Abstract
Capping of the 5′ ends of nascent RNA polymerase II transcripts is the first pre-mRNA processing event in all eukaryotic cells. Capping enzyme (CE) is recruited to transcription complexes soon after initiation by the phosphorylation of Ser-5 of the carboxyl-terminal domain of the largest subunit of RNA polymerase II. Here, we analyze the role of CE in promoter clearance and its functional interactions with different factors that are involved in promoter clearance. FCP1-mediated dephosphorylation of the carboxyl-terminal domain results in a drastic decrease in cotranscriptional capping efficiency but is reversed by the presence of DRB sensitivity-inducing factor (DSIF). These results suggest involvement of DSIF in CE recruitment. Importantly, CE relieves transcriptional repression by the negative elongation factor, indicating a critical role of CE in the elongation checkpoint control mechanism during promoter clearance. This functional interaction between CE and the negative elongation factor documents a dynamic role of CE in promoter clearance beyond its catalytic activities.
Eukaryotic mRNAs are modified at the 5′-end by the addition of a m7GpppN cap catalyzed by the sequential action of three enzymatic activities, RNA 5′-triphosphatase, guanylyl-transferase, and (guanine-N7) methyltransferase (1, 2). In yeast, these proteins are encoded by three different genes, whereas in human and other metazoans the RNA triphosphatase and guanylyltransferase are domains of a single polypeptide, and the methyltransferase activity resides in a separate protein (3). The initial pre-mRNA processing event is the 5′-capping of nascent RNA polymerase II (RNAPII) transcripts. The cap with its unique structural characteristics has important, positive impacts on multiple downstream steps in gene expression, including RNA stability, splicing, transport, and translation initiation (4, 5).
There is increasing evidence that capping occurs cotranscriptionally and is facilitated by the association of capping enzyme (CE) with transcribing RNAPII complexes through interaction with the phosphorylated carboxyl-terminal domain (CTD) of the RNAPII largest subunit, Rpb1. The CTD consists of a tandemly repeated YSPTSPS motif that undergoes extensive serine phosphorylation and dephosphorylation during the transcription cycle (6, 7). The CTD serves as a landing platform for, and regulator of, many transcription, splicing, polyadenylation, and termination factors (8-11). Several kinase activities have been implicated in CTD phosphorylation, and primarily one CTD-specific phosphatase, FCP1, serves to recycle RNAPII (12). Transcription begins with CTD phosphorylation at Ser-5 by the kinase activity of the Cdk7 subunit (equivalent to yeast Kin28) of transcription factor IIH. It is believed that during the formation of the transcription initiation complex, or soon after initiation, DRB sensitivity-inducing factor (DSIF) (13) is recruited to the transcription complex. Additionally, after initiation of transcription, the negative elongation factor (NELF) (13) is recruited through interaction with DSIF (5, 13, 14). This results in the arrest of the transcription complex before it enters into productive elongation, giving ample time for the recruitment and catalytic actions of the CE on nascent transcripts of ≈17-22 nt. DSIF/NELF mediated arrest is then relieved by means of phosphorylation of the CTD at Ser-2 by positive transcription elongation factor b (P-TEFb) (15), and the transcription complex resumes elongation (5).
Recruitment of the capping apparatus to the yeast RNAPII transcription complex in vivo requires the action of the transcription factor IIH-associated kinase Kin28 that phosphorylates Ser-5 of the CTD (16, 17). Ser-5 phosphorylation not only recruits the CE but also stimulates capping (7, 18-21). In addition, Spt5, the larger subunit of DSIF, interacts directly with the mammalian and fission yeast triphosphatase and guanylyl-transferase and stimulates RNA guanylylation (18, 19). These interactions demonstrate functional coupling of CEs with the transcription apparatus.
Recent studies showed that in yeast the CE complex is involved in transcriptional repression and may act at the level of reinitiation, consistent with a role in transcriptional regulation (22). In addition, the fission yeast Cdk9/Pch1, a putative counterpart of P-TEFb, functionally and physically interacts with the RNA triphosphatase, suggesting role(s) for CE, Spt5, and P-TEFb in an elongation checkpoint control (23). However, the effects of CE during promoter clearance and its functional relationships with the other factors operating during promoter clearance remain unclear. To investigate this issue, we developed a cotranscriptional capping assay using a highly purified reconstituted transcription system. Herein we report our findings that CE can be recruited to the transcription complex by means of an interaction with DSIF in addition to Ser-5 phosphorylated CTD. Surprisingly, we found that the recruitment of CE to the transcription complex disables NELF-mediated repression of transcription.
Materials and Methods
Purification of Proteins. FCP1 was purified as described (24). DSIF was expressed by using the baculovirus system and purified by Ni-NTA chromatography followed by S200 gel filtration. Human CE was expressed in Escherichia coli by using pHis (T)-hCAP1a, kindly provided by K. Mizumoto (Kitasato University, Tokyo), and purified by Ni-NTA. The catalytically inactive K294A mutant CE (25) was purified as wild-type protein. NELF was purified from human cell line-derived nuclear extract as described (26).
In Vitro Transcription Assays and Isolation of +16, +24, or +42 Ternary Complexes. In vitro transcription assays were reconstituted with highly purified general transcription factors and RNAPII on biotinylated linear DNA (EcoRI/ScaI fragment of the plasmid pML20-47 or PML5A) (27) essentially as described (24). Preinitiation complexes (PICs) formed as described (24) were incubated with RNasin (16 units) and a mixture of NTPs (50 μM ATP/5 μM CTP/50 μM 3′-Ome GTP/0.3 μM [α-32P]UTP) for 4 min at 30°C and chased with 1 μM UTP for 3 min to produce the +24 complexes (24). Stalled +24 complexes were washed with 0.05% Sarkosyl in TB60 buffer, equilibrated with TB60 buffer, and incubated with 5 μM ATP, GTP, and CTP with transcription factor IIS for 5 min at 30°C to produce the +42 complexes. The +42 complexes were washed with 0.05% Sarkosyl in TB60 and then with TB500 (TB buffer containing 500 mM KCl), and they were finally equilibrated with TB60. Stalled +16 complexes were isolated similarly by using plasmid pML5A.
Capping Assays. Ternary complexes or free RNAs were resuspended in 20 μl of TB60 and incubated with 50 μM GTP and CE for 5 min at 30°C. Reactions were terminated by adding stop buffer (24) and subjected to phenol-chloroform extraction, ethanol precipitation, and 17% PAGE in 8 M urea, followed by autoradiography.
P1 Nuclease and Alkaline Phosphatase (AP) Digestion. Ternary +24 complexes (not radiolabeled, washed with TB500, and equilibrated with TB60) were incubated with 0.3 μM [α-32P]GTP and CE (120 ng) in a 20-μl reaction for 30 min at 30°C and then terminated with stop buffer and subjected to phenol-chloroform extraction and ethanol precipitation. Purified RNAs were treated with P1 nuclease followed by calf intestine AP and analyzed by TLC and autoradiography (25).
Antibodies and Western Blot Analysis. The phosphorylation states of RNAPII CTD were determined with polyclonal antibody Gal4-CTD that detects the largest subunit (Rpb1) irrespective of phosphorylation states. Monoclonal H14 (Covance, Berkeley, CA) was used to probe Rpb1 of RNAPII containing phosphorylated Ser-5 in the CTD, and CE was detected with affinity purified polyclonal antibody. Transcription reactions were doubled and loaded directly onto 6% SDS-polyacrylamide gels in loading buffer for analysis by Western blotting by using enhanced chemiluminescence (Amersham Biosciences) detection.
Results and Discussion
Cotranscriptional Capping Reaction in a Purified System. Capping of nascent RNAPII transcripts occurs cotranscriptionally and is the first of many pre-mRNA processing events (5, 8, 28, 29). Recent studies indicate that components of the capping apparatus repress RNAPII transcription and also genetically interact with P-TEFb, suggesting an important role of capping in transcriptional control (22, 23). To further elucidate the role of CE during promoter clearance, and also to identify its functional interactions with different transcription factors operating during promoter clearance, we used a highly purified reconstituted transcription system. We isolated transcription complexes at different stages during promoter clearance and tested them as substrates for human CE.
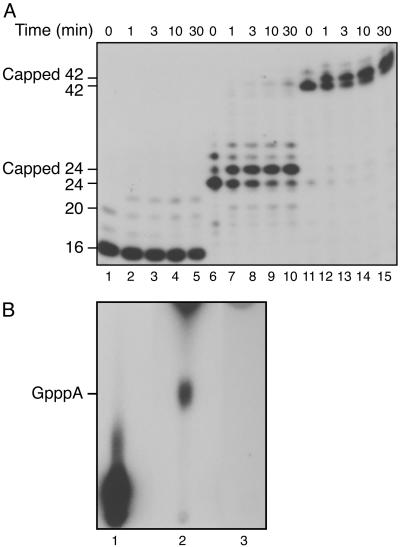
In the reconstituted transcription system, we used two different DNA templates that upon addition of a subset of ribonucleoside triphosphates allow initiation and subsequent stalling of the transcription complex at +16, +24, or +42 (24, 27). The stalled complexes were analyzed for the ability to support cap addition. Urea/PAGE analysis of the RNA produced demonstrated that the incubation of the stalled complexes with CE and GTP resulted in a slower migration of the RNA by ≈1 nt (Fig. 1A, lanes 6-15). This slower migration results from 5′ cap addition as confirmed by P1 nuclease and AP digestion, which yielded GpppA (Fig. 1B). RNAs in complexes stalled at 24 and 42 were efficiently capped within 1 min, whereas the majority of the stalled complexes at +16 were not capped, even after prolonged incubation (Fig. 1 A, lanes 1-5). Careful analysis of the gel profile demonstrates that RNAs of ≈20 nt, generated as by-products during formation of the +16 stalled complex through the slippage activity of RNAPII (30), were mostly capped (Fig. 1 A, lanes 1-5). Consistent with the high-resolution structure of the RNAPII elongation complex (31), the results indicate that the 5′-end of the 16 nucleotide RNA is hidden within the exit channel of the transcribing RNAPII complex and is thus unavailable to the capping apparatus, whereas transcripts of 20 nt had exited the channel and had become accessible to the CE (32).
Fig. 1.
Cotranscriptional capping in stalled transcription complexes. (A) Ternary complexes were incubated with 12 ng of CE and 50 μM GTP at 30°C. Capping reactions (lanes 1-5 for +16, lanes 6-10 for +24, and lanes 11-15 for +42) were stopped at the times shown. Products were subjected to phenol/chloroform extraction, ethanol precipitation, and PAGE/8 M urea, followed by autoradiography. The two small spots in lane 6 (above +24) are RNAs produced by slippage of RNAPII (30). (B) To confirm cap formation on RNA, +24 stalled complexes were incubated with [α-32P]GTP with and without CE (120 ng). RNAs (24 nt) were isolated, treated with P1 nuclease and AP, and analyzed by TLC. Lane 1, untreated RNA; lanes 2 and 3, capped and uncapped RNAs digested with P1 nuclease and AP. The position of authentic GpppA run in parallel is indicated.
DSIF Stimulates Cotranscriptional Capping. DSIF is a positive transcription elongation factor consisting of subunits Spt4 (p14) and Spt5 (p160) and interacts with NELF (14). NELF arrests transcribing RNAPII at an early stage of the transcription cycle well before the complex is converted into a mature elongation complex (5, 33). Spt5 is known to interact with several elongation factors and has been shown to bind to and stimulate human and yeast CE (19, 34-36).
By using the purified reconstituted transcription system, we analyzed the effect of DSIF on cotranscriptional capping, which was demonstrably greater than cap addition to free RNA (data not shown), in agreement with previous studies (29). However, because we used a highly purified reconstituted system to establish transcription initiation competent complexes, our findings permit the conclusion that stimulation of cotranscriptional cap addition apparently does not require additional factors that could have been present in the previous experiments using extracts to reconstitute transcription complexes (29).
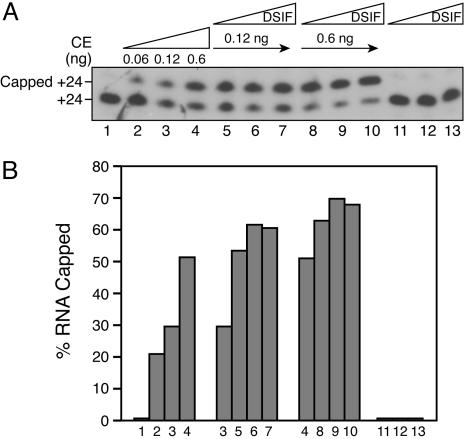
To assess the role of DSIF, stalled +24 complexes were incubated alone (Fig. 2A, lane 1) or with GTP and increasing amounts of CE, resulting in RNA capping (lanes 2-4). When +24 complexes were incubated with a limiting amount of CE, but in the presence of varying concentrations of DSIF, cotranscriptional capping was stimulated ≈2- to 3-fold (Fig. 2 A, compare lane 3 with lanes 5-7, and lane 4 with lanes 8-10; for quantitation, see Fig. 2B). These findings with stalled complexes are consistent with previous observations demonstrating that Spt5 stimulates cap addition to free RNA (19).
Fig. 2.
DSIF stimulates cotranscriptional capping. (A) Stalled +24 complexes were incubated alone (lane 1) or with increasing amounts of CE and GTP for 5 min (lanes 2-4). In parallel, +24 complexes that were first incubated with 10, 30, or 90 ng of DSIF for 10 min were further incubated with GTP in the absence (lanes 11-13) and presence (lanes 5-10) of CE for 5 min. (B) Quantitation of A.
FCP1-Mediated Dephosphorylation of CTD Ser-5 Decreases Cap Addition. CE binds to Ser-5 phosphorylated CTD (7, 21, 25, 37), and this interaction stimulates CE activity (18-20, 38, 39). To dissect the stimulatory effects of CTD phosphorylation, we assayed cotranscriptional cap addition in the purified system under Ser-5 phosphorylated and nonphosphorylated conditions. As the CTD is phosphorylated at Ser-5 by the kinase action of transcription factor IIH immediately after transcription initiation (5), all three stalled complexes (+16, 24, and 42) are phosphorylated (data not shown, see below). Complexes stalled at +24 were treated with the CTD-specific phosphatase FCP1 to dephosphorylate the CTD under conditions designed to keep the transcription complexes intact (refs. 12 and 24, data not shown). Although in vivo FCP1 preferentially dephosphorylates CTD Ser-2, in vitro under the conditions used FCP1 dephosphorylates both CTD Ser-2 and -5 (12, 40). Transcription complexes were then assayed for the ability to be capped by CE.
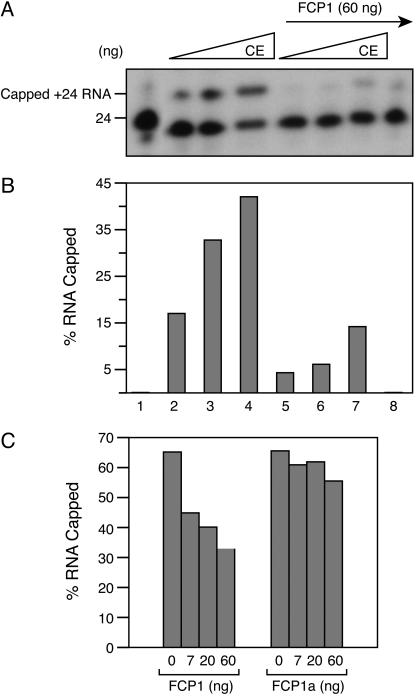
Addition of increasing amounts of CE resulted in more capped RNA (Fig. 3 A and B, lanes 2-4); however, pretreatment of the stalled +24 complexes with FCP1 diminished capping by nearly 70% (lanes 5-7, Fig. 3 A and B). In light of previous observations, we concluded that the removal of the CTD-Ser-5 phosphate by FCP1 results in inefficient recruitment of the capping apparatus to the ternary complex and loss of the stimulatory effect of Ser-5 phosphorylation on CE (18, 19). The observed effect was caused by dephosphorylation of the CTD, as treatment of the stalled complex with a catalytically inactive form of FCP1, FCP1a (12, 24), had no substantial effect on cap addition (Fig. 3C). It is important to note, however, that under the conditions of the assay, the decrease in cap addition by FCP1 treatment seems to be a kinetic phenomenon because prolonged incubation of the reaction resulted in a similar extent of capped RNA with or without FCP1 treatment (data not shown, and see below).
Fig. 3.
FCP1 inhibits capping. (A) Ternary +24 complexes were incubated alone (lane 1) or with 0.06, 0.12, and 0.6 ng of CE and GTP for 5 min (lanes 2-4). In parallel, +24 complexes were first incubated with 60 ng of FCP1 for 30 min at 30°C, washed with TB500, and then equilibrated with TB60. Finally, FCP1 treated complexes were incubated with 0.06, 0.12, and 0.6 ng of CE (lanes 5-7) and GTP for an additional 5 min. Lane 8: complexes treated with FCP1 only. (B) Quantitation of A. (C) Ternary +24 complexes were preincubated with FCP1 or catalytically inactive mutant FCP1a for 30 min at 30°C and then treated with 12 ng of CE and GTP for an additional 5 min.
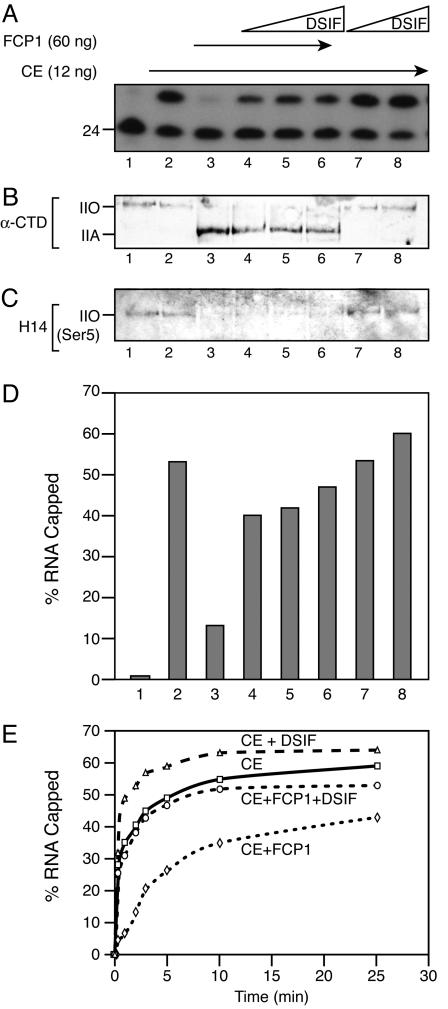
DSIF Overcomes FCP1-Mediated Decrease in Capping Activity. Studies have demonstrated that the larger subunit of DSIF, Spt5, interacts with the CE and stimulates capping (19, 34) and that DSIF is incorporated into transcription complexes during early steps of the transcription cycle. The results presented in Fig. 3 facilitated analysis of the role of DSIF on capping, independent of CTD Ser-5 phosphorylation. Ternary elongation complexes stalled at +24 were treated with excess FCP1 to achieve conversion of RNAPII to the nonphosphorylated A form (Fig. 4B). As expected, the +24 stalled complexes contain Ser-5 phosphorylated CTD (IIO form) resulting from the kinase activity of transcription factor IIH subsequent to transcription initiation (Fig. 4 B and C, lane 1). Addition of CE resulted in efficient RNA capping without altering the CTD phosphorylation state (Fig. 4 A-C, lane 2). FCP1 treatment of the complexes completely dephosphorylated the CTD and converted RNAPIIO to RNA-PIIA, resulting in loss of capping activity (Fig. 4 A-C, lane 3). Interestingly, when the +24 complexes were pretreated with FCP1 and then incubated with various amounts of DSIF followed by CE, capping was stimulated by DSIF, although the CTD remained dephosphorylated (Fig. 4 A-C, lanes 4-6). Thus, although DSIF treatment did not change the phosphorylation state of the transcription complexes, it stimulated capping (Fig. 4 A-C, lanes 7 and 8). Quantitation of the capping reactions is shown in Fig. 4D. These results suggest that the DSIF interaction with the CE stimulates the recruitment of CE to the ternary elongation complexes containing FCP1-dephosphorylated CTD.
Fig. 4.
DSIF can overcome FCP1-mediated capping inhibition. (A) Ternary +24 complexes alone (lane 1) or incubated with 0.6 ng of CE and GTP for 5 min at 30°C (lane 2). Ternary complexes were incubated with 60 ng of FCP1 for 30 min at 30°C, washed to remove FCP1, resuspended in TB60, and treated with CE and GTP in the absence (lane 3) or presence (lanes 4-6) of varying amounts of DSIF. As control, complexes not treated with FCP1 were incubated with DSIF and CE (lanes 7-8). (B and C) Capping reactions in A were subjected to 6% SDS/PAGE and analyzed by Western blot with GAL4-CTD polyclonal antibody, which recognizes both phosphorylated (IIO) and nonphosphorylated (IIA) Rpb1 and monoclonal antibody H14 to detect specifically Ser-5 phosphorylated CTD. (D) Quantitation of A. (E) Ternary +24 complexes were treated with FCP1 for 30 min, and treated and untreated complexes were incubated with 0.6 ng CE in the absence and presence of 30 ng of DSIF. Reactions were stopped at the indicated times, and the percentage of capped RNA was determined by SDS/PAGE and autoradiography.
Importantly, the stimulation of capping by DSIF observed with RNAPIIA did not reach the same levels as those observed with DSIF by using phosphorylated RNAPII. This finding is in agreement with previous studies demonstrating that CTD Ser-5 phosphorylation positively modulates cap addition (19, 20). However, the DSIF-mediated stimulation of capping with nonphosphorylated RNAPII suggests that the interaction between DSIF and CE provides an alternative pathway for the recruitment of CE. To further investigate this issue we performed a time course of cap addition within transcription complexes containing CTD Ser-5 phosphorylated or FCP1-dephosphorylated with and without addition of DSIF (Fig. 4E). We observed stimulation of capping by DSIF in each case, with a more pronounced effect during early times of the reaction. With prolonged incubation times, cap addition was observed on stalled complexes containing the nonphosphorylated RNAPII. These results collectively suggest that DSIF, as well as the CTD with phosphorylated Ser-5, likely affects the kinetics of capping, possibly by reducing the energy barrier for CE recruitment to transcription complexes.
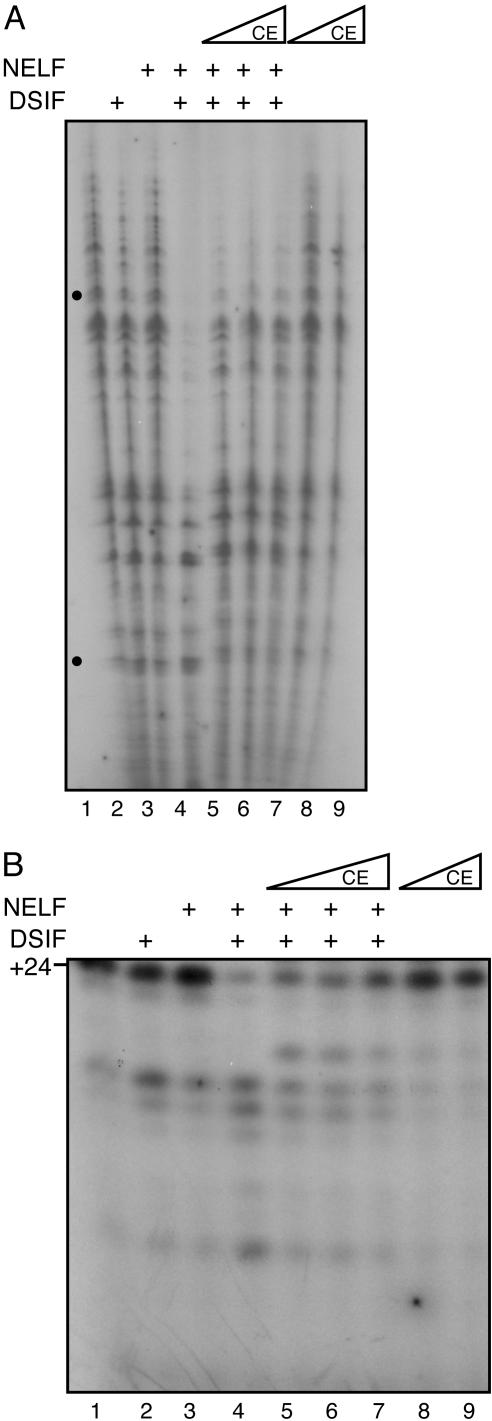
CE Overcomes NELF-Mediated Transcriptional Repression. NELF was originally identified as a factor that represses transcription in a DSIF-dependent manner (13, 14). This multisubunit factor interacts with RNAPII in the presence of DSIF and arrests transcription complexes at an early phase of the transcription cycle, during promoter clearance. The smallest subunit of NELF (NELF-E) also contains a functional RNA recognition motif required for transcriptional repression (14), and association of CE with nascent RNA (41) may interfere with NELF binding, contributing to release of repression. Recent studies showed that DSIF and NELF are both present in the promoter region during the early part of the transcription cycle (33, 36, 42). As transcription proceeds into the elongation phase, NELF dissociates, relieving arrest and allowing the conversion of the transcription complex into a mature elongation complex. On the other hand, DSIF remains bound to RNAPII and travels with the elongation complexes (33, 42). Capping likely occurs as soon as the nascent RNA protrudes from the RNAPII exit channel and is the first RNA processing event during promoter clearance, a step regulated by DSIF and NELF. We, therefore, investigated whether CE affects NELF function. Transcription initiation complexes were assembled with general transcription factors (GTFs) in the absence (Fig. 5A, lane 1) or in the presence of DSIF and/or NELF (lanes 2-4) and then chased with the 4 NTPs for 6 min. As expected, NELF alone had no significant effect on transcription (Fig. 5A, compare lanes 2 and 3 with 1). However, transcription was strongly inhibited when DSIF and NELF were present together (Fig. 5A, lane 4). Most interestingly, addition of CE to the transcription reaction inhibited by DSIF and NELF resulted in the reversal of NELF-mediated repression (Fig. 5A, lanes 5-7). CE alone had no significant effect on transcription either negatively or positively (lanes 8 and 9). These results suggest that CE alters NELF-mediated repression by interacting functionally with NELF or NELF-containing complexes during transcription.
Fig. 5.
CE can overcome NELF-mediated transcription repression. (A) PICs were assembled with purified GTFs on a linear DNA by incubation in TB60 for 20 min at 30°C. DSIF (30 ng), NELF, and CE were then added as shown, and incubations continued for 10 min. Transcription reactions were chased with a mixture of 600 μM ATP, CTP, and GTP; 0.3 μM [α-32P]UTP; 2 μM UTP and 16 units of RNasin for 6 min at 30°C. Reactions were stopped with stop buffer, extracted with phenol-chloroform, ethanol precipitated, and analyzed by 7% PAGE/8 M urea and autoradiography. PICs were chased with either NTPs alone (lane 1) or first incubated with DSIF (lane 2) or NELF (lane 3) or both (lane 4) and chased with NTPs. PICs incubated with both DSIF and NELF were further incubated with 0.12, 12, or 120 ng of CE (lanes 5-7) and chased with NTPs. For lanes 8 and 9, PICs were chased with NTPs in the presence of CE alone (12 and 120 ng). The dots indicate the approximate positions of transcripts of 42 and ≈200 nt. (B)Asin A, except that transcription reactions were chased with NTPs (50 μM ATP/5 μM CTP/0.3 μM [α-32P]UTP/50 μM 3′-OmeGTP/1 μM UTP/16 units of RNasin) for 5 min to produce +24 stalled transcripts, which were analyzed by 18% PAGE in 8M urea.
In another series of experiments, transcription-initiation complexes were chased with a subset of nucleotides (ATP, CTP, and UTP) and 3′-Ome GTP to restrict products to 24 nt and hold complexes in the promoter clearance stage. In agreement with the results shown in Fig. 5A, DSIF and NELF together efficiently repressed transcription and produced shorter transcripts (Fig. 5B, lane 4). However, addition of CE together with DSIF and NELF overcame the transcriptional repression by NELF and restored the 24-nt RNA products (Fig. 5B, lanes 5-7). The results shown in Fig. 5 indicate that CE can overcome NELF-mediated transcriptional repression.
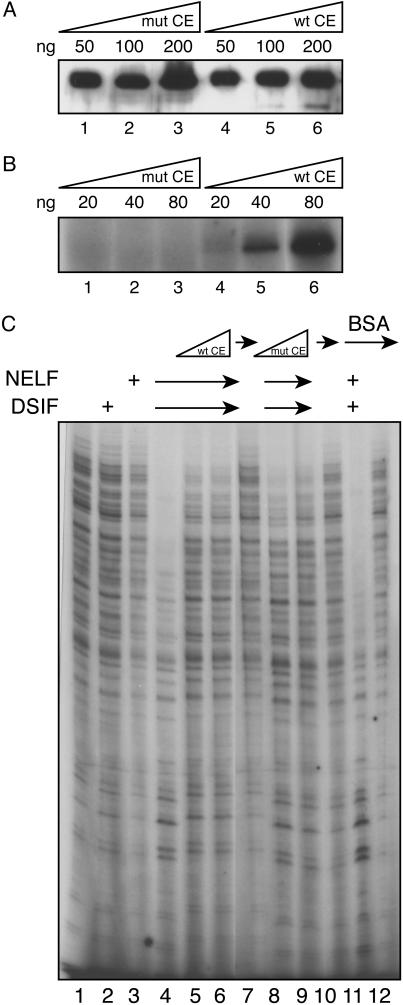
In the previous experiment, we used a mixture of NTPs that usually included GTP to chase transcription reactions, raising the possibility that the CE-mediated inhibition of NELF-mediated repression could be caused by cap addition. To investigate whether capping per se was responsible for overcoming NELF-mediated repression, we used the catalytically inactive K294A mutant CE (25). Transcription initiation complexes were assembled with GTFs in the presence of DSIF and NELF and then incubated with varying concentrations of wild-type or mutant CE. Reactions were chased with four NTPs for 6 min. Consistent with the results presented in Fig. 5, DSIF and NELF together strongly inhibited transcription (Fig. 6C, lane 4), and the wild-type CE reversed this effect (lanes 5 and 6). The catalytically inactive CE (Fig. 6B) could also relieve NELF-mediated transcriptional repression as efficiently as the wildtype enzyme (Fig. 6C, compare lane 4 with lanes 5 and 6 and with lanes 8 and 9), and BSA was unable to overcome NELF-mediated repression of transcription (lanes 11 and 12). We therefore concluded that the interaction of the CE with the transcription complex, most likely through DSIF, can rescue transcriptional repression by NELF.
Fig. 6.
Reversal of NELF-mediated transcription repression depends on CE but not on cap formation. (A) Western blot of K294A mutant CE and wild-type CE. (B) Guanylylation of CE. Wild-type and mutant CE were incubated in 20 μl of GTP-labeling buffer (25 mM Tris·HCl, pH 7.5/5 mM MgCl2/0.5 mM DTT/10 μCi (1 Ci = 37 GBq) [α-32P]GTP/0.1 μg of inorganic pyrophosphatase) at 37°C for 10 min and then analyzed by SDS/PAGE and autoradiography. (C) PICs were incubated with DSIF and NELF as in Fig. 5A and then chased with NTPs in the absence and presence of wild-type CE (12 and 120 ng, compare lanes 4-7), K294A inactive mutant CE (12 and 120 ng, compare lanes 8-10), or BSA as control (120 ng, compare lanes 11 and 12). Analysis by 7% PAGE/8 M urea is shown.
A direct test of the hypothesis that NELF is displaced from DSIF-NELF complexes destabilized by the interaction of CE with DSIF was not possible because stable DSIF-NELF complexes were not obtained, even in the presence of RNAPII (data not shown). We suggest that the functional interaction between DSIF, NELF, and CE, observed in the results presented above, may require a transcription complex, probably containing RNA. The RNA binding motif of NELF-E is required for transcriptional repression (14), and it is also possible that the RNA binding ability of the CE (41) interferes with the binding of NELF to the nascent transcript, leading to release of transcriptional repression. Additional studies on the interactions of CE, NELF, and DSIF, as well as the cap (guanine-N7) methyltransferase and RNAPII, should provide further insights into the critical events that occur during promoter clearance.
Abbreviations: RNAPII, RNA polymerase II; CE, capping enzyme; CTD, carboxyl-terminal domain; DSIF, DRB sensitivity-inducing factor; NELF, negative elongation factor; PIC, preinitiation complex; AP, alkaline phosphatase.
References
- 1.Furuichi, Y. & Shatkin, A. J. (2000) Adv. Virus. Res. 55, 135-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shatkin, A. J. (1976) Cell 9, 645-653. [DOI] [PubMed] [Google Scholar]
- 3.Shuman, S. (2001) Prog. Nucleic Acid Res. Mol. Biol. 66, 1-40. [DOI] [PubMed] [Google Scholar]
- 4.Maniatis, T. & Reed, R. (2002) Nature 416, 499-506. [DOI] [PubMed] [Google Scholar]
- 5.Orphanides, G. & Reinberg, D. (2002) Cell 108, 439-451. [DOI] [PubMed] [Google Scholar]
- 6.Greenleaf, A. (2003) Structure (London) 11, 900-902. [DOI] [PubMed] [Google Scholar]
- 7.Fabrega, C., Shen, V., Shuman, S. & Lima, C. D. (2003) Mol. Cell 11, 1549-1561. [DOI] [PubMed] [Google Scholar]
- 8.Shatkin, A. J. & Manley, J. L. (2000) Nat. Struct. Biol. 7, 838-842. [DOI] [PubMed] [Google Scholar]
- 9.Proudfoot, N. (2000) Trends Biochem. Sci. 25, 290-293. [DOI] [PubMed] [Google Scholar]
- 10.Ni, Z., Schwartz, B. E., Werner, J., Suarez, J. R. & Lis, J. T. (2004) Mol. Cell 13, 55-65. [DOI] [PubMed] [Google Scholar]
- 11.O'Brien, T., Hardin, S., Greenleaf, A. & Lis, J. T. (1994) Nature 370, 75-77. [DOI] [PubMed] [Google Scholar]
- 12.Cho, H., Kim, T. K., Mancebo, H., Lane, W. S., Flores, O. & Reinberg, D. (1999) Genes Dev. 13, 1540-1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamaguchi, Y., Takagi, T., Wada, T., Yano, K., Furuya, A., Sugimoto, S., Hasegawa, J. & Handa, H. (1999) Cell 97, 41-51. [DOI] [PubMed] [Google Scholar]
- 14.Yamaguchi, Y., Inukai, N., Narita, T., Wada, T. & Handa, H. (2002) Mol. Cell. Biol. 22, 2918-2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marshall, N. F. & Price, D. H. (1995) J. Biol. Chem. 270, 12335-12338. [DOI] [PubMed] [Google Scholar]
- 16.Rodriguez, C. R., Cho, E. J., Keogh, M. C., Moore, C. L., Greenleaf, A. L. & Buratowski, S. (2000) Mol. Cell. Biol. 20, 104-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schroeder, S. C., Schwer, B., Shuman, S. & Bentley, D. (2000) Genes Dev. 14, 2435-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pei, Y., Hausmann, S., Ho, C. K., Schwer, B. & Shuman, S. (2001) J. Biol. Chem. 276, 28075-28082. [DOI] [PubMed] [Google Scholar]
- 19.Wen, Y. & Shatkin, A. J. (1999) Genes Dev. 13, 1774-1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ho, C. K. & Shuman, S. (1999) Mol. Cell 3, 405-411. [DOI] [PubMed] [Google Scholar]
- 21.McCracken, S., Fong, N., Rosonina, E., Yankulov, K., Brothers, G., Siderovski, D., Hessel, A., Foster, S., Shuman, S. & Bentley, D. L. (1997) Genes Dev. 11, 3306-3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Myers, L. C., Lacomis, L., Erdjument-Bromage, H. & Tempst, P. (2002) Mol. Cell 10, 883-894. [DOI] [PubMed] [Google Scholar]
- 23.Pei, Y., Schwer, B. & Shuman, S. (2003) J. Biol. Chem. 278, 7180-7188. [DOI] [PubMed] [Google Scholar]
- 24.Mandal, S. S., Cho, H., Kim, S., Cabane, K. & Reinberg, D. (2002) Mol. Cell. Biol. 22, 7543-7552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yue, Z., Maldonado, E., Pillutla, R., Cho, H., Reinberg, D. & Shatkin, A. J. (1997) Proc. Natl. Acad. Sci. USA 94, 12898-12903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Narita, T., Yamaguchi, Y., Yano, K., Sugimoto, S., Chanarat, S., Wada, T., Kim, D. K., Hasegawa, J., Omori, M., Inukai, N., et al. (2003) Mol. Cell. Biol. 23, 1863-1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Samkurashvili, I. & Luse, D. S. (1998) Mol. Cell. Biol. 18, 5343-5354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Proudfoot, N. J., Furger, A. & Dye, M. J. (2002) Cell 108, 501-512. [DOI] [PubMed] [Google Scholar]
- 29.Moteki, S. & Price, D. (2002) Mol. Cell 10, 599-609. [DOI] [PubMed] [Google Scholar]
- 30.Pal, M. & Luse, D. S. (2002) Mol. Cell. Biol. 22, 30-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gnatt, A. L., Cramer, P., Fu, J., Bushnell, D. A. & Kornberg, R. D. (2001) Science 292, 1876-1882. [DOI] [PubMed] [Google Scholar]
- 32.Chiu, Y. L., Ho, C. K., Saha, N., Schwer, B., Shuman, S. & Rana, T. M. (2002) Mol. Cell 10, 585-597. [DOI] [PubMed] [Google Scholar]
- 33.Wu, C. H., Yamaguchi, Y., Benjamin, L. R., Horvat-Gordon, M., Washinsky, J., Enerly, E., Larsson, J., Lambertsson, A., Handa, H. & Gilmour, D. (2003) Genes Dev. 17, 1402-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pei, Y. & Shuman, S. (2002) J. Biol. Chem. 277, 19639-19648. [DOI] [PubMed] [Google Scholar]
- 35.Lindstrom, D. L., Squazzo, S. L., Muster, N., Burckin, T. A., Wachter, K. C., Emigh, C. A., McCleery, J. A., Yates, J. R., III, & Hartzog, G. A. (2003) Mol. Cell. Biol. 23, 1368-1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Andrulis, E. D., Guzman, E., Doring, P., Werner, J. & Lis, J. T. (2000) Genes Dev. 14, 2635-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cho, E. J., Takagi, T., Moore, C. R. & Buratowski, S. (1997) Genes Dev. 11, 3319-3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fong, N. & Bentley, D. L. (2001) Genes Dev. 15, 1783-1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cho, E. J., Rodriguez, C. R., Takagi, T. & Buratowski, S. (1998) Genes Dev. 12, 3482-3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin, P. S., Dubois, M. F. & Dahmus, M. E. (2002) J. Biol. Chem. 277, 45949-46956. [DOI] [PubMed] [Google Scholar]
- 41.Wen, Y., Yue, Z. & Shatkin, A. J. (1998) Proc. Natl. Acad. Sci. USA 95, 12226-12231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saunders, A., Werner, J., Andrulis, E. D., Nakayama, T., Hirose, S., Reinberg, D. & Lis, J. T. (2003) Science 301, 1094-1096. [DOI] [PubMed] [Google Scholar]