Abstract
Introduction
Cystic fibrosis is the most common autosomal recessive genetic disease in the Caucasian population. Extending knowledge about the molecular pathology on the one hand allows better delineation of the mutations in the CFTR gene and the other to dramatically increase the predictive power of molecular testing.
Methods
This study reports the results of a molecular screening of cystic fibrosis using DNA samples of patients enrolled from January 2009 to December 2013. Patients were referred to our laboratory for cystic fibrosis screening for infertile couples. In addition, we identified the gene mutations present in 76 patients affected by cystic fibrosis in the pediatric population of Basilicata.
Results
In the 964 infertile couples examined, 132 subjects (69 women and 63 men) resulted heterozygous for one of the CFTR mutations, with a recurrence of carriers of 6.85%. The recurrence of carriers in infertile couples is significantly higher from the hypothetical value of the general population (4%).
Conclusions
This study shows that in the Basilicata region of Italy the CFTR phenotype is caused by a small number of mutations.
Our aim is to develop a kit able to detect not less than 96% of CTFR gene mutations so that the relative risk for screened couples is superimposable with respect to the general population.
Keywords: Cystic fibrosis, Cystic fibrosis conductance transmembrane regulator, Screening in infertile couples, Mutation analysis, Polymerase Chain Reaction
Introduction
Cystic fibrosis (CF) is the most common autosomal recessive genetic disease for the Caucasian (white) population. In Italy, the disease occurs in 1/2500 to 1/3000 Caucasian newborns, with a carrier incidence ranging from 1/26 to 1/30 in the general population [1,2].
CF is a complex multisystem disease related to the buildup of thick, sticky mucus that can damage many of the body's organs (epithelia of the respiratory tract, exocrine pancreas, intestine, male genital tract, hepatobiliary system, and exocrine sweat glands). The pulmonary disease is present in 90% of patients and it is the major cause of morbidity and mortality in CF.
The CFTR gene provides instructions for making a glycoprotein called cystic fibrosis transmembrane conductance regulator (1,480 amino acid residues). This protein works as a channel across the membrane of cells producing mucus, sweat, saliva, tears, and digestive enzymes. The channel negatively transports charged particles called chloride ions into and out of the cells. The transport of chloride ions helps control the movement of water in tissues, which is necessary for the production of thin, freely flowing mucus. Mucus is a slippery substance that lubricates and protects the lining of the airways, digestive system, reproductive system and other organs and tissues [3]. In particular, more than 95% of men with CF are infertile as a result of azoospermia caused by congenital bilateral agenesis of the vas deferens (CBAVD); it occurs in men without pulmonary or gastrointestinal manifestations of CF.
Cystic fibrosis is caused by mutations in the CFTR gene (cystic fibrosis transmembrane regulator), detected for the first time in 1989. The CFTR gene is located on the long (q) arm of chromosome 7 (7q31.2) [4,5].
More than 1800 mutations in the CFTR gene have been identified [6]; many of which are so rare as to be called ‘private’ as they are only present within individual families. Moreover, the CFTR mutation detection rate varies by test method and ethnic background. The most common mutation, called F508del, is a deletion of one amino acid at position 508 in the CFTR protein. The resulting abnormal channel breaks down shortly after it is made, so it never reaches the cell membrane to transport chloride ions. The F508del mutation, accounts for two-thirds of all cystic fibrosis alleles worldwide. This mutation is particularly frequent in people of northern European ancestry (70% in Anglo-Saxon countries and 50% in the Mediterranean area).
The phenotypic variability is determined by the heterogeneity of mutations in the CFTR gene, but also by many other factors, such as modifier genes, epigenetic regulation, environment and timeliness in therapy [7,8].
Carrier screening for cystic fibrosis involves analysis for common mutations in the CFTR gene from people with no personal history, or family history, of the disease.
This analysis shows whether a person is a carrier, at risk (one in four) of having a baby with cystic fibrosis if their partner is also a carrier. Carrier screening has been recommended by the American College of Medical Genetics and American College of Obstetricians and Gynecologists (ACMG/ACOG) [9] and the Human Genetics Society of Australasia (HGSA) [10] and has been established in the USA, Australia, and parts of Europe [11]. Accurate identification of CF mutations results in more applicable programs for prevention, diagnosis, and treatment of CF. In a study carried out in some areas of Northern Italy, carrier screening was associated with a decrease in the incidence of CF [12].
Previous reports about CF patients born in the Basilicata region (southern Italy) have identified types and incidence of the most common mutations of the CFTR gene [13,14].
In this study are reported the results of a molecular screening of CF in patients enrolled from January 2009 to October 2013 using DNA samples. Patients were referred to our laboratory for CF screening for infertile couples [15]. In addition, we identified the gene mutations present in patients affected by cystic fibrosis in the pediatric population of Basilicata.
In this way the incidence and typology of CFTR mutations in Basilicata were characterized and related to previous reports on CF patients born in the same area.
Results
In the 964 infertile couples examined, 132 subjects (69 women and 63 men) were heterozygous for one of the mutations of CFTR, with a carrier occurrence of 6.85% (Table 1). The carrier occurrence in infertile couples is significantly higher than the hypothetical value of the general population (4%). The statistical analysis was performed according to the ‘hypothesis test for a proportion’. Our data indicated an expectation of 6.85% with a sample of 1,928 subjects. The Z-test revealed a value of 6.38% with a P value <0.0001. Nevertheless, mutation I148T, at first classified as a mutation, now is considered an ‘innocent’ polymorphism, able to be pathogenetic only if present in the same gene (on the same chromosome) with another polymorphism (3199del6). Hence it is necessary, in presence of polymorphism I148T, to detect the presence of polymorphism 3199del6. The polymorphism 3199del6 was not detected in all patients with the I148T variant.
Table 1.
Number of subjects tested who were carriers of the cystic fibrosis transmembrane regulator gene
| Mutation | Men | Women | Total |
|---|---|---|---|
| G551D |
1 |
2 |
3 |
| R553X |
0 |
1 |
1 |
| F508del |
35 |
32 |
67 |
| N1303K |
7 |
8 |
15 |
| I148T |
4 |
9 |
13 |
| G542X |
3 |
6 |
9 |
| DI507 |
2 |
0 |
2 |
| L1077P |
0 |
2 |
2 |
| D1152H |
1 |
6 |
7 |
| W1282X |
2 |
0 |
2 |
| 2183 AA>G |
3 |
0 |
3 |
| 1259insA |
0 |
1 |
1 |
| 4016insT |
1 |
0 |
1 |
| I507del |
1 |
0 |
1 |
| 2789+5G>A |
1 |
0 |
1 |
| 4382delA |
0 |
2 |
2 |
| G1244E |
1 |
0 |
1 |
| 621+3A>G |
1 |
0 |
1 |
| Total | 63 | 69 | 132 |
Among the infertile couples, three men, two of whom were brothers, were azoospermic with CBAVD. The molecular analysis of the CFTR gene revealed that the two brothers, aged 26 and 29 years old respectively, were both ‘compound heterozygotes G542X/D1152H’, while the third one, aged 37 years old, was ‘compound heterozygotes G542X/E831X’.
With regard to the 76 patients with cystic fibrosis and a positive sweat test, all have two CFTR-mutated genes (Figure 1).
Figure 1.
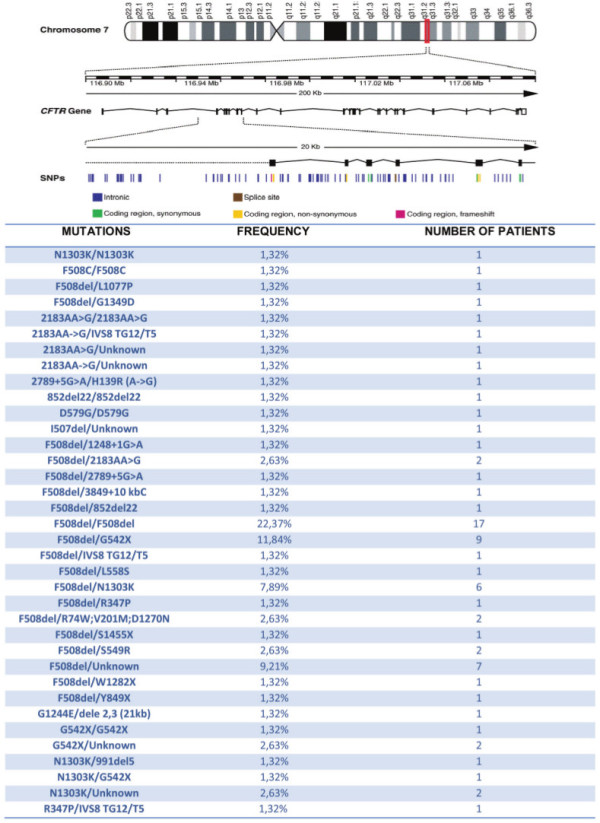
76 patients with cystic fibrosis and positive sweat test, all have two genes mutated. SNPs: Single nucleotide polymorphism.
Discussion
The aim of this study was to characterize, at molecular level, the most common CFTR mutations in the population of the Basilicata region. We have defined the type and frequency of mutations found and put them in comparison with data from previous studies (Table 2) [13,14]. In this study, we detected mutations that had not been revealed in the two previous studies [13,14]. In infertile couples from Basilicata, the occurrence of CF carriers is significantly higher than in the general population (6.85% vs. 4%). It was not clear if a screening for the general population would be viable or effective, so we invited the carriers in the families of affected people to participate in the research.
Table 2.
Comparison between the results obtained in this study and those obtained in a previous study
| Castaldo et al .[14] | Mutations observed in the present study | |
|---|---|---|
|
F508del |
55.8% (29) |
48.62% (141) |
|
N1303K |
3.8% (2) |
9.31% (27) |
|
G542X |
3.8% (2) |
8.96% (26) |
|
W1282X |
3.8% (2) |
1.03% (3) |
|
2183AA>G |
5.8% (3) |
2.76% (8) |
|
R1162X |
0 |
0 |
|
1717-1G>A |
1.9% (1) |
0 |
|
T338I |
0 |
0 |
|
R347P |
0 |
0.69% (2) |
|
711+5G>A |
0 |
0 |
|
852del22 |
5.8% (3) |
1.03% (3) |
|
4382delA |
0 |
0.69% (2) |
|
1259insA |
0 |
0.34% (1) |
|
4016insT |
0 |
0.34% (1) |
|
R553X |
0 |
0.34% (1) |
|
R1158X |
0 |
0 |
|
L1077P |
0 |
1.03% (3) |
|
I502T |
0 |
0 |
|
3849+10kbC>T |
1.9% (1) |
0.34% (1) |
|
D579G |
0 |
0.69% (2) |
|
G1244E |
3.8% (2) |
0 |
|
G1349D |
0 |
0.34% (1) |
|
2789+5G>A |
0 |
1.03% (3) |
|
711+1G>T |
0 |
0 |
|
L1065P |
0 |
0 |
|
2522insC |
0 |
0 |
|
E585X |
0 |
0 |
|
G85E |
0 |
0 |
|
G178R |
0 |
0 |
|
D1152H |
0 |
3.10% (9) |
|
I148T-3195del6 |
0 |
0 |
|
I148T (alone) |
0 |
4.48% (13) |
|
R334W |
0 |
0 |
|
DI507 |
0 |
0.69% (2) |
|
I1005R |
0 |
0 |
|
3272-26A>G |
0 |
0 |
|
2711delT |
0 |
0 |
|
L558S |
1.9% (1) |
0.34% (1) |
|
W1063X |
0 |
0 |
|
D110H |
0 |
0 |
|
S549R (A>C) |
1.9% (1) |
0.69% (2) |
|
2184insA |
0 |
0 |
|
3131del22 |
0 |
0 |
|
R709N |
0 |
0 |
|
A349V |
0 |
0 |
|
4015insA |
0 |
0 |
|
Y849X |
1.9% (1) |
0.34% (1) |
|
G551D |
0 |
1.03% (3) |
|
621+3A>G |
0 |
0.34% (1) |
|
E831X |
0 |
0 |
|
I507del |
0 |
0.69% (2) |
|
IVS8 TG12/t5 |
0 |
1.03% (3) |
|
H139R (A->G) |
0 |
0.34% (1) |
|
1248+1G>A |
0 |
0.34% (1) |
|
R74W;V201M;D1270N |
0 |
0.69% (2) |
|
S1455X |
0 |
0.34% (1) |
|
dele 2,3 (21kb) |
0 |
0.34% (1) |
|
991del5 |
0 |
0.34% (1) |
|
UNKNOWN |
7 %(4) |
4.83% (14) |
|
F508C |
0 |
0.69% (2) |
| TOTAL | 52 | 290 |
In CF, contrary to other genetic diseases (for example beta thalassemia), the identification of carriers is possible only through molecular research of CFTR mutations. A recent report has shown the costs of managing CF for the Italian National Health System [16]. The results reports a high cost for the chronic and evolutive aspects of the disease. As mentioned before, molecular screening of CF is highly recommended in the USA by the National Institutes of Health Consensus Development Conference Statement on genetic testing for cystic fibrosis [17]. The target of the screening could be men and women of reproductive age in the periconceptional period. A European consensus conference in 2009 has concluded that for complete information for couples, each national health system could implement a screening program. In Italy, the region of Veneto started a complete screening program for the general population some years ago. The results have shown a significant reduction of the incidence of the disease. More recently, an experimental periconceptional program of CF and thalassemia screening allowed for the identification of 94% of CFTR gene mutations, with respect to 80% in the rest of Italy. Moreover, this screening program involved a high number of couples [18].
The success of a genetic screening program is mainly the outcome of educational support informing the population about the disease, thereby offering the opportunity to make an informed decision about reproduction.
In this study, the aim was to establish the a priori risk of CF carrier status in the Basilicata population and to offer a reliable screening test to couples planning a pregnancy, allowing them to make informed decisions.
Conclusions
This study shows that, in the Basilicata region (Italy), it was observed that the CFTR phenotype is caused by a small number of mutations.
The knowledge of the frequency of mutations prevalent in CFTR genes in the province of Matera allows development of kits for their detection.
Our aim is to develop a kit able to detect not less than 96% of CTFR gene mutations so that the relative risk for the couples screened is superimposable with respect to the general population.
Extending knowledge about the molecular pathology, on the one hand, allows better delineation of the mutations in the CFTR gene, and on the other, dramatically increases the predictive power of molecular testing.
Methods
In this study, we investigated 964 infertile couples (1.928 subjects examined; 3.856 alleles studied) born and living in Basilicata, from 2009 to December 2013 (five years). Moreover, we evaluated gene mutation of CFTR in 76 children born in Basilicata (152 CF alleles examined) with a positive sweat test (>60mmol/L). Written informed consent was obtained from the subjects examined for publication of this study in compliance with the Helsinki Declaration.
We applied the following tests to each patient after a venous blood sample was collected (in EDTA-K3).
Molecular analysis of the CFTR gene was performed following these steps:
– DNA isolation, starting from 25μl of blood, using the Promega extraction kit (DNA IQ™ System, cod.C6701; Promega Italy S.r.l., Milan, Italy).
– Polymerase chain reaction (PCR) and reverse hybridization. The procedure includes two steps: PCR amplification using biotinylated primers and hybridization of amplification products to a test strip containing allele-specific oligonucleotide probes immobilized as an array of parallel lines. Bound biotinylated sequences are detected using streptavidin-alkaline phosphatase and color substrates. The amplification and the reverse hybridization on a strip were obtained with the use of commercial kits produced by Nuclear Laser Medicine S.r.l., Settala (MI), Italy (cod. AC023/AC025 and AC089): genetic tests aimed at checking 60 mutations in the CFTR genes. The mutations analyzed are listed in Table 3. The test has a sensitivity and a specificity of more than 99%. With a direct analysis of 60 mutations of the CTFR gene, with reverse dot blot, it is possible to detect 90% of the most common CF alleles in Southern Italy (the regions of Campania, Puglia, Basilicata and Molise).
Table 3.
List of 60 mutations in the cystic fibrosis transmembrane regulator gene (specificity 100%)
| F508del |
I507del |
F508C |
621+1G>T |
D110H |
E585X |
G1349D |
| I502T |
1706del17 |
1677delTA |
R117H |
H139R |
1898+1G>A |
4015delA |
| G542X |
1717-1G>A |
Q552X |
852del22 |
G178R |
1898+3A>G |
|
| G551D |
S549R(A>C) |
2183AA>G |
T338I |
991del5 |
1898+5G>T |
|
| N1303K |
4016insT |
3849+10kb C>T |
R347P |
R334W |
2184insA |
|
| G85E |
711+5G>A |
711+1G>T |
1259insA |
R347H |
2522insC |
|
| 2789+5G>A |
W1282X |
G1244E |
R1066H |
R352Q |
3120+1G>A |
|
| I148T |
3199del6 |
S912X |
R1158X |
1717-8G>A |
R1066C |
|
| R1162X |
4382delA |
D1152H |
L1077P |
D579G |
3272-26A>G |
|
| L1065P | R553X | PoliT: 5T, 7T, 9T | 1874insT | 3659delC | ||
– The patients who tested negative or with a single mutation detected by reverse dot blot and with a clinical suspicion of atypical cystic fibrosis were analyzed with a complete scanning of the codificant region, through amplification and direct sequencing of 27 exones of the CFTR gene. In patients negative for reverse dot blot and in whom there were no clinical signs of cystic fibrosis, sequencing of the CFTR gene was not necessary. DNA sequencing is not essential, since the detection of innocent polymorphisms is not important to control the disease. The sequence of oligonucleotides for each exone, with the annealing temperatures (T°A) and the length in base pair (bp) of the amplified product, are reported in Table 4. The amplification conditions for the 27 exones change according to the annealing temperature (T°A.), which depends on the oligonucleotides used, which in turn are specific for analysis of each exone. The amplification report is shown in Table 5.
Table 4.
Sequence of oligonucleotides for each exon, with the annealing temperatures (T°A) and the length in base pair (bp) of the amplified product
| Exon | Sequence of primers | T°A | bp |
|---|---|---|---|
| 1 |
GAGAAAGCCGCTAGAGCAAA(CF1F) |
55°C |
394 |
| TCCTTTACCCCAAACCCAAC(CF1R) | |||
| 2 |
TCCAAATCTGTATGGAGACCA(CF2F) |
55°C |
603 |
| TCAGTGTGAAAATGAGATGTTCC(CF2R) | |||
| 3 |
TCTGGCTGAGTGTTTGGTGT(CF3F) |
55°C |
399 |
| TTTGGAGTTGGATTCATCCTTT(CF3R) | |||
| 4 |
AAACTTGTCTCCCACTGTTGC(CF4F) |
55°C |
453 |
| GGCCTGTGCAAGGAAGTATT(CF4R) | |||
| 5 |
GTGCCTAGATGCTGGGAAAT(CF5F) |
55°C |
393 |
| AAAACTCCGCCTTTCCAGTT(CF5R) | |||
| 6a |
TGCTATGTGCTCCATGTAATGA(CF6AF) |
55°C |
415 |
| TGCATAGAGCAGTCCTGGTT(CF6AR) | |||
| 6b |
TGCCCATCTGTTGAATAAAAG(CF6BF) |
55°C |
411 |
| CCCATGAAAGTGAATTTGTGC(CF6BR) | |||
| 7 |
TTCCATTCCAAGATCCCTGA(CF7F) |
55°C |
404 |
| GCACATTTTTGCAAAGTTCA(CF7F) | |||
| 8 |
GAATCCTAGTGCTTGGCAAAT(CF8F) |
55°C |
404 |
| GATCCTCCTTCCAGTTCTACCA(CF8R) | |||
| 9 |
GGCCATGTGCTTTTCAAACT(CF9F) |
55°C |
389 |
| CTCCAAAAATACCTTCCAGCA(CF9R) | |||
| 10 |
TGAATCCTGAGCGTGATTTG(CF10F) |
55°C |
435 |
| TTCATGTGTTTGCAAGCTTCTT(CF10R) | |||
| 11 |
GAAGGAAGATGTGCCTTTCAA(CF11F) |
55°C |
395 |
| CCAAGATACGGGCACAGATT(CF11R) | |||
| 12 |
TCAGTGAATCGATGTGGTGAC(CF12F) |
55°C |
419 |
| ATGAGGCGGTGAGAAAAGGT(CF12R) | |||
| 13-1 |
TCATGCTATCAGAATTCACAAGG(CF13F1) |
56°C |
575 |
| GGGAGTCTTTTGCACAATGG(CF13R1) | |||
| 13-2 |
CTGGAGAGTTTGGGGAAAAA(CF13F2) |
56°C |
449 |
| AAATACCCCCAAGCGATGTA(CF13R2) | |||
| 14a |
CAATGGTGGCATGAACTGT(CF14AF) |
55°C |
437 |
| GTGGTTCTACTTGTTGATTTTTCAG(CF14AR) | |||
| 14b |
TGGCTTTCTTGTGAGGTTCA(CF4BF) |
55°C |
446 |
| TGCTTGGGAGAAATGAAACA(CF14BR) | |||
| 15 |
GTCGCCAAATAACGATTTCC(CF15F) |
55°C |
406 |
| AGGTTCAACAAAGGGCACAT(CF15R) | |||
| 16 |
TTTGGGTTCTGAATGCGTCT(CF16F) |
55°C |
388 |
| GGCCAGGTAAGCAGTTCTGA(CF16R) | |||
| 17a |
CTCACCAACATGTTTTCTTTGA(CF17AF) |
55°C |
399 |
| CCAAAATGAAGTCACATGGTCA(CF17AR) | |||
| 17b |
GAATGGCACCAGTGTGAAAA(CF17BF) |
55°C |
682 |
| CAATCTGTGTGCATCGGTTT(CF17BR) | |||
| 18 |
TGTGCCCTAGGAGAAGTGTG(CF18F) |
55°C |
335 |
| TGACAGATACACAGTGACCCTCA(CF18R) | |||
| 19 |
GCCCGACAAATAACCAAGTG(CF19F) |
55°C |
399 |
| GCAAGCAGTGTTCAAATCTCA(CF19R) | |||
| 20 |
CCAATTCCTTATGCCCAGTT(CF20F) |
55°C |
408 |
| TGGCTAAGTCCTTTTGCTCA(CF20R) | |||
| 21 |
TGATGGTAAGTACATGGGTGTTTC(CF21F) |
57°C |
578 |
| GGAGCCATACCAGTGAGGAG(CF21R) | |||
| 22 |
TCAAATGGTGGCAGGTAGTG(CF22F) |
55°C |
382 |
| TCACCATGAAGCAGGCATAA(CF22R) | |||
| 23 |
CCCATGGTTGAAAAGCTGAT(CF23F) |
55°C |
417 |
| TGAGTAAAGCTGGATGGCTG(CF23R) | |||
| 24 | GCCTTCTGTCCCAGATCTCA(CF24F) |
60°C | 362 |
| GAGCAAATGTCCCATGTCAA(CF24R) |
Table 5.
Amplification report
| Temperature | Time | Cycles |
|---|---|---|
| 95°C |
5’ |
1 |
| 95°C |
30” |
35 |
| T° A |
30” |
|
| 72°C |
20” |
|
| 72°C | 5’ | 1 |
Consent
Written informed consent was obtained from all of the patients (including legal guardians of the children) for publication of this case report and accompanying images. A copy of the written consent is available for review by the Editor-in-Chief of this journal. Our local institutional Ethics Committee approved this study.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
DD made substantial contributions to conception and design. MB, MM, AA and MG contributed to the acquisition, analysis and interpretation of data. DS, AAE and GG were involved in drafting the manuscript. DC, MLP, GD and EM gave final approval of the version to be published. All authors read and approved the final manuscript.
Contributor Information
Domenico Dell’Edera, Email: domenicodelledera68@gmail.com.
Michele Benedetto, Email: michelesi@hotmail.it.
Gemma Gadaleta, Email: gemma.gadaleta@uniba.it.
Domenico Carone, Email: d.carone@inwind.it.
Donatello Salvatore, Email: Donatello.Salvatore@libero.it.
Antonella Angione, Email: antonella.angione@regione.basilicata.it.
Massimiliano Gallo, Email: massimiliano.gallo@supporto.regione.basilicata.it.
Michele Milo, Email: michelemilo@alice.it.
Maria Laura Pisaturo, Email: pisaml@tiscali.it.
Giuseppe Di Pierro, Email: g.dipierro@alice.it.
Eleonora Mazzone, Email: eleonora.mazzone@gmail.com.
Annunziata Anna Epifania, Email: nunziaepifania69@libero.it.
Acknowledgements
The authors thank the ‘Association Gian Franco Lupo’ (ONLUS: a non-profit organization of social utility).
References
- Castellani C, Bonizzato A, Cabrini G, Mastella G. Newborn screening strategy for cystic fibrosis: a field study in an area with high allelic heterogeneity. Acta Paediatr. 1997;86:497–502. doi: 10.1111/j.1651-2227.1997.tb08920.x. [DOI] [PubMed] [Google Scholar]
- Davies JC, Alton EW, Bush A. Cystic fibrosis. BMJ. 2007;335:1255–1259. doi: 10.1136/bmj.39391.713229.AD. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Boeck K, Wilschanski M, Castellani C, Taylor C, Cuppens H, Dodge J, Sinaasappel M. Diagnostic Working Group. Cystic fibrosis: terminology and diagnostic algorithms. Thorax. 2006;61:627–635. doi: 10.1136/thx.2005.043539. Epub 2005 Dec 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riordan JR, Rommens JM, Kerem B, Alon N, Rozmahel R, Grzelczak Z, Zielenski J, Lok S, Plavsic N, Chou JL, Drumm ML, Iannuzzi MC, Collins FS, Tsui LC. Identification of the cystic fibrosis gene: cloning and characterization of complementary DNA. Science. 1989;245:1066–1073. doi: 10.1126/science.2475911. [DOI] [PubMed] [Google Scholar]
- Rommens JM, Iannuzzi MC, Kerem B, Drumm ML, Melmer G, Dean M, Rozmahel R, Cole JL, Kennedy D, Hidaka N, Zsiga M, Buchwald M, Riordan JR, Tsui L, Collins FS. Identification of the cystic fibrosis gene: chromosome walking and jumping. Science. 1989;245:1066–1073. doi: 10.1126/science.2475911. Erratum in: Science 1989 Sep 29;245:1437. [DOI] [PubMed] [Google Scholar]
- Cystic fibrosis mutation database. http://www.genet.sickkids.on.ca/app.
- Cutting GR. Modifier genes in Mendelian disorders: the example of cystic fibrosis. Ann N Y Acad Sci. 2010;1214:57–69. doi: 10.1111/j.1749-6632.2010.05879.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielenski J. Genotype and phenotype in cystic fibrosis. Respiration. 2000;67:117–133. doi: 10.1159/000029497. [DOI] [PubMed] [Google Scholar]
- Grody WW, Cutting GR, Klinger KW, Richards CS, Watson MS, Desnick RJ. Laboratory standards and guidelines for population-based cystic fibrosis carrier screening. Genet Med. 2001;3:149–154. doi: 10.1097/00125817-200103000-00010. [DOI] [PubMed] [Google Scholar]
- Cystic Fibrosis Population Screening Position Paper. Ed: Human Genetics Society of Australasia. 2010. http://www.hgsa.org.au/documents/item/27 (accessed June 20, 2013)
- Castellani C, Macek M Jr, Cassiman JJ, Duff A, Massie J, ten Kate LP, Barton D, Cutting G, Dallapiccola B, Dequeker E, Girodon E, Grody W, Highsmith EW, Kääriäinen H, Kruip S, Morris M, Pignatti PF, Pypops U, Schwarz M, Soller M, Stuhrman M, Cuppens H. Benchmarks for cystic fibrosis carrier screening: a European consensus document. J Cyst Fibros. 2010;9:165–178. doi: 10.1016/j.jcf.2010.02.005. [DOI] [PubMed] [Google Scholar]
- Castellani C, Picci L, Tamanini A, Girardi P, Rizzotti P, Assael BM. Association between carrier screening and incidence of cystic fibrosis. JAMA. 2009;302:2573–2579. doi: 10.1001/jama.2009.1758. [DOI] [PubMed] [Google Scholar]
- Castaldo G, Fuccio A, Cazeneuve C, Picci L, Salvatore D, Raia V, Scarpa M, Goossens M, Salvatore F. Detection of five rare cystic fibrosis mutations peculiar to Southern Italy: implications in screening for the disease and phenotype characterization for patients with homozygote mutations. Clin Chem. 1999;45:957–962. [PubMed] [Google Scholar]
- Castaldo G, Polizzi A, Tomaiuolo R, Cazeneuve C, Girodon E, Santostasi T, Salvatore D, Raia V, Rigillo N, Goossens M, Salvatore F. Comprehensive cystic fibrosis mutation epidemiology and haplotype characterization in a southern Italian population. Ann Hum Genet. 2005;69:15–24. doi: 10.1046/j.1529-8817.2004.00130.x. [DOI] [PubMed] [Google Scholar]
- Foresta C, Ferlin A, Gianaroli L, Dallapiccola B. Guidelines for the appropriate use of genetic tests in infertile couples. Eur J Hum Genet. 2002;10:303–312. doi: 10.1038/sj.ejhg.5200805. [DOI] [PubMed] [Google Scholar]
- Colombo C, Daccò V, Alicandro G, Loi S, Mazzi S, Lucioni C, Ravasio R. Cost of cystic fibrosis: analysis of treatment costs in a specialized center in northern Italy. Adv Ther. 2013;30:165–175. doi: 10.1007/s12325-013-0008-5. [DOI] [PubMed] [Google Scholar]
- National Institutes of Health Consensus Development Conference Statement on Genetic Testing for Cystic Fibrosis. Genetic testing for cystic fibrosis. Arch Intern Med. 1999;159:1529–1539. [PubMed] [Google Scholar]
- Coiana A, Faa' V, Carta D, Puddu R, Cao A, Rosatelli MC. Preconceptional identification of cystic fibrosis carriers in the Sardinian population: a pilot screening program. J Cyst Fibros. 2011;10:207–211. doi: 10.1016/j.jcf.2011.02.006. [DOI] [PubMed] [Google Scholar]


