Abstract
It is known that the DnaK and Trigger Factor (TF) chaperones cooperate in the folding of newly synthesized cytosolic proteins in Escherichia coli. We recently showed that despite a very narrow temperature range of growth and high levels of aggregated cytosolic proteins, E. coli can tolerate deletion of both chaperones, suggesting that other chaperones might be involved in this process. Here, we show that the secretion-dedicated chaperone SecB efficiently suppresses both the temperature sensitivity and the aggregation-prone phenotypes of a strain lacking both TF and DnaK. SecB suppression is independent of a productive interaction with the SecA subunit of the translocon. Furthermore, in vitro cross-linking experiments demonstrate that SecB can interact both co- and posttranslationally with short nascent chains of both secretory and cytosolic proteins. Finally, we show that such cotranslational substrate recognition by SecB is greatly suppressed in the presence of ribosome-bound TF, but not by DnaK. Taken together, our data demonstrate that SecB acts as a bona fide generalized chaperone.
Folding of newly synthesized cytosolic proteins in bacteria is principally orchestrated by three major chaperones, namely Trigger Factor (TF), DnaK (Hsp70), and GroEL (Hsp60). The occurrence of substrate sharing and cooperation among these chaperone machines in Escherichia coli has been extensively documented (1, 2). Recent studies have shown that the majority of the nascent polypeptides interact cotranslationally with the ATP-independent ribosome-associated TF and reach their native state without further folding assistance (3, 4). However, a substantial portion of nascent proteins has been shown to transit through the ATP-dependent DnaK and/or GroEL chaperone machines. DnaK, assisted by its DnaJ and GrpE cochaperone cohorts, interacts in a co- or posttranslational mode with nearly 15% of newly synthesized proteins (3, 5). In contrast, GroEL, together with its cochaperone partner GroES, interacts posttranslationally with ≈10% of the newly synthesized proteins (6, 7).
Recent studies have also highlighted the fact that TF and DnaK cooperate in the folding of newly synthesized proteins. For example, deletion of the tig gene, encoding TF, greatly enhances the number of newly synthesized polypeptides interacting with DnaK. In addition, the absence of TF at the ribosomal exit channel enables DnaK to interact with considerably shorter nascent chains (3, 5). Deuerling et al. (8) recently proposed that DnaK and TF possess overlapping substrate specificities. This proposal was based on the fact that the same population of cytosolic proteins aggregates when DnaK is depleted in the absence or presence of TF, although the amount of aggregated material is highly augmented in the absence of TF (8).
Genetic studies have previously shown that the simultaneous deletion of the dnaK and tig genes in E. coli causes synthetic lethality (3, 5). However, it was recently shown that E. coli can tolerate the deletion of both chaperones at temperatures <30°C (9, 10). In contrast to their respective isogenic single mutants, the strains lacking both DnaK/DnaJ and TF exhibit a very narrow range of growth and, in addition, accumulate high levels of aggregated proteins at the permissive temperatures. These findings indicate that protein folding in E. coli is not solely dependent on a productive interaction with either DnaK or TF and suggest that other cellular factors also play a role in this fundamental process. A candidate protein that could play a role in this process is SecB, a specialized chaperone known to assist preprotein translocation via the sec pathway (11). Because SecB can bind to nonnative polypeptide substrates independently of signal sequences and can also cooperate with the DnaK chaperone machine in the refolding of denatured protein in vitro, it has been suggested that SecB could also act as a generalized chaperone (12, 13).
In this study, we provide evidence supporting such a generalized role of SecB in protein folding. We first show that overproduction of SecB efficiently suppresses both the temperature-sensitive (ts) phenotype and the aggregation-prone phenotype of a strain lacking both DnaK/DnaJ and TF. Furthermore, by using an in vitro chemical-based cross-linking approach, we show that SecB efficiently associates with both secretory and cytosolic nascent polypeptides and that this interaction is greatly modulated by TF, but not by DnaK/DnaJ.
Materials and Methods
Bacterial Strains and Plasmid Constructs. All bacteria used in this study are E. coli K12 derivative strains. Strains MC4100, MG1655, and their isogenic Δtig::Cmr, ΔdnaKdnaJ::Knr, and Δtig::Cmr ΔdnaKdnaJ::Knr mutant derivatives have been recently described (9).
The 468-nt-long secB ORF was PCR-amplified from MG1655 genomic DNA by using primers SecB-For (5′-GGGAATTCACATGTCAGAACAAAACAACACTG-3′) and SecB-Rev (5′-GCGGATCCAAGCTTCAGGCATCCTGATGTTCTTC-3′). The amplified DNA fragment was digested with EcoRI and HindIII and ligated into either pSE380ΔNcoI (ColE1 ori, ptrc promoter) or p29SEN (pSC101 ori, ptrc promoter) digested with the same enzymes. Both plasmid vectors have been described (9). The mutation encoding SecB (E77K) was introduced by two-step PCR using the primers SecBE77K-For (5′-GTTCCTGTGTAAAGTTCAGCAGG-3′) and SecBE77K-Rev (5′-CCTGCTGAACTTTACACAGGAAC-3′) and wild-type secB DNA as template. The p29SEN-based TF expressing low copy plasmid was recently described (9). Plasmid pE01 was a kind gift of H. Tokuda (University of Tokyo, Tokyo) (14). Plasmid pC4Meth150PhoE has been described by Valent et al. (15). Plasmid pC4Meth150RpoB was constructed as follows. The first 438 nt of the rpoB ORF were amplified by PCR from MC4100 genomic DNA using primers 150RpoB-For (5′-CGCGGAATTCTAATATGGTTTACTCCTATACCGAG-3′)and150RpoB-Rev (5′-GCGCGGATCCGTCTGTCATGAGCGGAATTTCGCC-3′). The resulting PCR product was digested with EcoRI and HindIII and cloned into the pC4Met vector (16). All constructs were sequence-verified by using the appropriate primers.
Bacterial Viability Assay. Strains MC4100 Δtig ΔdnaKdnaJ or MG1655 Δtig ΔdnaKdnaJ were grown for 24 h at 20°C in LB broth (1% tryptone/0.5% yeast extract/0.5% NaCl, pH 7), electroporated with the indicated plasmids and incubated at the same temperature for 2 days on LB agar (1%) plates supplemented with ampicillin (always used at 100 μg/ml). Transformants were then grown for 24 h at 20°C in LB-ampicillin, and were serially diluted and spotted on LB-ampicillin agar plates supplemented with isopropyl-β-d-thiogalactopyranoside (IPTG) inducer when necessary. Plates were incubated at 20°C for 2 days and at 30°C, 34°C, 37°C, or 40°C for 18 h.
Isolation of Aggregated Proteins. Strain MC4100 Δtig ΔdnaKdnaJ, previously transformed with the pSE-based SecB-expressing plasmid, was grown for 24 h at 20°C, diluted 1:50 in LB-ampicillin with or without IPTG inducer, and grown to an optical density (OD600) of ≈0.4. The cultures were subsequently incubated for 3 h at 30°C or 2 h at 37°C. Identical densities of cells were pelleted, and aggregated proteins were isolated after repeated washing steps with 2% Nonidet P-40, as described by Tomoyasu et al. (17). This method efficiently separates protein aggregates from membrane proteins (17). Aggregation isolates were separated by SDS/12% PAGE and stained with Coomassie blue.
Preparation of Translation Lysates. The wild-type strain MC4100 and its Δtig, ΔdnaKdnaJ, or Δtig ΔdnaKdnaJ isogenic mutant derivatives were transformed with the pSE-based SecB-expressing vector and used to obtain the SecB-enriched translation lysates. The strains were grown overnight in LB-ampicillin and 0.4% glucose, diluted 100-fold in the same broth, and grown to an OD600 of 0.2. Overexpression of SecB was induced by the addition of 0.3 mM IPTG, cells were grown to an OD600 of 0.8, and translation lysates were prepared as described (18).
In Vitro Transcription, Translation, and Cross-Linking. Truncated mRNA was prepared as described (19). In vitro translation was carried out in the E. coli cell- and membrane-free S-135 extract as described (16). Bifunctional cross-linking was induced with 1 mM bis(sulfosuccinimidyl) suberate (BS3) for 10 min at 26°C and quenched at 4°C by adding 1/10 volume of quench buffer (1 M glycine/100 mM NaHCO3, pH 8.5). Ribosome-nascent chain complexes were collected as described (20) and analyzed either directly by SDS/15% PAGE or after immunoprecipitation as described (21), using 4-fold the amount used for direct analysis. The [35S]methionine-labeled bands were quantified by using imagequant software (Molecular Dynamics) and corrected for translation efficiency of the nascent chains as described (20).
Reagents, Enzymes, and Sera. Restriction enzymes and the Expand long template PCR kit were purchased from Roche Molecular Biochemicals. T4 DNA ligase was from Epicentre Technologies (Madison, WI). Megashort T7 transcription kit was from Ambion. [35S]methionine and protein A-Sepharose were from Amersham Pharmacia. BS3 was from Pierce. Sigma supplied all other chemicals. Antisera against L23, TF, and SecB were the kind gifts of R. Brimacombe (Max Planck Institute for Molecular Genetics, Berlin), W. Wickner (Dartmouth Medical School, Hanover, NH), and C. Kumamoto (Tufts University, Boston), respectively.
Results
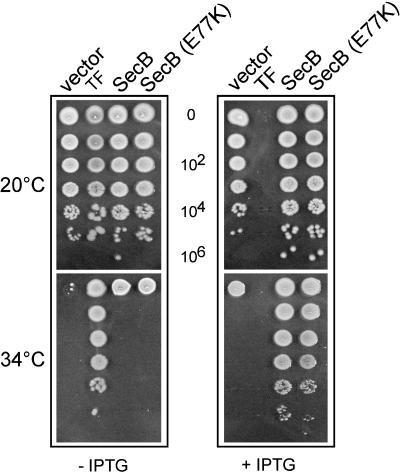
SecB Overexpression Efficiently Suppresses the Growth Defect of a Strain Lacking DnaK/DnaJ and TF. In a search for additional cellular factors that could assist folding of newly synthesized proteins, we found that plasmid pE01, which contains an E. coli genomic fragment encompassing the yibN, grxC, secB, and gpsA genes, could partially suppress the ts phenotype of the Δtig ΔdnaKdnaJ triple null mutant (data not shown). We asked whether SecB, a chaperone involved in the SecA/SecB-dependent secretion pathway (11, 22, 23), is responsible for the suppressive effect. To address this question, we PCR amplified and cloned the secB ORF under the control of an IPTG-inducible promoter on both a low (p29SEN) and a high (pSE380ΔNcoI) copy number plasmid and tested its ability to complement the ts phenotype of the Δtig ΔdnaKdnaJ mutant. Indeed, by using both plasmid constructs, we found that SecB expression alone efficiently suppresses the bacterial growth defect (Fig. 1). An ≈10-fold higher level of SecB was sufficient to detect suppression up to 34°-35°C (Fig. 1 and data not shown). The suppression was not background strain specific, as similar suppression results were also observed in the MG1655 Δtig ΔdnaKdnaJ mutant background (data not shown).
Fig. 1.
Overproduction of SecB efficiently suppresses the growth defect of a Δtig ΔdnaKdnaJ strain. Shown is in vivo complementation of the ts phenotype of the MC4100 Δtig ΔdnaKdnaJ triple mutant by the p29SEN-based IPTG-inducible constructs expressing the various proteins indicated at the top. Fresh transformants were then grown for 24 h at 20°C in LB-ampicillin, serially diluted, and spotted on LB-ampicillin agar plates without (-) or with (+)2mM IPTG inducer at the indicated temperatures. Note that TF overproduction is toxic in the Δtig ΔdnaKdnaJ triple mutant, as described (9).
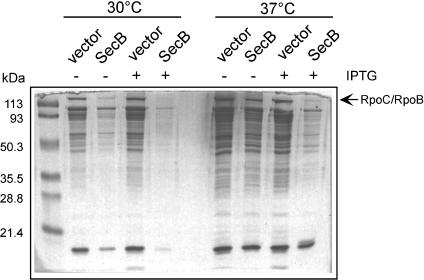
SecB Overexpression Suppresses Intracellular Protein Aggregation in the Absence of DnaK/DnaJ and TF. By using the methodology developed by Tomoyasu et al. (17), we next asked whether SecB overexpression protects proteins from aggregation in the absence of TF and DnaK. We found that SecB overexpression could indeed efficiently suppress the aggregation-prone phenotype of the MC4100 Δtig ΔdnaKdnaJ strain, at both the permissive (30°C) and nonpermissive (37°C) temperatures of growth (Fig. 2).
Fig. 2.
Overproduction of SecB rescues protein aggregation in the absence of TF and DnaK. Complementation of the aggregation-prone phenotype of the MC4100 Δtig ΔdnaKdnaJ triple mutant by the pSE380-based, IPTG-inducible constructs expressing SecB. The cultures were incubated for 3 h at permissive (30°C) and 2 h at nonpermissive (37°C) temperatures of growth. -, the absence of IPTG; + the presence of 1 mM IPTG. Identical densities of cells were pelleted, and aggregated proteins were isolated as described by Tomoyasu et al. (17). Aggregation samples were separated by SDS/12% PAGE and stained with Coomassie blue.
In Vivo Suppression by SecB Is Translocation-Independent. Because depletion of DnaK and DnaJ in a Δtig mutant seems to specifically lead to the aggregation of cytosolic proteins (3, 8), we asked whether suppression by SecB was independent of its translocation-specific function. To answer this question, we took advantage of a previously described mutation in secB (resulting in the E77K amino acid substitution in the SecB protein), which blocks productive interaction with SecA without significantly disrupting either the SecB oligomeric state or SecB/substrate complex formation (24, 25). We found that the SecB (E77K) expressing plasmid fully complements the growth defect (Fig. 1) as well as the aggregation-prone phenotype of the Δtig ΔdnaKdnaJ triple mutant (data not shown). The presence of endogenous wild-type SecB in the Δtig ΔdnaKdnaJ triple mutant does not positively influence SecB (E77K) suppression because the mutated protein suppresses the growth defect of a Δtig ΔsecB dnaJ triple mutant equally well (data not shown). In contrast to SecB, analogous experiments demonstrated that SecA, expressed from the same regulated promoter, does not suppress the Δtig ΔdnaKdnaJ ts phenotype (data not shown).
Taken together, our genetic data suggest that a functional interaction and substrate transfer between SecB and SecA is not necessary for the SecB-mediated suppression of the Δtig ΔdnaKdnaJ ts phenotype. Thus, our results emphasize the concept of SecB acting as a generalized chaperone, capable of assisting the folding of cytosolic proteins.
TF Modulates SecB Interaction with Nascent Chains of prePhoE Secretory Protein. The in vivo data presented above suggested that SecB and TF may compete for interaction with nascent polypeptide chains of both secretory and cytosolic proteins. We first examined whether TF modulates cotranslational interaction of SecB with a known SecB preprotein substrate. To do so, we performed in vitro chemical-based cross-linking experiments by using prePhoE, the cytosolic precursor form of the outer membrane protein PhoE as a model substrate. PrePhoE was previously shown to be a natural SecB substrate (26), but cotranslational interaction with SecB has not yet been detected.
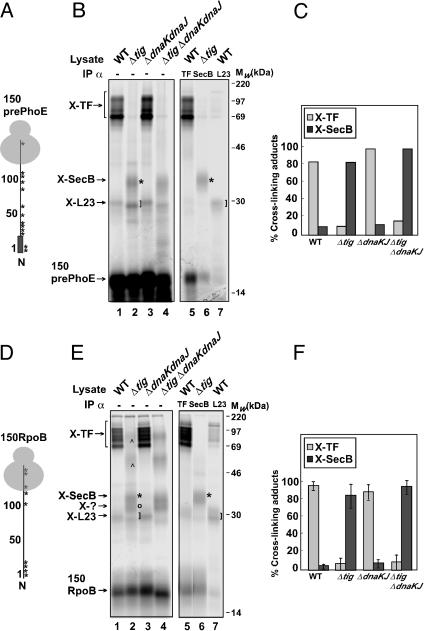
The N-terminal portion of prePhoE was translated from truncated mRNA to a length of 150 aa (Fig. 3A) in cell- and membrane-free E. coli extracts derived from wild-type, Δtig, ΔdnaKdnaJ, or Δtig ΔdnaKdnaJ mutant cells. All mutant cells overexpressed SecB resulting in lysates containing ≈20-fold higher levels of SecB than the wild type (data not shown). Translation of 150prePhoE in wild-type lysate and treatment of the translation products with the homobifunctional cross-linker BS3 resulted in the appearance of a cross-linked product of ≈29 kDa and a range of products from ≈70 to 150 kDa in size (Fig. 3B, lane 1). Subtracting the 17-kDa contribution of the nascent 150prePhoE chain from the ≈29-kDa product leaves a cross-linking partner of ≈12 kDa. In agreement with a recent study (20), this partner could be identified by immunoprecipitation as the ribosomal protein L23 (Fig. 3B, lane 7).
Fig. 3.
SecB and TF compete for nascent polypeptides of both secreted and cytosolic proteins. (A) Schematic representation of the 150prePhoE nascent chain with potential cross-linking sites depicted by asterisks. The signal sequence is represented by a thick line. (B) In vitro translation of 150prePhoE was carried out in cell- and membrane-free E. coli extracts from SecB-overproducing cells. After translation, samples were treated with 1 mM of the cross-linker BS3, and the ribosome-associated nascent chain complexes were purified over a high salt sucrose cushion. The pellet fractions were either separated directly by SDS/15% PAGE (lanes 1-4) or after immunoprecipitation with antiserum against TF, L23, or SecB (lanes 5-7, respectively). Left bracket, TF adducts; right brackets, L23 adducts; asterisk, SecB adducts. (C) TF and SecB cross-linking adducts from B (lanes 1-4) were quantified and corrected for efficiency of translation. The highest value for cross-linking efficiency was taken as 100%. (D) Schematic representation of 150RpoB nascent chain with potential cross-linking sites depicted by asterisks. (E) Nascent chains of 150RpoB were produced, cross-linked, and analyzed by SDS/PAGE and immunoprecipitation, as described in B. TF, L23, and SecB adducts are indicated as described in B. ⁁ or o, unknown adducts. (F) TF and SecB cross-linking adducts from E (lanes 1-4) were quantified as described in C. Error bars represent the SD of five independent experiments, using four different bifunctional cross-linking agents (see Results for more details).
The range of ≈70- to 150-kDa products corresponds to cross-linking partners of ≈53 to 135 kDa in size and could all be identified by immunoprecipitation as containing the chaperone TF (Fig. 3B, lanes 1 and 5). Although the reason why TF adducts migrate as multiple cross-linking species with different mobilities on SDS/PAGE is unclear, such behavior of TF adducts has been observed before (8, 27-29). In extracts prepared from ΔdnaKd-naJ cells, the same cross-linking products appeared as in extracts derived from wild-type cells, indicating that DnaK and DnaJ do not appreciably influence the interaction of TF with the nascent chain under our experimental conditions. Consistent with the immunoprecipitation data, when extracts were prepared from either Δtig or Δtig ΔdnaKdnaJ mutant cells, the cross-linking of 150prePhoE to TF was not observed (Fig. 3B, lanes 2 and 4). Interestingly, in the absence of TF, a cross-linking product of 34 kDa was clearly visible, corresponding to a cross-linking partner of 17 kDa (Fig. 3B, lanes 2 and 4). These results suggest that TF modulates the interaction of this component with nascent 150prePhoE.
We identified by immunoprecipitation the 17-kDa adduct as being the SecB chaperone (Fig. 3B, lane 6, marked with an asterisk). Thus, it seems that SecB can interact cotranslationally with the short nascent prePhoE substrate, especially in the absence of TF. In extracts prepared from either wild-type or ΔdnaKdnaJ cells, SecB was not found to be appreciably cross-linked, indicating that TF largely inhibits the interaction of SecB with nascent 150prePhoE (Fig. 3 B and C). Thus, the absence of ribosome-bound TF seems to considerably shorten the previously described minimal length required for SecB to interact cotranslationally with nascent polypeptide chains of the maltose-binding protein (30). In addition, we were also able to observe cross-linking of 150prePhoE to SecB when present at wild-type levels, albeit to a much lesser extent (data not shown). Again, such SecB association was only observed in Δtig extracts, demonstrating that competitive interaction of SecB and TF with nascent chains also occurs under normal growth conditions.
Furthermore, additional cross-linking products were observed only in the absence of both the TF and DnaK/DnaJ chaperone systems (Fig. 3B, lane 4). These unidentified bands could represent interactions of the leader sequence with some small, perhaps ribosomal protein partners.
TF Modulates SecB Interaction with Nascent Chains of the Cytosolic Protein RpoB. Because SecB can efficiently suppress the growth and the aggregation defects observed in the absence of TF and DnaK, we asked whether SecB could also interact cotranslationally with cytosolic proteins. To answer this question, we carried out similar in vitro cross-linking experiments by using an N-terminal portion of the 151-kDa cytosolic protein RpoB, the β subunit of the E. coli RNA polymerase, as a model substrate. We chose RpoB because it was previously shown to be a common substrate for both DnaK and TF. As a consequence, depletion of DnaK and DnaJ in a Δtig mutant leads to higher levels of aggregated RpoB polypeptides (3, 8). In addition, by using coimmunoprecipitation, Deuerling et al. (8) showed a direct interaction between RpoB and DnaK. We also observed an accumulation of aggregated RpoB in the Δtig ΔdnaKdnaJ triple mutant (Fig. 2 and ref. 9). Furthermore, the levels of RpoB aggregates were significantly reduced after SecB overproduction (Fig. 2).
Translation of 150RpoB and treatment with BS3 in wild-type and ΔdnaKdnaJ lysates resulted in the appearance of the same cross-linking patterns as for nascent PhoE (Fig. 3E, lanes 1 and 3). Immunoprecipitation experiments confirmed TF and L23 as 150RpoB cross-linking partners (Fig. 3E, lanes 5 and 7). Surprisingly, in the absence of TF, four other cross-linking products appeared, with sizes of ≈32-34, ≈55, and ≈100 kDa, corresponding to cross-linking partners of ≈15-17, ≈38, and ≈83 kDa, respectively (Fig. 3E, lanes 2 and 4). These results show that the presence of TF limits the interaction of all of these factors with nascent 150RpoB.
Interestingly, immunoprecipitation experiments revealed that the 17-kDa adduct of RpoB is SecB (Fig. 3E, lane 6, marked with an asterisk). These results indicate that SecB is also able to interact cotranslationally with cytosolic substrates and that this interaction is also modulated by TF. To prove that the cross-linking of SecB to the nascent 150RpoB is not critically dependent on the specificity of the cross-linker, we used several other cross-linking agents besides BS3.
We found that, in addition to BS3, the heterobifunctional cross-linkers 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide, N-hydroxysulfosuccinimidyl-4-azidobenzoate, and m-maleimidobenzoyl-N-hydroxysulfosuccinimide ester can also cross-link SecB to the 150RpoB nascent chain (data not shown). The adducts generated by the four different cross-linking agents mentioned above were quantified and corrected for translation efficiencies as described (20). The quantification results confirm that cross-linking to SecB is greatly enhanced in the absence of TF (Fig. 3F). In addition, similar to our results with 150prePhoE, we were able to observe low levels of SecB cross-linking to 150RpoB in Δtig extracts even when SecB was present at wild-type levels (data not shown).
The cross-linking partners of ≈15, ≈35, and ≈83 kDa that were specifically cross-linked to nascent 150RpoB in extracts lacking TF have not been identified yet.
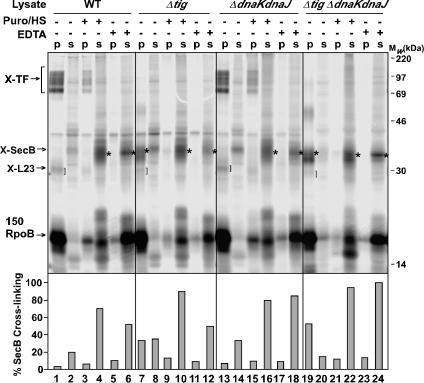
The Ribosome Is Not Required for Interaction of SecB with 150RpoB. As described above, SecB interacts with ribosome-associated nascent chains of 150RpoB in the absence of TF (Fig. 3E). To determine whether SecB association with nascent 150RpoB depends on the nascent chain being ribosome-associated, the ability of SecB to cross-link to released 150RpoB nascent chains was investigated. After in vitro translation of 150RpoB, one-third of the sample was mock-treated to keep the nascent chains ribosome-attached, one-third of the sample was treated with puromycin/high salt, and one-third was treated with EDTA before cross-linking with BS3. It is known that incorporation of either the aminoacyl tRNA analog puromycin or treatment of the ribosome nascent chain complexes with EDTA results in the release of the nascent chains (15). As expected, the released chains were found in the supernatant fraction, after appropriate centrifugation (see Materials and Methods), whereas in the absence of puromycin or EDTA, the nascent polypeptides were still ribosome-associated (Fig. 4 Top, lanes 1, 7, 13, and 19).
Fig. 4.
SecB interaction with 150RpoB is ribosome-independent. In vitro translation of nascent 150RpoB nascent chains was carried out in cell- and membrane-free E. coli extracts prepared from SecB-overproducing cells. After translation, the samples were divided into equal aliquots and treated with 0.2 mM puromycin and 0.4 M KOAc (Puro/HS) or with 25 mM EDTA, or were mock-treated with incubation buffer. After 10 min incubation at 26°C, the samples were treated with 1 mM of the cross-linker BS3, and the ribosome-associated nascent chain complexes were purified over a high-salt sucrose cushion. Both the supernatant (s) and pellet (p) fractions were separated by SDS/15% PAGE. Left bracket, TF adducts; right bracket, L23 adducts; asterisk, SecB adducts. Columns at the bottom show the quantification of SecB cross-linking adducts on the corresponding above lanes, corrected for translational efficiency. The highest value for cross-linking efficiency was taken as 100%.
As observed in previous experiments (Fig. 3E), TF was the dominant cross-linking partner of 150RpoB (Fig. 4 Top, lanes 1 and 13, indicated at the left), whereas cross-linking to SecB was only detected in the absence of TF (Fig. 4 Top, lanes 7 and 19, indicated by an asterisk). Upon addition of puromycin, most of the nascent chains were released from the ribosome (Fig. 4 Top, lanes 4, 10, 16, and 22). After such treatment, no detectable cross-linking of TF and L23 to 150RpoB was seen (Fig. 4 Top, lanes 4 and 16), indicating that the 150RpoB nascent chains were no longer ribosome-associated. Instead, a clear SecB adduct to 150RpoB was detected in the supernatant samples, independently of the presence or the absence of TF (Fig. 4 Top, lanes 4 and 10, indicated by an asterisk). Similar results were obtained when EDTA was used to disassemble the ribosome nascent chain complexes (Fig. 4, lanes 6, 12, 18, and 24). This conclusion was confirmed by immunoprecipitation using antisera against SecB (data not shown). Quantification of the cross-linked SecB adducts showed that cross-linking of SecB to released 150RpoB was approximately equivalent in all four extracts (Fig. 4 Bottom, lanes 4, 10, 16, and 22) and ≈40% more efficient than cross-linking of SecB to ribosome-associated 150RpoB (Fig. 4 Bottom, compare lanes 10 and 22 with lanes 7 and 19, respectively). These results indicate that, as observed with the secretory maltose-binding protein (30), interaction of SecB with nascent cytosolic substrates is not limited to the nascent chain's association with the ribosome but continues after nascent chain release.
Discussion
The SecB chaperone is known to promote the secretion of various E. coli proteins (11, 31) through its collaboration with the essential SecA member of the translocon (32). In this work, we have shown that SecB can also act as a generalized chaperone. This action was performed by first demonstrating that overproduction of SecB in vivo can suppress the growth defect, as well as the aggregation-prone phenotype, of an E. coli strain lacking both the TF and DnaK generalized chaperones. Furthermore, we showed that the suppression by SecB is independent of its translocation-dependent chores. Further evidence was obtained through the use of an in vitro chemical-based cross-linking method by showing that SecB can interact both co- and post-translationally with nascent chains of both secretory and cytosolic proteins. In both cases, only the cotranslational substrate recognition by SecB is actively inhibited by TF and not by DnaK.
The specialized function of SecB in the targeting of secretory proteins to the sec translocon has been well documented (11, 22, 23), whereas its active participation in the folding of cytosolic proteins has been suggested (13, 33). In agreement with a role for SecB as a generalized chaperone, previous in vitro studies indicated that SecB has no specific affinity for signal sequences (12, 13). In addition, peptide-binding scans revealed that SecB exhibits a preference for unstructured stretches of polypeptides, approximately nine amino acid residues in length, that contain both basic and aromatic residues (13, 34). Such stretches of amino acids, which are not specifically found in signal sequences, theoretically occur every 20-30 residues in proteins (13). In addition, SecB and DnaK share many potential binding sites in polypeptide substrates and could therefore interact with the same protein regions (13).
Functional in vivo overlapping among the SecB, DnaK, and GroEL chaperones has been previously observed. Specifically, it has been reported that DnaK can substitute in the export of several SecB-dependent secretory proteins (35) and that the DnaK and GroEL chaperone machines can assist protein export in some other cases as well (36, 37). Furthermore, similar to the DnaK system, SecB can prevent luciferase from aggregation and cooperate with DnaK/DnaJ/GrpE in the refolding of luciferase in vitro (13). Finally, an increased cellular level of SecB has been observed in strains with defective alleles of the dnaJ, dnaK, groEL, or groES genes (38).
Recently, we reported that of many other known chaperone and protease genes tested, only GroEL/GroES overproduction could provide enough assistance to support growth of our Δtig ΔdnaKdnaJ strain as well as prevent aggregation under these conditions (9). Therefore, it is conceivable that TF, DnaK, GroEL, and SecB may compete for the same pool of both secretory and cytosolic proteins, although with a considerable advantage for TF, being more abundant and localized near the ribosome exit channel (28). In agreement with this concept, our in vitro data revealed a strong exclusion by TF on SecB for binding to the nascent polypeptides of both secretory and cytosolic proteins. However, TF did not interfere with SecB binding to the same nascent chains once they were released from the ribosome. Similarly, it has been previously shown that in the absence of ribosome-bound TF, the total amount of nascent polypeptides bound to DnaK is increased (3, 5); in addition, DnaK is able to interact with considerably shorter nascent chains (5).
Because SecB is an abundant protein (its intracellular concentration having been estimated between 4 and 20 μM; refs. 39 and 40) with a high affinity for unfolded substrates (12, 33, 41, 42), it may interact both co- and posttranslationally with a large number of newly synthesized secretory and cytosolic polypeptides. Upon binding and release from SecB, polypeptide substrates may subsequently reach their native state without further assistance, be transferred to other chaperones, or be targeted for translocation or proteolysis. Consistent with this concept, proteomic analyses of a secB mutant strain indicate that both secretory and cytosolic proteins aggregate in the absence of SecB (J. W. de Gier, University of Stockholm, Stockholm, personal communication). Thus, specific interaction of the SecB/substrate complex with SecA at the inner membrane would result in protein translocation only when a signal sequence is also present. Part of the SecB-bound cytosolic polypeptides may partition before interacting with SecA, whereas a substantial amount may remain associated with SecB. In such a model, SecA must somehow discriminate between SecB bound to cytosolic versus secretory polypeptide substrates, so that the translocon is not saturated with cytosolic proteins. Such a SecA-dependent quality control mechanism has recently been illustrated by Eser and Ehrmann (43). These authors showed that SecA can partition substrates with or without a signal sequence and either target them for translocation (those with a signal sequence) or directly assist their folding in the cytoplasm (those lacking a signal sequence).
In addition to SecB, our in vitro cross-linking experiments revealed the unexpected presence of other, thus far unidentified, putative protein factors potentially involved in nascent polypeptide folding. Interestingly, these factors could only be cross-linked specifically to the nascent cytosolic model substrate and not to the nascent secretory model substrate. The intriguing possibility is that these factors represent additional chaperones also capable of assisting de novo protein folding in vivo. Future studies to identify these putative chaperones are warranted.
Acknowledgments
We thank Debbie Ang for a critical reading of the manuscript and David N. Chaperon for plasmid gifts. F.S., C.G., and P.G. were supported by Swiss National Science Foundation Grant FN-31-65403 and the Canton of Geneva. R.S.U., J.L., and N.H. were supported by the Council for Chemical Sciences of the Netherlands Organization for Scientific Research.
Abbreviations: TF, Trigger Factor; ts, temperature-sensitive; IPTG, isopropyl-β-d-thiogalactopyranoside; BS3, bis(sulfosuccinimidyl) suberate.
References
- 1.Hartl, F. U. & Hayer-Hartl, M. (2002) Science 295, 1852-1858. [DOI] [PubMed] [Google Scholar]
- 2.Bukau, B., Deuerling, E., Pfund, C. & Craig, E. A. (2000) Cell 101, 119-122. [DOI] [PubMed] [Google Scholar]
- 3.Deuerling, E., Schulze-Specking, A., Tomoyasu, T., Mogk, A. & Bukau, B. (1999) Nature 400, 693-696. [DOI] [PubMed] [Google Scholar]
- 4.Valent, Q. A., Kendall, D. A., High, S., Kusters, R., Oudega, B. & Luirink, J. (1995) EMBO J. 14, 5494-5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Teter, S. A., Houry, W. A., Ang, D., Tradler, T., Rockabrand, D., Fischer, G., Blum, P., Georgopoulos, C. & Hartl, F. U. (1999) Cell 97, 755-765. [DOI] [PubMed] [Google Scholar]
- 6.Houry, W. A., Frishman, D., Eckerskorn, C., Lottspeich, F. & Hartl, F. U. (1999) Nature 402, 147-154. [DOI] [PubMed] [Google Scholar]
- 7.Ewalt, K. L., Hendrick, J. P., Houry, W. A. & Hartl, F. U. (1997) Cell 90, 491-500. [DOI] [PubMed] [Google Scholar]
- 8.Deuerling, E., Patzelt, H., Vorderwulbecke, S., Rauch, T., Kramer, G., Schaffitzel, E., Mogk, A., Schulze-Specking, A., Langen, H. & Bukau, B. (2003) Mol. Microbiol. 47, 1317-1328. [DOI] [PubMed] [Google Scholar]
- 9.Genevaux, P., Keppel, F., Schwager, F., Langendijk-Genevaux, P. S., Hartl, F. U. & Georgopoulos, C. (2004) EMBO Rep. 5, 195-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vorderwulbecke, S., Kramer, G., Merz, F., Kurz, T. A., Rauch, T., Zachmann-Brand, B., Bukau, B. & Deuerling, E. (2004) FEBS Lett. 559, 181-187. [DOI] [PubMed] [Google Scholar]
- 11.Randall, L. L. & Hardy, S. J. (2002) Cell. Mol. Life Sci. 59, 1617-1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Randall, L. L., Topping, T. B. & Hardy, S. J. (1990) Science 248, 860-863. [DOI] [PubMed] [Google Scholar]
- 13.Knoblauch, N. T., Rudiger, S., Schonfeld, H. J., Driessen, A. J., Schneider-Mergener, J. & Bukau, B. (1999) J. Biol. Chem. 274, 34219-34225. [DOI] [PubMed] [Google Scholar]
- 14.Shimizu, H., Nishiyama, K. & Tokuda, H. (1997) Mol. Microbiol. 26, 1013-1021. [DOI] [PubMed] [Google Scholar]
- 15.Valent, Q. A., de Gier, J. W., von Heijne, G., Kendall, D. A., ten Hagen-Jongman, C. M., Oudega, B. & Luirink, J. (1997) Mol. Microbiol. 25, 53-64. [DOI] [PubMed] [Google Scholar]
- 16.Urbanus, M. L., Scotti, P. A., Froderberg, L., Saaf, A., de Gier, J. W., Brunner, J., Samuelson, J. C., Dalbey, R. E., Oudega, B. & Luirink, J. (2001) EMBO Rep. 2, 524-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tomoyasu, T., Mogk, A., Langen, H., Goloubinoff, P. & Bukau, B. (2001) Mol. Microbiol. 40, 397-413. [DOI] [PubMed] [Google Scholar]
- 18.De Vrije, T., Batenburg, A. M., Jordi, W. & De Kruijff, B. (1989) Eur. J. Biochem. 180, 385-392. [DOI] [PubMed] [Google Scholar]
- 19.Scotti, P. A., Urbanus, M. L., Brunner, J., de Gier, J. W., von Heijne, G., van der Does, C., Driessen, A. J., Oudega, B. & Luirink, J. (2000) EMBO J. 19, 542-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ullers, R. S., Houben, E. N., Raine, A., ten Hagen-Jongman, C. M., Ehrenberg, M., Brunner, J., Oudega, B., Harms, N. & Luirink, J. (2003) J. Cell Biol. 161, 679-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luirink, J., High, S., Wood, H., Giner, A., Tollervey, D. & Dobberstein, B. (1992) Nature 359, 741-743. [DOI] [PubMed] [Google Scholar]
- 22.Muller, M., Koch, H. G., Beck, K. & Schafer, U. (2001) Prog. Nucleic Acid Res. Mol. Biol. 66, 107-157. [DOI] [PubMed] [Google Scholar]
- 23.Kim, J. & Kendall, D. A. (2000) Cell Stress Chaperones 5, 267-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fekkes, P., de Wit, J. G., van der Wolk, J. P., Kimsey, H. H., Kumamoto, C. A. & Driessen, A. J. (1998) Mol. Microbiol. 29, 1179-1190. [DOI] [PubMed] [Google Scholar]
- 25.Kimsey, H. H., Dagarag, M. D. & Kumamoto, C. A. (1995) J. Biol. Chem. 270, 22831-22835. [DOI] [PubMed] [Google Scholar]
- 26.Kusters, R., de Vrije, T., Breukink, E. & de Kruijff, B. (1989) J. Biol. Chem. 264, 20827-20830. [PubMed] [Google Scholar]
- 27.Beck, K., Wu, L. F., Brunner, J. & Muller, M. (2000) EMBO J. 19, 134-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kramer, G., Rauch, T., Rist, W., Vorderwulbecke, S., Patzelt, H., Schulze-Specking, A., Ban, N., Deuerling, E. & Bukau, B. (2002) Nature 419, 171-174. [DOI] [PubMed] [Google Scholar]
- 29.Eisner, G., Koch, H. G., Beck, K., Brunner, J. & Muller, M. (2003) J. Cell Biol. 163, 35-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Randall, L. L., Topping, T. B., Hardy, S. J., Pavlov, M. Y., Freistroffer, D. V. & Ehrenberg, M. (1997) Proc. Natl. Acad. Sci. USA 94, 802-807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kumamoto, C. A. & Beckwith, J. (1983) J. Bacteriol. 154, 253-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hartl, F. U., Lecker, S., Schiebel, E., Hendrick, J. P. & Wickner, W. (1990) Cell 63, 269-279. [DOI] [PubMed] [Google Scholar]
- 33.Hardy, S. J. & Randall, L. L. (1991) Science 251, 439-443. [DOI] [PubMed] [Google Scholar]
- 34.Randall, L. L. (1992) Science 257, 241-245. [DOI] [PubMed] [Google Scholar]
- 35.Wild, J., Altman, E., Yura, T. & Gross, C. A. (1992) Genes Dev. 6, 1165-1172. [DOI] [PubMed] [Google Scholar]
- 36.Qi, H. Y., Hyndman, J. B. & Bernstein, H. D. (2002) J. Biol. Chem. 277, 51077-51083. [DOI] [PubMed] [Google Scholar]
- 37.Wild, J., Rossmeissl, P., Walter, W. A. & Gross, C. A. (1996) J. Bacteriol. 178, 3608-3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Muller, J. P. (1996) J. Bacteriol. 178, 6097-6104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Watanabe, M. & Blobel, G. (1989) Proc. Natl. Acad. Sci. USA 86, 2728-2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Woodbury, R. L., Topping, T. B., Diamond, D. L., Suciu, D., Kumamoto, C. A., Hardy, S. J. & Randall, L. L. (2000) J. Biol. Chem. 275, 24191-24198. [DOI] [PubMed] [Google Scholar]
- 41.Fekkes, P., den Blaauwen, T. & Driessen, A. J. (1995) Biochemistry 34, 10078-10085. [DOI] [PubMed] [Google Scholar]
- 42.Stenberg, G. & Fersht, A. R. (1997) J. Mol. Biol. 274, 268-275. [DOI] [PubMed] [Google Scholar]
- 43.Eser, M. & Ehrmann, M. (2003) Proc. Natl. Acad. Sci. USA 100, 13231-13234. [DOI] [PMC free article] [PubMed] [Google Scholar]