Rx(E/D)DTD” of bacterial, organellar, and archaeal GluRSs, and the conserved motif “Rf(E/D)DTN′” of eukaryotic GluRSs and of all GlnRSs. In the T. thermophilus GluRS/tRNAGlu/GoA structure, the corresponding Thr-43 side chain is hydrogen-bonded with the 5′-hydroxyl group of A76 (31), which, by directly contacting Glu, could help the enzyme to select its amino acid substrate. Thus, the absence of this crucial residue or of any functional equivalent at this position in all YadB proteins suggests the mode of interaction of tRNAAsp with YadB differs from that of tRNAGlu with GluRS. This finding could also be linked to the unforeseen feature of YadB, to activate Glu in the absence of tRNA.
The evolutionary linkage between YadB and members of the GluRS/GlnRS subfamily was investigated by maximum-parsimony, maximum-likelihood, and neighbor-joining analyses of the aligned sequences by using the class I LysRS from Pyrococcus horikoshii, the aaRS closest to the GluRS/GlnRS subfamily (53, 54), as an outgroup for the constructions. The maximum-likelihood phylogenetic tree could not be fully resolved and thus was useless as a general guiding tree. However, the maximum-parsimony evolutionary tree and the neighbor-joining distance tree have very similar topologies and show that YadB proteins represent a monophylogenic group that branches between the α- and β-type GlxRSs (Fig. 5), confirming their correct identification as homologues of E. coli YadB and supporting the existence of the three YadB subgroups identified above from a specific sequence motif.
Questions and Perspectives. The presence of YadB proteins in almost all bacteria that possess a GlnRS sheds a new light to the puzzling fact that only a minority of bacteria did integrate eukaryotic GlnRS (13). Indeed, a single substitution in the anticodon of tRNAAsp (GUC) is sufficient to convert it into a Glu anticodon (UUC). Such substitution prevents aspartylation of tRNAAsp variant with a U34 mutation by E. coli AspRS (55) but should not affect its glutamylation by YadB because it lacks the anticodon-binding domain. Therefore, glutamylation by YadB of tRNAs variants with Glu or Gln anticodons could have offered an alternative pathway to incorporate Glu into protein, during the transition from ND-GluRSs to discriminating GluRSs in parallel with the acquisition of a GlnRS. For this to happen, however, Glu-tRNAAsp must have been recognized by an elongation factor, which is not the case with the extant E. coli EF-Tu.
The recognition of tRNAAsp by YadB could support a model according to which pairs of ancestral aaRSs, of class I and class II, respectively, would bind opposite sides of the same tRNA molecule (54). Such a model is based in part on evidence for interaction of extant AspRS and ArgRS with the same tRNA (56), on the requirement for the binding of both class I and class II LysRSs to tRNAPyl for its aminoacylation (57), and on the observation that tRNAGlu and tRNAAsp possess the same G73 identity determinant (58). The model would imply that the ancestral AspRS and GluRS would have formed a pair of class I and class II primitive aaRSs interacting with the same tRNA. In favor of this view is the linkage of Asp and Glu with their codons (55). Docking experiments of YadB with the minor groove side of the acceptor arm of tRNAAsp bound to modern AspRS were unsuccessful (results not shown); however, this fact does not exclude the possibility that ancestral AspRS and YadB (a miniGluRS) formed a ternary complex with tRNAAsp. From another viewpoint, the similar affinities of tRNAAsp for YadB lacking the anticodon-binding domain (but bearing an SC fold; see Figs. 1 and 2) and AspRS (Table 2) are intriguing and suggest a stronger binding of the amino acid-accepting half and part of the anticodon stem of tRNAAsp with YadB than with AspRS.
The fate of glutamylated tRNAAsp formed by YadB remains an open question. The present data exclude a direct role in classical protein synthesis. This aminoacylated tRNAAsp, however, could be an amino acid donor in non-ribosome-mediated peptide synthesis or in another metabolic pathway. Experiments are needed to unravel these different possibilities.
Acknowledgments
We thank G. Eriani for E. coli AspRS and tRNAAsp and J. Perona and R. Chênevert for gifts of pure E. coli GlnRS and GoA, respectively. This work was supported by Natural Sciences and Engineering Research Council of Canada Grant OGP0009597 and Fonds pour la Formation de Chercheurs et l'Aide à la Recherche du Québec Grant 2003-ER-2481 (to J.L.), by the Centre National de la Recherche Scientifique, the French Association de la Recherche contre le Cancer, French Ministry of Industry Grant ASG (to C.C.), the Genopoles Network of the French Ministry of Research, and the Université Louis Pasteur (Strasbourg). D.Y.D. and M. B. received doctoral fellowships, respectively, from Fonds pour la Formation de Chercheurs et l'Aide à la Recherche du Québec and from Ministère de la Recherche et de la Technologie.
Abbreviations: aaRS, aminoacyl-tRNA synthetase; GluRS, glutamyl-tRNA synthetase; GlnRS, glutaminyl-tRNA synthetase; AspRS, aspartyl-tRNA synthetase; ND-GluRS, nondiscriminating GluRS; GlxRS, GluRS and GlnRS superfamily; EF-Tu, elongation factor Tu; SC, stem-contact; GoA, glutamol-AMP.
See Commentary on page 7493.
References
- 1.Woese, C. R., Olsen, G. J., Ibba, M. & Söll, D. (2000) Microbiol. Mol. Biol. Rev. 64, 202-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schimmel, P. & Ribas De Pouplana, L. (2000) Trends Biochem. Sci. 25, 207-209. [DOI] [PubMed] [Google Scholar]
- 3.Francklyn, C. S., Perona, J. J., Pütz, J. & Hou, Y. M. (2002) RNA 8, 1363-1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fujita, N., Mori, H., Yura, T. & Ishihama, A. (1994) Nucleic Acids Res. 22, 1637-1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blattner, F. R., Plunkett, G., III, Bloch, C. A., Perna, N. T., Burland, V., Riley, M., Collado-Vides, J., Glasner, J. D., Rode, C. K., Mayhew, G. F., et al. (1997) Science 277, 1453-1474. [DOI] [PubMed] [Google Scholar]
- 6.Nureki, O., Vassylyev, D. G., Katayanagi, K., Shimizu, T., Sekine, S., Kigawa, T., Miyazawa, T., Yokoyama, S. & Morikawa, K. (1995) Science 267, 1958-1965. [DOI] [PubMed] [Google Scholar]
- 7.Ibba, M. & Söll, D. (2000) Annu. Rev. Biochem. 69, 617-650. [DOI] [PubMed] [Google Scholar]
- 8.Gagnon, Y., Lacoste, L., Champagne, N. & Lapointe, J. (1996) J. Biol. Chem. 271, 14856-14863. [DOI] [PubMed] [Google Scholar]
- 9.Lapointe, J., Duplain, L. & Proulx, M. (1986) J. Bacteriol. 165, 88-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schön, A., Kannangara, C. G., Gough, S. & Söll, D. (1988) Nature 331, 187-190. [DOI] [PubMed] [Google Scholar]
- 11.Wilcox, M. & Nirenberg, M. (1968) Proc. Natl. Acad. Sci. USA 61, 229-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ruan, B. F., Ahel, I., Ambrogelly, A., Becker, H. D., Bunjun, S., Feng, L., Tumbula-Hansen, D., Ibba, M., Korencic, D., Kobayashi, H., et al. (2001) Acta Biochim. Pol. 48, 313-321. [PubMed] [Google Scholar]
- 13.Lamour, V., Quevillon, S., Diriong, S., N′Guyen, V. C., Lipinski, M. & Mirande, M. (1994) Proc. Natl. Acad. Sci. USA 91, 8670-8674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Handy, J. & Doolittle, R. F. (1999) J. Mol. Evol. 49, 709-715. [DOI] [PubMed] [Google Scholar]
- 15.Brown, J. R. & Doolittle, W. F. (1999) J. Mol. Evol. 49, 485-495. [DOI] [PubMed] [Google Scholar]
- 16.Doolittle, R. F. & Handy, J. (1998) Curr. Opin. Genet. Dev. 8, 630-636. [DOI] [PubMed] [Google Scholar]
- 17.Rould, M. A., Perona, J. J., Söll, D. & Steitz, T. A. (1989) Science 246, 1135-1142. [DOI] [PubMed] [Google Scholar]
- 18.Siatecka, M., Rozek, M., Barciszewski, J. & Mirande, M. (1998) Eur. J. Biochem. 256, 80-87. [DOI] [PubMed] [Google Scholar]
- 19.Russell, R. R. & Pittard, A. J. (1971) J. Bacteriol. 108, 790-798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lapointe, J. & Delcuve, G. (1975) J. Bacteriol. 122, 352-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Madore, E., Florentz, C., Giegé, R., Sekine, S., Yokoyama, S. & Lapointe, J. (1999) Eur. J. Biochem. 266, 1128-1135. [DOI] [PubMed] [Google Scholar]
- 22.Campanacci, V., Dubois, D. Y., Becker, H. D., Kern, D., Spinelli, S., Valencia, C., Pagot, F., Salomoni, A., Grisel, S., Vincentelli, R., et al. (2004) J. Mol. Biol. 337, 273-283. [DOI] [PubMed] [Google Scholar]
- 23.Pütz, J., Wientges, J., Sissler, M., Giegé, R., Florentz, C. & Schwienhorst, A. (1997) Nucleic Acids Res. 25, 1862-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Donis-Keller, H. (1979) Nucleic Acids Res. 7, 179-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keith, G., Desgres, J. & de Murcia, G. (1990) Anal. Biochem. 191, 309-313. [DOI] [PubMed] [Google Scholar]
- 26.Grosjean, H., Keith, G. & Droogmans, L. (2004) in Methods in Molecular Biology, ed. Gott, J. M. (Humana Press, Totowa, NJ).
- 27.Rudinger, J., Hillenbrandt, R., Sprinzl, M. & Giegé, R. (1996) EMBO J. 15, 650-657. [PMC free article] [PubMed] [Google Scholar]
- 28.Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. (1990) J. Mol. Biol. 215, 403-410. [DOI] [PubMed] [Google Scholar]
- 29.Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F. & Higgins, D. G. (1997) Nucleic Acids Res. 25, 4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hall, T. A. (1999) Nucleic Acids Symp. Ser. 41, 95-98. [Google Scholar]
- 31.Sekine, S., Nureki, O., Dubois, D. Y., Bernier, S., Chenevert, R., Lapointe, J., Vassylyev, D. G. & Yokoyama, S. (2003) EMBO J. 22, 676-688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Felsenstein, J. (1996) Methods Enzymol. 266, 418-427. [DOI] [PubMed] [Google Scholar]
- 33.Dayhoff, M. O., Eck, R. V. & Park, C. M. (1972) in Atlas of Protein Sequence and Structure, ed. Dayhoff, M. O. (National Biomedical Research Foundation, Washington, DC), Vol. 5, pp. 89-99. [Google Scholar]
- 34.Strimmer, K. & von Haeseler, A. (1996) Mol. Biol. Evol. 13, 964-969. [Google Scholar]
- 35.Jones, D. T., Taylor, W. R. & Thornton, J. M. (1992) Comput. Appl. Biosci. 8, 275-282. [DOI] [PubMed] [Google Scholar]
- 36.Page, R. D. W. (1996) Comput. Appl. Biosci. 12, 357-358. [DOI] [PubMed] [Google Scholar]
- 37.Kern, D. & Lapointe, J. (1980) J. Biol. Chem. 255, 1956-1961. [PubMed] [Google Scholar]
- 38.Desjardins, M., Garneau, S., Desgagnes, J., Lacoste, L., Yang, F., Lapointe, J. & Chênevert, R. (1998) Bioorg. Chem. 26, 1-13. [Google Scholar]
- 39.Sekiya, S., Mori, M., Takahashi, N. & Nishimura, S. (1980) Nucleic Acids Res. 8, 3809-3827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Csonka, L. N. (1989) Microbiol. Rev. 53, 121-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McLaggan, D., Naprstek, J., Buurman, E. T. & Epstein, W. (1994) J. Biol. Chem. 269, 1911-1917. [PubMed] [Google Scholar]
- 42.Becker, H. D. & Kern, D. (1998) Proc. Natl. Acad. Sci. USA 95, 12832-12837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lariviere, F. J., Wolfson, A. D. & Uhlenbeck, O. C. (2001) Science 294, 165-168. [DOI] [PubMed] [Google Scholar]
- 44.Stover, C. K., Pham, X. Q., Erwin, A. L., Mizoguchi, S. D., Warrener P., Hickey M. J., Brinkman F. S., Hufnagle W. O., Kowalik D. J., Lagrou M., et al. (2000) Nature 406, 959-964. [DOI] [PubMed] [Google Scholar]
- 45.Parkhill, J., Wren, B. W., Thomson, N. R., Titball, R. W., Holden M. T., Prentice M. B., Sebaihia M., James K. D., Churcher C., Mungall K. L., et al. (2001) Nature 413, 523-527. [DOI] [PubMed] [Google Scholar]
- 46.Heidelberg, J. F., Eisen, J. A., Nelson, W. C., Clayton, R. A., Gwinn, M. L., Dodson, R. J., Haft, D. H., Hickey E. K., Peterson J. D., Umayam L., et al. (2000) Nature 406, 477-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kang, P. J. & Craig, E. A. (1990) J. Bacteriol. 172, 2055-2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Webb, C., Moreno, M., Wilmes-Riesenberg, M., Curtiss, R., III, & Foster, J. W. (1999) Mol. Microbiol. 34, 112-123. [DOI] [PubMed] [Google Scholar]
- 49.Brown, L., Gentry, D., Elliott, T. & Cashel, M. (2002) J. Bacteriol. 184, 4455-4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jude, F., Kohler, T., Branny, P., Perron, K., Mayer, M. P., Comte, R. & van Delden, C. (2003) J. Bacteriol. 185, 3558-3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Webster, T., Tsai, H., Kula, M., Mackie, G. A. & Schimmel, P. (1984) Science 226, 1315-1317. [DOI] [PubMed] [Google Scholar]
- 52.Sugiura, I., Nureki, O., Ugaji-Yoshikawa, Y., Kuwabara, S., Shimada, A., Tateno, M., Lorber, B., Giegé, R., Moras, D., Yokoyama, S. & Konno, M. (2000) Structure (London) 8, 197-208. [DOI] [PubMed] [Google Scholar]
- 53.Terada, T., Nureki, O., Ishitani, R., Ambrogelly, A., Ibba, M., Söll, D. & Yokoyama, S. (2002) Nat. Struct. Biol. 9, 257-262. [DOI] [PubMed] [Google Scholar]
- 54.Ribas de Pouplana, L. & Schimmel, P. (2001) Cell 104, 191-193. [DOI] [PubMed] [Google Scholar]
- 55.Nameki, N., Tamura, K., Himeno, H., Asahara, H., Hasegawa, T. & Shimizu, M. (1992) Biochem. Biophys. Res. Commun. 189, 856-862. [DOI] [PubMed] [Google Scholar]
- 56.Sissler, M., Eriani, G., Martin, F., Giegé, R. & Florentz, C. (1997) Nucleic Acids Res. 25, 4899-4906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Polycarpo, C., Ambrogelly, A., Ruan, B., Tumbula-Hansen, D., Ataide, S. F., Ishitani, R., Yokoyama, S., Nureki, O., Ibba, M. & Söll, D. (2003) Mol. Cell 12, 287-294. [DOI] [PubMed] [Google Scholar]
- 58.Giegé, R., Sissler, M. & Florentz, C. (1998) Nucleic Acids Res. 26, 5017-5035. [DOI] [PMC free article] [PubMed] [Google Scholar]