Abstract
Accumulation of somatic mutations is thought to contribute to the aging process. Genomic instability has been shown to increase during aging, suggesting an aberrant function of DNA double-strand break (DSB) repair. Surprisingly, DSB repair has not been examined with respect to cellular senescence. Therefore, we have studied the ability of young, presenescent, and senescent normal human fibroblasts to repair DSBs in transfected DNA by using a fluorescent reporter substrate. We have found that the efficiency of end joining is reduced up to 4.5 fold in presenescent and senescent cells, relative to young cells. Sequence analysis of end junctions showed that the frequency of precise ligation was higher in young cells, whereas end joining in old cells was associated with extended deletions. These results indicate that end joining becomes inefficient and more error-prone during cellular senescence. Furthermore, the ability to use microhomologies for end joining was compromised in senescent cells, suggesting that young and senescent cells may use different end joining pathways. We hypothesize that inefficient and aberrant end joining is a likely mechanism underlying the age-related genomic instability and higher incidence of cancer in the elderly.
Accumulation of somatic mutations is considered to be a contributing cause of aging. Age-associated decline in the function of base excision repair has been documented in nuclear protein extracts from tissues of aged mice (1, 2), in rat neurons (3), and in human skin (4). Studies using transgenic mice carrying chromosomally integrated lacZ (5) or lacI (6, 7) reporter genes have shown that point mutations in the reporter gene accumulate with age (5, 7) and that the mutation rate is higher in old animals (7). Whereas young animals accumulate mainly point mutations, old animals begin to accumulate large genomic rearrangements (8), which is consistent with cytogenetic studies, showing that cells isolated from old animals contain high numbers of abnormal chromosomes (9–11). A recent study in yeast showed a dramatic increase in genomic rearrangements during yeast aging (12). Rearrangements result primarily from errors in repair of double-strand breaks (DSBs), which suggests that changes in DSB repair contribute to the aging process. The relevance of DSB repair to the aging process is further underscored by the finding that the number of DSBs is increased in tissues of old mice (13).
DSBs are repaired in two ways: by homologous recombination and by nonhomologous end joining (NHEJ). Homologous recombination typically copies the missing information from the sister chromatid into the break site, resulting in exact reconstitution of the original sequence. In contrast, NHEJ fuses the two broken ends, with little regard for homology, leading to deletions and other rearrangements. Growing evidence suggests that there are multiple pathways for NHEJ. The best understood pathway involves the heterodimeric Ku protein, DNA-dependent protein kinase, and the DNA ligase IV-XRCC4 complex (14). However, end joining occurs in cell-free extracts lacking Ku or DNA-PK, indicating the existence of additional Ku-independent pathways of NHEJ (15, 16). In addition, extracts of cells from Fanconi anemia, a disease characterized by chromosomal instability and predisposition to cancer, have a deficiency in an NHEJ process that appears to be independent of DNA-PK/Ku (17). Finally, genetic analysis of end joining in yeast confirms the existence of Ku-independent pathways (18).
The role of DSBs in aging is supported by studies showing that mutations in the genes involved in DSB repair and recombination lead to accelerated aging and cellular senescence. The premature aging syndrome, Werner syndrome, is caused by a mutation in the WRN gene that encodes a helicase involved in DSB repair by homologous recombination (19, 20) and NHEJ (21), and possibly telomere maintenance (22). Cultured somatic cells from Werner patients display an increased rate of genome rearrangements (23) and extensive deletions during NHEJ (24). Ku86 knockout mice, which are completely defective in the classic pathway of NHEJ, display various symptoms of accelerated aging (25). Disruption of the mouse ERCC1 gene results in DNA repair deficiency and senescence (26), which is consistent with the idea that DSB repair plays an important role in the aging process. The tumor suppressor p53 protein provides a further link between DSB repair and senescence. p53 is a major regulator in the DNA-damage response (27, 28), it is also essential for induction of cellular senescence (29), and has been implicated in regulating homologous recombination (30–32) and NHEJ (33–35). Thus, p53 plays a major role in both the induction of senescence (36) and the regulation of DSB repair (33–35, 37).
Here, we examined the status of end joining during cellular senescence of normal human fibroblasts, by using a fast-readout plasmid-rejoining assay. To date, it is unclear whether plasmid rejoining in human cells reflects the process controlled by DNA-PK/Ku or alternative NHEJ pathways. Ku-deficient yeast strains show a dramatic decrease in plasmid rejoining (38, 39), whereas several studies in Ku- and XRCC4-deficient rodent cell lines failed to demonstrate a significant defect (40, 41). In contrast, LIG4-null human cells lines and cells from LIG4 syndrome patients are markedly impaired in plasmid rejoining (42), arguing that this assay monitors the DNA-PK/Ku/XRCC4/LIG4 pathway in human cells. Notably, knockout studies suggest that humans and rodents may differ in their use of NHEJ pathways, because murine knockouts of the components of DNA-PK complex are viable, whereas no human patient has been described with a mutation of Ku86 or Ku70, and the Ku86 locus was shown to be essential in human somatic tissue culture cells (43).
The plasmid rejoining assay has proven to be an important tool for examining both the efficiency and fidelity of the end-joining process, especially in the analysis of human primary cells with limited lifespan, where the establishment of stably integrated clones is not feasible. Plasmid end-joining assays in cells have been used to demonstrate the defect in the fidelity of DNA repair in Fanconi anemia (44), the reduction in the efficiency of joining (45), and change in the spectrum of NHEJ products (46) in Bloom syndrome cells, the abnormal processing of ends in Ataxia telangiectasia (47) and BRCA1-mutant cells (48), and the extensive deletions that occur at plasmid ends during end joining in Werner syndrome cells (24). Thus, NHEJ activity, as measured by the plasmid assay, has yielded important insights into the physiology of human disease.
We report here for the first time, to our knowledge, that the efficiency of end joining is reduced in presenescent and senescent cells. The end junctions in presenescent and senescent cells are associated with extensive deletions. These results indicate that end joining becomes inefficient and more error-prone during cellular senescence, and suggest a mechanism for age-related genomic instability.
Materials and Methods
Construction of Plasmids. GFP-Pem1 plasmid was kindly provided by L. Li, University of Texas, M. D. Anderson Cancer Center, Houston. This plasmid consists of a modified pEGFP-N1 (Clontech), in which the enhanced GFP (EGFP) gene is interrupted by the Pem1 intron. The Pem1 intron is efficiently spliced and does not affect GFP expression. To construct GFP-Pem1-Ad2 (NHEJ substrate), the HindIII site in the EGFP polylinker was removed by XhoI/SalI digestion and self-ligation of GFP-Pem1 plasmid. The adenoviral major late mRNA leader sequence (adenoviral exon 2) was then inserted into the HindIII site within the Pem1 intron. Plasmid pSP65-Ad, which contains wild-type adenoviral exon 2, was kindly provided by S. Berget, Baylor College of Medicine. The entire adenoviral exon 2, including splice sites, was PCR-amplified with primers containing HindIII sites and inverted I-SceI sites. The construct was verified by sequencing, and GFP activity was analyzed by fluorescent microscopy and fluorescence-activated cell sorter (FACS) analysis.
End-Joining Assay, Transfection, and FACS Analysis. The experimental strategy for the NHEJ assay is depicted in Fig. 5, which is published as supporting information on the PNAS web site. The NHEJ reporter plasmid was digested with HindIII or I-SceI (Roche Applied Science) for 6 h and purified by using a Qiagen gel extraction kit. Aliquots were analyzed by gel electrophoresis to confirm complete digestion. Normal human fibroblasts were transfected by using an Amaxa Nucleofector according to manufacturer's instructions. In a typical reaction, 1 × 106 cells were transfected with 0.5 μg of predigested NHEJ reporter substrate along with 0.1 μg of pDsRed2-N1 (Clontech) to serve as transfection control. Expression of GFP and DsRed was monitored by fluorescence microscopy (Nikon, Eclipse TE2000-U). Maximum expression of both GFP and DsRed was observed 48–72 h after transfection. After transfection, cells were incubated for 72 h. Cells were harvested, resuspended in 0.5 ml of PBS, pH 7.4 (GIBCO, Invitrogen), and analyzed by FACS (Beckman Coulter, EPICS XL-MCL, by using system ii, version 3.0).
Further information on cell lines and growth conditions and plasmid rescue may be found in Supporting Text, which is published as supporting information on the PNAS web site.
Results
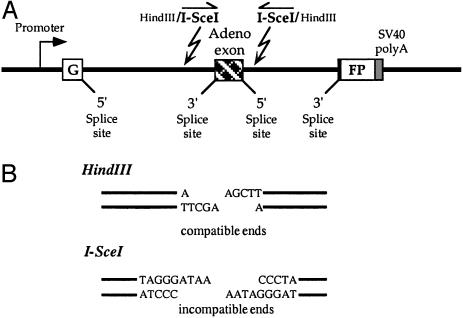
Fluorescence-Based Assay for End Joining in Human Cells. To examine the changes in end joining during cellular senescence, we constructed a reporter substrate that allows detection of a wide spectrum of NHEJ events (Fig. 1A). The substrate consists of a GFP gene, engineered to contain a 2.4-kb intron from Rattus norvegicus Pem gene (GFP-Pem1). The Pem intron is efficiently spliced out and does not affect GFP expression. Upon induction of a DSB within the intron, a wide spectrum of NHEJ events can restore GFP activity because the intron can tolerate large insertions and deletions. Subsequent analysis of NHEJ events showed that deletions large enough to disrupt the GFP ORF represented <10% of the end-joining events, indicating that the GFP activity of NHEJ reporter substrate provides a representative measure of end-joining efficiency.
Fig. 1.
Reporter substrate for analysis of NHEJ. (A) The reporter substrate consists of GFP with an artificially engineered intron, interrupted by an adenoviral exon, flanked by restriction sites for induction of DSBs. In this construct, the GFP gene is inactive; however, upon digestion with HindIII or I-SceI enzymes and successful NHEJ, the construct becomes GFP+. (B) Restriction sites used to introduce DSBs. Digestion with HindIII generates compatible cohesive ends. Because I-SceI has a nonpalindromic 18-bp recognition site, cleavage of the two inverted I-SceI sites generates incompatible ends.
Because we wanted the starting substrate to be GFP-, we inserted exon 2 from adenovirus into the middle of the Pem1 intron. This exon is efficiently spliced into the middle of the GFP ORF, killing GFP activity. On both sides of the killer exon, we introduced recognition sites for I-SceI (in an inverted orientation) and HindIII (Fig. 1B). Cleavage with either of those endonucleases removes the killer exon, and successful DSB repair reconstitutes GFP activity.
To examine the changes in the efficiency of end joining during cellular senescence, we used two normal human diploid fibroblast cell lines, WI-38 and HCA2. WI-38 cells are fetal lung fibroblasts, and HCA2 cells are foreskin fibroblasts. Both cell lines have been extensively characterized with regard to cellular senescence. The cells were passaged to senescence, and aliquots were frozen at each passage for use in subsequent experiments. HCA2 cells senesced at a population doubling (PD) of 70, and WI-38 senesced at PD 72. A shorter lifespan (50 ± 10 PD) has been reported for WI-38 cells (American Type Culture Collection); however, the growth condition that we developed for these cells (see Materials and Methods) consistently resulted in a longer lifespan. Cells at PD 25–32 were defined as young, cells at PD 58–62 (≈10 PDs before the arrest of cell division) were defined as presenescent, and cells that had stopped proliferation (PD 70+) were defined as senescent.
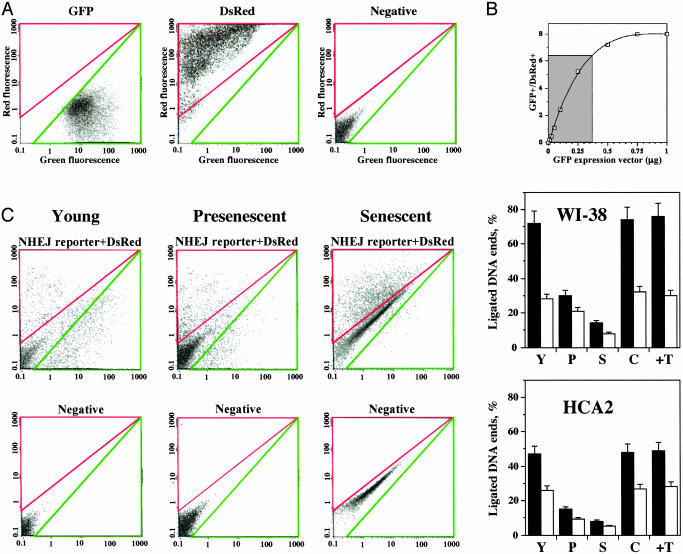
Cells were transfected with the I-SceI- or HindIII-digested NHEJ reporter substrate and the DsRed expression vector to normalize for transfection efficiency. Transfections were performed by using Amaxa electroporation, which, at high DNA concentrations (5 μg), yields 90–95% transfection efficiency in the young cells and 70–75% transfection efficiency in presenescent and senescent cells (data not shown). Cells were incubated for 72 h to allow expression of GFP (green fluorescence) and DsRed (red fluorescence), and were analyzed by flow cytometry. Cells were analyzed on a red-versus-green fluorescence plot. The gating for GFP+ or DsRed+ cells was determined in each experiment by using cells transfected with GFP-Pem1 (green), or pDsRed2-N1 (red), or pHPRT (negative control) plasmids (Fig. 2A). To analyze the products of end joining, genomic DNA was extracted, the repaired NHEJ reporter substrates were rescued by transformation into Escherichia coli, and the fidelity of NHEJ events was examined by sequencing.
Fig. 2.
Efficiency of NHEJ declines during cellular senescence. (A) Calibration of the parameters for FACS analysis. Cells were analyzed on a red-versus-green fluorescence plot. The gating for the analysis of green and red cells was set up by using cells transfected with 5 μg of GFP or 5 μg of DsRed vectors, and the cells were transfected with a negative control plasmid expressing the hypoxanthine phosphoribosyltransferase gene (to exclude autofluorescent cells). GFP and DsRed- cells possess autofluorescence and fall along the green/red diagonal, whereas GFP+ and DsRed+ cells appear in separate populations shifted off the diagonal. (B) Relationship between the amount of transfected DNA and the number of GFP+ cells. Cells were transfected with various amounts of circular GFP-Pem1 vector (from 0.01 to 1 μg) mixed with 0.1 μg of pDsRed. After 72 h, the percentages of red and green cells were determined by FACS analysis. The ratio of GFP+ to DsRed+ cells was plotted as a function of the amount of GFP-Pem1 vector. The shaded area indicates the range of GFP+/DsRed+ obtained upon transfection with digested NHEJ substrate. (C)Efficiency of NHEJ in young, presenescent, and senescent WI-38 and HCA2 fibroblasts. Cells were cotransfected with 0.5 μgof HindIII- or I-SceI-digested NHEJ reporter substrate and 0.1 μg of the DsRed expression vector. The numbers of green (GFP+) and red (DsRed+) cells were determined by FACS analysis, and typical FACS traces are shown. The ratio of GFP+ to DsRed+ was used as a measure of NHEJ efficiency. The percent of ligated DNA ends was determined by using the curve shown in B. Hind III, filled bars; I-SceI, open bars. All experiments were repeated at least five times and the SD are shown. Y, young cells; P, presenescent; S, senescent; C, confluent; T, nTERT-immortalized.
To determine the relationship between the number of repaired NHEJ substrates and percent of GFP+ cells, we transfected normal human fibroblasts with increasing concentrations of GFP-Pem1 vector mixed with 0.1 μg of pDsRed to normalize for transfection efficiency. The percentages of GFP+ and DsRed+ cells were determined by FACS analysis. The ratio of GFP+ to DsRed+ cells was plotted as a function of the amount of GFP-Pem1 vector (Fig. 2B). The number of GFP+ cells reached saturation when 0.75 μg or more of circular GFP-Pem1 was transfected. Therefore, in all experiments, we used 0.5 μg of digested NHEJ substrate and 0.1 μg of the DsRed expression vector. The various ratios of GFP+/DsRed+ cells obtained upon transfection with the digested NHEJ substrate are shown in the shaded area in Fig. 2B, where there is a close to linear dependence on the amount of input DNA. The observed ratio is less than expected for 0.5 μg of GFP vector, because the efficiency of NHEJ is <100%. To calculate the percentage of ligated NHEJ substrate, the GFP+/DsRed+ ratio obtained from 0.5 μg of linear NHEJ substrate was first converted to an equivalent amount of circular GFP expression vector by using the standard curve in Fig. 2B. The percentage of ligated ends was then calculated as equivalent circular DNA over 0.5 μg × 100%.
Efficiency of End Joining Is Reduced in Presenescent and Senescent Cells. Young, presenescent, and senescent cells were cotransfected with the NHEJ reporter substrate, digested with either I-SceI or HindIII, and with the DsRed expression vector, as described above, and the percentages of GFP+ and DsRed+ cells were determined by FACS analysis. The percentage of GFP+ cells corresponds to the frequency of NHEJ events. To normalize for the efficiency of transfection, the ratio of GFP+ to DsRed+ cells was used as a measure of NHEJ efficiency. The percentage of ligated NHEJ substrates was calculated as described above.
We observed a striking (up to 4.5 fold) reduction in the efficiency of end joining in senescent cells compared with young cells (Fig. 2C). NHEJ of cohesive HindIII ends was more efficient than that of incompatible I-SceI ends, and WI-38 cells were, generally more efficient at NHEJ than were HCA2 cells. However, in each experimental group, the efficiency of NHEJ was reduced in presenescent cells compared with young cells, and was further reduced in terminally senescent cells. When circularized NHEJ plasmids and DsRed plasmids from human fibroblasts were rescued in E. coli, the ratio of NHEJ plasmids to DsRed plasmids was lower in presenescent and senescent cells, which is in agreement with the FACS data (Fig. 6, which is published as supporting information on the PNAS web site).
To rule out the possibility that the differences in NHEJ efficiency between young, presenescent, and senescent cells were due to different rates of cell division, we tested NHEJ in young, confluent cells. Cells were grown to confluence and kept there for 7 days, electroporated with the NHEJ substrate, and were then plated at high density to prevent resumption of cell division. The incorporation of 3H thymidine 24 h after plating indicated that fewer than 1% of cells in the culture were dividing (data not shown). The efficiency of end joining in young nondividing cells was equivalent to that of young cells (Fig. 2C), arguing that the decline in NHEJ during cellular senescence does not depend on cell division. This conclusion is further supported by the result with presenescent cells, which have a significantly reduced efficiency of NHEJ but only a slightly longer doubling time than young cells (18–20 h in presenescent cells versus 15–16 h in the young cells, data not shown), again suggesting that cell division is not required for NHEJ.
To rule out the possibility that the time cells spent in culture, or “cell culture shock,” rather than onset of cellular senescence, might be responsible for the decline of NHEJ, we used HCA2-hTERT and WI-38-hTERT cells, immortalized by expression of the catalytic component of human telomerase. At the time of testing, both HCA2-hTERT and WI-38-hTERT cell lines had undergone >100 PD, and had spent a longer time in culture than the senescent cells. The efficiency of NHEJ in hTERT-immortalized cells was equivalent to that of young cells (Fig. 2C). Thus, cell culture stress is not responsible for the observed decline of NHEJ during senescence. Collectively, these data indicate that the decrease in the efficiency of NHEJ is due to the process of cellular senescence.
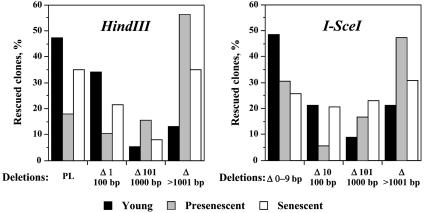
End Joining in Presenescent and Senescent Cells Is Associated with Larger Deletions. To determine whether the fidelity of NHEJ changes during cellular senescence, we rescued the repaired NHEJ substrates in E. coli, sequenced the junctions, and compared the deletion sizes among young, presenescent, and senescent cells. The plasmid rescue procedure is described in the supporting information on the PNAS web site. A total of 222 independent clones were analyzed (71 clones from young, 75 clones from presenescent, and 76 clones from senescent cells, and in each category, half of the clones were obtained from WI-38 cells and half from HCA2 cells). The complete list of deletions in the rescued products is shown in Table 1, which is published as supporting information on the PNAS web site. In 38 of the 222 clones, the ends had been precisely ligated, whereas the remainder contained deletions of various lengths, ranging from 1 nucleotide to 5 kb (the maximum deletion that can be tolerated to maintain the E. coli origin of replication and antibiotic resistance in NHEJ reporter plasmid). To facilitate comparison, the clones were grouped into four classes according to deletion size, as shown in Fig. 3, where deletion size was calculated as a sum of deletions from both ends. In the young cells, NHEJ of both HindIII- and I-SceI-induced breaks was preferentially precise, or associated with small deletions, whereas, as cells progressed to senescence, end-joining products contained larger deletions (Fig. 3). χ2 tests indicate that for both HindIII- and I-SceI-induced breaks, the distribution of deletion sizes in NHEJ products from young cells is significantly different from presenescent cells [P(χ2)<0.05], and possibly different from senescent cells [P(χ2)<0.1]. Overall, the average deletion size was 316 bp in the young cells, 1,247 bp in presenescent cells, and 718 bp in senescent cells. The average deletion size in presenescent and senescent cells was significantly larger than in the young cells (P < 0.001, t test). To rule out that the average deletion size is influenced by few very large deletions, we also determined the median deletion size, which was 10 bp in the young cells, 1,074 bp in presenescent cells, and 83 bp in senescent cells. The median deletion size in presenescent and senescent cells was significantly larger than in the young cells (P < 0.001, t test). Thus, NHEJ in presenescent and senescent cells is associated with larger deletions, suggesting that the fidelity of NHEJ declines during cellular senescence.
Fig. 3.
Distribution of deletion sizes among young, presenescent, and senescent cells. A total of 222 clones were analyzed. The clones were grouped according to the deletion sizes. PL, precise ligation. The complete list of deletions in the rescued products is shown in Table 1.
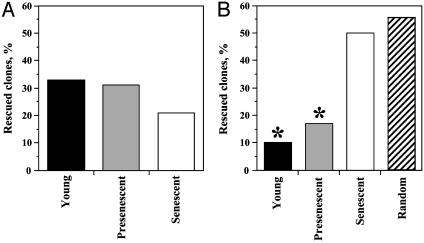
Senescent Cells Do Not Use Microhomologies for End Joining. Analysis of the products of NHEJ that had lost one or more nucleotides from the ends revealed three major classes of junctions: junctions with no homology, junctions at regions of microhomology, and junctions containing insertions (1–410 bp). We compared the percentage of each type of junction among the clones rescued from young, presenescent, and senescent cells. There was no significant difference between young and presenescent (P = 0.86, t test) and between young and senescent cells (P = 0.25, t test) in the frequency of insertions (Fig. 4A). However, the frequency NHEJ without microhomology increased as cells progressed to senescence (Fig. 4B). Junctions with no homology constituted 10% from the total junctions with deletions rescued from young cells, 17% from presenescent cells, and 50% from senescent cells. We calculated that the probability of finding microhomology by chance is 44%; i.e., if the ends were joined randomly, 56% of the junctions would not contain microhomology. To calculate this number, we classified the 1,357,800 junction scenarios resulting from all possible deletions at the DSB in the PEM1 intron (1.5 kb from either end) as either containing a micrhoomology, or not containing a microhomology. We identified 76,1867 (56%) of the junctions as not having a microhomology. Subsequent analysis of all possible deletions of smaller portions of the PEM1 intron yielded the same percentage of junctions without microhomology. Previous theoretical calculations suggest that this proportion is independent of deletion size, assuming an equal usage of the four nucleotides (49).
Fig. 4.
The frequency of NHEJ events with insertions (A) and without microhomologies (B) among the clones rescued from young, presenescent, and senescent cells. The hatched bar represents the theoretically calculated number of junctions with zero nucleotides of homology if the ends were joined at random. The t test was used to estimate the significance of the differences between the expected frequency of randomly occurring microhomologies and the experimentally observed frequency. *, Statistically significant difference from random.
The frequency of junctions without microhomology in young and presenescent cells was significantly different from random (P < 0.001, t test). However, the frequency of junctions without microhomology in senescent cells was not statistically different from random (P = 0.64, t test), indicating that the ability to use microhomologies for NHEJ may be compromised in senescent cells.
Discussion
Decline of End Joining During Senescence. Despite a large body of data that implicates the involvement of DSB repair in aging and cellular senescence (8, 13, 19, 25, 26, 36), age-associated changes in DSB repair have received little attention. Here, we present a systematic study of end joining with respect to cellular senescence. We have developed a sensitive fluorescence-based assay for NHEJ in human cells that detects a wide range of NHEJ products. By using this assay, we showed that the efficiency of end joining declines during cellular senescence in human diploid fibroblasts. NHEJ begins to decline before the terminal growth arrest in presenescent cells and continues as the cells become nondividing. This senescence-associated decline in end joining is likely to be a general phenomenon, because it was shown in two different normal fibroblast cell lines (lung and foreskin) by using two types of DSBs.
The efficiency of end joining was not compromised in young, nondividing cells, which rules out the possibility that the decline of NHEJ is caused by differences in rates of cell division. In addition, hTERT-immortalized cells, which had spent more time in culture than senescent cells, had levels of NHEJ similar to that of young cells, demonstrating that the reduced efficiency of NHEJ in presenescent and senescent cells is not due to cell-culture stress.
In addition to the reduced efficiency of end joining, we have demonstrated that the fidelity of NHEJ is compromised in presenescent and senescent cells. Older cells do not perform precise ligation of cohesive ends efficiently, and tend to generate larger deletions at the junctions. Such a shift toward larger deletions might be explained by a competition between exonucleases and NHEJ activities at the sites of DSBs. The drop in efficiency of end joining in senescent cells may allow more time for exonuclease degradation of the ends, leading to larger deletions. The efficiency of NHEJ was more severely affected in terminally senescent cells, whereas the deletion size was greater in presenescent cells. This apparent discrepancy might be due to higher exonuclease activity in actively dividing presenescent cells versus growth-arrested senescent cells. In this regard, it is interesting to note that the exonuclease activity responsible for processing broken DNA ends is repressed in G1-arrested Saccharomyces cerevisiae cells (G. Ira and J.E. Haber, personal communication), a situation that may be similar to that in G0-arrested senescent cells. Another difference in the way senescent cells process DNA ends was that the ability to use microhomologies for end joining was compromised in senescent cells compared with young cells. Interestingly, extracts from Bloom's syndrome cells are unable to use microhomologies for NHEJ (46), suggesting that specific proteins, possibly RecQ helicases, lose their function in senescent cells. Because more than one NHEJ pathway is likely to operate in human cells, it is possible that a choice of NHEJ pathway changes during cellular aging. Future experiments aimed at comparing protein requirements for NHEJ in young and senescent cells will help to elucidate the specific NHEJ pathways that are used by cells at different points of their lifespan.
The Role of Age-Related Decline of NHEJ in Tumorogenesis. Cancer incidence increases exponentially near the end of human life (50). Chromosomal abnormalities are a hallmark of most tumors, and genomic instability is believed to be a prerequisite for tumorigenesis (51). It has been observed that tissues of old animals accumulate genomic rearrangements (7–11). Our results suggest a mechanism for this age-related pathology. Reduced efficiency of NHEJ may lead to persistent DSBs. Indeed, a recent study (52) that used an alkaline extraction assay has shown that the number of DSBs increases during senescence. Formation of extended deletions at the sites of DSB repair in presenescent and senescent cells may lead to the loss of genetic material and cell transformation. In addition to larger deletions at the site of a break, the persistence of DSBs due to lower efficiency of NHEJ could lead to the joining of inappropriate ends, giving rise to genomic rearrangements. We hypothesize that inefficient and aberrant NHEJ is a likely mechanism underlying the age-related genomic instability and higher incidence of cancer in the elderly.
Contribution of Inefficient NHEJ to Organismal Aging. The finding that end joining declines during cellular senescence may be directly relevant to aging, because senescent cells accumulate in older organisms (53–56). The studies that have analyzed DSB repair in aging have shown that cells from old mice contain more DSBs (13). Furthermore, analysis of x-ray-induced DSBs in lymphocytes from donors of different ages showed an age-associated decline in the repair efficiency (57, 58). Thus, inefficient end joining in presenescent and senescent cells may contribute to the decline of DSB repair observed in aging organisms.
In the absence of efficient DSB repair, cells are more likely to die by apoptosis, as was shown for XRCC4- and Lig4-deficient mice, which die during the final days of gestation due to massive apoptosis (59, 60). Accelerated cell death has been implicated in several diseases of an old age (61, 62), and hence, an increase in cell death might be another mechanism by which inefficient NHEJ contributes to age-related pathologies.
Replicative senescence is an important mechanism for suppressing the development of malignant tumors in vivo. Because senescent cells are incapable of self-renewal, it has been proposed that cellular senescence might cause aging phenotypes such as immune failure, poor wound healing, skin atrophy, the decline of gastrointestinal function, etc. Furthermore, changes in cellular physiology and function that take place in senescent cells are thought to promote age-related diseases. For example, atherosclerosis has been proposed to be initiated by secreted proteins produced by senescent cells (63, 64), and senescent cells have been proposed to act in progression of cancer by stimulating the growth of neighboring premalignant cells (65). Our finding that senescent cells have inefficient and error-prone NHEJ provides an additional mechanism by which cellular senescence may contribute to the development of cancer in the elderly.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grants GM38219 and EY11731 (to J.H.W.), the Ellison Medical Foundation Senior Scholar Award in Aging (to O.M.P.-S.), a Human Frontier of Science Fellowship, and the Ellison Medical Foundation New Scholar Award in Aging (to V.G.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: DSB, double-strand break; NHEJ, nonhomologous end joining; FACS, fluorescence-activated cell sorter; PD, population doubling.
References
- 1.Cabelof, D. C., Raffoul, J. J., Yanamadala, S., Ganir, C., Guo, Z. & Heydari, A. R. (2002) Mutat. Res. 500, 135-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Intano, G. W., McMahan, C. A., McCarrey, J. R., Walter, R. B., McKenna, A. E., Matsumoto, Y., MacInnes, M. A., Chen, D. J. & Walter, C. A. (2002) Mol. Cell. Biol. 22, 2410-2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rao, K. S., Annapurna, V. V. & Raji, N. S. (2001) Ann. N.Y. Acad. Sci. 928, 113-120. [DOI] [PubMed] [Google Scholar]
- 4.Xu, G., Snellman, E., Bykov, V. J., Jansen, C. T. & Hemminki, K. (2000) Mutat. Res. 459, 195-202. [DOI] [PubMed] [Google Scholar]
- 5.Vijg, J., Dolle, M. E. T., Martus, H.-J. & Boerrigter, M. E. T. I. (1997) Mech. Ageing Dev. 98, 189-202. [DOI] [PubMed] [Google Scholar]
- 6.Kohler, S. W., Provost, G. S., Fieck, A., Kretz, P. L., Bullock, W. O., Sorge, J. A., Putman, D. L. & Short, J. M. (1991) Proc. Natl. Acad. Sci. USA 88, 7958-7962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stuart, G. R. & Glickman, B. W. (2000) Genetics 155, 1359-1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vijg, J. & Dolle, M. E. T. (2002) Mech. Ageing Dev. 123, 907-915. [DOI] [PubMed] [Google Scholar]
- 9.Curtis, H. & Crowley, C. (1963) Radiat. Res. 19, 337-344. [PubMed] [Google Scholar]
- 10.Ramsey, M. J., Moore, D. H., Briner, J. F., Lee, D. A., Olsen, L. A., Senft, J. R. & Tucker, J. D. (1995) Mutat. Res. 338, 95-106. [DOI] [PubMed] [Google Scholar]
- 11.Tucker, J. D., Spruill, M. D., Ramsey, M. J., Director, A. D. & Nath, J. (1999) Mutat. Res. 425, 135-141. [DOI] [PubMed] [Google Scholar]
- 12.McMurray, M. A. & Gottschling, D. E. (2003) Science 301, 1908-1911. [DOI] [PubMed] [Google Scholar]
- 13.Singh, N. P., Ogburn, C. E., Wolf, N. S., van Belle, G. & Martin, G. M. (2001) Biogerontology 2, 261-270. [DOI] [PubMed] [Google Scholar]
- 14.Jackson, S. P. (2002) Carcinogenesis 23, 687-696. [DOI] [PubMed] [Google Scholar]
- 15.Wang, H., Perrault, A. R., Takeda, Y., Qin, W. & Iliakis, G. (2003) Nucleic Acids Res. 31, 5377-5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feldmann, E., Schmiemann, V., Goedecke, W., Reichenberger, S. & Pfeiffer, P. (2000) Nucleic Acids Res. 28, 2585-2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lundberg, R., Mavinakere, M. & Campbell, C. (2001) J. Biol. Chem. 276, 9543-9549. [DOI] [PubMed] [Google Scholar]
- 18.Lee, S. E., Pellicioli, A., Vaze, M. B., Sugawara, N., Malkova, A., Foiani, M. & Haber, J. E. (2003) Mol. Cell. Biol. 23, 8913-8923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Opresko, P. L., Cheng, W. H., von Kobbe, C., Harrigan, J. A. & Bohr, V. A. (2003) Carcinogenesis 24, 791-802. [DOI] [PubMed] [Google Scholar]
- 20.Saintigny, Y., Makienko, K., Swanson, C., Emond, M. J. & Monnat, R. J. J. (2002) Mol. Cell. Biol. 22, 6971-6978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yannone, S. M., Roy, S., Chan, D. W., Murphy, M. B., Huang, S., Campisi, J. & Chen, D. J. (2001) J. Biol. Chem. 276, 38242-38248. [DOI] [PubMed] [Google Scholar]
- 22.Opresko, P. L., von Kobbe, C., Laine, J. P., Harrigan, J. A., Hickson, I. D. & Bohr, V. A. (2002) J. Biol. Chem. 277, 41110-41119. [DOI] [PubMed] [Google Scholar]
- 23.Fukuchi, K., Martin, G. M. & Monnat, R. J. (1989) Proc. Natl. Acad. Sci. USA 86, 5893-5897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oshima, J., Huang, S., Pae, C., Campisi, J. & Schiestl, R. H. (2002) Cancer Res. 62, 547-551. [PubMed] [Google Scholar]
- 25.Vogel, H., Lim, D. S., Karsenty, J., Finegold, M. & Hasty, P. (1999) Proc. Natl. Acad. Sci. USA 96, 10770-10775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weeda, G., Donker, I., de Wit, J., Morreau, H., Janssens, R., Vissers, C. J., Nigg, A., van Steeg, H., Bootsma, D. & Hoeijmakers, J. H. (1997) Curr. Biol. 7, 427-439. [DOI] [PubMed] [Google Scholar]
- 27.Hanawalt, P. C., Ford, J. M. & Lloyd, D. R. (2003) Mutat. Res. 554, 107-114. [DOI] [PubMed] [Google Scholar]
- 28.Levine, A. J. (1997) Cell 88, 323-331. [DOI] [PubMed] [Google Scholar]
- 29.Shay, J. W., Pereira-Smith, O. M. & Wright, W. E. (1991) Exp. Cell Res. 196, 33-39. [DOI] [PubMed] [Google Scholar]
- 30.Yang, Q., Zhang, R., Wang, X. W., Spillare, E. A., Linke, S. P., Subramanian, D., Griffith, J. D., Li, J. L., Hickson, I. D., Shen, J. C., Loeb, L. A., et al. (2002) J. Biol. Chem. 277, 31980-31987. [DOI] [PubMed] [Google Scholar]
- 31.Janz, C., Susse, S. & Wiesmuller, L. (2002) Oncogene 21, 2130-2140. [DOI] [PubMed] [Google Scholar]
- 32.Lee, S. H., Cavallo, L. & Griffith, J. (1997) J. Biol. Chem. 272, 7532-7539. [DOI] [PubMed] [Google Scholar]
- 33.Bill, C. A., Yu, Y., Miselis, N. R., Little, J. B. & Nickoloff, J. A. (1997) Mutat. Res. 385, 21-29. [DOI] [PubMed] [Google Scholar]
- 34.Okorokov, A. L., Warnock, L. & Milner, J. (2002) Carcinogenesis 23, 549-557. [DOI] [PubMed] [Google Scholar]
- 35.Tang, W., Willers, H. & Powell, S. N. (1999) Cancer Res. 59, 2562-2565. [PubMed] [Google Scholar]
- 36.Itahana, K., Dimri, G. & Campisi, J. (2001) Eur. J. Biochem. 268, 2784-2791. [DOI] [PubMed] [Google Scholar]
- 37.Xia, S. J., Shammas, M. A. & Shmookler Reis, R. J. (1997) Mol. Cell. Biol. 17, 7151-7158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boulton, S. J. & Jackson, S. P. (1996) Nucleic Acids Res. 24, 4639-4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Milne, G. T., Jin, S., Shannon, K. B. & Weaver, D. T. (1996) Mol. Cell. Biol. 16, 4189-4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kabotyanski, E. B., Gomelsky, L., Han, J. O., Stamato, T. D. & Roth, D. B. (1998) Nucleic Acids Res. 26, 5333-5342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liang, F. & Jasin, M. (1996) J. Biol. Chem. 271, 14405-14411. [DOI] [PubMed] [Google Scholar]
- 42.Smith, J., Riballo, E., Kysela, B., Baldeyron, C., Manolis, K., Masson, C., Lieber, M. R., Papadopoulo, D. & Jeggo, P. (2003) Nucleic Acids Res. 31, 2157-2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li, G., Nelsen, C. & Hendrickson, E. A. (2002) Proc. Natl. Acad. Sci. USA 99, 832-837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Escarceller, M., Buchwald, M., Singleton, B. K., Jeggo, P. A., Jackson, S. P., Moustacchi, E. & Papadopoulo, D. (1998) J. Mol. Biol. 279, 375-385. [DOI] [PubMed] [Google Scholar]
- 45.Runger, T. M. & Kraemer, K. H. (1989) EMBO J. 8, 1419-1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Langland, G., Elliott, J., Li, Y., Creaney, J., Dixon, K. & Groden, J. (2002) Cancer Res. 62, 2788-2770. [PubMed] [Google Scholar]
- 47.Runger, T. M., Poot, M. & Kraemer, K. H. (1992) Mutat. Res. 293, 47-54. [DOI] [PubMed] [Google Scholar]
- 48.Baldeyron, C., Jacquemin, E., Smith, J., Jacquemont, C., De Oliveira, I., Gad, S., Feunteun, J., Stoppa-Lyonnet, D. & Papadopoulo, D. (2002) Oncogene 21, 1401-1410. [DOI] [PubMed] [Google Scholar]
- 49.Roth, D. B., Porter, T. N. & Wilson, J. H. (1985) Mol. Cell. Biol. 5, 2599-2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.DePinho, R. A. (2000) Nature 408, 248-254. [DOI] [PubMed] [Google Scholar]
- 51.Nowell, P. C. (1976) Science 194, 23-28. [DOI] [PubMed] [Google Scholar]
- 52.Chevanne, M., Caldini, R., Tombaccini, D., Mocali, A., Gori, G. & Paoletti, F. (2003) Biogerontology 4, 97-104. [DOI] [PubMed] [Google Scholar]
- 53.Dimri, G. P., Lee, X., Basile, G., Acosta, M., Scott, G., Roskelley, C., Medrano, E. E., Linskens, M., Rubelj, I., Pereira-Smith, O., et al. (1995) Proc. Natl. Acad. Sci. USA 92, 9363-9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pendergrass, W. R., Lane, M. A., Bodkin, N. L., Hansen, B. C., Ingram, D. K., Roth, G. S., Yi, L., Bin, H. & Wolf, N. (1999) J. Cell. Physiol. 180, 123-130. [DOI] [PubMed] [Google Scholar]
- 55.Choi, J., Shendrik, I., Peacocke, M., Peehl, D., Buttyan, R., Ikeguchi, E. F., Katz, A. E. & Benson, M. C. (2000) Urology 56, 160-166. [DOI] [PubMed] [Google Scholar]
- 56.Ding, G., Franki, N., Kapasi, A. A., Reddy, K., Gibbons, N. & Singhal, P. C. (2001) Exp. Mol. Pathol. 70, 43-53. [DOI] [PubMed] [Google Scholar]
- 57.Singh, N. P., Danner, D. B., Tice, R. R., Brant, L. & Schneider, E. L. (1990) Mutat. Res. 237, 123-130. [DOI] [PubMed] [Google Scholar]
- 58.Mayer, P. J., Lange, C. S., Bradley, M. O. & Nichols, W. W. (1989) Mutat. Res. 219, 95-100. [DOI] [PubMed] [Google Scholar]
- 59.Gao, Y., Sun, Y., Frank, K. M., Dikkes, P., Fujiwara, Y., Seidl, K. J., Sekiguchi, J. M., Rathbun, G. A., Swat, W., Wang, J., et al. (1998) Cell 95, 891-902. [DOI] [PubMed] [Google Scholar]
- 60.Barnes, D. E., Stamp, G., Rosewell, I., Denzel, A. & Lindahl, T. (1998) Curr. Biol. 1998, 1395-1398. [DOI] [PubMed] [Google Scholar]
- 61.Martin, L. J. (2001) Int. J. Mol. Med. 7, 455-478. [PubMed] [Google Scholar]
- 62.Thompson, C. B. (1995) Science 267, 1456-1462. [DOI] [PubMed] [Google Scholar]
- 63.Chang, E. & Harley, C. B. (1995) Proc. Natl. Acad. Sci. USA 92, 11190-11194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vasile, E., Tomita, Y., Brown, L., Kocher, O. & Dvorak, H. F. (2001) FASEB J. 15, 458-466. [DOI] [PubMed] [Google Scholar]
- 65.Krtolica, A., Parrinello, S., Lockett, S., Desprez, P. Y. & Campisi, J. (2001) Proc. Natl. Acad. Sci. USA 98, 12072-12077. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.