Abstract
Transient or reversible protein–protein interactions are commonly used to ensure efficient targeting of signaling enzymes to their cellular substrates. These interactions include direct binding to substrate, interaction with an accessory or scaffold protein, and positioning at subcellular locations in proximity to substrates. The existence of specialized targeting mechanisms raises the possibility of designing inhibitors that do not block enzyme activity per se, but rather interfere with targeting of the enzyme to one or more of its substrates within the cell. Here, we identify small organic molecules that specifically block targeting of the protein phosphatase calcineurin to its substrate nuclear factor of activated T cells (NFAT, also termed NFATc) and show that they are effective inhibitors of calcineurin-NFAT signaling.
Transient or weak protein–protein interactions are widely used in intracellular signaling pathways for docking signaling proteins to their downstream partners or substrates (1–5), targeting proteins to signaling complexes (6, 7), or transmitting activating conformational changes within larger complexes (8). Peptide inhibitors of these interactions are providing new tools in cell biology, for example to map the physiological coupling of receptors to specific G proteins and effectors (9, 10), and to dissect the mechanisms by which α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptor trafficking and recruitment of signaling proteins to N-methyl-d-aspartate (NMDA) receptor complexes alter neuronal synaptic signaling (11–15). In addition, studies with peptide inhibitors have indicated that blockade of specific protein–protein interactions has therapeutic promise for treating inflammatory and autoimmune diseases (16, 17), blunting development of pathological myocardial hypertrophy (18, 19), killing cancer cells selectively (20), and limiting ischemic damage in stroke (21).
Small organic molecules are particularly attractive as inhibitors of intracellular protein–protein interactions because their virtually limitless structural diversity can be exploited to achieve tight binding to the target protein surface and because effective inhibitors can be tailored chemically to improve stability, to eliminate nonspecific side effects, and to enhance delivery in vivo or in cell culture. Whereas small organic inhibitors have shown promise in interfering with binding of extracellular protein ligands to their protein partners and with assembly of certain dimeric or multimeric protein complexes, their utility in preventing specific enzyme–substrate interactions that direct intracellular signaling remains to be established (22–24).
We have used calcineurin-nuclear factor of activated T cells (NFAT) signaling as a test case. Calcineurin, a phosphoprotein phosphatase, is poised at a branch point of calcium/calmodulin signaling and controls the function of diverse effector proteins, ranging from transcription factors to enzymes, transmembrane ion channels, and proteins involved in apoptosis (25, 26). Its effectors include the four NFAT-family proteins, transcription factors that activate cytokine gene expression in T cells (27–30) and that also participate in the genetic programs of muscle fiber-type specialization, osteoclast differentiation, cardiac valve development, and myocardial hypertrophy (29–32). The conventional method of blocking calcineurin-NFAT signaling is to apply the immunosuppressive compounds cyclosporin A (CsA) and FK506, which, in the form of CsA–cyclophilin or FK506–FKBP12 complexes, inhibit the enzymatic activity of calcineurin toward all its physiological substrates (33). However, calcineurin employs a range of targeting mechanisms (1, 34–43) that offer conceptually novel possibilities for disrupting calcineurin-substrate signaling. In particular, a protein–protein interaction of calcineurin with NFAT-family proteins controls the efficiency of NFAT dephosphorylation in vitro and in cells (1, 16, 44, 45). Here, we identify inhibitors of calcineurin-NFAT signaling that act at this protein–protein contact rather than at the calcineurin catalytic site.
Materials and Methods
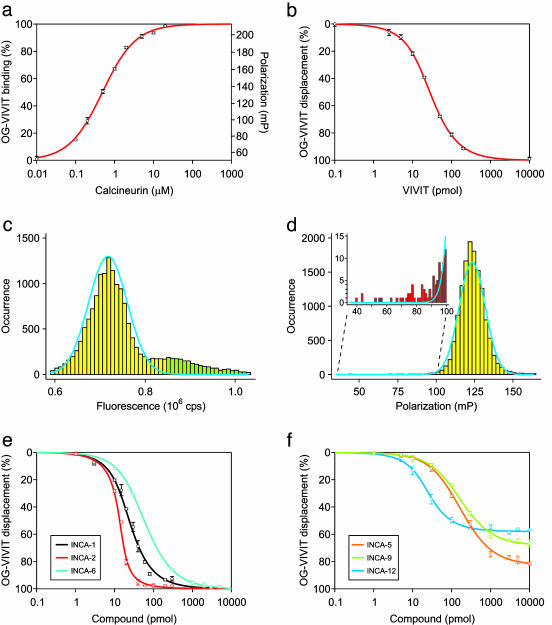
Fluorescence Polarization Assay. Fluorescence measurements were made on samples arrayed in 384-well plates by using an Analyst plate reader (Molecular Devices) to monitor the interaction between the catalytic domain of human calcineurin Aα (16) and an Oregon Green-labeled VIVIT peptide (OG-VIVIT). See Supporting Materials and Methods, which is published as supporting information on the PNAS web site, for a full technical description of the assay. Experimental polarization data from simple and competitive binding experiments were fitted to analytic expressions for two-state (Fig. 1a) or three-state (Fig. 1 b and e) equilibrium models as described in Supporting Materials and Methods. The initial screen for inhibitors examined 16,320 compounds in the DiverSet E library (ChemBridge, San Diego). Additional stocks of compounds were obtained from Chem-Bridge, AsInEx (Moscow), and Maybridge (Tintagel, U.K.).
Fig. 1.
Fluorescence polarization assay of binding at the NFAT recognition site on calcineurin. (a) Binding of 30 nM fluorescent VIVIT (OG-VIVIT) as a function of calcineurin added to the 10-μl assay volume. The Kd estimated by fitting a two-state equilibrium binding model to the experimental data was 0.55 μM. (b) Displacement of OG-VIVIT from calcineurin by unlabeled VIVIT peptide. The IC50 for VIVIT under the conditions of this experiment was ≈25 pmol. The Kd for unlabeled VIVIT estimated by fitting a competitive three-state binding model to the experimental data was 0.42 μM. (c) Histogram of total fluorescence for 16,320 samples in the high-throughput assay (cps, counts per second). The blue curve depicts a Gaussian fit to the main peak of the fluorescence intensity distribution. Compounds that caused total fluorescence to fall either below the 1st or above the 99th percentile of the Gaussian distribution (green bars) were excluded from further analysis. (d) Histogram of fluorescence polarization for 13,445 samples that met the criterion in c. The blue curve represents a Gaussian fit to the fluorescence polarization distribution. (Inset) An excess of samples (red bars) in the leftmost tail of the distribution over the number expected from the Gaussian distribution (blue curve) could reflect the presence of compounds that displaced VIVIT from calcineurin. (e) Displacement of OG-VIVIT from calcineurin by three INCA inhibitors. The IC50 values for INCA-1, INCA-2, and INCA-6 under the conditions of this experiment were in the range of 10–60 pmol, and the Kd values from the fitted curves were 0.44, 0.11, and 0.76 μM, respectively. (f) Displacement of OG-VIVIT by other representative INCA compounds, illustrating incomplete displacement of labeled peptide even at high concentrations of inhibitor. The value that would correspond to 100% displacement was obtained with samples of free peptide. The smooth curves are intended solely as an aid in grouping individual data sets.
Ligand-Binding Experiments. Binding of selected compounds to calcineurin was confirmed in T2-filtered NMR titration experiments by observing resonances of the free compounds in the presence of varied concentrations of calcineurin. Additionally, binding was observed in gel filtration experiments in which 50-μl samples containing compound and calcineurin were centrifuged in Micro Bio-Spin P-30 columns (Bio-Rad) for 4 min at 1,000 × g. The detailed conditions of these assays are stated in Supporting Materials and Methods.
Cellular and Biochemical Assays. Dephosphorylation of NFAT, nuclear import of NFAT, and induction of cytokine mRNAs in Cl.7W2 T cells were assessed as described (1, 16). Calcineurin activity in cell lysates was measured in a standard assay (46) by using 32P-labeled RII peptide substrate, with the adaptations noted in Supporting Materials and Methods.
Results
To identify specific inhibitors of the calcineurin-NFAT interaction, we developed a fluorescence polarization assay taking advantage of the high affinity of VIVIT peptide (16) for the NFAT recognition site on calcineurin. The fluorescent VIVIT peptide bound to calcineurin with dissociation constant (Kd) 0.50 ± 0.03 μM (Fig. 1a), and its binding was inhibited by unlabeled VIVIT with Kd 0.48 ± 0.05 μM (Fig. 1b). We screened a library of 16,320 small organic compounds for inhibitors of the calcineurin-VIVIT interaction. Library compounds that were themselves fluorescent, or that strongly quenched fluorescence, were identified by statistical analysis of the fluorescence intensity distribution and excluded (Fig. 1c). Fluorescence polarization data from samples containing the remaining 13,445 compounds were analyzed to identify candidate inhibitors that caused a decrease in the polarization signal (Fig. 1d), and the inhibitory activity of 11 of these compounds was verified in more extensive competitive binding assays (Fig. 1 e and f and data not shown). We term these compounds inhibitors of NFAT-calcineurin association (INCA).
Three compounds, INCA-1, INCA-2, and INCA-6, displaced VIVIT completely from calcineurin at low micromolar concentrations (Fig. 1e). The estimated dissociation constants were 0.50 ± 0.09 μM, 0.12 ± 0.03 μM, and 0.80 ± 0.11 μM, respectively. The remaining eight compounds at micromolar concentrations also displaced VIVIT, but inhibition reached a plateau at 50–90% displacement (Fig. 1f and data not shown). At least for INCA-5, INCA-7, INCA-12, and INCA-19, the plateau was not caused by limited aqueous solubility (data not shown). Two plausible physical explanations are that inhibitors in the second group only partially occlude the VIVIT binding site, or that these inhibitors bind to a nearby site and alter the geometry of the VIVIT binding site.
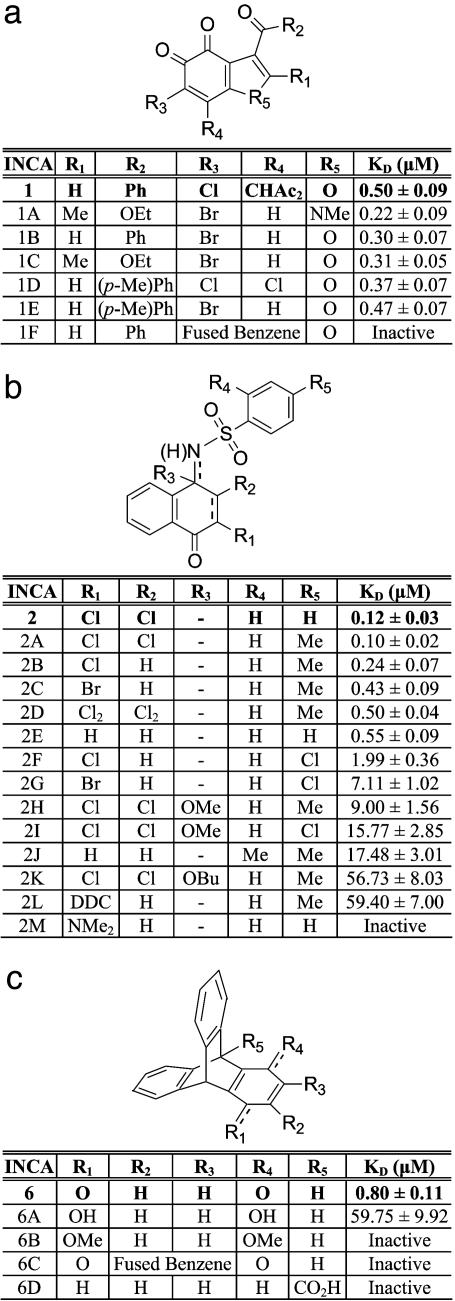
Further analysis focused on INCA-1, INCA-2, and INCA-6 because of their high affinities and their ability to displace the fluorescent probe completely from its binding site. To gain insight into the structure-activity relationships of INCA compounds, we examined a number of structural analogues of these compounds in competitive binding experiments (Fig. 2). In many cases, the inhibitory effectiveness was only marginally affected by conservative changes in ring substituents. However, certain changes caused moderate to dramatic losses of potency. For example, expansion of the ring system of INCA-1 (INCA-1F), or reduction of the vicinal keto groups of INCA-1 to hydroxyl groups or their replacement by halogen substituents (not shown), resulted in inactive compounds. Introduction of bulky substituents at R1 in INCA-2 (INCA-2L and INCA-2M) or of Cl at R5 (INCA-2F and INCA-2G) was detrimental to binding. Full reduction of the quinonimine of INCA-2 (not shown), reduction of the imino linkage with introduction of an alkyl ether at R3 (INCA-2H and INCA-2K), or reduction of INCA-6 to the hydroquinone or its dimethoxy derivative (INCA-6A and INCA-6B) caused a pronounced decrease in or loss of inhibitory activity. Expansion of the INCA-6 quinone ring to a naphthoquinone (INCA-6C) abolished activity.
Fig. 2.
Structure-activity relationships for three families of INCA compounds. INCA-1 (a), INCA-2 (b), and INCA-6 (c), with entries in boldface type, were identified in the high-throughput screen. Kd estimates are from fluorescence polarization data similar to those shown in Fig. 1e, analyzed using the three-state equilibrium binding model. In INCA-2D, the ring is disubstituted at positions R1 and R2, and a single bond connects the corresponding ring carbons. INCA-2H, INCA-2I, and INCA-2K are N-substituted amines whereas the other INCA-2 analogues are N-substituted imines. INCA-6 and INCA-6C are quinones, and the other INCA-6 analogues are substituted triptycenes. Ac, acetyl; Bu, butyl; DDC, 4,4-dimethyl-2,6-dioxo-cyclohexyl; Et, ethyl; Me, methyl; Ph, phenyl.
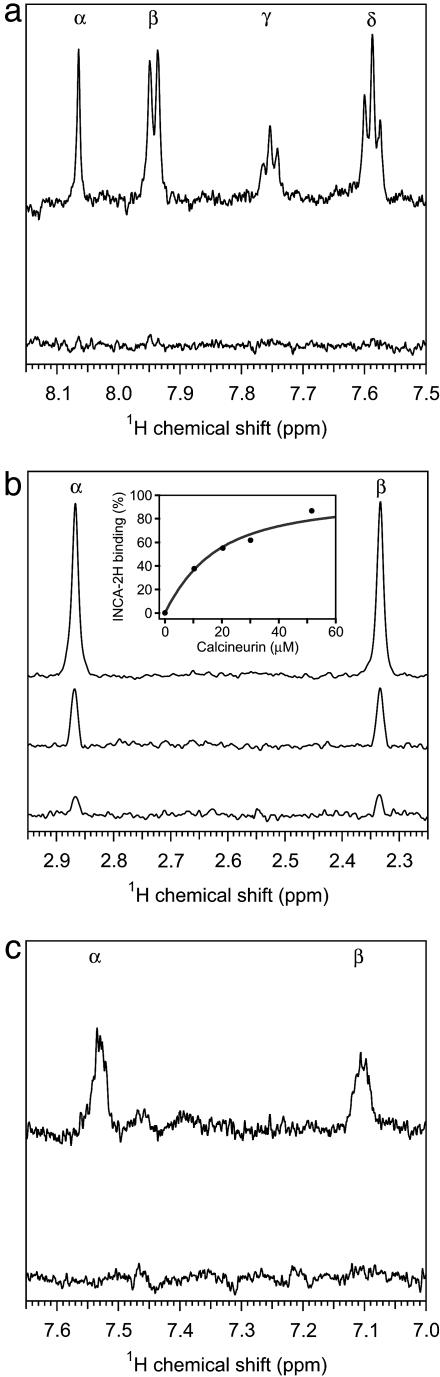
We used NMR titration spectroscopy to investigate binding of the compounds to calcineurin. In T2-filtered experiments, the proton resonances characteristic of the individual INCA compounds diminished or disappeared in the presence of calcineurin (Fig. 3), providing strong evidence that the compounds interact directly with calcineurin. The NMR titration data for INCA-1, INCA-2, and INCA-6 (Fig. 7, which is published as supporting information on the PNAS web site, and not shown) were consistent with dissociation constants at least an order of magnitude below the concentrations of calcineurin (5–20 μM) used in the experiments. The Kd derived from NMR titration curves for the intermediate-affinity compound INCA-2H was 11.9 ± 1.5 μM (Fig. 3b Inset). These estimates are in excellent agreement with Kd values determined by displacement of fluorescent VIVIT (Fig. 2).
Fig. 3.
Evidence for direct interaction between INCA compounds and calcineurin. (a–c) T2-filtered 1H NMR spectra of 10 μM INCA compound in the presence of different concentrations of calcineurin. (a) INCA-1, resonances from protons at R1 (α) and at the ortho (β), para (γ), and meta (δ) positions of R2 in the presence of 0 μM (upper trace) or 20 μM (lower trace) calcineurin. (b) INCA-2H, methyl resonances from R3 (α) and R5 (β) in the presence of 0 μM (top trace), 30 μM (middle trace), or 52 μM (bottom trace) calcineurin. (Inset) Bound INCA-2H, determined from the intensity integral of the methyl resonances, is plotted as a function of total calcineurin concentration present in the sample. Modeling the situation as a two-state equilibrium gave the fitted curve with Kd 11.9 μM. (c) INCA-6, resonances from the benzene ring protons (α, β) in the presence of 0 μM (upper trace) or 20 μM (lower trace) calcineurin.
Using miniaturized size exclusion chromatography, we also obtained direct visual evidence of the binding of INCA compounds to calcineurin (Fig. 8, which is published as supporting information on the PNAS web site). Under conditions where calcineurin, bovine IgG, and BSA eluted in the void volume of the column, INCA-1, INCA-2, or INCA-6 that had been preincubated with calcineurin coeluted with the protein whereas the compounds preincubated alone or with IgG or BSA were retained in the column.
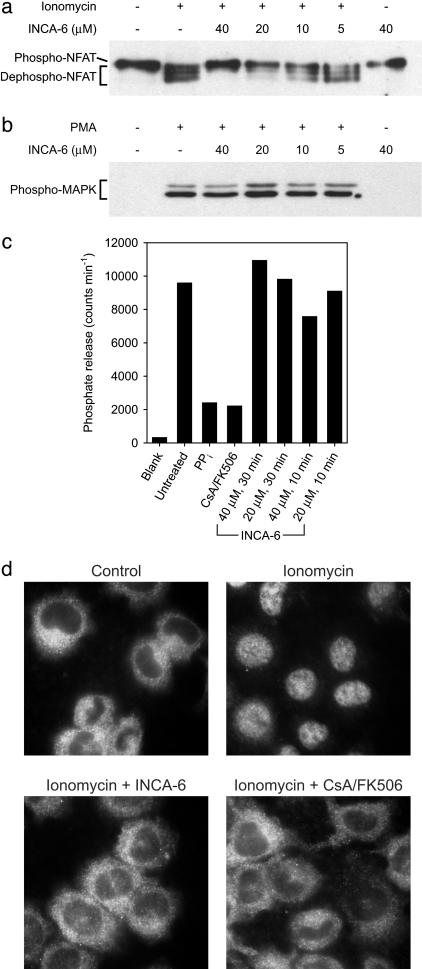
The ability of INCA-6 to inhibit NFAT activation was examined in cellular assays. Cl.7W2 T cells were used for these experiments to avoid the known nonspecific toxicity of quinones for primary T cells (see Comment 1 in Supporting Materials and Methods). Stimulation of cells with the calcium ionophore ionomycin caused dephosphorylation of NFAT, and the dephosphorylation depended on calcium-calmodulin-calcineurin signaling (Fig. 4a and not shown). Pretreatment with INCA-6 resulted in a concentration-dependent blockade of NFAT dephosphorylation that was partial with 10 μM INCA-6, nearly complete with 20 μM INCA-6, and total with 40 μM INCA-6 (Fig. 4a). INCA-6 at these concentrations did not cause a general impairment of intracellular signaling because activation of the protein kinase C–mitogen-activated protein (MAP) kinase signaling pathway was not blocked (Fig. 4b).
Fig. 4.
INCA-6 inhibits activation of NFAT in Cl.7W2 T cells. (a) Western blot of phosphorylated NFAT1 (Phospho-NFAT) and the faster-migrating dephosphorylated forms (Dephospho-NFAT). Cells were stimulated with ionomycin in the presence of the indicated concentrations of INCA-6. (b) Western blot of the activated form of p44/p42 mitogen-activated protein (MAP) kinase (Phospho-MAPK), phosphorylated at Thr-202 and Tyr-204. Cells were stimulated with PMA in the presence of the indicated concentrations of INCA-6. (c) Activity of calcineurin in lysates of cells that had been incubated with medium alone (Untreated) or with INCA-6 as indicated. The controls confirm that the phosphatase detected is calcineurin because selective inhibition of calcineurin by treating cells with a combination of CsA and FK506 (CsA/FK506) reduces phosphate release to the same extent as nonselective inhibition of phosphatases in the lysate with sodium pyrophosphate (PPi). (d) Cellular localization of NFAT1 detected by immunocytochemistry. Cells were stimulated with ionomycin in the absence of inhibitor, or in the presence of 20 μM INCA-6 or a combination of CsA and FK506 (CsA/FK506).
Cellular metabolism of quinones like INCA-6 can produce reactive oxygen species as byproducts, and calcineurin can be inactivated by oxidation (47–50). To assess whether treatment of intact cells with INCA-6 resulted in the inactivation of calcineurin, we incubated Cl.7W2 cells with 20 μM and 40 μM INCA-6 as for the NFAT dephosphorylation experiments, lysed the cells, and assayed calcineurin activity in the cell lysates. In three independent experiments, calcineurin activity in lysates from INCA-6-treated cells ranged from 57% to 122% of the control value (Fig. 4c) and was not consistently lower with the higher concentration of INCA-6 or with a longer time of treatment. We conclude that the block of NFAT dephosphorylation by INCA-6 is not explained by nonspecific inactivation of calcineurin (see Comment 2 in Supporting Materials and Methods).
Preventing the dephosphorylation of NFAT should also inhibit recruitment of NFAT from the cytoplasm to the cell nucleus. Cl.7W2 T cells were stimulated with ionomycin, alone or in the presence of INCA-6 or a combination of CsA and FK506, and NFAT was localized by immunocytochemistry. Like the established immunosuppressive drugs, INCA-6 completely blocked nuclear import of NFAT (Fig. 4d).
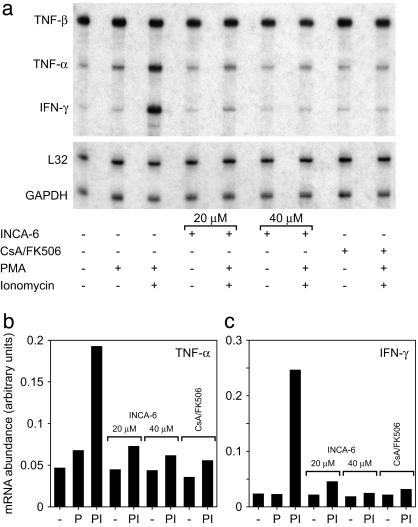
Finally, INCA-6 prevented induction of cytokine mRNAs that are downstream targets of NFAT (Fig. 5, Fig. 9, which is published as supporting information on the PNAS web site, and data not shown). Treatment of Cl.7W2 T cells with the phorbol ester phorbol 12-myristate 13-acetate (PMA) and ionomycin caused rapid induction of the mRNAs for tumor necrosis factor (TNF)-α, IFN-γ, granulocyte–macrophage/colony-stimulating factor (GM-CSF), lymphotactin (Ltn), macrophage inflammatory protein (MIP)-1α, and MIP-1β. Consistent with previous work (51–55), the increase in levels of these mRNAs was blocked by inhibiting calcineurin with a combination of CsA and FK506. Cytokine mRNA induction was likewise inhibited by 20 μM or 40 μM INCA-6, with 40 μM INCA-6 reducing mRNA levels to those in unstimulated cells. Levels of TNF-β, RANTES (regulated upon activation, normal T cell expressed and secreted), and macrophage migration inhibitory factor (MIF) mRNAs, which are not downstream targets of NFAT, were not increased at early times by treatment with PMA and ionomycin and were unaffected by INCA-6 (Figs. 5 and 9 and data not shown).
Fig. 5.
INCA-6 inhibits the induction of NFAT-dependent cytokine mRNAs in Cl.7W2 T cells. (a) RNase protection assay for transcripts of the genes encoding TNF-α, TNF-β, and IFN-γ, and of the housekeeping genes L32 and GAPDH. Cells were preincubated with INCA-6 or a combination of CsA and FK506 where indicated, and further stimulated with PMA or a combination of PMA and ionomycin. (b and c) Quantitation of radiolabel in protected probes for TNF-α and IFN-γ mRNAs, presented in the same order as the lanes in a. -, unstimulated; P, stimulated with PMA alone; PI, stimulated with PMA and ionomycin.
Discussion
Here, we have used our knowledge of the calcineurin-NFAT interaction in a stepwise strategy to identify small organic molecules that inhibit NFAT signaling by interfering with protein–protein recognition. After defining the principal site of calcineurin-NFAT interaction, we isolated a peptide ligand (VIVIT) with increased affinity for the site and verified that the peptide achieved selective inhibition of calcineurin-NFAT signaling and cytokine gene induction in cells (1, 16). This optimized ligand proved to be the key to a sensitive calcineurinpeptide binding assay. To identify nonpeptide inhibitors, we established that fluorescent VIVIT peptide is a specific probe for binding at the targeted site, used the probe in a fluorescence polarization assay to screen a library of organic compounds, and showed that compounds identified in this screen are effective inhibitors of calcineurin-NFAT signaling in vitro and in cells.
The INCA compounds we have identified interfere selectively with the interaction between calcineurin and its substrate NFAT without preventing dephosphorylation of other substrates. This substrate-selective enzyme inhibition represents a conceptual and practical advance over inhibition with CsA or FK506, which indiscriminately block all signaling downstream of calcineurin. Current biochemical techniques are increasingly efficient at pinpointing the protein–protein interactions that channel intracellular signaling, and considerable effort is being directed toward producing comprehensive maps of protein–protein interactions in yeast, worm, and mammalian cells (56–61). These efforts will provide increasing opportunities for selective interference with enzyme-substrate recognition or with recognition of particular protein partners in other signaling pathways.
Supplementary Material
Acknowledgments
We acknowledge the assistance of staff and use of facilities of the Institute of Chemistry and Cell Biology, Harvard Medical School. This research was supported by Grants GM038608 (to G.W.), AI40127 and CA42471 (to A.R.), and AI43726 (to P.G.H.) from the National Institutes of Health. J.A. was supported by a grant from Fundació la Caixa (00/012-00). Purchase and maintenance of NMR equipment used for this research were supported by National Institutes of Health Grant RR000995.
Abbreviations: NFAT, nuclear factor of activated T cells; CsA, cyclosporin A; INCA, inhibitor of NFAT-calcineurin association; PMA, phorbol 12-myristate 13-acetate; TNF, tumor necrosis factor.
References
- 1.Aramburu, J., García-Cozár, F., Raghavan, A., Okamura, H., Rao, A. & Hogan, P. G. (1998) Mol. Cell 1, 627-637. [DOI] [PubMed] [Google Scholar]
- 2.Schulman, B. A., Lindstrom, D. L. & Harlow, E. (1998) Proc. Natl. Acad. Sci. USA 95, 10453-10458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown, N. R., Noble, M. E. M., Endicott, J. A. & Johnson, L. N. (1999) Nat. Cell Biol. 1, 438-443. [DOI] [PubMed] [Google Scholar]
- 4.Takeda, D. Y., Wohlschlegel, J. A. & Dutta, A. (2001) J. Biol. Chem. 276, 1993-1997. [DOI] [PubMed] [Google Scholar]
- 5.Nash, P., Tang, X., Orlicky, S., Chen, Q., Gertler, F. B., Mendenhall, M. D., Sicheri, F., Pawson, T. & Tyers, M. (2001) Nature 414, 514-521. [DOI] [PubMed] [Google Scholar]
- 6.Tsunoda, S., Sierralta, J., Sun, Y., Bodner, R., Suzuki, E., Becker, A., Socolich, M. & Zuker, C. S. (1997) Nature 388, 243-249. [DOI] [PubMed] [Google Scholar]
- 7.Scott, K. & Zuker, C. S. (1998) Nature 395, 805-808. [DOI] [PubMed] [Google Scholar]
- 8.Buck, E., Li, J., Chen, Y., Weng, G., Scarlata, S. & Iyengar, R. (1999) Science 283, 1332-1335. [DOI] [PubMed] [Google Scholar]
- 9.Chang, M. S. S., Tam, J. P. & Sanders-Bush, E. (August 29, 2000) Sci. STKE., 10.1126/stke.2000.47.pl1. [DOI] [PubMed]
- 10.Gilchrist, A., Li, A. & Hamm, H. E. (February 5, 2002) Sci. STKE., 10.1126/stke.2002.118.pl1. [DOI] [PubMed]
- 11.Li, P., Kerchner, G. A., Sala, C., Wei, F., Huettner, J. E., Sheng, M. & Zhuo, M. (1999) Nat. Neurosci. 2, 972-977. [DOI] [PubMed] [Google Scholar]
- 12.Sattler, R., Xiong, Z., Lu, W.-Y., Hafner, M., MacDonald, J. F. & Tymianski, M. (1999) Science 284, 1845-1848. [DOI] [PubMed] [Google Scholar]
- 13.Xia, J., Chung, H. J., Wihler, C., Huganir, R. L. & Linden, D. J. (2000) Neuron 28, 499-510. [DOI] [PubMed] [Google Scholar]
- 14.Daw, M. I., Chittajallu, R., Bortolotto, Z. A., Dev, K. K., Duprat, F., Henley, J. M., Collingridge, G. L. & Isaac, J. T. (2000) Neuron 28, 873-886. [DOI] [PubMed] [Google Scholar]
- 15.Kim, C. H., Chung, H. J., Lee, H. K. & Huganir, R. L. (2001) Proc. Natl. Acad. Sci. USA 98, 11725-11730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aramburu, J., Yaffe, M. B., López-Rodríguez, C., Cantley, L. C., Hogan, P. G. & Rao, A. (1999) Science 285, 2129-2133. [DOI] [PubMed] [Google Scholar]
- 17.May, M. J., D'Acquisto, F., Madge, L. A., Glockner, J., Pober, J. S. & Ghosh, S. (2000) Science 289, 1550-1554. [DOI] [PubMed] [Google Scholar]
- 18.Akhter, S. A., Luttrell, L. M., Rockman, H. A., Iaccarino, G., Lefkowitz, R. J. & Koch, W. J. (1998) Science 280, 574-577. [DOI] [PubMed] [Google Scholar]
- 19.van Rooij, E., Doevendans, P. A., de Theije, C. C., Babiker, F. A., Molkentin, J. D. & de Windt, L. J. (2002) J. Biol. Chem. 277, 48617-48626. [DOI] [PubMed] [Google Scholar]
- 20.Chen, Y.-N. P., Sharma, S. K., Ramsey, T. M., Jiang, L., Martin, M. S., Baker, K., Adams, P. D., Bair, K. W. & Kaelin, W. G., Jr. (1999) Proc. Natl. Acad. Sci. USA 96, 4325-4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aarts, M., Liu, Y., Liu, L., Besshoh, S., Arundine, M., Gurd, J. W., Wang, Y.-T., Salter, M. W. & Tymianski M. (2002) Science 298, 846-850. [DOI] [PubMed] [Google Scholar]
- 22.Cochran, A. G. (2000) Chem. Biol. 7, R85-R94. [DOI] [PubMed] [Google Scholar]
- 23.Toogood, P. L. (2002) J. Med. Chem. 45, 1543-1558. [DOI] [PubMed] [Google Scholar]
- 24.Gadek, T. R. & Nicholas, J. B. (2003) Biochem. Pharmacol. 65, 1-8. [DOI] [PubMed] [Google Scholar]
- 25.Rusnak, F. & Mertz, P. (2000) Physiol. Rev. 80, 1483-1521. [DOI] [PubMed] [Google Scholar]
- 26.Aramburu, J., Rao, A. & Klee, C. B. (2000) Curr. Top Cell Regul. 36, 237-295. [DOI] [PubMed] [Google Scholar]
- 27.Rao, A., Luo, C. & Hogan, P. G. (1997) Annu. Rev. Immunol. 15, 707-747. [DOI] [PubMed] [Google Scholar]
- 28.Serfling, E., Berberich-Siebelt, F., Chuvpilo, S., Jankevics, E., Klein-Hessling, S., Twardzik, T. & Avots, A. (2000) Biochim. Biophys. Acta 1498, 1-18. [DOI] [PubMed] [Google Scholar]
- 29.Crabtree, G. R. & Olson, E. N. (2002) Cell 109, S67-S79. [DOI] [PubMed] [Google Scholar]
- 30.Hogan, P. G., Chen, L., Nardone, J. & Rao, A. (2003) Genes Dev. 17, 2205-2232. [DOI] [PubMed] [Google Scholar]
- 31.Horsley, V. & Pavlath, G. K. (2002) J. Cell Biol. 156, 771-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schiaffino, S. & Serrano, A. (2002) Trends Pharmacol. Sci. 23, 569-575. [DOI] [PubMed] [Google Scholar]
- 33.Liu, J., Farmer, J. D., Jr., Lane, W. S., Friedman, J., Weissman, I. & Schreiber, S. L. (1991) Cell 66, 807-815. [DOI] [PubMed] [Google Scholar]
- 34.Liu, J. P., Sim, A. T. & Robinson, P. J. (1994) Science 265, 970-973. [DOI] [PubMed] [Google Scholar]
- 35.Lai, M. M., Hong, J. J., Ruggiero, A. M., Burnett, P. E., Slepnev, V. I., De Camilli, P. & Snyder, S. H. (1999) J. Biol. Chem. 274, 25963-25966. [DOI] [PubMed] [Google Scholar]
- 36.Cousin, M. A. & Robinson, P. J. (2001) Trends Neurosci. 24, 659-665. [DOI] [PubMed] [Google Scholar]
- 37.Coghlan, V. M., Perrino, B. A., Howard, M., Langeberg, L. K., Hicks, J. B., Gallatin, W. M. & Scott, J. D. (1995) Science 267, 108-111. [DOI] [PubMed] [Google Scholar]
- 38.Tavalin, S. J., Colledge, M., Hell, J. W., Langeberg, L. K., Huganir, R. L. & Scott, J. D. (2002) J. Neurosci. 22, 3044-3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cameron, A. M., Steiner, J. P., Roskams, A. J., Ali, S. M., Ronnett, G. V. & Snyder, S. H. (1995) Cell 83, 463-472. [DOI] [PubMed] [Google Scholar]
- 40.Cameron, A. M., Nucifora, F. C., Jr., Fung, E. T., Livingston, D. J., Aldape, R. A., Ross, C. A. & Snyder, S. H. (1997) J. Biol. Chem. 272, 27582-27588. [DOI] [PubMed] [Google Scholar]
- 41.Frey, N., Richardson, J. A. & Olson, E. N. (2000) Proc. Natl. Acad. Sci. USA 97, 14632-14637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shin, D. W., Pan, Z., Bandyopadhyay, A., Bhat, M. B., Kim, D. H. & Ma, J. (2002) Biophys. J. 83, 2539-2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Santana, L. F., Chase, E. G., Votaw, V. S., Nelson, M. T. & Greven, R. (2002) J. Physiol. 544, 57-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chow, C. W., Rincón, M. & Davis, R. J. (1999) Mol. Cell. Biol. 19, 2300-2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu, J., Arai, K. & Arai, N. (2001) J. Immunol. 167, 2677-2687. [DOI] [PubMed] [Google Scholar]
- 46.Fruman, D. A., Pai, S. Y., Klee, C. B., Burakoff, S. J. & Bierer, B. E. (1996) Methods 9, 146-154. [DOI] [PubMed] [Google Scholar]
- 47.Wang, X., Culotta, V. C. & Klee, C. B. (1996) Nature 383, 434-437. [DOI] [PubMed] [Google Scholar]
- 48.Furuke, K., Shiraishi, M., Mostowski, H. S. & Bloom, E. T. (1999) J. Immunol. 162, 1988-1993. [PubMed] [Google Scholar]
- 49.Reiter, T. A. & Rusnak, F. (2002) J. Biol. Inorg. Chem. 7, 823-834. [DOI] [PubMed] [Google Scholar]
- 50.Rusnak, F. & Reiter, T. (2000) Trends Biochem. Sci. 25, 527-529. [DOI] [PubMed] [Google Scholar]
- 51.Tsai, E. Y., Jain, J., Pesavento, P. A., Rao, A. & Goldfeld, A. E. (1996) Mol. Cell. Biol. 16, 459-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Macián, F., García-Rodríguez, C. & Rao, A. (2000) EMBO J. 19, 4783-4795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kiani, A., García-Cozár, F. J., Habermann, I., Laforsch, S., Aebischer, T., Ehninger, G. & Rao, A. (2001) Blood 98, 1480-1488. [DOI] [PubMed] [Google Scholar]
- 54.Peng, S. L., Gerth, A. J., Ranger, A. M. & Glimcher, L. H. (2001) Immunity 14, 13-20. [DOI] [PubMed] [Google Scholar]
- 55.Lee, D. U., Avni, O., Chen, L. & Rao, A. (2004) J. Biol. Chem. 279, 4802-4810. [DOI] [PubMed] [Google Scholar]
- 56.Ashman, K., Moran, M. F., Sicheri, F., Pawson, T. & Tyers, M. (October 9, 2001) Sci. STKE., 10.1126/stke.2001.103.pe33. [DOI] [PubMed]
- 57.Legrain, P., Wojcik, J. & Gauthier, J.-M. (2001) Trends Genet. 17, 346-352. [DOI] [PubMed] [Google Scholar]
- 58.Tucker, C. L., Gera, J. F. & Uetz, P. (2001) Trends Cell Biol. 111, 102-106. [DOI] [PubMed] [Google Scholar]
- 59.Walhout, A. J. M. & Vidal, M. (2001) Nat. Rev. Mol. Cell Biol. 2, 55-62. [DOI] [PubMed] [Google Scholar]
- 60.Tong, A. H. Y., Drees, B., Nardelli, G., Bader, G. D., Brannetti, B., Castagnoli, L., Evangelista, M., Ferracuti, S., Nelson, B., Paoluzi, S., et al. (2002) Science 295, 321-324. [DOI] [PubMed] [Google Scholar]
- 61.Uetz, P. (2002) Curr. Opin. Chem. Biol. 6, 57-62. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.