Abstract
Ubiquitin-(Ub) like proteins (Ubls) are conjugated to their targets by an enzymatic cascade involving an E1 activating enzyme, an E2 conjugating enzyme, and in some cases an E3 ligase. ISG15 is a Ubl that is conjugated to cellular proteins after IFN-α/β stimulation. Although the E1 enzyme for ISG15 (Ube1L/E1ISG15) has been identified, the identities of the downstream components of the ISG15 conjugation cascade have remained elusive. Here we report the purification of an E2 enzyme for ISG15 and demonstrate that it is UbcH8, an E2 that also functions in Ub conjugation. In vitro assays with purified Ub E2 enzymes and in vivo RNA interference assays indicate that UbcH8 is a major E2 enzyme for ISG15 conjugation. These results indicate that the ISG15 conjugation pathway overlaps or converges with the Ub conjugation pathway at the level of a specific E2 enzyme. Furthermore, these results raise the possibility that the ISG15 conjugation pathway might use UbcH8-competent Ub ligases in vivo. As an initial test of this hypothesis, we have shown that a UbcH8-competent Ub ligase conjugates ISG15 to a specific target in vitro. These results challenge the concept that Ub and Ubl conjugation pathways are strictly parallel and nonoverlapping and have important implications for understanding the regulation and function of ISG15 conjugation in the IFN-α/β response.
IFN-α/β play an essential role in innate immunity and are induced during many types of viral infections (1). Many genes are transcriptionally induced by IFN-α/β, including ISG15 (IFN-stimulated gene, 15 kDa) (2, 3). The ISG15 protein is a 15-kDa ubiquitin (Ub)-like protein (Ubl), consisting of two Ub-related domains, ≈30% (N-terminal domain) and 36% (C-terminal domain) identical to Ub. ISG15 becomes conjugated to a diverse set of cellular proteins after IFN-α/β stimulation (4). Although the biochemical consequences of ISG15 conjugation and the fate of the conjugated proteins are not known, it does not appear that ISG15 targets proteins for proteasomal degradation (5, 6).
Conjugation of Ub to target proteins requires the cooperative activities of at least three classes of enzymes (7). The ATP-dependent E1 enzyme activates Ub by C-terminal adenylation, followed by formation of a high-energy thioester bond between the terminal carboxylate of Ub and the active-site cysteine of E1. Ub is then transferred to the active-site cysteine of one of a number of related E2 enzymes. E3 enzymes then promote transfer of Ub from the E2 to the substrate, resulting in a stable amide bond between ε-amino groups of lysine side chains and Ub. E3 enzymes are the primary determinants of substrate specificity and can be divided into two classes based on mechanism. HECT E3s accept Ub from the E2 enzyme, again in the form of a thioester adduct, and transfer Ub from their active-site cysteine to the bound substrate (8). RING E3s consist of several subclasses and are either single or multisubunit enzymes that serve as docking proteins for both protein substrates and activated E2 enzymes, with transfer of Ub being from the E2 to the substrate (9).
Conjugation pathways for several Ubls have been at least partially characterized, and the emerging view is that each pathway is parallel and distinct from the Ub pathway (5, 10). That is, each Ubl has unique and dedicated E1 and E2 enzymes, related to their cognate enzymes in the Ub system but functioning only with that particular Ubl. The only exception to this is E1Apg7, which activates two Ubls (Apg12 and Aut7), both of which play key roles in the autophagy process in budding yeast (11). None of the Ub E2 enzymes have been found to function with Ubls, and none of the Ubl E2 enzymes have been found to function with Ub. The best characterized E3s for Ubls are those for small ubiquitin-like modifier (SUMO) family members (12-14), and these are specific for SUMO conjugation and function only with the SUMO-specific E2, Ubc9.
We previously identified the E1 enzyme for ISG15, Ube1L/E1ISG15, a single-subunit enzyme capable of activating ISG15 but not Ub (15). To gain further insight into the function and regulation of ISG15 conjugation, we sought here to identify the E2 enzyme that functions in ISG15 conjugation. Surprisingly, we found that UbcH8, an E2 that functions in Ub conjugation pathways (16-22), is a major E2 enzyme for the ISG15 conjugation pathway. Therefore, our results indicate that the ISG15 conjugation pathway overlaps or converges with the Ub conjugation pathway at the level of a specific E2 enzyme. Such a convergence or overlap of a Ubl conjugation pathway with a Ub conjugation pathway is so far unprecedented. Furthermore, these results raise the possibility that the ISG15 conjugation pathway might use UbcH8-competent Ub ligases in vivo, and we present in vitro data that support this possibility. These findings have important implications for our understanding of both Ub/Ubl conjugation pathways and IFN-α/β-induced ISG15 conjugation.
Methods
Preparation of A549 Cell Extracts. Crude cell extracts from A549 cells, either untreated or treated with 1,000 units/ml IFN-β (Berlex Biosciences, Richmond, CA), were prepared as described (15). The extracts were centrifuged at 12,000 × g for 10 min, and the supernatant was passed through a glutathione-Sepharose column to remove cellular proteins that bind directly to this matrix. The flow-through was applied to a SP Sepharose column to remove activities that interfered with optimum ISG15 conjugation. The flow-through from this column was concentrated to 20 mg/ml by using Centricon-10 filters (Amicon) and then used for the purification of the E2 enzyme for ISG15 as described in the legend to Fig. 1B.
Fig. 1.
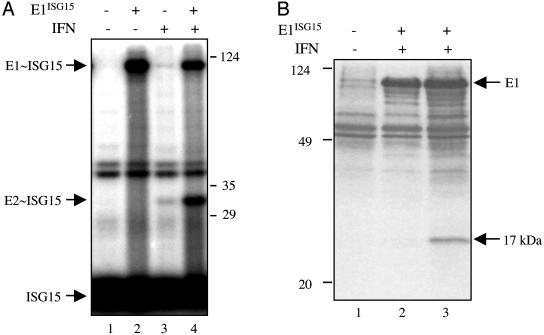
Identification of the E2 enzyme for ISG15 conjugation. (A) Extracts from IFN-β-treated (lanes 3 and 4) or untreated (lanes 1 and 2) A549 cells were incubated with 32P-ISG15 protein in the absence (lanes 1 and 3) or presence (lanes 2 and 4) of E1ISG15 for 30 min at 25°C. The protein products were resolved by electrophoresis on a 10% polyacrylamide gel. Positions of molecular mass markers are shown on the right. (B)Affinity purification of the E2 enzyme for ISG15. Cell extract (15 mg of protein) from IFN-β-treated (lanes 1 and 3) or untreated (lane 2) A549 cells were incubated with GST-ISG15 (2 mg) in either the absence (lane 1) or presence (lanes 2 and 3) of E1ISG15 (500 μg) in a final volume of 1 ml for 30 min at 25°C under the thioester reaction conditions described in Methods. The reaction products were affinity selected on glutathione-Sepharose, and thioester bonds were cleaved by DTT treatment. The eluted proteins were subjected to electrophoresis on 15% polyacrylamide gels, followed by Coomassie blue staining. The 17-kDa protein in lane 3 was digested with trypsin, and the smallest tryptic peptide was sequenced by automated Edman degradation at the W. M. Keck Foundation Biotechnology Resource Laboratory at Yale University.
Protein Expression and Purification. E1Ub and E1ISG15 were expressed as GST fusion proteins by using baculovirus vectors (Pharmingen and Invitrogen) in High Five insect cells (Invitrogen), and Ub and ISG15 were expressed as GST fusion proteins by using the pGEX2TK vector (Amersham Pharmacia) in Escherichia coli strain BL21. UbcH5b, UbcH7, and UbcH8 were expressed in E. coli strain BL21 as GST fusions by using the pGEX4T1 vector (Amersham Pharmacia Biosciences). These proteins were purified by glutathione affinity chromatography and, where indicated, the GST fusions were further purified by size exclusion chromatography after thrombin cleavage to remove GST. Rsp5, Rsp5 C-A, and FLAG-tagged WBP2 protein were expressed in E. coli as a GST fusion protein by using the pGEX-6p vector (Amersham Pharmacia). After glutathione affinity chromatography, the GST moiety of the latter three fusions was removed by digestion with PreScission protease (Amersham Pharmacia). Proteins were stored at -80 at concentrations of 0.5-15 mg/ml.
Thioester and Ub and ISG15 Conjugation Assays. Ub and ISG15 thioester assays (Fig. 2) used 200 ng of purified E1Ub or E1ISG15, 2 μg of purified E2 enzyme, and 400 ng/106 cpm of 32P-labeled Ub or ISG15. Ub and ISG15 were labeled as described (15). Reactions contained 25 mM Tris (pH 7.5), 50 mM NaCl, 10 mM MgCl2, 5 mM ATP, and 0.1 mM DTT and were incubated at 25° for 10 min. Reactions were stopped with SDS/PAGE loading buffer lacking DTT and analyzed by SDS/PAGE and autoradiography.
Fig. 2.
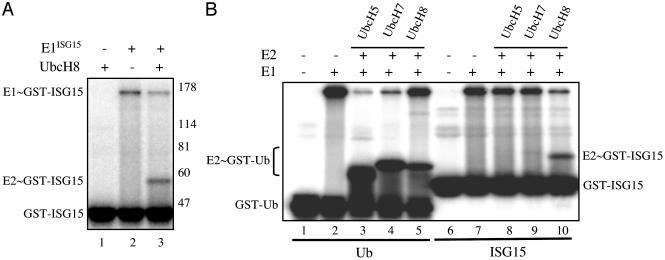
UbcH8 functions as a major E2 enzyme for ISG15 in vitro. (A) 32P-GST-ISG15 was incubated in the presence (lanes 1 and 3) or absence (lane 2) of UbcH8 and in the presence (lanes 2 and 3) or absence (lane 1) of E1ISG15. (B) Either 32P-GST-Ub (lanes 1-5) or 32P-GST-ISG15 (lanes 6-10) was incubated in the absence (lanes 1 and 6) or presence of E1Ub (lanes 2-5) or E1ISG15 (lanes 7-10) and in the absence or presence of the indicated E2 proteins.
Substrate conjugation assays (Fig. 4) using unlabeled Ub or ISG15 contained 200 ng of Ub or ISG15, 100 ng of E1Ub or E1ISG15, 1.5 μg of UbcH8, 100 ng of Rps5p protein, and 2 μg of purified FLAG-WBP2. Reactions contained 25 mM Tris (pH 7.5), 50 mM NaCl, 10 mM MgCl2, 5 mM ATP, and 0.1 mM DTT, and were incubated at 25° for 30 min. Reactions were stopped with DTT-containing SDS/PAGE loading buffer and analyzed by immunoblotting by using anti-FLAG antibody (Sigma). Reactions containing 32P-labeled Ub or ISG15 (400 ng/106 cpm) were done under the same conditions. Reactions were stopped by dilution into buffer containing 10 mM EDTA. FLAG-WBP2 was immunoprecipitated with anti-FLAG antibody, and the immunoprecipitates were analyzed by SDS/PAGE and autoradiography.
Fig. 4.
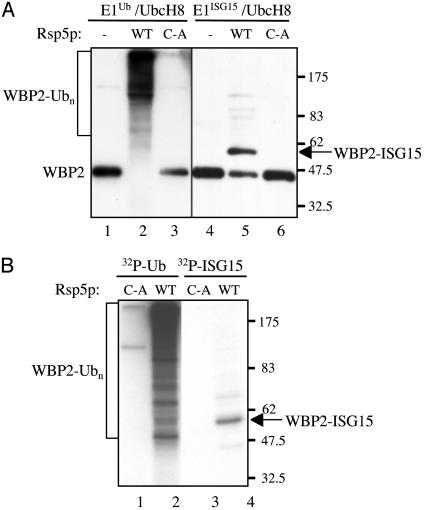
A Ub E3 functions with ISG15 in vitro. (A) Purified FLAG-WBP2 protein was incubated with either Ub, E1Ub, and UbcH8 (lanes 1-3) or ISG15, E1ISG15, and UbcH8 (lanes 4-6), in the absence (lanes 1 and 4) or presence of Rsp5p (lanes 2 and 5) or in the presence the C-A mutant of Rsp5p (lanes 3 and 6). Reactions products were analyzed by immunoblotting with anti-FLAG antibody. (B) Conjugation reactions were performed as in A, in the presence of 32P-Ub (lanes 1 and 2) or 32P-ISG15 (lanes 3 and 4). Reaction products were immunoprecipitated with anti-FLAG antibody and analyzed by SDS/PAGE and autoradiography.
Results
Conjugation of ISG15 to target proteins occurs in IFN-β-treated human A549 human lung cells (4, 15), indicating that these cells contain the ISG15-conjugating enzymes. To test for the presence of an E2 enzyme for ISG15, an extract from these cells was incubated with 32P-ISG15 in the absence or presence of added E1ISG15 (Fig. 1A, lanes 3 and 4). A labeled 32-kDa product was observed. Several lines of evidence indicated that this 32-kDa species was likely to correspond to the thioester-linked complex of ISG15 with its E2 enzyme. First, this species was sensitive to DTT, as expected for thioester-linked complexes (data not shown). Second, the molecular mass of the protein that was conjugated to the 15-kDa ISG15 was ≈17 kDa, similar to the molecular masses of the E2s for Ub and other Ubls (7). Third, formation of the labeled 32-kDa species was stimulated ≈20-fold by the addition of E1ISG15, indicating that it was likely the downstream product of a reaction involving ISG15 and E1ISG15. The labeled 32-kDa species was not observed by using extracts from A549 cells that had not been treated with IFN-β (lanes 1 and 2), indicating that the E2 enzyme activity was itself induced by IFN-β.
To purify the E2 enzyme for ISG15, GST-ISG15 and E1ISG15 were incubated with an extract from IFN-β-treated A549 cells, resulting in the formation of GST-ISG15∼E1 and GST-ISG15∼E2 complexes (“∼” is used to indicate a thioester bond). The complexes were affinity selected by using glutathione-Sepharose. The thioester bonds between ISG15 and E1 and E2 were cleaved with DTT, and the eluted material was subjected to denaturing gel electrophoresis (Fig. 1B, lane 3). Coomassie blue staining detected a protein species of 17 kDa. The putative E2 species was detected neither in the absence of added E1ISG15 (lane 1) nor when using extracts from cells not treated with IFN-β (lane 2). The identity of the 17-kDa species was determined by Edman degradation sequencing of the shortest peptide generated by trypsin digestion. Its sequence, AEEFTLR, was identical to a sequence in UbcH8 (amino acids 149-155), an E2 for Ub (16).
To test whether UbcH8 functions as an E2 for ISG15 in vitro, GST-32P-ISG15 was incubated with purified UbcH8 in the absence or presence of E1ISG15 (Fig. 2A). ISG15 was covalently linked to UbcH8 only in the presence of E1ISG15 (lane 3), and the amount of ISG15 linked to E1ISG15 decreased concomitantly with the increase in the amount of ISG15 linked to UbcH8 (compare lanes 2 and 3). The UbcH8∼ISG15 complex was sensitive to DTT (data not shown). These results indicate that ISG15 was transferred from E1ISG15 to UbcH8, and that UbcH8 functions as an E2 for ISG15. To determine whether other E2s that are closely related to UbcH8 can also function as E2s for ISG15, we tested the activity of UbcH5b (23) (56% similarity to UbcH8) and UbcH7 (24) (72% similarity to UbcH8), both of which function efficiently as E2s for Ub (Fig. 2B, lanes 2-5). No E2∼ISG15 complexes were observed with UbcH5b (compare lanes 8 and 10), and a low amount of complex was observed with UbcH7 (≈5% of that of UbcH8, as determined by a long exposure of the gel of Fig. 2B). Therefore, our in vitro results suggest that UbcH8 is a major E2 enzyme for ISG15. Interestingly, like ISG15, the UbcH8 gene is transcriptionally induced by IFN-α/β (25), consistent with our results that the E2 activity for ISG15 was detected only in A549 cells after IFN-β treatment (Fig. 1).
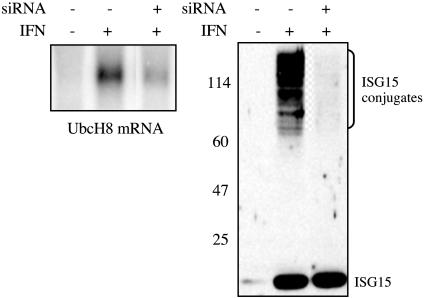
To determine whether UbcH8 is a major E2 for ISG15 in vivo, we carried out RNA interference experiments (26) in IFN-β-treated HeLa cells. We used a double-stranded short interfering RNA (siRNA) directed against a UbcH8 mRNA sequence that is not identical to a sequence in any other predicted human mRNA, including the closely related UbcH7 mRNA. This siRNA reduced the amount of UbcH8 mRNA by 80-90% and inhibited IFN-β-induced ISG15 conjugation to a similar extent (Fig. 3). Another siRNA against UbcH8 had the same effect on UbcH8 mRNA and ISG15 conjugation, and Northern and RT-PCR analyses showed that UbcH7 mRNA was not affected by either siRNA (data not shown). Northern analysis (Fig. 3) also verified that UbcH8 mRNA was induced by IFN-β. We therefore conclude that UbcH8 serves as a major E2 enzyme for ISG15 conjugation in vivo.
Fig. 3.
UbcH8 functions as a major E2 enzyme for ISG15 in vivo. Cells were either mock transfected (-siRNA) or transfected with an siRNA (20 nM final concentration) directed against bases 28-49 of the ORF of human UbcH8 mRNA (+siRNA lanes). After 24 h, the cells were left untreated (-IFN lanes) or were treated with IFN-β (1,000 units/ml; +IFN lanes). After another 24 h, the cells were collected. RNA was analyzed for UbcH8 mRNA by Northern analysis (Left), and proteins were analyzed by immunoblotting with ISG15 antiserum (Right). Each lane of the Northern blot contained 12 μg of total RNA, and the presence of equal amounts of RNA in each lane was confirmed by ethidium bromide staining of 28S ribosomal RNA (not shown). The same results were obtained by using a second siRNA that was directed against bases 239-258 of the ORF of UbcH8 mRNA, and an siRNA directed against a sequence in the mRNA for GFP did not decrease either UbcH8 mRNA or ISG15 conjugation (not shown).
The finding that UbcH8 functions as a major ISG15 E2 enzyme raises the possibility that ISG15 conjugation might use UbcH8-competent Ub E3 enzymes. UbcH8 has been reported to function as an E2 for several Ub E3 enzymes, including both HECT E3s (16) and RING E3s (17-22). As an initial test of this hypothesis, we determined whether a UbcH8-competent Ub E3 was capable of conjugating ISG15 to a substrate protein in vitro. Conjugation reactions were performed by using the Saccharomyces cerevisiae Rsp5p E3 ligase, which efficiently catalyzes the in vitro ubiquitination of human WBP2 (27). Two assays established that purified Rsp5p could function with UbcH8 to conjugate ISG15 to WBP2 in vitro. In the first assay, purified FLAG-WBP2 was incubated with Ub or ISG15 in the presence of UbcH8 and the cognate E1 enzyme, and the reaction products were analyzed by SDS/PAGE and immunoblotting by using anti-FLAG antibody (Fig. 4A). Rsp5p catalyzed efficient polyubiquitination of WBP2 in the presence of E1Ub and UbcH8, whereas the active-site mutant (C-A) was inactive (lanes 1-3). Wild-type Rsp5p also catalyzed the conjugation of ISG15 to WBP2 in the presence of E1ISG15 and UbcH8 (lanes 4-6). The molecular weight of the predominant WBP2-ISG15 conjugate corresponded to the addition of a single molecule of ISG15. In the second assay, conjugation reactions contained 32P-Ub or 32P-ISG15, and the reaction products were immunoprecipitated with anti-FLAG antibody and analyzed by SDS/PAGE and autoradiography. A broad distribution of polyubiquitinated species was seen in the Ub-containing reaction in the presence of wild-type Rsp5p but not in the presence of the C-A mutant (Fig. 4B). The ISG15-containing reaction again produced a predominant species corresponding in size to a single molecule of ISG15 conjugated to WBP2, and this was not seen in the presence of the C-A mutant. Consistent with these findings, Rsp5p formed the expected ISG15-thioester catalytic intermediate (data not shown). Therefore, a HECT domain ligase competent to function with UbcH8 is capable of conjugating ISG15 in vitro.
Discussion
Results to date have suggested that Ubl conjugation pathways are parallel to but distinct from pathways for Ub conjugation (5), with each Ubl using E1, E2, and in some cases E3 enzymes related to their cognate enzymes in the Ub pathway but dedicated to conjugation of that specific Ubl. Indeed, for the NEDD8 Ubl, there are selectivity determinants that prevent ubiquitinating enzymes from conjugating NEDD8 and that prevent NEDD8-specific enzymes from conjugating Ub (28-31). In contrast, our results indicate that the pathways for Ub and ISG15 conjugation converge or overlap at the level of a specific E2 enzyme, UbcH8. This was established by purifying the E2 that functions with E1ISG15 in extracts from IFN-β-treated cells and identifying this E2 as UbcH8 by protein sequencing. Transfer of ISG15 from E1ISG15 to UbcH8 was confirmed in vitro with purified components, and experiments using UbcH8-specific siRNAs demonstrated that UbcH8 is a major E2 for the ISG15 pathway in vivo.
For UbcH8 to be charged with both Ub and ISG15, UbcH8 must have the ability to interact appropriately with both E1Ub and E1ISG15 and with both Ub and ISG15. The sequences of the E1s for Ub and ISG15 and of Ub and ISG15 themselves reveal similarities that provide insights into the cross talk between the two pathways. Thus, E1ISG15 is the most similar to E1Ub among all known human Ubl E1 enzymes (≈44% identity). A significant difference between E1ISG15 and E1Ub, however, is that E1ISG15 demonstrates specificity for the E2 enzyme UbcH8 over other Ub E2s, whereas the E1 enzyme for Ub functions with multiple E2 enzymes, including UbcH8 (7). The molecular basis for the selectivity of E1ISG15 for UbcH8 over other closely related E2s, particularly UbcH7, remains to be determined. With respect to the similarity between Ub and ISG15, ISG15 is the only known Ubl whose C-terminal six residues are identical to those of Ub, suggesting that this C-terminal tail may be crucial for a functional interaction with UbcH8. Indeed, the structural model of a Ubc∼Ub thioester complex revealed that the C-terminal six residues of Ub position themselves in a shallow cleft in the E2 protein that is highly conserved among E2 proteins (32). It is also apparent that the N-terminal Ub-like domain of ISG15 does not interfere with the ability of ISG15 to function with UbcH8.
We have shown that a UbcH8-competent Ub ligase (Rsp5p) conjugates ISG15 to a specific target in vitro, indicating that there are no inherent structural features of ISG15 that interfere with its utilization by at least this type of Ub ligase. Therefore, the ISG15 conjugation pathway, which utilizes an E2 enzyme of the Ub pathway, might use a subset of UbcH8-competent Ub E3 enzymes in vivo. Although the putative Ub ligases that function with ISG15 in vivo have not been identified, our results suggest several possible models for the involvement of Ub E3s in the ISG15 conjugation pathway. In the “converging pathways” model, a given UbcH8-competent E3 enzyme would have the capacity to catalyze either Ub or ISG15 conjugation, depending on whether it was presented with UbcH8∼ISG15 or a Ub-charged E2 protein. The E3 would be presented with high levels of UbcH8∼ISG15 after IFN-α/β stimulation, because ISG15, E1ISG15, and UbcH8 are all transcriptionally induced by IFN-α/β. In the absence of IFN-α/β stimulation, the E3 might function with Ub and UbcH8-related E2 enzymes, such as UbcH7 or UbcH5. This model implies possible alternative modification (Ub or ISG15) of substrates, depending on IFN-α/β stimulation. Perhaps consistent with this model, an earlier study that examined total cellular ISG15 conjugates suggested that proteins that are conjugated to ISG15 are not simultaneously modified by Ub (33). The rationale of incorporating UbcH8 into the ISG15 conjugation pathway in this model might be to take advantage of a preexisting set of E3s and switch them from performing Ub conjugation to ISG15 conjugation. Candidate E3s in this model include both HECT and single-subunit RING E3s, which are so far the only families of E3s that contain members capable of using UbcH8 (16-22) and possibly those E3s that interact with the closely related E2 UbcH7.
A second model, the “induced ligase” model, invokes E3s that, like the other components of the ISG15 conjugation pathway, are also induced by IFN-α/β. As in the first model, such E3s might have an inherent capacity to catalyze Ub and ISG15 conjugation. Their capacity to catalyze Ub conjugation might be limited, however, if they are expressed only under conditions where ISG15, E1ISG15, and UbcH8 are abundant. Distinguishing between these models for the ISG15 conjugation pathway will obviously require identification of the E3 enzymes that are used for conjugation of ISG15 to specific targets in vivo.
The ultimate biochemical function of ISG15 modification remains unknown. ISG15 modification does not appear to promote proteasome-mediated degradation (5, 6). Consistent with this, the predominant ISG15-WBP2 conjugate produced in vitro contained a single ISG15 molecule, under conditions where Ub was conjugated in polyubiquitin chains. Although there has been only limited characterization of in vivo ISG15 substrates and their conjugates, it appears that most substrates may have only one or two molecules of conjugated ISG15 (6, 34). In the alternative modification model, one function of ISG15 may be to stabilize proteins that would otherwise be targeted for Ub-mediated degradation. Such a function has previously been suggested for SUMO (10). Regardless of whether ISG15 protects against proteasomal degradation, ISG15 might alter the activities of its target proteins, perhaps by altering their localization or association with other proteins. In either case, ISG15 conjugation may be the basis for some of the profound biological effects of IFN-α/β. The integral role of ISG15 in the IFN-α/β response pathway is further highlighted by the fact that at least one virus, influenza B virus, has evolved to short-circuit the ISG15 conjugation pathway (15).
Interaction or cross-regulation of the Ub conjugation pathway with other Ubl pathways has been seen previously, for example in the alternative modification of a given substrate, at the same lysine residue, by Ub or SUMO (35), and in the regulation of SCF ubiquitin ligase activity by modification with NEDD8/RUB1 (36). The apparent convergence or overlap of the Ub and ISG15 conjugation systems described here is so far unprecedented and challenges the concept that Ub and Ubl proteins use parallel but separate conjugation pathways.
Acknowledgments
This work was supported by National Institutes of Health Grants AI17772 (to R.M.K.), CA72943 (to J.M.H.), and GM69530 (to B.A.S.); a Pew Scholar in Biomedical Sciences Award (to B.A.S.); American Lebanese Syrian Associated Charities/St. Jude Children's Research Hospital (to B.A.S.); and a National Institutes of Health National Research Service Award (to M.L.K.).
Abbreviations: Ub, ubiquitin; Ubl, Ub-like; SUMO, small ubiquitin-like modifier; siRNA, short interfering RNA.
References
- 1.Levy, D. E. & Garcia-Sastre, A. (2001) Cytokine Growth Factor Rev. 12, 143-156. [DOI] [PubMed] [Google Scholar]
- 2.Haas, A. L., Ahrens, P., Bright, P. M. & Ankel, H. (1987) J. Biol. Chem. 262, 11315-11323. [PubMed] [Google Scholar]
- 3.Farrell, P. J., Broeze, R. J. & Lengyel, P. (1979) Nature 279, 523-525. [DOI] [PubMed] [Google Scholar]
- 4.Loeb, K. R. & Haas, A. L. (1992) J. Biol. Chem. 267, 7806-7813. [PubMed] [Google Scholar]
- 5.Schwartz, D. C. & Hochstrasser, M. (2003) Trends Biochem. Sci. 28, 321-328. [DOI] [PubMed] [Google Scholar]
- 6.Malakhov, M. P., Kim, K. I., Malakhova, O. A., Jacobs, B. S., Borden, E. C. & Zhang, D. E. (2003) J. Biol. Chem. 278, 16608-16613. [DOI] [PubMed] [Google Scholar]
- 7.Pickart, C. M. (2001) Annu. Rev. Biochem. 70, 503-533. [DOI] [PubMed] [Google Scholar]
- 8.Scheffner, M., Nuber, U. & Huibregtse, J. M. (1995) Nature 373, 81-83. [DOI] [PubMed] [Google Scholar]
- 9.Weissman, A. M. (2001) Nat. Rev. Mol. Cell Biol. 2, 169-178. [DOI] [PubMed] [Google Scholar]
- 10.Jentsch, S. & Pyrowolakis, G. (2000) Trends Cell Biol. 10, 335-342. [DOI] [PubMed] [Google Scholar]
- 11.Huang, W. P. & Klionsky, D. J. (2002) Cell Struct. Funct. 27, 409-420. [DOI] [PubMed] [Google Scholar]
- 12.Jackson, P. K. (2001) Genes Dev. 15, 3053-3058. [DOI] [PubMed] [Google Scholar]
- 13.Pichler, A., Gast, A., Seeler, J. S., Dejean, A. & Melchior, F. (2002) Cell 108, 109-120. [DOI] [PubMed] [Google Scholar]
- 14.Kagey, M. H., Melhuish, T. A. & Wotton, D. (2003) Cell 113, 127-137. [DOI] [PubMed] [Google Scholar]
- 15.Yuan, W. & Krug, R. M. (2001) EMBO J. 20, 362-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumar, S., Kao, W. H. & Howley, P. M. (1997) J. Biol. Chem. 272, 13548-13554. [DOI] [PubMed] [Google Scholar]
- 17.Chin, L. S., Vavalle, J. P. & Li, L. (2002) J. Biol. Chem. 277, 35071-35079. [DOI] [PubMed] [Google Scholar]
- 18.Tanaka, K., Suzuki, T., Chiba, T., Shimura, H., Hattori, N. & Mizuno, Y. (2001) J. Mol. Med. 79, 482-494. [DOI] [PubMed] [Google Scholar]
- 19.Niwa, J., Ishigaki, S., Doyu, M., Suzuki, T., Tanaka, K. & Sobue, G. (2001) Biochem. Biophys. Res. Commun. 281, 706-713. [DOI] [PubMed] [Google Scholar]
- 20.Urano, T., Saito, T., Tsukui, T., Fujita, M., Hosoi, T., Muramatsu, M., Ouchi, Y. & Inoue, S. (2002) Nature 417, 871-875. [DOI] [PubMed] [Google Scholar]
- 21.Moynihan, T. P., Ardley, H. C., Nuber, U., Rose, S. A., Jones, P. F., Markham, A. F., Scheffner, M. & Robinson, P. A. (1999) J. Biol. Chem. 274, 30963-30968. [DOI] [PubMed] [Google Scholar]
- 22.Zhang, Y., Gao, J., Chung, K. K., Huang, H., Dawson, V. L. & Dawson, T. M. (2000) Proc. Natl. Acad. Sci. USA 97, 13354-13359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jensen, J. P., Bates, P. W., Yang, M., Vierstra, R. D. & Weissman, A. M. (1995) J. Biol. Chem. 270, 30408-30414. [DOI] [PubMed] [Google Scholar]
- 24.Nuber, U., Schwarz, S., Kaiser, P., Schneider, R. & Scheffner, M. (1996) J. Biol. Chem. 271, 2795-2800. [DOI] [PubMed] [Google Scholar]
- 25.Nyman, T. A., Matikainen, S., Sareneva, T., Julkunen, I. & Kalkkinen, N. (2000) Eur. J. Biochem. 267, 4011-4019. [DOI] [PubMed] [Google Scholar]
- 26.Elbashir, S. M., Harborth, J., Lendeckel, W., Yalcin, A., Weber, K. & Tuschl, T. (2001) Nature 411, 494-498. [DOI] [PubMed] [Google Scholar]
- 27.Salvat, C., Wang, G., Dastur, A., Lyon, N. & Huibregtse, J. M. (2004) J. Biol. Chem., in press. [DOI] [PubMed]
- 28.Bohnsack, R. N. & Haas, A. L. (2003) J. Biol. Chem. 278, 26823-26830. [DOI] [PubMed] [Google Scholar]
- 29.Whitby, F. G., Xia, G., Pickart, C. M. & Hill, C. P. (1998) J. Biol. Chem. 273, 34983-34991. [DOI] [PubMed] [Google Scholar]
- 30.Burch, T. J. & Haas, A. L. (1994) Biochemistry 33, 7300-7308. [DOI] [PubMed] [Google Scholar]
- 31.Walden, H., Podgorski, M. S., Huang, D. T., Miller, D. W., Howard, R. J., Minor, D. L., Jr., Holton, J. M. & Schulman, B. A. (2003) Mol. Cell 12, 1427-1437. [DOI] [PubMed] [Google Scholar]
- 32.Hamilton, K. S., Ellison, M. J., Barber, K. R., Williams, R. S., Huzil, J. T., McKenna, S., Ptak, C., Glover, M. & Shaw, G. S. (2001) Structure (Cambridge, U.K. 9, 897-904. [DOI] [PubMed] [Google Scholar]
- 33.Liu, M., Li, X. L. & Hassel, B. A. (2003) J. Biol. Chem. 278, 1594-1602. [DOI] [PubMed] [Google Scholar]
- 34.Hamerman, J. A., Hayashi, F., Schroeder, L. A., Gygi, S. P., Haas, A. L., Hampson, L., Coughlin, P., Aebersold, R. & Aderem, A. (2002) J. Immunol. 168, 2415-2423. [DOI] [PubMed] [Google Scholar]
- 35.Muller, S., Hoege, C., Pyrowolakis, G. & Jentsch, S. (2001) Nat. Rev. Mol. Cell Biol. 2, 202-210. [DOI] [PubMed] [Google Scholar]
- 36.del Pozo, J. C. & Estelle, M. (2000) Plant Mol. Biol. 44, 123-128. [DOI] [PubMed] [Google Scholar]